Abstract
The functional abnormality of circadian regulation genes is involved in the development and progression of hepatocellular carcinoma (HCC). However, the association between functional single nucleotide polymorphisms (SNPs) in circadian gene NPAS2 and the overall survival of HCC patients treated with transcatheter arterial chemoembolization (TACE) has never been investigated. Six functional SNPs in the NPAS2 gene were genotyped using the Sequenom iPLEX genotyping system in a cohort of 448 unresectable Chinese patients with HCC treated with TACE. Multivariate Cox proportional hazards model and Kaplan–Meier curves were used for the prognosis analysis. We found that two SNPs, rs1053096 and rs2305160, in the NPAS2 gene showed significant associations with overall death risk in HCC patients in the recessive model (hazard ratio [HR] = 1.48; 95% confidence interval [CI], 1.13–1.94; P = 0.004) and in the dominant model (HR = 1.63; 95% CI, 1.29–2.07; P < 0.001), respectively. Moreover, we observed a cumulative effect of these two SNPs on HCC overall survival, indicating a significant trend of increasing death risk with increasing number of unfavorable genotypes (P for trend < 0.001). Compared with the patients without any unfavorable genotypes, the HRs for patients with one and two unfavorable genotypes were 1.41 (95% CI, 1.10–1.82; P = 0.007) and 2.09 (95% CI, 1.46–2.97, P < 0.001), respectively. The haplotype and diplotype analyses further characterized the association between NPAS2 genotype and survival of HCC patients. Our results for the first time suggest that NPAS2 gene polymorphisms may serve as an independent prognostic marker for HCC patients treated with TACE.
Keywords: Hepatocellular carcinoma, NPAS2, prognosis, single nucleotide polymorphism, transcatheter arterial chemoembolization
Hepatocellular carcinoma is the sixth most common cancer and the third leading cause of cancer-related deaths worldwide. Almost half of these cases and deaths were estimated to occur in China.(1) According to the data from the National Central Cancer Registry of China, the crude incidence of HCC was 28.71/100 000, and the crude mortality was 26.04/100 000 in 2009.(2) Unfortunately, only a few patients with HCC are diagnosed at an early stage, when the disease is amenable to treatment approaches such as surgical resection, liver transplantation, or local ablation. Most HCC patients, who are diagnosed at advanced stages, had to receive palliative or conservative therapy only.(3) Transcatheter arterial chemoembolization is the current standard of care for patients with intermediate or advanced HCC.(4) It has been shown that TACE significantly improves the OS of unresectable HCC patients.(5) But there is still a lack of effective and specific biomarkers for prediction of TACE treatment responses.
Circadian rhythm is the 24-h cycle in the behavioral, physiological, and biochemical processes.(6) The molecular mechanisms of circadian oscillation are based on a positive/negative transcriptional–translational feedback loop generated by at least nine core circadian genes.(7) Among them, NPAS2, which is located on chromosome 2 at 2q11.2, is one of the most important circadian genes. The NPAS2 gene encodes a member in the basic helix–loop–helix–PAS class of transcription factors. NPAS2 forms heterodimers with BMAL1 and then activates the expressions of the circadian genes PER and CRY, which are required for maintaining biological rhythms in many organisms.(8) An animal study showed that the loss of normal function of the NPAS2 gene may cause defects in several aspects of the circadian system, such as patterns of sleep and behavior.(9) Moreover, evidence has suggested that NPAS2 may play an important role in tumorigenesis by affecting expression of cancer-related genes and could be considered as a novel tumor suppressor.(10) In addition, genetic association studies have shown that a missense SNP (rs2305160) in NPAS2 gene is associated with the risk of breast cancer,(11) prostate cancer,(12) and non-Hodgkin's lymphoma.(13) More interestingly, potential involvement of circadian genes in cancer progression has recently been reported, indicating that high levels of NPAS2 expression are significantly associated with tumor grade, OS, and disease-free survival in breast cancer patients.(14) Our group has tested six functional SNPs in the NPAS2 gene in a cohort of 411 resected Chinese colorectal cancer patients, but we did not observe any association between these SNPs and clinical outcomes.(15)
Recently, there has been growing interest in the impact of circadian genes on HCC. For example, one study reported that circadian disruption accelerates hepatocellular carcinogenesis in mice.(16) Another study found that the downregulation of PER, CRY2, and TIM genes causes the disturbance of circadian rhythm in HCC patients, which may benefit the selective survival of cancerous cells and promote carcinogenesis.(17) Of more interest, the overexpression of the BMAL1 gene and low expression of the PER1 gene were correlated with liver metastasis in colorectal cancer.(18) Recently, our group reported that a functional polymorphism (rs2640908) in the PER3 gene is associated with prognosis in HCC treated with TACE.(19) All these data strongly indicated that the functional abnormality of circadian regulation genes is involved in the development and progression of HCC. It has been reported that NPAS2 significantly affects the cell cycle and DNA damage response, which plays an important role in the response of cancer cells to chemotherapeutic reagents.(10) Moreover, the PAS domain of NPAS2 protein mediates the responses of cells to environmental stress, such as ischemia.(20) These data suggest that NPAS2 may play a role in the response of HCC cells to TACE treatment, an important therapeutic regimen for unresectable HCC through both cytotoxicity and ischemic embolism. However, the association between functional SNPs of NPAS2 and OS of HCC patients treated with TACE has not been reported.
To evaluate whether functional SNPs of NPAS2 could be a potential biomarker to predict HCC prognosis, we assessed the effects of six functional SNPs in the NPAS2 gene on survival in a cohort of 448 unresectable Chinese HCC patients. To the best of our knowledge, this is the first study to investigate the predictive role of SNPs in the NPAS2 gene for clinical prognosis of unresectable HCC patients treated with TACE.
