Abstract
Osteosarcoma typically arises in tissues of mesenchymal origin, and is the most malignant bone tumor characterized by high local aggressiveness, with poor therapeutic outcome. Busulfan has been widely used to treat CML. So far, there are no reports on the therapeutic effect of busulfan on osteosarcoma. Here, we showed that busulfan dose-dependently reduced the cell viability and proliferation, and induced cell apoptosis, senescence, and reactive oxygen species levels in two osteosarcoma cell lines. Moreover, a series of loss-of-function and gain-of-function experiments further indicated that busulfan may have its anti-osteosarcoma effect by upregulating the microRNA-200 (miR-200) family which subsequently downregulated its target genes ZEB1 and ZEB2. Furthermore, treatment with busulfan potentially inhibited the growth of implanted osteosarcoma in nude mice. Taken together, our data suggest that busulfan may have an anti-osteosarcoma effect through downregulating ZEB1 and ZEB2 through activating the miR-200 family, highlighting a possibility of using busulfan as a novel therapy for osteosarcoma.
Keywords: Busulfan, microRNAs, osteosarcoma, ZEB1, ZEB2
Osteosarcoma is characterized by high local aggressiveness and rapid metastasizing potential, resulting in an early onset tumor with poor survival.(1) A wide spectrum of osteosarcoma has been found histologically. However, the histological appearance is not correlated with clinical outcome.(1) The histological heterogeneity of osteosarcoma may indicate an origin of mesenchymal stem cells, along their path of differentiation to the osteoblastic lineage.(2) The origin and etiology of osteosarcoma is further complicated by its extreme rearranged genome, lack of precursor lesions, and high genetic instability. Osteosarcoma is the most common primary bone tumor that frequently occurs in the pediatric age, and it appears to be the second highest cause of cancer-related death.(1) Therefore, there is an urgent need for studying the tumor biology in order to increase our comprehension to treat it efficiently. A number of human osteosarcoma cell lines have been established, among which U2OS(3) and MG-63(4) cells lines have been extensively characterized and widely used for osteosarcoma research.
MicroRNAs (miRNAs) are non-coding small RNAs, usually 18–25 nucleotides in length, that inhibit translation and cleave mRNA by base-pairing to the 3′-UTR of the target gene. MicroRNAs plays an important role in regulation of gene expression at post-transcriptional levels, and were implicated in tumorigenesis and cancer progression.(1,5–12)
The chemical systematic name of busulfan is butane-1,4-diyl dimethanesulfonate. Busulfan has been used as a palliative treatment of CML and other myeloproliferative disorders since the 1950s.(13,14) Busulfan has been potentially displaced by imatinib nowadays in the treatment of CML.
So far, there have been no reports on the therapeutic effect of busulfan on osteosarcoma. Here, we showed that busulfan dose-dependently reduced the cell viability and proliferation, and induced cell apoptosis, senescence, and reactive oxygen species (ROS) levels in two osteosarcoma cell lines. Moreover, further analyses showed that busulfan effected its anti-osteosarcoma effect by upregulating the miR-200 family, which subsequently downregulated its target genes ZEB1 and ZEB2, manifested by a series of loss-of-function and gain-of-function experiments. Furthermore, treatment with busulfan potentially inhibited the growth of implanted osteosarcoma in nude mice. Taken together, our data suggest that busulfan may produce its anti-osteosarcoma effect by downregulating ZEB1 and ZEB2 through activating the miR-200 family, highlighting a possibility of using busulfan as a novel therapy for osteosarcoma.
Materials and Methods
Cell lines and culture
Human osteosarcoma U2OS and MG63 cells lines were purchased from ATCC and maintained in DMEM supplemented with 10% FBS (PAA Laboratories, Morningside, Australia). U2OS and MG63 cells were seeded in 24-well plates at 6 × 104 cells per well for in vitro experiments. Busulfan (Sigma-Aldrich, USA) stock solution was prepared in DMSO (Sigma-Aldrich) and further diluted in complete cell culture medium. Medium containing DMSO, but not busulfan, was used as a control.
Cell viability assay and proliferation assay
The viability of cells was assayed by the Trypan blue exclusion test. Trypan blue solution (0.4%) was purchased from Life Technologies (USA). Cell viability was calculated as the number of viable cells divided by the total number of cells within the grids on the hemocytometer. Trypan blue taken cells are considered non-viable.(15)
For assay of cell proliferation, pretreated cells were seed into 96-well plates at 4000 cells per well and subjected to a Cell Proliferation Kit (MTT; Roche, USA), according to the manufacturer's instructions.
