Abstract
This study found that long-term exposure of chronic myelogenous leukemia (CML) K562 cells to BCR/ABL thyrosine kinase inhibitors (TKI) caused drug-resistance in association with an increase in levels of DNA methyltransferases (DNMT) and a decrease in levels of microRNA miR-217. These observations are clinically relevant; an increase in levels of DNMT3A in association with downregulation of miR-217 were noted in leukemia cells isolated from individuals with BCR/ABL TKI-resistant Philadelphia chromosome positive acute lymphoblastic leukemia (Ph+ ALL) and CML. Further studies with TKI-resistant K562 cells found that forced expression of miR-217 inhibited expression of DNMT3A through a miR-217-binding site within the 3′-untranslated region of DNMT3A and sensitized these cells to growth inhibition mediated by the TKI. Of note, long-term exposure of K562 cells to dasatinib (10 nM) together with 5-Aza-2′-deoxycytidine (5-AzadC) (0.1 μM) potently inhibited proliferation of these cells in association with upregulation of miR-217 and downregulation of DNMT3A in vitro. In addition, a decrease in levels of DNMT3A and an increase in levels of miR-217 were noted in K562 tumors growing in immune-deficient mice that were treated with the combination of 5-AzadC and dasatinib. Taken together, Ph+ leukemia cells acquire TKI resistance via downregulation of miR-217 and upregulation of DNMT3A. Inhibition of DNMT3A by forced expression of miR-217 or 5-AzadC may be useful to prevent drug resistance in individuals who receive TKI.
Keywords: 5-AzadC, chronic myelogenous leukemia, dasatinib, DNA methyltransferases, miR-217
More potent second-line BCR/ABL tyrosine kinase inhibitors (TKI) such as nilotinib and dasatinib are approved as the first line chemotherapeutic agent for individuals with chronic myelogenous leukemia (CML) and Philadelphia chromosome positive acute lymphoblastic leukemia (Ph+ ALL).1–3 However, patients treated with the second-line TKI also experience resistance to TKI.4,5 Thus, the elucidation of the molecular mechanism of TKI resistance and novel treatment strategies to prevent and overcome TKI resistance are urgently needed.
Our previous studies showed that long-term exposure of chronic eosinophilic leukemia (CEL) cells to imatinib significantly increased levels of DNA methyltransferases (DNMT) and polycomb group (PcG) proteins in parallel with induction of hypermethylation of the promoter region of the phosphatase and tensin homolog deleted on chromosome 10 (PTEN), and these cells became resistant to imatinib-mediated growth inhibition.6 Expression of PTEN recovered when these cells were cultured in the presence of DNMT inhibitor 5-Aza-2′-deoxycytidine (decitabine, 5-AzadC), suggesting that anti-epigenetic agents could be useful for individuals who receive tyrosine kinase inhibitors in overcoming drug resistance.7 5-AzadC acts as a hypometylating agent in low concentrations; in contrast, higher concentrations of 5-AzadC can cause DNA-damage and apoptosis in cancer cells.8,9 5-AzadC and azacytidine are approved for the treatment of myelodysplastic syndromes.10 Epigenetic mechanisms are attributed to heritable changes in gene expression that occur in a cell without changes in their genomic sequence.11 Epigenetic mechanisms regulating gene expression include DNA methylation and histone modification. DNMT such as DNMT1, DNMT3A and DNMT3B are key epigenetic regulators involved in transcriptional repression.12–14
Recent studies show that microRNA (miRNA) negatively regulate epigenetic regulators such as DNMT and histone deacetylase (HDAC) in leukemia cells.15–18 miRNA are noncoding RNA of 19–25 nucleotides in length that regulate gene expression by inducing translational inhibition or cleavage of their target mRNA through base pairing at partially or fully complementary sites.19 The mature miRNA are processed, after a complex but well-established biogenesis, from a larger stem loop precursor (pri-miRNA) and negatively regulate gene expression at the post-transcriptional level. When mature miRNA bind to the 3′-untranslated region (3′-UTR) of the target mRNA with entire complementarity, the miRNA can lead to degradation of its target mRNA with partial complementarity, in this case inducing a translational repression of its target gene.20–22 For example, enforced expression of miR-29b in acute myeloid leukemia cells resulted in marked reduction of the expression of DNA methyltransferases DNMT1, DNMT3A and DNMT3B at both RNA and protein levels. This, in turn, led to a decrease in global DNA methylation and re-expression of p15 (INK4b) via promoter DNA hypomethylation. miRNA are also involved in the control of DNA methylation by targeting the DNA methylation machinery. The miR-29 family directly targets DNMT3A and 3B in lung cancer,16 thereby leading to downregulation of these genes, reduction of global DNA methylation, and re-expression of the hypermethylated DNA, and silenced tumor suppressor genes FHIT and WWOX.
In this study, we examined the relationship between TKI resistance and miRNA and found that long-term exposure of CML K562 cells to ABL TKI such as dasatinib and nilotinib decreased the levels of miR-217 and increased the levels of DNMT1 and DNMT3A, as well as resulting in acquisition of TKI resistance. Low concentration of 5-Aza-2′-deoxycytidine in combination with dasatinib potently inhibited the proliferation of ph+ leukemia cells in vitro and in vivo.
Materials and Methods
Cells
Bcr/Abl-expressing K562 cells were obtained from the American Type Culture Collection (Manassas, VA, USA). Dasatinib-resistant K562 (designated as K562DR) and nilotinib-resistant K562 (designated as K562NR) cell lines were established by culturing with increasing concentrations of dasatinib (10 nM) or nilotinib (100 nM) for 12 months. Leukemic bone marrow-mononuclear cells (BM-MNC) were isolated from individuals with Ph+ ALL (case # 1, n = 1) and CML (case # 2–7, n = 7) after obtaining informed consent with Kochi University Institutional Review Board approval. The characteristics of the patients are listed in Table1.
Table 1.