Materials and Methods
Study group
A total of 578 Han Chinese patients who had unresectable HCC were recruited from an ongoing study at the Department of Pain Management, Tangdu Hospital affiliated to The Fourth Military Medical University in Xi'an and the Eastern Hepatobiliary Surgery Hospital affiliated to the Secondary Military Medical University in Shanghai, China. These patients were recruited between December 2006 and November 2011. As described previously,(19) HCC diagnosis was based on the National Comprehensive Cancer Network clinical practice guidelines in oncology. All cases had no previous history of other cancers or cancer-related treatments. There were no recruitment restrictions on age, gender, and TNM stage. After recruitment, all patients received TACE as the first-line treatment within a week after diagnosis. A combination of oxaliplatin, fluorouracil, and doxorubicin was used for the majority of unresectable HCC patients. In the present study, 130 cases were excluded, including 83 patients with incomplete clinical information or lost during follow-up, 35 patients with distant metastases or Child–Pugh score C, and 12 patients with poor DNA quality. Finally, a total of 448 patients were included in the primary cohort and successfully genotype-assayed. Written informed consent was obtained from each participant before enrolment. Our study was approved by the hospital ethics committees both in Shanghai and Xi'an.
Demographic and clinical data
Demographic variables, including age, sex, ethnicity, residential region, and family history of cancer, were collected by well-trained interviewers. Detailed clinical information was collected through medical chart review or consulting with treating physicians, including the date of diagnosis, history of hepatitis virus infection, tumor number, tumor size, lymph node invasion and/or distant metastases, PVTT, Child–Pugh score, serum AFP, treatment protocol, and response. Clinical staging was determined according to the TNM classification system. The follow-up information was updated at 3-month intervals through onsite interview, direct calling, or medical chart review by a trained clinical specialist. The latest follow-up data in this analysis were obtained in January 2013. For each patient, 5 mL venous blood was collected for genomic DNA extraction using the EZNA Blood DNA Midi Kit (Omega Bio-Tek, Norcross, GA, USA) in the laboratory.
Single nucleotide polymorphism selection and genotyping
Functional SNPs in the NPAS2 gene were selected using a set of web-based SNP selection tools (http://snpinfo.niehs.nih.gov/snpinfo/snpfunc.htm) according to the previous description.(15) Finally, a total of six SNPs were selected, which include one non-synonymous SNP (rs2305160), one in transcription factor binding sites (rs3811558), two in splice sites (rs1562313 and rs9223), and two in miRNA-binding sites (rs2305158 and rs1053096). Genotyping was carried out using the Sequenom iPLEX genotyping system (Sequenom, San Diego, CA, USA). Laboratory persons who carried out the genotyping were blinded to patient information. The average call rate for the SNP array was 98.4%. Strict quality control measures were implemented during genotyping with over 99.0% concordance with the main genotyping results.
Immunohistochemistry
Tissue samples from another cohort of 243 surgical HCC patients were used for evaluation of NPAS2 expression by immunohistochemistry as previously described(21) with rabbit anti-human NPAS2 (1:200; Santa Cruz Biotechnology, Dallas, TX, USA) as primary antibody. The visualization signal was developed with the Histostain Plus kit (Life Technologies, Grand Island, NY, USA). The nuclear NPAS2 immunostaining score was evaluated for all samples under double-blinded conditions as previously described.(21)
Statistical analysis
All statistical analyses were carried out using the spss 16.0 statistical package (SPSS, Chicago, IL, USA). The survival time was defined as the period from the date of first treatment to the date of death or last follow-up. The three genetic models (additive, dominant, and recessive) were applied to assess the association of single SNPs with clinical outcome of HCC patients, and the best-fitting model was defined as that with the smallest P-value for the association analysis. Hazard ratios and 95% CIs were estimated by multivariate Cox proportional hazards model, with adjustment for age, sex, HBV infection, PVTT, TNM stage, serum AFP, and treatment, where appropriate. Overall survival was analyzed by plotting Kaplan–Meier curves and the survival probability distributions were compared using the log–rank test. The Mann–Whitney U-test was used to compare the difference in NPAS2 immunostaining score between different genotypes. All statistical tests were two-sided and P ≤ 0.05 was considered to be statistically significant.
Results
Distribution of patients' characteristics and prognosis analysis
Patients' characteristics and clinical data are summarized in Table 1. The patients included 391 men (87.3%) and 57 women (12.7%) with a median age of 53 years (range, 20–80 years); 383 (85.5%) patients were HBsAg positive. The minority (29.9%) of patients had PVTT and almost all patients (96.2%) had a Child–Pugh score A. The percentage of patients with TNM stage I and II was 49.3%. Two hundred and seventeen (48.4%) patients had significantly increased serum AFP (≥400 μg/L). Among all patients, 366 (81.7%) received only TACE treatment whereas the remaining 82 (18.3%) received TACE and other non-surgical treatments, such as radio frequency ablation, percutaneous ethanol injection, gamma knife radiosurgery, and/or immunoradiotherapy. During the median follow-up of 12.1 months (range, 2.0–70.1 months), a total of 339 (75.7%) patients died, and 8.9% patients were lost during follow-up. The median survival time for all patients was 11.1 months. Furthermore, multivariate analysis indicated significant poor OS in HCC patients with PVTT (HR = 1.92; 95% CI, 1.49–2.46), advanced TNM stage III and IV (HR = 1.56; 95% CI, 1.23–1.98), and higher serum AFP (HR = 1.51; 95% CI, 1.20–1.89). But patients who received TACE and other non-surgical treatments showed a significantly decreased risk of death (HR = 0.42; 95% CI, 0.30–0.59) compared with those treated by TACE alone. There was no association between HCC outcome and age (P = 0.31), sex (P = 0.17), HBV infection status (P = 0.30), or Child–Pugh score (P = 0.06).