Apoptosis assay
Cells were labeled with annexin V–FITC and propidium iodide, and then examined with an apoptosis detecting kit (Invitrogen, Burlington, Canada) for apoptosis. Samples were analyzed by flow cytometry and the results were analyzed by CellQuest software (Becton Dickinson, San Jose, CA, USA) as previously described.(16) In addition, TUNEL staining was carried out with an assay (ApopTag Peroxidase In Situ Apoptosis Detection Kit; Millipore, Billerica, MA, USA), according to the manufacturer's instruction.
Senescence β-Gal staining
The senescence of osteosarcoma cells was analyzed by a Senescence β-Galactosidase Staining kit (Cell Signaling, Danvers, MA, USA), according to the protocol from the manufacturer.(17)
Reactive oxygen species assay
An OxiSelect Intracellular ROS Assay kit was purchased from Cell Biolabs. Cells were cultured in a 96-well culture plate and assayed according to the manufacturer's instructions.(18) The fluorescence was quantified by a fluorescent microplate reader (Gemini).
RNA extraction, reverse transcription, and quantitative PCR
Total RNA was extracted using Trizol (Invitrogen), according to the manufacturer's instructions. For mRNA analysis, cDNA was randomly primed from 2.0 μg total RNA using the Omniscript reverse transcription kit (Qiagen, Hilden, Germany). Real-time PCR was subsequently carried out in triplicate with a 1:4 dilution of cDNA using the Quantitect SyBr green PCR system (Qiagen) on a Rotorgene 6000 series PCR machine (Qiagen). Data were collected and analyzed using the Rotorgene software accompanying the PCR machine. Relative expression levels were determined using the comparative quantification feature of the Rotorgene software. All mRNA quantification data were normalized to GAPDH. For miRNA analysis, quantitative PCR was carried out as above, using TaqMan miRNA assays according to the manufacturer's instructions (Applied Biosystems, Foster City, CA, USA). Sequences of PCR primers and RNA oligonucleotides for real-time PCR were generated by Shanghai Sangon Company as previously described.(17)
Transfections and infections
All miRNA mimics, antisense oligonucleotides (ASO) and plasmids were produced by Genema (Shanghai, China). Cells were transfected with synthetic miRNA (mimics), miRNA ASO, pcDNA3.1-ZEB1, pcDNA3.1-ZEB2, and pcDNA3.1-p-pre-miR-200a/b-miR-429 (p-pre-miR-200a/b-miR-429) using Lipofectamine 2000 (Invitrogen) transfection reagents, according to the manufacturer's instructions. Cells were analyzed 48 h after treatment.
To trace the osteosarcoma cells in vivo, we infected the U2OS cells with a recombinant lentivirus expressing luciferase and GFP under the control of cytomegalovirus promoter at MOI 100 and resulted in nearly 100% infection efficiency based on green fluorescence. The reporter-carrying osteosarcoma cells were termed U2OS-LUC.
Mouse manipulation and induction of intracranial tumor and busulfan injection
All mouse experiments were approved by the general principles contained in the Guide for the Care and Use of Laboratory Animals published by Nanjing University (Nanjing, China). Ten-week-old male NOD/SCID mice were used for experiments. U2OS-LUC cells (105) were orthotopically injected into the tibia of the mice to develop tumors. Tumor growth was monitored by bioluminescence imaging. Busulfan was i.v. injected at a dose of 4 mmol/kg body weight every other day, into the tail vein of the mice.
Imaging of osteosarcoma by bioluminescence
Bioluminescence was measured with the IVIS imaging system (Xenogen, Alameda, CA, USA). All of the images were taken 10 min after i.p. injection of luciferin (Sigma) at 150 mg/kg body weight, as a 60-s acquisition and 10 of binning. During image acquisition, mice were sedated continuously by inhalation of 3% isoflurane. Image analysis and bioluminescent quantification was carried out using Living Image software (Xenogen).
Statistical analysis
Each experiment condition contained five repeats. Five animals were used in each group in the in vivo study. All values are depicted as mean ± standard deviation and were considered significant if P < 0.05. Student's t-test was used for comparison between experimental and control groups. One-way anova was used for comparison between three groups.
