Abstract
To test the hypothesis that population subdivision into small demes promotes both rapid speciation and evolutionary changes in gene arrangement by inbreeding and drift, we estimated rates of speciation and rates of chromosomal evolution in 225 genera of vertebrates. Rates of speciation were estimated by considering the number of living species in each genus and the fossil record of each genus as well as information about extinction rates. Speciation rate was strongly correlated with rate of chromosomal evolution and average rates of speciation in lower vertebrate genera were one-fifth those in mammalian genera. Genera with high karyotypic diversity and rapid speciation rates may generally have small effective population size (Ne), whereas large Ne values may be associated with karyotypically uniform genera and slow rates of speciation. Speciation and chromosomal evolution seem fastest in those genera with species organized into clans or harems (e.g., some primates and horses) or with limited adult vagility and juvenile dispersal, patchy distribution, and strong individual territoriality (e.g., some rodents). This is consistent with the above hypothesis regarding the evolutionary importance of demes.
Full text
PDF
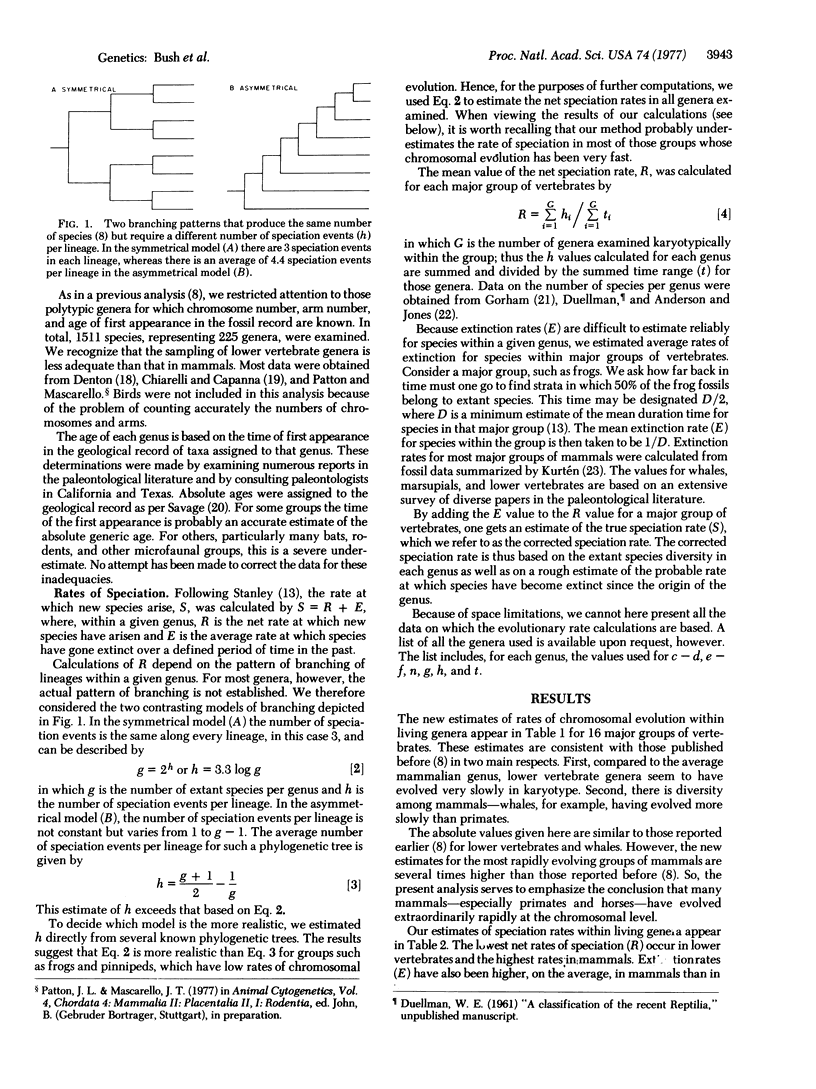



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Arnason U The role of chromosomal rearrangement in mammalian speciation with special reference to Cetacea and Pinnipedia. Hereditas. 1972;70(1):113–118. [PubMed] [Google Scholar]
- Bickham J. W., Baker R. J. Chromosome homology and evolution of emydid turtles. Chromosoma. 1976 Feb 23;54(3):201–219. doi: 10.1007/BF00293451. [DOI] [PubMed] [Google Scholar]
- Cappana E., Cristaldi M., Perticone P., Rizzoni M. Identification of chromosomes involved in the 9 Robertsonian fusions of the Apennine mouse with a 22-chromosome karyotype. Experientia. 1975 Mar 15;31(3):294–296. doi: 10.1007/BF01922545. [DOI] [PubMed] [Google Scholar]
- Feist J. D., McCullough D. R. Behavior patterns and communication in feral horses. Z Tierpsychol. 1976 Aug;41(4):337–371. doi: 10.1111/j.1439-0310.1976.tb00947.x. [DOI] [PubMed] [Google Scholar]
- Gropp A., Winking H., Zech L., Müller H. Robertsonian chromosomal variation and identification of metacentric chromosomes in feral mice. Chromosoma. 1972;39(3):265–288. doi: 10.1007/BF00290787. [DOI] [PubMed] [Google Scholar]
- Jotterand M. Le polymorphisme chromosomique des Mus (Leggadas) africains. Cytogénétique, zoogéographie, évolution. Rev Suisse Zool. 1972;79(1):287–359. [PubMed] [Google Scholar]
- Levin D. A., Wilson A. C. Rates of evolution in seed plants: Net increase in diversity of chromosome numbers and species numbers through time. Proc Natl Acad Sci U S A. 1976 Jun;73(6):2086–2090. doi: 10.1073/pnas.73.6.2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M., Fuerst P. A., Chakraborty R. Testing the neutral mutation hypothesis by distribution of single locus heterozygosity. Nature. 1976 Aug 5;262(5568):491–493. doi: 10.1038/262491a0. [DOI] [PubMed] [Google Scholar]