Table 1.
Distribution of patient characteristics and prognosis analysis in Chinese patients with hepatocellular carcinoma treated with transcatheter arterial chemoembolization (TACE) (n = 448)
| Variable | Total, n (%) (n = 448) | Deaths, n (%) (n = 339) | Adjusted HR† (95% CI) | P-value‡ |
|---|---|---|---|---|
| Age, years | ||||
| ≤53 | 222 (49.6) | 168 (49.6) | Ref. | |
| >53 | 226 (50.4) | 171 (50.4) | 1.13 (0.89–1.42) | 0.310 |
| Sex | ||||
| Female | 57 (12.7) | 40 (11.8) | Ref. | |
| Male | 391 (87.3) | 299 (88.2) | 1.27 (0.90–1.77) | 0.170 |
| HBV infection | ||||
| Negative | 65 (14.5) | 48 (14.2) | Ref. | |
| Positive | 383 (85.5) | 291 (85.8) | 0.84 (0.61–1.17) | 0.300 |
| PVTT | ||||
| No | 314 (70.1) | 221 (65.2) | Ref. | |
| Yes | 134 (29.9) | 118 (34.8) | 1.92 (1.49–2.46) | <0.001 |
| Child–Pugh score | ||||
| A | 431 (96.2) | 324 (95.6) | Ref. | |
| B | 17 (3.8) | 15 (4.4) | 1.64 (0.97–2.75) | 0.060 |
| TNM stage | ||||
| I + II | 221 (49.3) | 142 (41.9) | Ref. | |
| III + IV | 227 (50.7) | 197 (58.1) | 1.56 (1.23–1.98) | <0.001 |
| Serum AFP, μg/L | ||||
| <400 | 231 (51.6) | 157 (46.3) | Ref. | |
| ≥400 | 217 (48.4) | 182 (53.7) | 1.51 (1.20–1.89) | <0.001 |
| Treatment | ||||
| TACE | 366 (81.7) | 299 (88.2) | Ref. | |
| TACE + other | 82 (18.3) | 40 (11.8) | 0.42 (0.30–0.59) | <0.001 |
Adjusted by age, sex, hepatitis B virus (HBV) infection, portal vein tumor thrombus (PVTT), tumor stage, serum α-fetoprotein (AFP), and treatment, where appropriate. ‡Significant P-values (<0.05) are in bold. CI, confidence interval; HR, hazard ratio; Ref., reference; TACE, transcatheter arterial chemoembolization.
Association between SNPs and clinical outcome in HCC patients
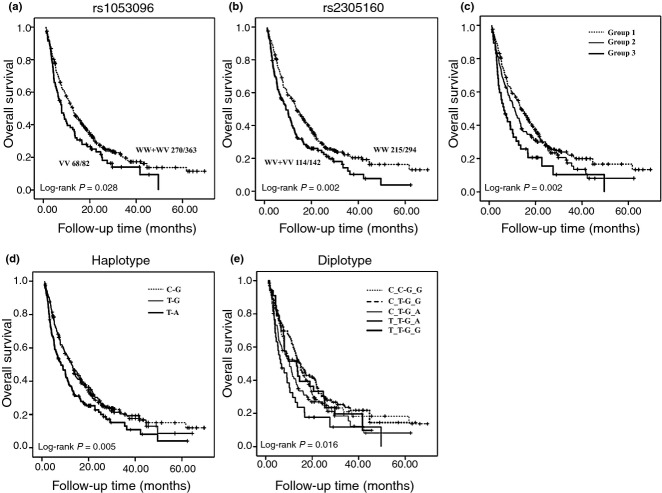
We assessed the association between each individual SNP and clinical outcome using the multivariate Cox proportional hazard model with adjustment for age, sex, HBV infection, PVTT, TNM stage, serum AFP, and treatment under dominant, additive, and recessive models (Tables 2, S1). We found that rs1053096 was significantly associated with HCC OS (Table 2). Compared to the WW and WV of rs1053096, the VV conferred a significant increased risk of death (HR = 1.48; 95% CI, 1.13–1.94; P = 0.004) in HCC patients. In addition, compared to the WW of rs2305160, patients carrying at least one variant allele (WV + VV) had a significant increased risk of death (HR = 1.63; 95% CI, 1.29–2.07; P < 0.001) (Table 2). Kaplan–Meier curve analysis showed that there was no significant difference in OS among patients with different genotypes in each SNP except rs2305160 (Fig. S1). Further analysis showed that patients with VV genotype in rs1053096 had better OS than those with WW or WV genotype (Fig. 1a), whereas patients with WW genotype in rs2305160 had worse OS than those with WW or WV genotype (Fig. 1b). To further evaluate the predictive effect on HCC outcome of SNPs in the NPAS2 gene, we carried out a stratified analysis (Table 3). The death risk conferred by rs1053096 remained significant in patients who were more than 53 years old (HR = 1.64; 95% CI, 1.11–2.41, P = 0.01), in male patients (HR = 1.44; 95% CI, 1.08–1.92, P = 0.01), in patients who were HBsAg positive (HR = 1.46; 95% CI, 1.10–1.94, P = 0.01), in patients without PVTT (HR = 1.45; 95% CI, 1.04–2.01, P = 0.03), in patients with TNM stage I and II disease (HR = 1.53; 95% CI, 1.01–2.32, P = 0.05), and in patients treated by TACE alone (HR = 1.56; 95% CI, 1.17–2.08, P = 0.003). Similar results were observed with rs2305160. In particular, P-values were <0.001 in male patients (HR = 1.69; 95% CI, 1.31–2.17), in HBsAg-positive patients (HR = 1.57; 95% CI, 1.22–2.03), those with stage III and IV disease (HR = 1.94; 95% CI, 1.41–2.67), in patients with increased serum AFP (HR = 1.76; 95% CI, 1.28–2.42), and in patients treated with TACE alone (HR = 1.76; 95% CI, 1.37–2.26).