Results
Busulfan reduced osteosarcoma cell viability in a dose-dependent manner
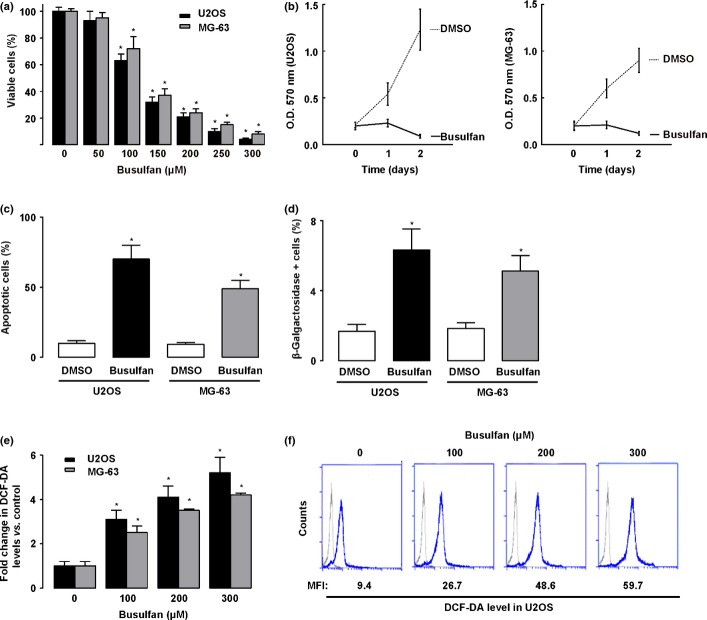
Two osteosarcoma cell lines (U2OS and MG-63) were treated with busulfan for 48 h in culture, and the viable cells were detected by Trypan blue exclusion test. We found that busulfan reduced the cancer cell viability in a dose-dependent manner (Fig. 1a). As a concentration of 150 μM potentially decreased cell viability by more than 70% in both lines, it was thus applied in the following experiments.
Figure 1.
Measurements of cell viability, proliferation, apoptosis, senescence, and reactive oxygen species (ROS) levels in busulfan-treated osteosarcoma cells. (a) Percentage of viable cells (U2OS and MG-63) 24 h after different doses of busulfan. (b) Busulfan (100 μM) significantly inhibited the proliferation of U2OS and MG-63 cells. (c) Busulfan increased the apoptosis of U2OS and MG-63 cells. (d) Quantification of β-Gal positive cells in busulfan-treated U2OS and MG-63 cells. (e,f) 2'-7'-Dichlorodihydrofluorescein diacetate (DCF-DA) levels in U2OS after the different doses of busulfan treatment by representative cytometry histogram (e) and by quantification (f). MFI, mean fluorescence intensity. *P < 0.05.
Busulfan reduced osteosarcoma cell proliferation and induced apoptosis, senescence, and ROS levels
Next, we examined the changes in the proliferation of osteosarcoma cells by busulfan treatment in an MTT assay. Our data showed that the proliferation of both U2OS and MG-63 cells were significantly inhibited by busulfan treatment (Fig. 1b). The apoptosis of the cells were then evaluated by flow cytometry, using an apoptosis detection kit. We found that busulfan treatment induced much higher apoptosis of U2OS cells (36%) or of MG-63 (25%), compared with controls (Fig. 1c). As cellular senescence may also suppress tumorigenesis,(19,20) we then examined the cancer cell senescence after busulfan treatment and β-Gal activity as an indicator for cell senescence. Our data showed that busulfan treatment significantly increased the number of β-galactosidase-positive cells, suggesting that busulfan may induce the senescence of osteosarcoma cells (Fig. 1d). Cancer cells usually show greater levels of ROS than normal cells.(19,20) This high level of ROS in cancer cells likely suppresses tumor growth through sustained activation of cell-cycle inhibitor. The use of 2′-7′-dichlorodihydrofluorescein diacetate is well established as a technique for measuring the ROS in a cell.(19,20) Thus, we used it to examine the ROS levels in the osteosarcoma cells. We found that busulfan significantly increased the ROS level in a dose-dependent manner in both U2OS and MG-63 cells (Fig. 1e,f).