Characteristics of patients
| Patient # | Disease | Treatment | Drug response | Time to relapse |
|---|---|---|---|---|
| 1 | Ph+ ALL | Dasatinib 100 mg | Relapse | 3 months |
| 2 | CML | Dasatinib 50 mg | Loss of CHR | 4 months |
| 3 | CML | Dasatinib 75 mg | MMR | – |
| 4 | CML | Dasatinib 100 mg | MMR | – |
| 5 | CML | Nilotinib 600 mg | MMR | – |
| 6 | CML | Imatinib 100 mg | MMR | – |
| 7 | CML | Dasatinib 100 mg | MMR | – |
| 8 | CML | Dasatinib 100 mg | MMR | – |
CHR, complete hematologic response; CML, chronic myeloid leukemia; MMR, major molecular response; Ph+ ALL, Philadelphiachromosome-positive acute lymphoblastic leukemia.
Chemical
Nilotinib was provided by Novartis (Basel, Switzerland). Dasatinib was provided by Bristol-Myers Squibb (NY, USA). DNMT inhibitor 5-AzadC was purchased from Sigma (St. Louis, MO, USA). These reagents were dissolved in 100% DMSO to a stock concentration 10 mM and stored at −80°C.
MTT assay
Cells (3 × 105/mL) were cultured with various concentrations of the indicated agents. After 24–96 h, cell viability was measured by MTT assay as previously described.7 All experiments were performed in triplicate and repeated at least three times.
RNA isolation and reverse transcription-polymerase chain reaction
RNA isolation and cDNA preparation were performed as described previously.7 We measured expression of 18S for normalization as previously described.7 Real-time PCR was carried out by using a Power SYBR Green PCR Master Mix (Applied Biosystems, Warrington, UK) as described previously.7 Primers for PCR are shown in Table2.
Table 2.
PCR primers
| Gene | Direction | Primer |
|---|---|---|
| DNMT1 | Forward | 5′-ACCGCTTCTACTTCCTCGAGGCCTA-3′ |
| Reverse | 5′-GTTGCAGTCCTCTGTGAACACTGTGG-3′ | |
| DNMT3A | Forward | 5′-CACACAGAAGCATATCCAGGAGTG-3′ |
| Reverse | 5′-AGTGGACTGGGAAACCAAATACCC-3′ | |
| DNMT3B | Forward | 5′-AATGTGAATCCAGCCAGGAAAGGC-3′ |
| Reverse | 5′-ACTGGATTACACTCCAGGAACCGT-3′ | |
| p21 | Forward | 5′-CGTGGTGGTGGTGAGCTAGA-3′ |
| Reverse | 5′-CTGTCTGCACCTTCGCTCCT-3′ | |
| BAX | Forward | 5′-AATCCCCGATTCATCTACCC-3′ |
| Reverse | 5′-AGGGCAGAAGGCACTAATCA-3′ | |
| 18S | Forward | 5′-AAACGGCTACCACATCCAAG-3′ |
| Reverse | 5′-CCTCCAATGGATCCTCGTTA-3′ |
Mice
Female immune deficient BALB/c nude mice of 4 weeks of age were purchased from JAPAN SLC (Shizuoka, Japan), and were maintained in pathogen-free conditions with irradiated chow. Animals were bilaterally injected s.c. with 1 × 107 K562 cells/tumor in 0.1 mL Matrigel (Collaborative Biomedical Products, Bedford, MA, USA) and 0.1 mL RPMI-1640 (Sigma). When K562 cells formed palpable tumors, mice were divided randomly into four groups receiving control (n = 9), or either dasatinib (n = 9) or 5-AzadC (n = 9), or a combination of both dasatinib and 5-AzadC (n = 9). Dasatinib (10 mg/kg) was given to mice i.p. three times over 2 weeks. The dose of these agents was determined by our preliminary studies (data not shown). 5-AzadC (0.1 mg/kg) was given to mice i.p. from Monday to Friday for 2 weeks.23 Treatment with 0.1 mg/kg/d 5-AzadC was well-tolerated and caused a moderate, reversible leukocytopenia.23 PBS was given to the untreated control mice. After 2 weeks, tumor tissue was removed from mice assigned to each group and washed to make a single cell. Collected 5 × 106 cells were implanted s.c. into the second recipient mice. When cells formed palpable tumors, each drug was given to mice i.p. over 2 weeks. These experiments were repeated five times. Body weight and tumors were measured twice a week. Tumor sizes were calculated using the formula a × b × c, where “a” is the length, “b” is the width and “c” is the height in millimeters. At the end of the experiment, animals were killed by CO2 asphyxiation and tumor weights were measured after their careful resection. Tumor tissue was collected for analysis. The experiments were approved by the Review Board of Kochi University.
Methylation analysis by methylation-specific PCR
DNA (300 ng) extracted from K562 tumors established in nude mice was used for bisulfite treatment done using the EZ DNA Methylation Kit (Zymo Research, Orange, CA, USA) according to the supplier's protocol. The primer sets used to amplify the promoter region of the p53, p16, p21waf1 and Bax genes are shown in Table3. Amplification was carried out in a Mycycler thermal cycler (Bio-Rad, Tokyo, Japan) at 94°C for 1 min, cycled at 98°C for 10 s, 60°C for 15 s and 68°C for 30 s (30 cycles).
Table 3.