Table 2.
Association between single nucleotide polymorphisms (SNPs) in the NPAS2 gene and clinical outcome in Chinese patients with hepatocellular carcinoma treated with transcatheter arterial chemoembolization (n = 448)
| SNP | Genotype and best-fitting model | Death/total 339/448 | Death HR† (95%CI) | P-value‡ |
|---|---|---|---|---|
| rs9223 | WW | 200/270 | Ref. | |
| WV | 122/156 | 1.03 (0.82–1.30) | 0.790 | |
| VV | 17/21 | 1.03 (0.62–1.70) | 0.910 | |
| Dominant | 1.03 (0.83–1.28) | 0.780 | ||
| rs1053096 | WW | 105/143 | Ref. | |
| WV | 165/220 | 1.08 (0.85–1.39) | 0.520 | |
| VV | 68/82 | 1.55 (1.14–2.12) | 0.005 | |
| Recessive | 1.48 (1.13–1.94) | 0.004 | ||
| rs1562313 | WW | 198/260 | Ref. | |
| WV | 115/157 | 0.87 (0.69–1.09) | 0.230 | |
| VV | 25/30 | 0.90 (0.59–1.37) | 0.620 | |
| Dominant | 0.87 (0.7–1.09) | 0.220 | ||
| rs2305158 | WW | 226/298 | Ref. | |
| WV | 94/128 | 1.12 (0.88–1.43) | 0.360 | |
| VV | 19/22 | 1.63 (1.01–2.63) | 0.040 | |
| Dominant | 1.18 (0.94–1.49) | 0.150 | ||
| rs2305160 | WW | 215/294 | Ref. | |
| WV | 107/133 | 1.61 (1.26–2.05) | ||
| <0.001 | ||||
| VV | 7/9 | 2.15 (1.00–4.64) | 0.050 | |
| Dominant | 1.63 (1.29–2.07) | <0.001 | ||
| rs3811558 | WW | 98/132 | Ref. | |
| WV | 163/208 | 1.05 (0.82–1.35) | 0.710 | |
| VV | 71/101 | 1.07 (0.78–1.46) | 0.670 | |
| Dominant | 1.06 (0.83–1.34) | 0.660 |
Adjusted for age, sex, hepatitis B virus infection, portal vein tumor thrombus, tumor stage, serum α-fetoprotein, and treatment.
Significant P-values (<0.05) are in bold. CI, confidence interval; HR, hazard ratio; Ref., reference; VV, homozygous variant; WV, heterozygous variant; WW, homozygous wild-type.
Figure 1.
Kaplan–Meier plots of overall survival in Chinese patients with hepatocellular carcinoma treated with transcatheter arterial chemoembolization. Overall survival analysis of patients with single nucleotide polymorphisms (SNPs) rs1053096 (a) and rs2305160 (b) in the NPAS2 gene. VV, homozygous variant; WV, heterozygous variant; WW, homozygous wild-type. (c) Cumulative effect of unfavorable genotypes. Group 1, no unfavorable genotypes; Group 2, one unfavorable genotype; Group 3, two unfavorable genotypes. (d) Haplotype analysis of combined SNPs rs1053096 and rs2305160. (e) Diplotype analysis of combined SNPs rs1053096 and rs2305160. The differences between indicated groups in each analysis were compared by the log–rank test.
Table 3.
Associations between single nucleotide polymorphisms rs1053096 and rs2305160 in the NPAS2 gene and clinical outcome in Chinese patients with hepatocellular carcinoma treated with transcatheter arterial chemoembolization (TACE) (n = 448), stratified by host characteristics
| rs1053096 | rs2305160 | |||||||
|---|---|---|---|---|---|---|---|---|
| Variable | Genotype | Genotype | ||||||
| Death/total | HR† (95% CI) | P-value‡ | Death/total | HR† (95% CI) | P-value‡ | |||
| Age, years | ||||||||
| ≤53 | WW+WV | 133/179 | Ref. | WW | 111/154 | Ref. | ||
| VV | 35/42 | 1.32 (0.91–1.93) | 0.150 | WV+VV | 54/64 | 1.70 (1.21–2.40) | 0.002 | |
| >53 | WW+WV | 137/184 | Ref. | WW | 104/140 | Ref. | ||
| VV | 33/40 | 1.64 (1.11–2.41) | 0.010 | WV+VV | 60/78 | 1.51 (1.08–2.10) | 0.020 | |
| Sex | ||||||||
| Female | WW+WV | 32/48 | Ref. | WW | 28/39 | Ref. | ||
| VV | 8/9 | 1.75 (0.78–3.90) | 0.170 | WV+VV | 11/15 | 1.47 (0.66–3.30) | 0.350 | |
| Male | WW+WV | 238/315 | Ref. | WW | 187/255 | Ref. | ||
| VV | 60/73 | 1.44 (1.08–1.92) | 0.010 | WV+VV | 103/127 | 1.69 (1.31–2.17) | <0.001 | |
| HBV infection | ||||||||
| Negative | WW+WV | 41/56 | Ref. | WW | 34/47 | Ref. | ||
| VV | 7/8 | 1.39 (0.58–3.30) | 0.460 | WV+VV | 13/17 | 2.46 (1.18–5.12) | 0.020 | |
| Positive | WW+WV | 229/307 | Ref. | WW | 181/247 | Ref. | ||
| VV | 61/74 | 1.46 (1.10–1.94) | 0.010 | WV+VV | 101/125 | 1.57 (1.22–2.03) | <0.001 | |
| PVTT | ||||||||
| No | WW+WV | 173/254 | Ref. | WW | 140/207 | Ref. | ||
| VV | 47/57 | 1.45 (1.04–2.01) | 0.030 | WV+VV | 75/99 | 1.59 (1.19–2.12) | 0.002 | |
| Yes | WW+WV | 97/109 | Ref. | WW | 75/87 | Ref. | ||
| VV | 21/25 | 1.39 (0.85–2.28) | 0.180 | WV+VV | 39/43 | 1.52 (1.00–2.31) | 0.050 | |
| TNM stage | ||||||||
| I + II | WW+WV | 112/180 | Ref. | WW | 84/138 | Ref. | ||