Busulfan upregulated miR-200 family and modulated its target genes in osteosarcoma cells
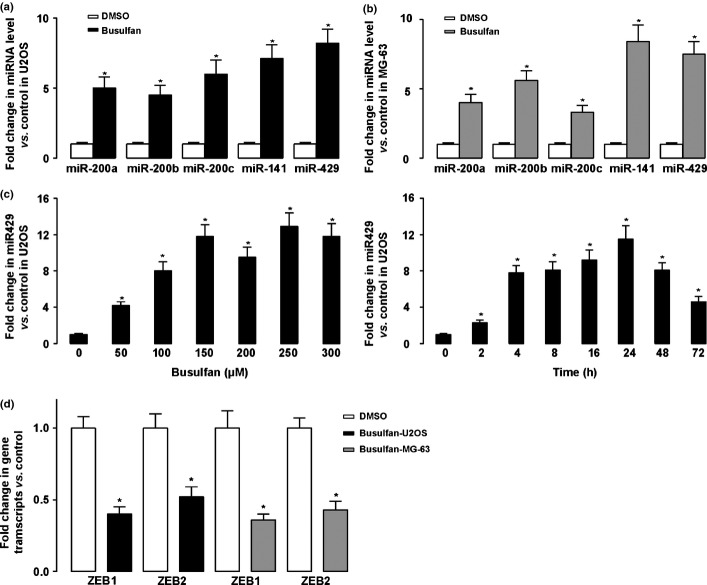
All five members of the miR-200 family (miR-200a, miR-200b, miR-200c, miR-141, and miR-429) can inhibit epithelial–mesenchymal transition through activating E-cadherin transcription repressors ZEB1 and ZEB2. Thus, we measured the miRNA expression levels in U2OS and MG-63 cells. We found that the five members in the miRNA-200 family were all upregulated after busulfan treatment in both cell lines (Fig. 2a).
Figure 2.
Busulfan upregulated the microRNA-200 (miR-200) family and modulated its target genes in osteosarcoma. (a,b) Messenger RNA from the miR-200 family (miR-200a, miR-200b, miR-200c, miR-141, and miR-429) was assayed by quantitative RT-PCR and normalized to control, showing that all were upregulated after busulfan treatment in U2OS (a) and MG-63 cells (b). (c) Dose-dependent and time-dependent effect of busulfan on the activation of miR-429. Busulfan (150 μM) seemed to be efficient for activation of miR-429. Moreover, efficient induction of miR-429 was detected 24 h after busulfan treatment. (d) Changes in mRNA of ZEB1 and ZEB2 in both lines were assayed by quantitative RT-PCR and normalized to control. *P < 0.05
Next, we examined the expression levels of the miRNA-200 family dose- and time-dependent on busulfan treatment. We found that and 150 μM busulfan seemed to be efficient for activation of the miRNA-200 family. Further increases in dosage did not further increase the levels of the miRNA-200. Moreover, the efficient induction of the miRNA-200 family was detected 24 h after busulfan treatment (with data for miR-249 shown in Fig. 2b).
As ZEB1 and ZEB2 were target genes of the miRNA-200 family, we then assayed the ZEB1 and ZEB2 mRNA expression 24 h after busulfan treatment. We found that expression of both ZEB1 and ZEB2 was significantly decreased by busulfan treatment in both cell lines (Fig. 2c). Taken together, our data suggest that busulfan upregulated the miRNA-200 family and modulated its target genes in osteosarcoma cells.