Methylation analysis by methylation-specific PCR primers
| Gene | Direction | Primer |
|---|---|---|
| p21 M | Forward | 5′-TACGCGAGGTTTCGGGATC-3′ |
| Reverse | 5′-CCCTAATATACAACCGCCCCG-3′ | |
| p21 U | Forward | 5′-GACCCCGAACCGCGACCGTAA-3′ |
| Reverse | 5′-ACAACCCTAATATACAACCACCCCA-3′ | |
| BAX M | Forward | 5′-ATTAAATTTTTCGAGGGAGC-3′ |
| Reverse | 5′-GCTAAACGTACGTCCTTCAC-3′ | |
| BAX U | Forward | 5′-GGGATTAAATTTTTTGAGGGAGT-3′ |
| Reverse | 5′-CACTAAACATACATCCTTCACAT-3′ | |
| p53 M | Forward | 5′-TTCGGTAGGCGGATTATTTG-3′ |
| Reverse | 5′-AAATATCCCCGAAACCCAAC-3′ | |
| p53 U | Forward | 5′-TTGGTAGGTGGATTATTTGTTT-3′ |
| Reverse | 5′-CCAATCCAAAAAAACATATCAC-3′ | |
| p16 M | Forward | 5′-TTATTAGAGGGTGGGGCGGATCGC-3′ |
| Reverse | 5′-GACCCCGAACCGCGACCGTAA-3′ | |
| p16 U | Forward | 5′-TTATTAGAGGGTGGGGTGGATTGT-3′ |
| Reverse | 5′-CAACCCCAAACCACAACCATAA-3′ |
M, methylation; U, unmethylation.
Western blot analysis
Western blot analysis was performed as described previously.7 Protein concentrations were quantitated using a Bio-Rad assay (Bio-Rad Laboratories, Hercules, CA, USA). Proteins were resolved on a 10% SDS polyacrylamide gel, and transferred to an immobilon polyvinylidene difluoride membrane (Amersham, Arlington Heights, IL, USA). The membrane was probed sequentially with the antibodies. Anti-p21waf1 (Santa Cruz Biotechnology, Santa Cruz, CA, USA), Anti-Bax (Santa Cruz Biotechnology) and Anti-GAPDH (Abcam, Cambridge, UK, USA) antibodies were used.
Expression of miRNA
Expression of miRNA was analyzed using Mir-X miRNA qRT-PCR SYBR Kit (638314; Clontech Laboratories, Mountain View, CA, USA) according to the supplier's protocol. Levels of miRNA gene were normalized using the U6 (638314; Clontech Laboratories) and relative quantities were determined using the delta CT method. Primers for PCR are shown in Table2.
miR-217 vector
Lntiviral miR-217 expression vector and control vector were purchased from Biosettia (San Diego, CA, USA). These plasmids were transfected into K562DR cells by using FuGENE HD (Promega KK, Tokyo, Japan). After 48 h, medium containing puromycin (10 μg/mL) was replaced to select for stably transduced cells.
Small interfering RNA and transfections
Control small interfering (si)RNA and two siRNA against DNMT3A were purchased from Santa Cruz Biotechnology and Sigma (Deisenhofen, Germany), respectively. K562DR cells were transiently transfected with either control or DNMT3A siRNA (300 nM) by Amaxa electroporator Nucleofector II (Wako Pure Chemical Industries, Osaka, Japan), using the Nucleofector Kit V (program T-016) as previously described.7
Luciferase reporter assay for targeting DNMT3A 3′-UTR
For luciferase reporter experiments, a DNMT3A 3′-UTR segment of 898 bp were amplified by PCR from human genomic DNA (636401, Clontech, Heidelberg, Germany). Primers complementary to the published human DNMT3A 3′-UTR sequence containing NheI and XhoI restriction sites for the forward (GCGGCTAGCAGTCAGGGACTTGGCTCTCC) and reverse (GCGCTCGAGCCTGCATGAACATTAGGTTGG) primers, respectively, were synthesized. The PCR product and pGL4.10 [Luc2] vector (E6651, Promega, Madison, WI, USA) were digested with NheI (1241A, Takara Bio) and XhoI (1094A, Takara Bio) restriction endonucleases. The PCR product was ligated into the pGL4.10 [Luc2] vector using T4 DNA ligase (2011A, Takara Bio).We also generated the DNMT3A 3′-UTR mutant vector with 4 bp deletions (CAUG) in the binding site of miR-217 by using the PrimeSTAR Mutagenesis Basal Kit (Takara, Osaka, Japan). These plasmids were transfected into K562DR cells by using FuGENE HD (Promega KK). After 48 h, cell lysate luciferase activity was measured using the Dual-Luciferase assay system (Promega). Lysate luciferase activity was normalized to that of Renilla luciferase, which was used as a control.
Statistical analysis
When comparing two groups, Student's t-test was used. All statistical analyses were carried out using spss software (Version 11.03; SPSS Inc., Tokyo, Japan) and the results were considered to be significant when the P-value was <0.05, and highly significant when the P-value was <0.01.