| VV | 29/38 | 1.53 (1.01–2.32) | 0.050 | WV+VV | 53/76 | 1.23 (0.86–1.75) | 0.250 | |
| III + IV | WW+WV | 158/183 | Ref. | WW | 131/156 | Ref. | ||
| VV | 39/44 | 1.35 (0.94–1.93) | 0.100 | WV+VV | 61/66 | 1.94 (1.41–2.67) | <0.001 | |
| Serum AFP, μg/L | ||||||||
| <400 | WW+WV | 128/195 | Ref. | WW | 105/158 | Ref. | ||
| VV | 29/35 | 1.40 (0.93–2.12) | 0.110 | WV+VV | 46/65 | 1.46 (1.01–2.10) | 0.040 | |
| ≥400 | WW+WV | 142/168 | Ref. | WW | 110/136 | Ref. | ||
| VV | 39/47 | 1.42 (0.99–2.05) | 0.060 | WV+VV | 68/77 | 1.76 (1.28–2.42) | <0.001 | |
| Treatment | ||||||||
| TACE | WW+WV | 239/298 | Ref. | WW | 184/234 | Ref. | ||
| VV | 59/66 | 1.56 (1.17–2.08) | 0.003 | WV+VV | 107/123 | 1.76 (1.37–2.26) | <0.001 | |
| TACE + other | WW+WV | 31/65 | Ref. | WW | 31/60 | Ref. | ||
| VV | 9/16 | 0.85 (0.39–1.83) | 0.670 | WV+VV | 7/19 | 0.68 (0.29–1.59) | 0.380 | |
Adjusted by age, sex, hepatitis B virus (HBV) infection, portal vein tumor thrombus (PVTT), tumor stage, serum α-fetoprotein (AFP), and treatment, where appropriate.
Significant P-values (<0.05) are in bold. CI, confidence interval; HR, hazard ratio; Ref., reference; VV, homozygous variant; WV, heterozygous variant; WW, homozygous wild-type.
Cumulative effect of unfavorable genotypes on HCC OS
To further assess the cumulative effect of genetic variants on HCC OS, we carried out a combination analysis by including the two SNPs with a significant association in the single SNP analysis. The unfavorable genotypes were defined as VV for rs1053096 and WV + VV for rs2305160. When we combined these unfavorable genotypes, there was a significant trend of increasing death risk with increasing number of unfavorable genotypes. Compared with the reference (group 1 without any unfavorable genotypes), the HRs for patients with one (group 2) and two (group 3) unfavorable genotypes were 1.41 (95% CI, 1.10–1.82; P = 0.007) and 2.09 (95% CI, 1.46–2.97, P < 0.001), respectively. A significant dose–response trend was also observed (P < 0.001) (Table 4). The Kaplan–Meier analysis showed that there was a significant trend for decreased OS with the increasing number of unfavorable genotypes (log–rank, P = 0.002; Fig. 1c).
Table 4.
Cumulative effect of unfavorable genotypes on overall survival in Chinese patients with hepatocellular carcinoma (n = 448)
| Group† | Death/total | HR‡ (95% CI) | P-value§ |
|---|---|---|---|
| Group 1 (0) | 188/259 | Ref. | |
| Group 2 (1) | 101/127 | 1.41 (1.10–1.82) | 0.007 |
| Group 3 (2) | 39/47 | 2.09 (1.46–2.97) | <0.001 |
| P for trend | <0.001 |
Patients were grouped by the number of unfavorable genotypes (0–2) in single nucleotide polymorphisms rs1053096 (VV) and rs2305160 (WV+VV) in the NPAS2 gene.
Hazard ratios (HRs) were adjusted by age, sex, hepatitis B virus infection, portal vein tumor thrombus, tumor stage, serum α-fetoprotein, and treatment.
Significant P-values (<0.05) are in bold. CI, confidence interval; Ref. reference.
Haplotype and diplotype of NPAS2 gene and HCC OS
We carried out haplotype and diplotype analyses to further characterize the impact of NPAS2 genotypes on the OS of HCC patients. There were four haplotypes in terms of rs1053096 and rs2305160 (CG, 56.0%; TG, 26.7%; TA, 16.4%; and CA, 0.9%) and seven diplotypes (CC-GG, 30.5%; CT-GG, 29.3%; CT-GA, 19.9%; TT-GA, 8.8%; TT-GG, 7.6%; TT-AA, 2.1%; and CC-GA, 1.8%). The frequency of haplotypes and diplotypes <5% were combined as another group. When compared to the CG haplotype, there was a 1.59-fold increase of death risk for the TA haplotype (95% CI, 1.28–1.97; P < 0.001) (Table 5). Kaplan–Meier curves indicated that significant difference of OS existed among the three haplotypes (log–rank, P = 0.005; Fig. 1d). Patients with the CT-GA diplotype had a significantly increased risk of death (HR = 1.50; 95% CI, 1.08–2.08; P = 0.01), and those with TT-GA had a 1.94-fold increased risk of death (95% CI, 1.29–2.92; P = 0.001), compared with those carrying the CC-GG diplotype (Table 5). The P-value was also significant by Kaplan–Meier analysis (log–rank, P = 0.016; Fig. 1e). In addition, we also carried out haplotype and diplotype analyses for six SNPs in the NPAS2 gene, but no significant result was observed (data not shown).