Downregulating miRNA-200 family or upregulating ZEB1 and ZEB2 reversed anti-osteosarcoma effect of busulfan
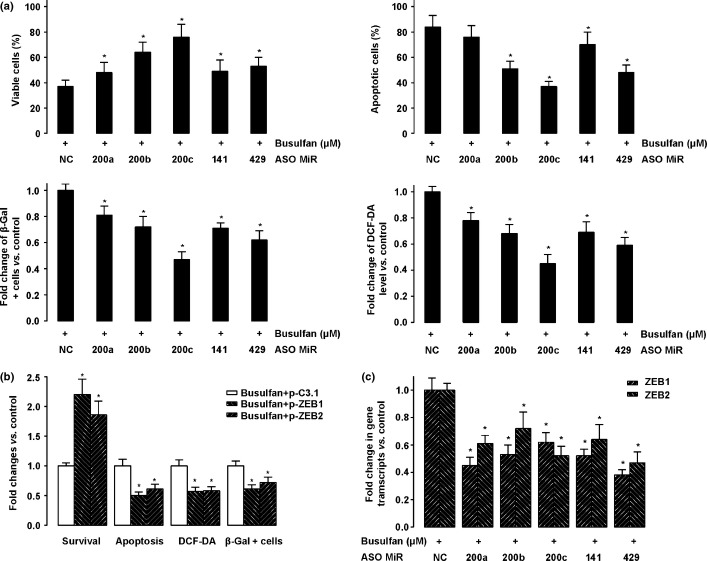
To define whether busulfan produced its anti-osteosarcoma effect through downregulation of the miRNA-200 family and by modulation of expression of its downstream targets ZEB1 and ZEB2, we inhibited the expression of miR-200a, miR-200b, miR-200c, miR-141, and miR-429, or overexpressed ZEB1 and ZEB1, by miRNA antisense oligonucleotide transfection or plasmid transfection (p-ZEB1 or p-ZEB2), respectively. We found that downregulation of all five members in the miR-200 family reversed the effect induced by busulfan in terms of cell viability, proliferation, level of ROS apoptosis, and cell senescence (Fig. 3a). Similarly, we found that upregulation of ZEB1 or ZEB2 expression also reversed the anti-osteosarcoma effect of busulfan (Fig. 3b), without affecting the expression levels of the miR-200 family (data not shown). Of note, downregulation of all five members in the miR-200 family resulted in upregulation of ZEB1 and ZEB2 (Fig. 3c). Taken together, these loss-of-function experiments thus indicate that busulfan may have its anti-osteosarcoma effect by upregulating ZEB1 and ZEB2 through activating the miRNA-200 family.
Figure 3.
Downregulating the microRNA-200 (miR-200) family or upregulating ZEB1 and ZEB2 reversed the anti-osteosarcoma effect of busulfan. (a) U2OS cells were transfected with antisense oligonucleotides (ASO) of miR-200a, miR-200b, miR-200b, miR-141, and miR-429 to downregulate the expression of miRNAs. The percentage of viable cells, apoptosis, β-gal-positive cells and 2'-7'-Dichlorodihydrofluorescein diacetate (DCF-DA) levels were assayed after 24 h. All data were normalized to negative control (NC). (b) MG-63 cells were transfected with pcDNA3.1-ZEB1 or pcDNA3.1-ZEB2, two overexpressing plasmids. The percentage of viable cells, apoptosis, β-gal-positive cells and DCF-DA levels were assayed after 24 h. (c) Downregulation of all five members in the miR-200 family by ASO resulted in upregulation of ZEB1 and ZEB2. *P < 0.05.
Upregulating miRNA-200 family, or inhibiting ZEB1 and ZEB2, partly mimicked the effect of busulfan
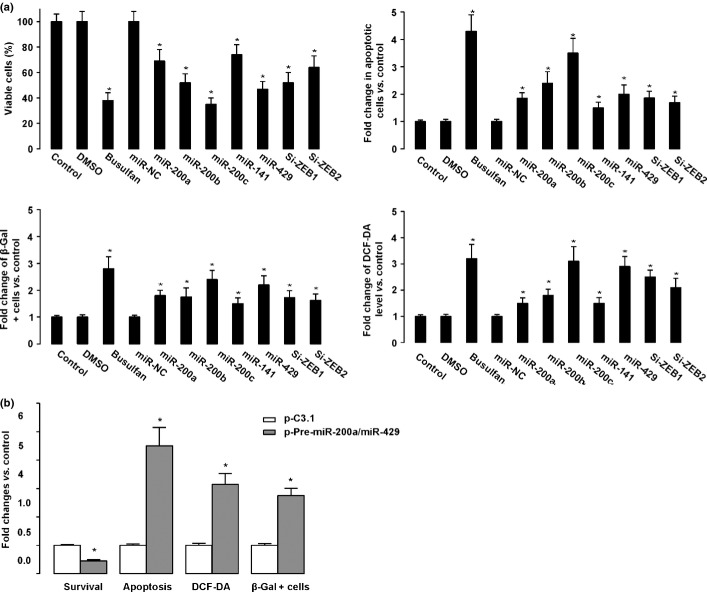
In order to find out whether upregulating the miRNA-200 family, or inhibiting ZEB1 and ZEB2, may produce the anti-osteosarcoma effect without the need of busulfan, we overexpressed miR-200a, miR-200b, miR-200c, miR-141, and miR-429, or inhibited ZEB1 and ZEB2, alone (without treatment of busulfan) in osteosarcoma cells. The anti-osteosarcoma effects of busulfan were partially mimicked by these approaches (Fig. 4). These gain-of-function experiments thus strengthened our findings and further showed that busulfan may produce its anti-osteosarcoma effect by upregulating ZEB1 and ZEB2 through activating the miRNA-200 family.