Results
Long-term exposure of dasatinib and nilotinib increased levels of DNMT3A in K562 cells
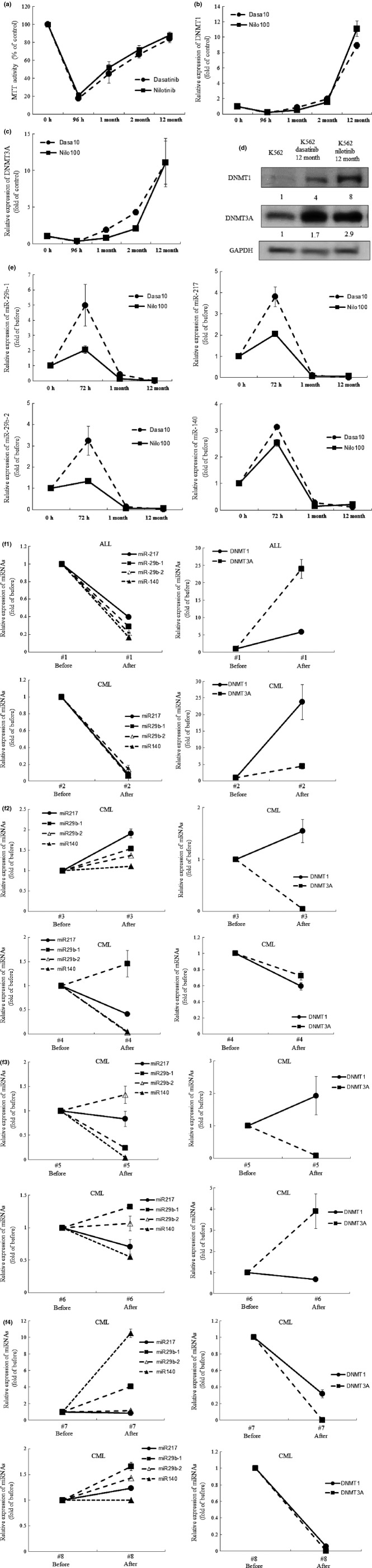
Exposure of CEL cells to low concentration of nilotinib (10 nM, 96 h) or dasatinib (1 nM, 96 h) increased the levels of DNMT proteins.6 Similarly, exposure of K562 cells to low concentration of dasatinib (1 nM, 48–96 h) or nilotinib (10 nM, 48–96 h) increased the levels of DNMT (DNMT1, DNMT3A and DNMT3B) (data not shown). This dose of dasatinib (1 nM, 96 h) or nilotinib (10 nM, 96 h) was not able to inhibit the proliferation of these cells (data not shown). However, exposure of K562 cells to 10-fold higher concentration of dasatinib (10 nM, 96 h) or nilotinib (100 nM, 96 h) inhibited the proliferation of K562 cells and decreased levels of DNMT1 and DNMT3A (Fig.1a,b), but not DNMT3B (data not shown). Intriguingly, when K562 cells were cultured in the presence of this dose of TKI (dasatinib 10 nM, nilotinib 100 nM) for a long period, up to 12 months, these cells lost sensitivity to TKI. In parallel, levels of DNMT1 and DNMT3A were elevated (Fig.1c,d). We next compared the DNA methylation status on the promoter regions of cell cycle regulator and proapoptotic genes between K562 and dasatinib-resistant K562 (K562DR) cells by using methylation-specific PCR. It was found that DNA methylaton on the promoter region of p21waf1 and BAX was increased in K562DR cells as compared with that in K562 cells (Fig. S1).
Figure 1.
(Next page) Thyrosine kinase inhibitors (TKI) increases levels of DNA methyltransferases (DNMT). (a) MTT assay. K562 cells were plated in 96-well plates and cultured with dasatinib (10 nM) or nilitinib (100 nM). At the indicated time point, their proliferation was measured by MTT assay. Results represent the mean ± SD of three experiments performed in triplicate. (b, c) Real-time RT-PCR. RNA was extracted from K562 cells. cDNA were synthesized and subjected to real-time RT-PCR to measure the levels of the indicated gene. Results represent the mean ± SD of three experiments performed in triplicate. The statistical significance was assessed using a paired t-test. **P < 0.01; *P < 0.05. (d) Western blot analysis. These cells were harvested, and subjected to western blot analysis to monitor the levels of the indicated proteins. Each lane was loaded with 30 μg of whole protein lysate. Band intensities were quantified using ImageJ software (Wayne Rasband, NIH). (e) Expression of miRNA. Expression of miRNA of K562 cells treated with dasatinib or nilotinib (0–12 months) was analyzed using an Mir-X miRNA qRT-PCR SYBR Kit to measure the levels of the indicated gene. Results represent mean ± SD of duplicate cultures. (f) Real-time RT-PCR. RNA was extracted from bone marrow mononuclear cells isolated from Ph+ ALL (n = 1) and CML (n = 7) patients before and after treatment with TKI. cDNA were synthesized and subjected to real-time RT-PCR to measure the levels of the indicated genes. Results represent mean ± SD of duplicate cultures. **P < 0.01; *P < 0.05.
Long-term exposure of dasatinib and nilotinib decreased levels of miRNA in leukemia cells
We next measured levels of miRNA in K562 cells after exposure to TKI, as previous studies showed that miRNA regulated expression of DNMT.15–18 Interestingly, exposure of K562 cells to a cytotoxic dose of dasatinib (10 nM) or nilotinib (100 nM) for a short period (72 h) increased levels of miRNA, including miR-29b-1, miR-29b-2, miR-140 and miR-217 (Fig.1e). In contrast, long-term exposure of K562 cells to dasatnib (10 nM, 12 months) or nilotinib (100 nM, 12 months) decreased the levels of these miRNA (Fig.1e). Furthermore, we compared the levels of DNMT and miRNA in leukemia cells isolated from individuals with dasatinib-resistant Ph+ ALL and CML who were not able to take dasatinib every day because of thrombocytopenia, resulting in loss of complete hematologic response (CHR), with those in cells obtained at initial diagnosis (case #1 and 2 [Fig.1f]). Notably, the levels of DNMT1 and DNMT3A increased and the levels of miRNA decreased in BM-MNC isolated from case #1 at the time of relapse and case #2 at the time when the patient lost CHR (Fig.1f). In contrast, the levels of DNMT3A were not elevated in BM-MNC isolated from individuals with CML who remained on major molecular response by treatment with TKI, except for one case, case #6 (case #3–8 [Fig.1f]).24 These observations suggested the association of dysregulation of DNMT3A by miRNA and TKI resistance.