Table 5.
Haplotype and diplotype analysis of rs1053096 and rs2305160 single nucleotide polymorphisms in the NPAS2 gene in Chinese patients with hepatocellular carcinoma (n = 448)
| Group | Frequency,% | Death, n (%) | HR† (95% CI) | P-value‡ |
|---|---|---|---|---|
| Haplotype | ||||
| C-G | 56.0 | 359 (54.7) | Ref. | |
| T-G | 26.7 | 177 (27.0) | 1.04 (0.87–1.24) | 0.680 |
| T-A | 16.4 | 114 (17.4) | 1.59 (1.28–1.97) | <0.001 |
| Other groups | 0.9 | 6 (0.9) | 0.81 (0.36–1.83) | 0.610 |
| Diplotype | ||||
| C_C-G_G | 30.5 | 97 (29.6) | Ref. | |
| C_T-G_G | 29.3 | 91 (27.7) | 0.88 (0.66–1.18) | 0.390 |
| C_T-G_A | 19.9 | 68 (20.7) | 1.50 (1.08–2.08) | 0.010 |
| T_T-G_A | 8.8 | 32 (9.8) | 1.94 (1.29–2.92) | 0.001 |
| T_T-G_G | 7.6 | 27 (8.2) | 1.16 (0.75–1.80) | 0.490 |
| Other groups | 3.9 | 13 (4.0) | 1.22 (0.67–2.21) | 0.510 |
Adjusted by age, sex, hepatitis B virus infection, portal vein tumor thrombus, tumor stage, serum α-fetoprotein, and treatment.
Significant P-values (<0.05) are in bold. CI, confidence interval; HR, hazard ratio; Ref., reference.
Association between genotypes and expression of NPAS2
To assess the potential impact of genotypes of SNPs on the expression of NPAS2, we examined the genotype and expression of NPAS2 in another 243 HCC cases. As shown in Table S2, we found that patients who had the VV genotype of rs1053096 showed a significantly higher expression level of NPAS2 when compared with those who had the WW and WV genotypes (P = 0.03). In addition, no significant difference of NPAS2 expression was found between patients carrying different genotypes of other SNPs.
Discussion
In the present study, we evaluated the effects of six functional SNPs in NPAS2 on the prognosis of a cohort of 448 unresectable Chinese patients with HCC treated with TACE. The most important finding was that two SNPs (rs1053096 and rs2305160) are significantly associated with HCC OS. Additionally, we identified an accumulative death risk with increasing number of unfavorable genotypes. Furthermore, we observed that several haplotypes and diplotypes were significantly associated with HCC death risk, indicating the potential interactions between these SNPs on the OS of HCC patients. To the best of our knowledge, this is the first study to report that NPAS2 gene polymorphisms may serve as an independent prognostic marker for HCC patients treated with TACE.
Previous reports have suggested that NPAS2 may directly regulate other genes in the circadian regulatory system.(22) For example, the heterodimer of transactivators NPAS2 with BMAL1 regulate the transcription of PER1, PER2, CRY1, and CRY2.(23) Decreased expressions of PER1, PER2, PER3, and CRY2 in HCCs were observed.(17) It was reported in both in vitro and in vivo studies that overexpression of PER1 and PER2 inhibited cancer cell growth,(24) and induced apoptosis.(25) Yi et al.(26) reported that overexpression of NPAS2 increased PER1 and PER2 expression and consequently inhibited the growth of cancer cells. This could be one molecular explanation for the observed association between NPAS2 expression and OS among breast cancer patients.(14) A genome-wide ChIP-on-chip study identified 16 transcriptional targets of NPAS2, and nine of these genes were involved in cancer development.(26) Furthermore, Hoffman et al.(10) reported that NPAS2 is also involved in DNA damage response and cell-cycle regulation, and therefore plays an important role in tumorigenesis by affecting the expression of cancer-related genes, and could be considered a novel tumor suppressor. All these data strongly support the idea that NPAS2 is involved in the development and progression of human cancers, including HCC.
The functional prediction indicates that several microRNA binding sites exist near the sequence of rs1053096, which is located in the 3′-UTR region of the NPAS2 gene, suggesting the potential regulation on NPAS2 expression by rs1053096. Previous evidence showed that SNP sequences in 3′-UTRs of many other genes were strongly involved in the expression regulation of mRNA either by providing mutated binding sites for proteins and microRNAs to alter mRNA stability, or by forming hairpin loop structures to stabilize and thus slow down mRNA degradation. In this study, we found that the genotype of rs1053096 was significantly associated with the expression level of NPAS2. Therefore, the SNP rs1053096 in the NPAS2 gene could possibly affect the expression of NPAS2 and ultimately regulate its biological functions in cancer cells. In our study, for the first time, we found that SNP rs1053096 was associated with the prognosis of HCC patients, indicating that variants in this SNP site may affect the functions of NPAS2 in HCC cells. However, the underlying mechanisms need to be studied further. Moreover, the prognostic value of rs1053096 in other types of cancer warrant detailed investigation.