Figure 4.
Upregulating the microRNA-200 (miR-200) family, or inhibiting ZEB1 and ZEB2, partly mimicked the effect of busulfan in osteosarcoma cell lines. (a) U2OS cells were transfected with mimics of miR-200a, miR-200b, miR-200b, miR-141, and miR-429 to upregulate miRNAs. The percentage of viable cells, apoptosis, β-gal-positive cells, and 2'-7'-Dichlorodihydrofluorescein diacetate (DCF-DA) levels were assayed after 24 h. All data were normalized to negative control. (b) MG-63 cells were transfected with siRNA for ZEB1 and ZEB2 to downregulate the expression of ZEB1 and ZEB2. The percentage of viable cells, apoptosis, β-gal-positive cells, and DCF-DA levels were assayed after 24 h. All data were normalized to negative control (NC). *P < 0.05.
Busulfan inhibited osteosarcoma growth in vivo in a mouse model
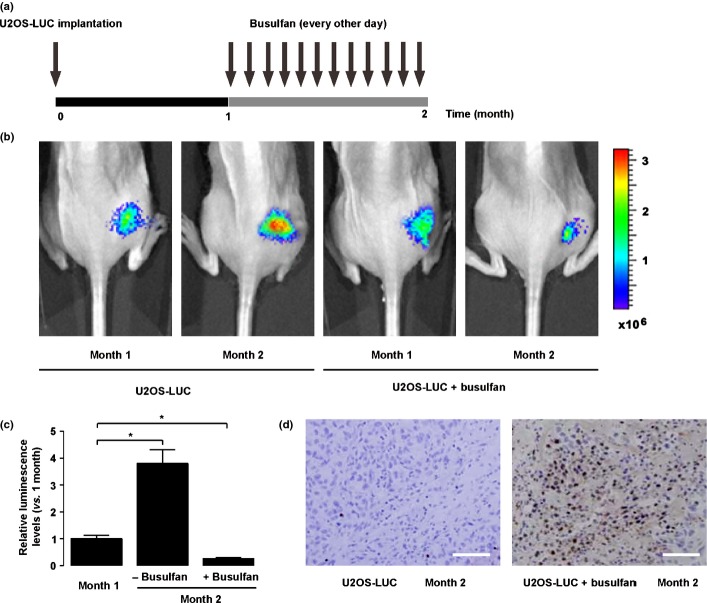
In order to evaluate the therapeutic effect of busulfan on an osteosarcoma in vivo, we generated a mouse model by orthotopically injecting luciferase-transfected U2OS (U2OS-LUC) into the tibia of nude mice, as it is one of the most common locations of primary osteosarcoma in humans. Tumor growth was monitored by bioluminescence imaging (Fig. 5a). The bioluminescence levels in the mice without busulfan treatment increased by 3.8 ± 0.5% fold, whereas the bioluminescence levels in the mice that received busulfan treatment decreased by 74.3 ± 11.6%, in 1 month (Fig. 5b,c), suggesting that the tumor growth was significantly inhibited in the mice that received busulfan. The TUNEL assay further confirmed that busulfan induced significant apoptosis of osteosarcoma cells in vivo (Fig. 5d). Taken together, busulfan inhibited osteosarcoma growth in vivo.
Figure 5.
Busulfan inhibited osteosarcoma growth in vivo in a mouse model. (a) In order to evaluate the therapeutic effect of busulfan on osteosarcoma in vivo, we generated a mouse model by orthotopically injecting luciferase-transfected U2OS (U2OS-LUC) into the tibia of nude mice. One month later, busulfan was i.v. injected into the mice every other day for another month. (b,c) Tumor growth was monitored by bioluminescence imaging. We found a continuous growth of the implanted tumor in the control mice, but the tumor growth was significantly inhibited in the mice that received busulfan, shown by representative images (b), and by quantification (c). (d) TUNEL assay further confirmed that busulfan induced significant apoptosis of osteosarcoma cells in vivo. Scale bar = 40 μm.