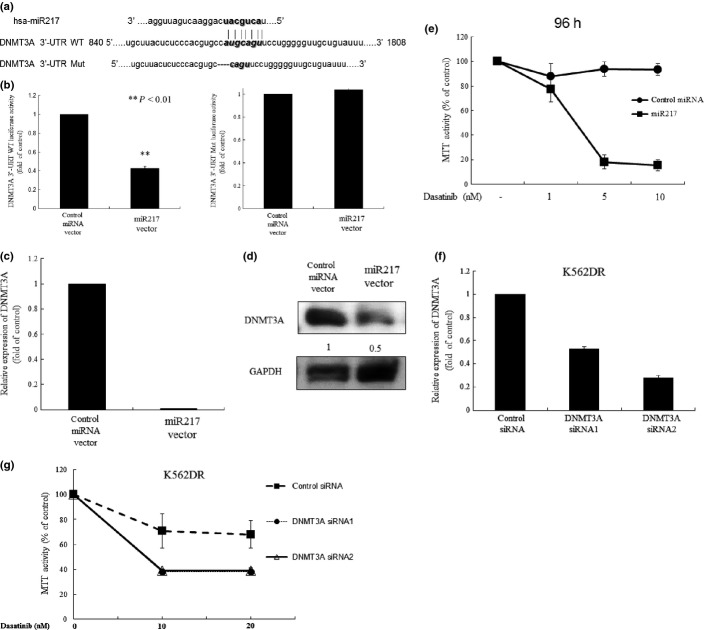
Forced expression of miR-217 in K562DR cells sensitized these cells to thyrosine kinase inhibitors via downregulation of DNMT3A
Recent studies show that miR-29b and miR-140 regulate expression of DNMT, including DNMT1 and DNMT3A.15–18 However, the role of miR-217 in the regulation of DNMT remains unknown. Computational miRNA target analysis from miRNA databases identified homology between miR-217 and the 3′-UTR of the human DNMT3A gene, but not DNMT1 genes, prompting us to elucidate the link between miR-217 and DNMT3A (Fig.2a). We explored the effect of miR-217 on transcriptional activity of DNMT3A by using DNMT3A 3′-UTR luciferase reporter vector (Fig.2b). The luciferase activity in miR-217 stably expressing K562DR cells were less than that in control miRNA transfected cells (Fig.2b, Fig. S2). We further deleted four nucleotides (CAUG) from the miR-217 binding site of DNMT3A 3′-UTR and transfected this mutant construct in miR-217 stably expressing K562DR cells (Fig.2b). This mutant abrogated the miR-217/DNMT3A interaction, as evidenced by unchanged luciferase activity (Fig.2b). These results suggest that miR-217 negatively regulates expression of DNMT3A in leukemia cells. In fact, downregulated levels of DNMT3A mRNA and protein were noted in miR-217 stably expressing K562DR cells (Fig.2c,d). Intriguingly, forced expression of miR-217 in K562DR cells sensitized these cells to dasatinib (5 or 10 nM, 96 h), as measured by the MTT assay (Fig.2e). To explore the involvement of the elevated levels of DNMT3A in the acquisition of drug resistance of K562DR cells, we suppressed DNMT3A by siRNA and tested the sensitivity of these cells to dasatinib. Both DNMT3A siRNA1 and DNMT3A siRNA2 effectively downregulated the levels of DNMT3A in K562DR cells (Fig.2f). As expected, DNMT3A-depleted K562DR cells regained sensitivity to dasatinib (Fig.2g).
Figure 2.
Forced expression of miR-217 sensitizes dasatinib-resistant K562 (K562DR) cells to dasatinib. (a) The sequence of miR-217 and its potential matching site in the DNMT3A 3′-UTR. DNMT3A 3′-UTR mutant vector with deletion of 4 bp (CAUG) in the binding site of miR-217 was generated by using the PrimeSTAR Mutagenesis Basal Kit. (b) Reporter gene assay. miR-217 overexpressed K562DR cells were transfected with either DNMT3A 3′-UTR WT or mutant luciferase reporter vector. The pRL-SV40-Luciferase (Renilla luciferase) vector was co-transfected for normalization. After 48 h, cells were harvested and subjected to the reporter assay. (c) Real-time RT-PCR. cDNA were synthesized and subjected to real-time RT-PCR to measure the levels of the indicated gene. Results represent the mean ± SD of three experiments performed in triplicate. The statistical significance was assessed using a paired t-test. **P < 0.01; *P < 0.05. (d) Western blot analysis. K562DR cells were transfected with control or miR-217 expression vector. These cells were subjected to western blot analysis to monitor the levels of the indicated proteins. Each lane was loaded with 30 μg of whole protein lysate. Band intensities were quantified using ImageJ software (Wayne Rasband, NIH). (e) MTT assay. K562DR cells were transfected with control or miR-217 expression vector. These cells were plated in 96-well plates and cultured with dasatinib (1, 5 or 10 nM). After 96, their proliferation was measured by MTT assay. Results represent the mean ± SD of three experiments performed in triplicate. Downregulation of DNMT3A in K562DR cells sensitizes these cells to dasatinib. (f) Real-time RT-PCR. K562DR cells were transiently transfected with either scrambled control or DNMT3A siRNA. RNA was extracted from these cells. cDNA were synthesized and subjected to real-time RT-PCR to measure the levels of the indicated gene. Results represent the mean ± SD of three experiments performed in triplicate. The statistical significance was assessed using a paired t-test. **P < 0.01; *P < 0.05. (g) MTT assay. K562DR cells were transiently transfected with either scrambled control or DNMT3A siRNA. These cells were plated in 96-well plates and cultured with dasatinib (10 nM). At the indicated time point, their proliferation was measured by MTT assay. Results represent the mean ± SD of three experiments performed in triplicate. Mut, mutation; WT, wild type.