Another SNP, rs2305160, which is a non-synonymous SNP in the NPAS2 gene, has previously been reported to be a susceptibility biomarker for human cancers in earlier case–control studies.(11–13) The current results showed that the homozygous variant of rs2305160 is significantly associated with poor OS in HCC patients. It may have several potential impacts on NPAS2 functions. First, an amino acid change caused by this polymorphism may alter the NPAS2 protein structure. Second, Zhu et al.(11) has predicted that rs2305160 is located in the PAS sensory domain, therefore, it may affect protein sensory capacity directly or interfere with NPAS2/BMAL1 heterodimerization, which is essential for normal sensory function. Further functional assay is still needed to verify our hypothesis.
In our study, the mortality rate in unresectable HCC patients after TACE treatment is similar to previous reports in Chinese patients.(27,28) The TACE technique combines transarterial embolization with regional chemotherapy. The major advantage of this treatment is the high concentration of chemo-agents in tumor tissue, which bring toxicity to cancer cells. TACE shows great application to HCC patients by restricting the location of chemo-agents to HCC tissue, therefore improving the treatment response. The final clinical outcome is usually determined by the balance between tumor progression and treatment response. On the basis of these reports, we put forward the hypothesis that rs1053096 and rs2305160 of the NPAS2 gene may influence the treatment response of TACE through regulating cell proliferation, apoptosis, and DNA damage response by controlling transcription and affecting the protein sensory capacity of NPAS2. Further validations of this hypothesis using independent populations, high-density mapping, and functional characterizations are warranted.
Our study has one specific strength. The patient group was mainly enrolled from Xi'an and adjacent areas. This region is highly attractive for carrying out population-based research because of the demographic stability and low mobility rate. The homogenous patient characteristics and treatments, as well as the extremely low rate of patient loss during follow-up, greatly reduced the confounding effects of the heterogeneous therapeutic methods in most cancer clinical outcome studies. Our study also has limitations. Our sample size may not be large enough to detect minimal associations and interactions, and the generalizability of our findings identified from this Chinese patient group needs to be further validated in other ethnic populations.
In conclusion, our study reported the first epidemiological evidence supporting a role for rs1053096 and rs2305160 in the prognosis prediction of Chinese HCC patients treated with TACE. If validated, these two SNPs may potentially be developed as a simple non-invasive biomarker to help clinicians identify specific patients who will benefit from TACE.
Acknowledgments
This work was supported by the Program for New Century Excellent Talents in University (Grant Nos. 81201583 and 81171966) from the National Natural Science Foundation of China, the International S&T Cooperation Program of China (Grant No. s2011gr0239), and the National Key Scientific and Technological Project (Grant No. 2011ZX09307-001-04) from the Ministry of Science and Technology of China.
Glossary
Abbreviations
- AFP
α-fetoprotein
- CI
confidence interval
- HBsAg
hepatitis B surface antigen
- HBV
hepatitis B virus
- HCC
hepatocellular carcinoma
- HR
hazard ratio
- OS
overall survival
- PVTT
portal vein tumor thrombus
- SNP
single nucleotide polymorphism
- TACE
transcatheter arterial chemoembolization
- VV
homozygous variant
- WV
heterozygous variant
- WW
homozygous wild-type
Disclosure Statement
The authors have no conflict of interest.
Supporting Information
Additional supporting information may be found in the online version of this article:
Fig. S1. Kaplan–Meier survival curves of patients with hepatocellular carcinoma by single nucleotide polymorphisms in the NPAS2 gene. VV, homozygous variant; WV, heterozygous variant; WW, homozygous wild-type.
Table S1. Association of rs1053096 and rs2305160 with clinical outcome in patients with hepatocellular carcinoma.
Table S2. NPAS2 scores by subgroups with different single nucleotide polymorphism genotypes.
References
- 1.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.Chen WQ, Zheng RS, Zhang SW. Liver cancer incidence and mortality in China, 2009. Chin J Cancer. 2013;32:162–9. doi: 10.5732/cjc.013.10027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Forner A, Llovet JM, Bruix J. Hepatocellular carcinoma. Lancet. 2012;379:1245–55. doi: 10.1016/S0140-6736(11)61347-0. [DOI] [PubMed] [Google Scholar]
- 4.Murata S, Mine T, Ueda T, et al. Transcatheter arterial chemoembolization based on hepatic hemodynamics for hepatocellular carcinoma. ScientificWorldJournal. 2013;2013:479–805. doi: 10.1155/2013/479805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bruix J, Sherman M. Management of hepatocellular carcinoma. Hepatology. 2005;42:1208–36. doi: 10.1002/hep.20933. [DOI] [PubMed] [Google Scholar]
- 6.Kondratov RV, Gorbacheva VY, Antoch MP. The role of mammalian circadian proteins in normal physiology and genotoxic stress responses. Curr Top Dev Biol. 2007;78:173–216. doi: 10.1016/S0070-2153(06)78005-X. [DOI] [PubMed] [Google Scholar]