Discussion
Busulfan was used as a palliative treatment for CML. Busulfan is now widely used as an alternative to total body irradiation in conditioning therapy for hematopoietic stem cell transplantation. However, there are no reports on the therapeutic effect of busulfan on osteosarcoma. Here, we found that busulfan induced osteosarcoma cell death in vitro. Although a previous report has shown that some osteosarcoma cell lines resisted cell death induced by 1000 μM busulfan,(21) we showed that at a concentration of 100 μM, busulfan induced significant cell death in some osteosarcoma cell lines. This discrepancy may largely result from the different cell lines that had been used in the experiments. In our study, U2OS and MG-63 were used; in the previous report, the authors used OST and MNNG-HOS cell lines.
Here we showed that busulfan reduced cell viability and proliferation, and induced cell apoptosis, senescence, and ROS levels by upregulating five members in the miR-200 family, and subsequently inhibiting their target genes, ZEB1 and ZEB2. The miR-200 family has been shown to modulate cellular motility and control “stemness” and apoptosis.(5–12) Our further analyses showed that busulfan produced its anti-osteosarcoma effect by upregulating the miRNA-200 family, which subsequently downregulated its target genes ZEB1 and ZEB2, in a series of loss-of-function and gain-of-function experiments. Furthermore, treatment with busulfan potentially inhibited the growth of implanted osteosarcomas in nude mice.
It has been previously shown that knockdown of ZEB2 by siRNA suppressed cell proliferation, migration, and invasion, as well as induced cell apoptosis in glioma cells. Furthermore, ZEB downregulation was accompanied by decreased expression of CDK4/6, cyclin D1, cyclin E, E2F1, and c-myc, while p15 and p21 were upregulated. Several apoptosis-related regulators such as caspase-3, -6, and -9, and cleaved poly(ADP-ribose) polymerase were activated after ZEB siRNA treatment.(22) Therefore, these mechanisms may also explain what we have found in the current study.
Our finding is consistent with previous reports on the abnormal expression of miRNAs in osteosarcoma.(5–12) Moreover, overexpression of miR-181a, miR-181b, and miR-181c, and decreased expression of miR-16, miR-29b, and miR-142-5p have been also reported to be related to osteosarcoma.(11) further investigation is needed to elucidate the exact roles of these miRNAs during the pathogenesis of osteosarcoma.
Reactive oxygen species play double-edged roles in cancer. They facilitate the growth of cancer cells through activation of receptor tyrosine kinases,(23) supported by the data from knockout animals lacking antioxidant enzymes or repair enzymes.(17) However, high ROS can suppress tumor growth through the sustained activation of cell-cycle inhibitor.(17) In our study, busulfan-induced ROS seem to contribute to tumor suppression.
Taken together, our data suggest that busulfan may produce its anti-osteosarcoma effect by downregulating ZEB1 and ZEB2 through activating the miRNA-200 family, highlighting the possibility of using busulfan as a novel therapy for osteosarcoma.
Acknowledgments
This work was supported by internal funding from the 169th Hospital of Hunan Normal University, the Third Military Medical University, and Jinling Hospital, Nanjing University School of Medicine.
Disclosure Statement
The authors have no conflict of interest.