- Reig O. A., Kiblisky P. Chromosome multiformity in the genus Ctenomys (Rodentia, Octodontidae). A progress report. Chromosoma. 1969;28(2):211–244. doi: 10.1007/BF00331531. [DOI] [PubMed] [Google Scholar]
- Selander R. K. Behavior and genetic variation in natural populations. Am Zool. 1970 Feb;10(1):53–66. doi: 10.1093/icb/10.1.53. [DOI] [PubMed] [Google Scholar]
- Stanley S. M. A theory of evolution above the species level. Proc Natl Acad Sci U S A. 1975 Feb;72(2):646–650. doi: 10.1073/pnas.72.2.646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struhsaker T. T. Correlates of ecology and social organization among African cercopithecines. Folia Primatol (Basel) 1969;11(1):80–118. doi: 10.1159/000155259. [DOI] [PubMed] [Google Scholar]
- Thaeler C. S., Jr Karyotypes of sixteen populations of the Thomomys talpoides complex of pocket gophers (Rodentia-Geomyidae). Chromosoma. 1968;25(2):172–183. doi: 10.1007/BF00327176. [DOI] [PubMed] [Google Scholar]
- Wahrman J., Goitein R., Nevo E. Mole rat Spalax: evolutionary significance of chromosome variation. Science. 1969 Apr 4;164(3875):82–84. doi: 10.1126/science.164.3875.82. [DOI] [PubMed] [Google Scholar]
- White M. J. Models of speciation. New concepts suggest that the classical sympatric and allopatric models are not the only alternatives. Science. 1968 Mar 8;159(3819):1065–1070. doi: 10.1126/science.159.3819.1065. [DOI] [PubMed] [Google Scholar]
- Wilson A. C., Bush G. L., Case S. M., King M. C. Social structuring of mammalian populations and rate of chromosomal evolution. Proc Natl Acad Sci U S A. 1975 Dec;72(12):5061–5065. doi: 10.1073/pnas.72.12.5061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson A. C., Carlson S. S., White T. J. Biochemical evolution. Annu Rev Biochem. 1977;46:573–639. doi: 10.1146/annurev.bi.46.070177.003041. [DOI] [PubMed] [Google Scholar]
- Wilson A. C., Sarich V. M., Maxson L. R. The importance of gene rearrangement in evolution: evidence from studies on rates of chromosomal, protein, and anatomical evolution. Proc Natl Acad Sci U S A. 1974 Aug;71(8):3028–3030. doi: 10.1073/pnas.71.8.3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright S. Evolution in Mendelian Populations. Genetics. 1931 Mar;16(2):97–159. doi: 10.1093/genetics/16.2.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurster-Hill D. H., Gray C. W. The interrelationships of chromosome banding patterns in procyonids, viverrids, and felids. Cytogenet Cell Genet. 1975;15(5):306–331. doi: 10.1159/000130528. [DOI] [PubMed] [Google Scholar]
- Zieg J., Silverman M., Hilmen M., Simon M. Recombinational switch for gene expression. Science. 1977 Apr 8;196(4286):170–172. doi: 10.1126/science.322276. [DOI] [PubMed] [Google Scholar]