- 7.Lee C, Etchegaray JP, Cagampang FR, Loudon AS, Reppert SM. Posttranslational mechanisms regulate the mammalian circadian clock. Cell. 2001;107:855–67. doi: 10.1016/s0092-8674(01)00610-9. [DOI] [PubMed] [Google Scholar]
- 8.DeBruyne JP, Weaver DR, Reppert SM. CLOCK and NPAS2 have overlapping roles in the suprachiasmatic circadian clock. Nat Neurosci. 2007;10:543–5. doi: 10.1038/nn1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dudley CA, Erbel-Sieler C, Estill SJ, et al. Altered patterns of sleep and behavioral adaptability in NPAS2-deficient mice. Science. 2003;301:379–83. doi: 10.1126/science.1082795. [DOI] [PubMed] [Google Scholar]
- 10.Hoffman AE, Zheng T, Ba Y, Zhu Y. The circadian gene NPAS2, a putative tumor suppressor, is involved in DNA damage response. Mol Cancer Res. 2008;6:1461–8. doi: 10.1158/1541-7786.MCR-07-2094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhu Y, Stevens RG, Leaderer D, et al. Non-synonymous polymorphisms in the circadian gene NPAS2 and breast cancer risk. Breast Cancer Res Treat. 2008;107:421–5. doi: 10.1007/s10549-007-9565-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chu LW, Zhu Y, Yu K, et al. Variants in circadian genes and prostate cancer risk: a population-based study in China. Prostate Cancer Prostatic Dis. 2008;11:342–8. doi: 10.1038/sj.pcan.4501024. [DOI] [PubMed] [Google Scholar]
- 13.Zhu Y, Leaderer D, Guss C, et al. Ala394Thr polymorphism in the clock gene NPAS2: a circadian modifier for the risk of non-Hodgkin's lymphoma. Int J Cancer. 2007;120:432–5. doi: 10.1002/ijc.22321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yi C, Mu L, de la Longrais IA, et al. The circadian gene NPAS2 is a novel prognostic biomarker for breast cancer. Breast Cancer Res Treat. 2009;120:663–9. doi: 10.1007/s10549-009-0484-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhou F, He X, Liu H, et al. Functional polymorphisms of circadian positive feedback regulation genes and clinical outcome of Chinese patients with resected colorectal cancer. Cancer. 2011;118:937–46. doi: 10.1002/cncr.26348. [DOI] [PubMed] [Google Scholar]
- 16.Filipski E, Subramanian P, Carriere J, Guettier C, Barbason H, Levi F. Circadian disruption accelerates liver carcinogenesis in mice. Mutat Res. 2009;680(1–2):95–105. doi: 10.1016/j.mrgentox.2009.10.002. [DOI] [PubMed] [Google Scholar]
- 17.Lin YM, Chang JH, Yeh KT, et al. Disturbance of circadian gene expression in hepatocellular carcinoma. Mol Carcinog. 2008;47:925–33. doi: 10.1002/mc.20446. [DOI] [PubMed] [Google Scholar]
- 18.Oshima T, Takenoshita S, Akaike M, et al. Expression of circadian genes correlates with liver metastasis and outcomes in colorectal cancer. Oncol Rep. 2011;25:1439–46. doi: 10.3892/or.2011.1207. [DOI] [PubMed] [Google Scholar]
- 19.Zhao B, Lu J, Yin J, et al. A functional polymorphism in PER3 gene is associated with prognosis in hepatocellular carcinoma. Liver Int. 2012;32:1451–9. doi: 10.1111/j.1478-3231.2012.02849.x. [DOI] [PubMed] [Google Scholar]
- 20.Bersten DC, Sullivan AE, Peet DJ, Whitelaw ML. bHLH-PAS proteins in cancer. Nat Rev Cancer. 2013;13:827–41. doi: 10.1038/nrc3621. [DOI] [PubMed] [Google Scholar]
- 21.Shi H, Zhou Y, Liu H, et al. Expression of CIAPIN1 in human colorectal cancer and its correlation with prognosis. BMC Cancer. 2010;10:477. doi: 10.1186/1471-2407-10-477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Levi F, Okyar A, Dulong S, Innominato PF, Clairambault J. Circadian timing in cancer treatments. Annu Rev Pharmacol Toxicol. 2010;50:377–421. doi: 10.1146/annurev.pharmtox.48.113006.094626. [DOI] [PubMed] [Google Scholar]
- 23.Chen-Goodspeed M, Lee CC. Tumor suppression and circadian function. J Biol Rhythms. 2007;22:291–8. doi: 10.1177/0748730407303387. [DOI] [PubMed] [Google Scholar]
- 24.Lee CC. Tumor suppression by the mammalian Period genes. Cancer Causes Control. 2006;17:525–30. doi: 10.1007/s10552-005-9003-8. [DOI] [PubMed] [Google Scholar]
- 25.Hua H, Wang Y, Wan C, et al. Circadian gene mPer2 overexpression induces cancer cell apoptosis. Cancer Sci. 2006;97:589–96. doi: 10.1111/j.1349-7006.2006.00225.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yi CH, Zheng T, Leaderer D, Hoffman A, Zhu Y. Cancer-related transcriptional targets of the circadian gene NPAS2 identified by genome-wide ChIP-on-chip analysis. Cancer Lett. 2009;284:149–56. doi: 10.1016/j.canlet.2009.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shi M, Lu LG, Fang WQ, et al. Roles played by chemolipiodolization and embolization in chemoembolization for hepatocellular carcinoma: single-blind, randomized trial. J Natl Cancer Inst. 2013;105:59–68. doi: 10.1093/jnci/djs464. [DOI] [PubMed] [Google Scholar]
- 28.Yuen MF, Chan AO, Wong BC, et al. Transarterial chemoembolization for inoperable, early stage hepatocellular carcinoma in patients with Child-Pugh grade A and B: results of a comparative study in 96 Chinese patients. Am J Gastroenterol. 2003;98:1181–5. doi: 10.1111/j.1572-0241.2003.07404.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Kaplan–Meier survival curves of patients with hepatocellular carcinoma by single nucleotide polymorphisms in the NPAS2 gene. VV, homozygous variant; WV, heterozygous variant; WW, homozygous wild-type.
Table S1. Association of rs1053096 and rs2305160 with clinical outcome in patients with hepatocellular carcinoma.
Table S2. NPAS2 scores by subgroups with different single nucleotide polymorphism genotypes.