References
- 1.Yang J, Zhang W. New molecular insights into osteosarcoma targeted therapy. Curr Opin Oncol. 2013;25:398–406. doi: 10.1097/CCO.0b013e3283622c1b. [DOI] [PubMed] [Google Scholar]
- 2.Tsuchiya H, Tomita K, Mori Y, et al. Caffeine-assisted chemotherapy and minimized tumor excision for nonmetastatic osteosarcoma. Anticancer Res. 1998;18(1B):657–66. [PubMed] [Google Scholar]
- 3.Ponten J, Saksela E. Two established in vitro cell lines from human mesenchymal tumours. Int J Cancer. 1967;2:434–47. doi: 10.1002/ijc.2910020505. [DOI] [PubMed] [Google Scholar]
- 4.Billiau A, Edy VG, Heremans H, et al. Human interferon: mass production in a newly established cell line, MG-63. Antimicrob Agents Chemother. 1977;12(1):11–5. doi: 10.1128/aac.12.1.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim VN, Han J, Siomi MC. Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol. 2009;10:126–39. doi: 10.1038/nrm2632. [DOI] [PubMed] [Google Scholar]
- 6.Shukla GC, Singh J, Barik S. MicroRNAs: processing, maturation, target recognition and regulatory functions. Mol Cell Pharmacol. 2011;3(3):83–92. [PMC free article] [PubMed] [Google Scholar]
- 7.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136(2):215–33. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6(11):857–66. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 9.Miao J, Wu S, Peng Z, Tania M, Zhang C. MicroRNAs in osteosarcoma: diagnostic and therapeutic aspects. Tumour Biol. 2013;34(4):2093–8. doi: 10.1007/s13277-013-0940-7. [DOI] [PubMed] [Google Scholar]
- 10.Zhou G, Shi X, Zhang J, Wu S, Zhao J. MicroRNAs in osteosarcoma: from biological players to clinical contributors, a review. J Int Med Res. 2013;41:1–12. doi: 10.1177/0300060513475959. [DOI] [PubMed] [Google Scholar]
- 11.Jones KB, Salah Z, Del Mare S, et al. miRNA signatures associate with pathogenesis and progression of osteosarcoma. Cancer Res. 2012;72(7):1865–77. doi: 10.1158/0008-5472.CAN-11-2663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liang W, Gao B, Fu P, Xu S, Qian Y, Fu Q. The miRNAs in the pathgenesis of osteosarcoma. Front Biosci. 2013;18:788–94. doi: 10.2741/4142. [DOI] [PubMed] [Google Scholar]
- 13.Goldman JM. Chronic myeloid leukemia: a historical perspective. Semin Hematol. 2010;47:302–11. doi: 10.1053/j.seminhematol.2010.07.001. [DOI] [PubMed] [Google Scholar]
- 14.Deininger MW. Chronic myeloid leukemia: an historical perspective. Hematology Am Soc Hematol Educ Program. 2008;1:418. doi: 10.1182/asheducation-2008.1.418. [DOI] [PubMed] [Google Scholar]
- 15.Strober W. Trypan blue exclusion test of cell viability. Curr Protoc Immunol. 2001 doi: 10.1002/0471142735.ima03bs21. Appendix 3: Appendix 3B. doi: 10.1002/0471142735.ima03bs21. [DOI] [PubMed] [Google Scholar]
- 16.Lu J, Wen M, Huang Y, et al. C2ORF40 suppresses breast cancer cell proliferation and invasion through modulating expression of M phase cell cycle genes. Epigenetics. 2013;8:571–83. doi: 10.4161/epi.24626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yu J, Liu C, Zheng G, et al. Pseudolaric acid B induced cell cycle arrest, autophagy and senescence in murine fibrosarcoma l929 cell. Int J Med Sci. 2013;10:707–18. doi: 10.7150/ijms.5726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Eruslanov E, Kusmartsev S. Identification of ROS using oxidized DCFDA and flow-cytometry. Methods Mol Biol. 2010;594:57–72. doi: 10.1007/978-1-60761-411-1_4. [DOI] [PubMed] [Google Scholar]
- 19.Ramsey MR, Sharpless NE. ROS as a tumour suppressor? Nat Cell Biol. 2006;8(11):1213–5. doi: 10.1038/ncb1106-1213. [DOI] [PubMed] [Google Scholar]
- 20.Liu Y, Sharpless NE. Tumor suppressor mechanisms in immune aging. Curr Opin Immunol. 2009;21(4):431–9. doi: 10.1016/j.coi.2009.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lanvers-Kaminsky C, Bremer A, Dirksen U, Jurgens H, Boos J. Cytotoxicity of treosulfan and busulfan on pediatric tumor cell lines. Anticancer Drugs. 2006;17(6):657–62. doi: 10.1097/01.cad.0000215059.93437.89. [DOI] [PubMed] [Google Scholar]
- 22.Qi S, Song Y, Peng Y, et al. ZEB2 mediates multiple pathways regulating cell proliferation, migration, invasion, and apoptosis in glioma. PLoS ONE. 2012;7(6):e38842. doi: 10.1371/journal.pone.0038842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Irani K, Xia Y, Zweier JL, et al. Mitogenic signaling mediated by oxidants in Ras-transformed fibroblasts. Science. 1997;275(5306):1649–52. doi: 10.1126/science.275.5306.1649. [DOI] [PubMed] [Google Scholar]