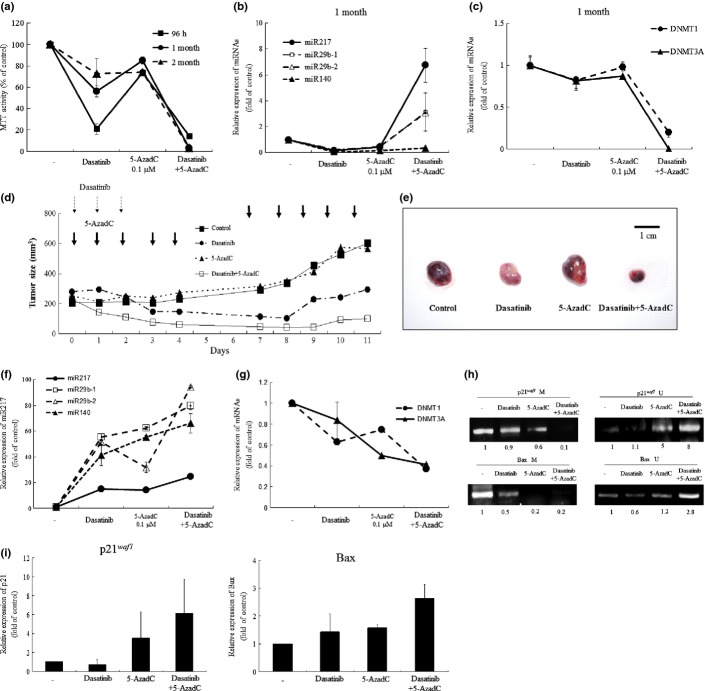
5-AzadC decreased levels of DNA methyltransferase and prevented dasatinib resistance in K562 cells
We next examined whether DNMT inhibitor 5-AzadC prevented resistance to dasatinib in K562 cells. K562 cells were cultured with either dasatinib (10 nM, 96 h – 2 months) and/or 5-AzadC (0.1 μM, 96 h – 2 months). The culture media were replaced and drugs were added every 5 days. 5-AzadC only slightly inhibited the proliferation of K562 cells. K562 cells lost their sensitivity to dasatinib after 2 months culture in the presence of dasatinib (Fig.3a). Notably, when K562 cells were cultured with a combination of dasatinib (10 nM) and 5-AzadC (0.1 μM), their proliferation was potently inhibited even after 2 months in parallel with upregulation of miR-29b-1, miR-29b-2 and miR-217 and downregulation of DNMT (Fig.3b,c).
Figure 3.
The effect of DNA methyltransferases (DNMT) inhibitor 5-AzadC on BCR/ABL tyrosine kinase inhibitors-resistance. (a) MTT assay. K562 cells were plated in 96-well plates and cultured with either dasatinib (10 nM) and/or 5-AzadC (0.1 μM). At the indicated time point, their proliferation was measured by MTT assay. Results represent the mean ± SD of three experiments performed in triplicate. (b, c) Real-time RT-PCR. RNA was extracted from K562 cells. cDNA were synthesized and subjected to real-time RT-PCR to measure the levels of the indicated gene. Results represent the mean ± SD of three experiments performed in triplicate. The statistical significance was assessed using a paired t-test. **P < 0.01; *P < 0.05. The combination of dasatinib and 5-AzadC blocks leukemic growth in the murine xenograft model. (d) When K562 tumors were palpable, mice were randomized into four groups (n = 9) and treatment was initiated. Dasatinib (10 mg/kg) was given to mice i.p. three times a week during a 2-week experimental period. 5-AzadC (0.1 mg/kg) was given to mice i.p. from Monday to Friday for 2 weeks. Each point represents the mean of nine tumors. (e) Tumor size at autopsy. After 2 weeks of treatment, tumors were removed. This experiment was repeated three times. (f, g) Real-time RT-PCR. RNA was extracted from K562 tumors. cDNA were synthesized and subjected to real-time RT-PCR to measure the levels of the indicated gene. Results represent the mean ± SD of three experiments performed in triplicate. The statistical significance was assessed using a paired t-test. **P < 0.01; *P < 0.05. The effects of dasatinib and 5-AzadC in K562 tumors. Methylation-specific PCR. (h) DNA was extracted from K562 tumor cells. DNA with methylated CpG was processed using the EZ DNA Methylatiohn Kit. The recovered DNA was amplified by PCR on methylation of the p21waf1, and BAX promoter. Band intensities were quantified using ImageJ software (Wayne Rasband, NIH). Real time RT-PCR. (i) RNA was extracted from tumor cells. cDNA were synthesized and subjected to real-time RT-PCR to measure the levels of p21waf1 and BAX. Results represent the mean ± SD of the three experiments performed. **P < 0.01; *P < 0.05.
The combination of dasatinib and 5-AzadC inhibited leukemic growth in vivo
To explore the therapeutic potential of the combination of dasatinib and 5-AzadC on proliferation of CML cells, BALB/c nude mice bearing CML tumors were treated with dasatinib (10 mg/kg, 1 time per day of on days 1–3) and/or 5-AzadC (0.1 mg/kg, 1 time per day of on days 1−5, 8−12). Dasatinib (10 mg/kg, 1 time per day of on days 1−3) alone inhibited the proliferation of K562 xenografts for 8 days (103 ± 80 mm3 [Fig.3d]); however, tumors started to regrow after 9 days (192 ± 178 mm3, Fig.3d). 5-AazadC (0.1 mg/kg) alone was not able to inhibit the proliferation of K562 xenografts over the 9 days compared with the control tumors (488 ± 259 vs 471 ± 112 mm3 [Fig.3d]). When mice were treated with both compounds at the same concentration, tumor growth was significantly inhibited even after 9 days (Fig.3d), resulting in a decrease in the size of K562 tumors at autopsy (Fig.3e). None of the treated mice showed signs of illness or significant weight loss (data not shown). Real-time PCR revealed that a combination of dasatinib and 5-AzadC decreased levels of DNMT1 and DNMT3A and increased levels of miR-217 in K562 tumor cells as compared with those in cells removed from mice treated by either dasatinib or 5-AzadC alone (Fig.3f,g). Moreover, we examined the DNA methylation status on the promoter region of cell cycle regulator genes p21waf1, p53 and p16, and proapoptotic gene BAX in K562 tumor cells removed from mice (Fig.3h) (data not shown). Notably, DNA methylation of the promoter region of p21waf1 and BAX, but not p53 and p16, was significantly reduced in parallel with upregulation of p21waf1 and BAX both at mRNA and protein levels after treatment with both dasatinib and 5-AzadC (Fig.3i and Fig. S3) (data not shown).
Discussion
We previously reported that long-term exposure of leukemia cells to imatinib induced activation of AKT, ERK and STAT5 signaling via epigenetic silencing of the PTEN gene.7 Similarly, long-term exposure of the K562 cells to TKI such as dasatinib and nilotinib failed to inactivate AKT and ERK signaling, although BCR/ABL was effectively dephosphorylated in these cells (Fig. S4), suggesting BCR/ABL-independent activation of these signal pathways in the TKI-resistant cells. The present study shows that long-term exposure of K562 cells to TKI increases the levels of DNMT1 and DNMT3A in parallel with a decrease in the levels of miRNA, including miR-217 and miR-29b, and these cells became resistant to growth inhibition mediated by TKI (Fig.1a–e). Notably, the levels of DNMT1 and DNMT3A were elevated in leukemia cells isolated from a dasatinib-resistant Ph+ ALL patient (case #1) and a CML patient who lost CHR (case #2) in association with a decrease in levels of miR-217, miR-29b-1, miR-29b-2 and miR-140 as compared with those in cells isolated at initial diagnosis (Fig.1f). In contrast, reverse correlation of levels of DNMT3A and these miRNA were not noted in remission cases, except for one case. These observations suggest that downregulation of miRNA and upregulation of DNMT3A may contribute to TKI resistance in Ph+ leukemia cells.
Further study using the lentiviral gene expression system demonstrated the link between miR-217 and expression of DNMT3A in K562DR cells; miR-217 inhibited expression of DNMT3A through a miR-217-binding site within the 3′-UTR of DNMT3A (Fig.2a,b). In addition, forced expression of miR-217 by lentiviral transduction restored the sensitivity of K562DR cells to dasatinib in association with the downregulation of DNMT3A (Fig.2c–e). These observations indicate the crucial role of miR-217 in prevention of drug resistance in CML cells via downregulation of DNMT3A. In fact, downregulation of DNMT3A by an siRNA in K562DR cells sensitized these cells to dasatinib (Fig.2f,g).
The drug resistance against TKI was counteracted by treatment with an anti-epigenetic agent HDACI, highlighting a potential therapeutic strategy.7 We also previously showed that HDACI successfully overcame imatinib resistance in CEL cells in association with restoration of PTEN expression.7
The present study found that combination of dasatinib (10 nM) and 5-AzadC (0.1 μM) potently inhibited proliferation of K562 cells in association with upregulation of miR-217 and downregulation of DNMT3A in vitro and in vivo (Fig.3a–g). In addition, DNA methylation on promoter regions of p21waf1 and BAX was significantly reduced in association with a decrease in levels of DNMT1 and DNTM3A mRNA in K562 cells after exposure to 5-AzadC (Fig.3f–i). Consequently, the expression of p21waf1 and BAX increased both at mRNA and protein levels (Fig.3h,i). The p21waf1 is a cyclin-dependent kinase inhibitor that is frequently epigenetically silenced in various types of human cancer. Others have shown that the levels of DNMT protein are inversely correlated with the levels of p21waf1 in breast cancer cells.25 BAX protein regulates apoptosis in a cellular pathway that involves both Bcl-2 and p53. BAX functions as a tumor suppressor gene in glioma cells.26 K562 cells carry a monoallelic insertion mutation in exon 5 of p53, resulting in a frameshift mutation and consequent expression of a truncated nonfunctional p53 protein of 148 amino acids.27 Exposure of K562 cells to 5-AzadC did not increase levels of p53 proteins. Thus, the anti-leukemia effect of the combination of dasatinib and 5-AzadC is not dependent on p53 in K562 cells. Upregulation of p21waf1 and BAX by treatment with 5-AzadC might sensitize K562 cells to dasatinib-mediated growth inhibition. Low concentration of 5-AzadC may be useful for maintaining the anti-leukemia effect of dasatinib long term.
Taken together, long-term exposure of K562 cells to TKI decreased levels of miR-217, resulting in upregulation of DNMT3A, which likely plays a role in the acquisition of drug resistance. Inhibition of DNMT3A by forced expression of miR-217 or 5-AzadC may be useful to prevent drug resistance in individuals who receive TKI.
Acknowledgments
This work was supported in part by The Kochi University President's Discretionary Grant (to TI) and the Japanese Leukemia Research Fund (to CN).
Disclosure Statement
The authors have no conflict of interest.
Funding information
The Kochi University President's Discretionary Grant. Japanese Leukemia Research Fund.
Supporting Information
Additional supporting information may be found in the online version of this article:
DNA methylation of the p21waf1, and BAX promoter in K562 and dasatinib-resistnt K562 (K562DR) cells.
The expression of miR-217.
The effects of dasatinib and 5-AzadC in K562 tumors.
Long-term exposure of the K562 cells to tyrosine kinase inhibitors failed to inactivate AKT and ERK signaling.
References
- 1.Kantarjian H, Giles F, Wunderle L, et al. Nilotinib in imatinib-resistant CML and Philadelphia chromosome-positive ALL. N Engl J Med. 2006;354:2542–51. doi: 10.1056/NEJMoa055104. [DOI] [PubMed] [Google Scholar]
- 2.Kantarjian H, Pasquini R, Hamerschlak N, et al. Dasatinib or high-dose imatinib for chronic-phase chronic myeloid leukemia after failure of first-line imatinib: a randomized phase 2 trial. Blood. 2007;109:5143–50. doi: 10.1182/blood-2006-11-056028. [DOI] [PubMed] [Google Scholar]
- 3.Kantarjian HM, Giles FJ, Bhalla KN, et al. Nilotinib is effective in patients with chronic myeloid leukemia in chronic phase following imatinib resistance or intolerance: 24-month follow-up results. Blood. 2011;117:1141–5. doi: 10.1182/blood-2010-03-277152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cannella L, Breccia M, Stefanizzi C, Napoleone L, Santopietro M, Alimena G. Dasatinib overcomes imatinib and nilotinib failure in Philadelphia chromosome positive chronic myeloid leukemia with different mechanisms of resistance. Leuk Lymphoma. 2009;50:848–50. doi: 10.1080/10428190902829425. [DOI] [PubMed] [Google Scholar]
- 5.Giles FJ, Abruzzese E, Rosti G, et al. Nilotinib is active in chronic and accelerated phase chronic myeloid leukemia following failure of imatinib and dasatinib therapy. Leukemia. 2010;24:1299–301. doi: 10.1038/leu.2010.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nishioka C, Ikezoe T, Yang J, Udaka K, Yokoyama A. Imatinib causes epigenetic alterations of PTEN gene via upregulation of DNA methyltransferases and polycomb group proteins. Blood Cancer J. 2011;1:e48. doi: 10.1038/bcj.2011.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nishioka C, Ikezoe T, Yang J, Yokoyama A. Long-term exposure of leukemia cells to multi-targeted tyrosine kinase inhibitor induces activations of AKT, ERK and STAT5 signaling via epigenetic silencing of the PTEN gene. Leukemia. 2010;24:1631–40. doi: 10.1038/leu.2010.145. [DOI] [PubMed] [Google Scholar]
- 8.Jabbour E, Issa JP, Garcia-Manero G, Kantarjian H. Evolution of decitabine development: accomplishments, ongoing investigations, and future strategies. Cancer. 2008;112:2341–51. doi: 10.1002/cncr.23463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Qin T, Jelinek J, Si J, Shu J, Issa JP. Mechanisms of resistance to 5-aza-2′-deoxycytidine in human cancer cell lines. Blood. 2009;113:659–67. doi: 10.1182/blood-2008-02-140038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Oki Y, Kondo Y, Yamamoto K, et al. A phase I/II study of decitabine in patients with myelodysplastic syndrome: a multi-center study in Japan. Cancer Sci. 2012;103:1839–47. doi: 10.1111/j.1349-7006.2012.02386.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jones PA, Baylin SB. The epigenetic of cancer. Cell. 2007;128:683–92. doi: 10.1016/j.cell.2007.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bachman KE, Park BH, Rhee I, et al. Histone modifications and silencing prior to DNA methylation of a tumor suppressor gene. Cancer Cell. 2003;3:89–95. doi: 10.1016/s1535-6108(02)00234-9. [DOI] [PubMed] [Google Scholar]
- 13.Fuks F, Burgers WA, Brehm A, Hughes-Davies L, Kouzarides T. DNA methyltransferase Dnmt1 associates with histone deacetylase activity. Nat Genet. 2000;24:88–91. doi: 10.1038/71750. [DOI] [PubMed] [Google Scholar]
- 14.Fuks F, Burgers WA, Godin N, Kasai M, Kouzarides T. Dnmt3a binds deacetylases and is recruited by a sequence-specific repressor to silence transcription. EMBO J. 2001;20:2536–44. doi: 10.1093/emboj/20.10.2536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Garzon R, Liu S, Fabbri M, et al. MicroRNA-29b induces global DNA hypomethylation and tumor suppressor gene reexpression in acute myeloid leukemia by targeting directly DNMT3A and 3B and indirectly DNMT1. Blood. 2009;113:6411–8. doi: 10.1182/blood-2008-07-170589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fabbri M, Garzon R, Cimmino A, et al. MicroRNA-29 family reverts aberrant methylation in lung cancer by targeting DNA methyltransferases 3A and 3B. Proc Natl Acad Sci USA. 2007;104:15805–10. doi: 10.1073/pnas.0707628104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Takata A, Otsuka M, Yoshikawa T, et al. MiRNA-140 acts as a liver tumor suppressor by controlling NF-κB activity via directly targeting Dnmt1 expression. Hepatology. 2013;57:162–70. doi: 10.1002/hep.26011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Menghini R, Casagrande V, Cardellini M, et al. MicroRNA 217 modulates endothelial cell senescence via silent information regulator 1. Circulation. 2009;120:1524–32. doi: 10.1161/CIRCULATIONAHA.109.864629. [DOI] [PubMed] [Google Scholar]
- 19.Garzon R, Fabbri M, Cimmino A, Calin GA, Croce C. MicroRNA expression and function in cancer. Trends Mol Med. 2006;12:580–7. doi: 10.1016/j.molmed.2006.10.006. [DOI] [PubMed] [Google Scholar]
- 20.Esquela-Kerscher A, Slack FJ. Oncomirs – microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259–69. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 21.He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5:522–31. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- 22.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–97. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 23.Sharma SV, Lee DY, Li B, et al. A chromatin-mediated reversible drug-tolerant state in cancer cell subpopulations. Cell. 2010;141:69–80. doi: 10.1016/j.cell.2010.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Baccarani M, Deininger MW, Rosti G, et al. European LeukemiaNet recommendations for the management of chronic myeloid leukemia: 2013. Blood. 2013;122:872–84. doi: 10.1182/blood-2013-05-501569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nass SJ, Ferguson AT, El-Ashry D, Nelson WG, Davidson NE. Expression of DNA methyl-transferase (DMT) and the cell cycle in human breast cancer cells. Oncogene. 1999;18:7453–61. doi: 10.1038/sj.onc.1203138. [DOI] [PubMed] [Google Scholar]
- 26.Chou D, Miyashita T, Mohrenweiser HW, et al. The BAX gene maps to the glioma candidate region at 19q13.3, but is not altered in human gliomas. Cancer Genet Cytogenet. 1009;88:136–40. doi: 10.1016/0165-4608(95)00341-x. [DOI] [PubMed] [Google Scholar]
- 27.Thakur BK, Dittrich T, Chandra P, et al. Involvement of p53 in the cytotoxic activity of the NAMPT inhibitor FK866 in myeloid leukemic cells. Int J Cancer. 2013;132:766–74. doi: 10.1002/ijc.27726. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
DNA methylation of the p21waf1, and BAX promoter in K562 and dasatinib-resistnt K562 (K562DR) cells.
The expression of miR-217.
The effects of dasatinib and 5-AzadC in K562 tumors.
Long-term exposure of the K562 cells to tyrosine kinase inhibitors failed to inactivate AKT and ERK signaling.