Abstract
Malignant rhabdoid tumor (MRT) is a rare and highly lethal cancer that mainly affects infants and young children. The majority of MRT are characterized by loss of function of SMARCB1 on chromosome 22q11.2. However, little is known about genetic changes other than SMARCB1 alterations that are responsible for the development and/or progression of MRT. To explore additional gene targets in MRT, we analyzed 21 MRT specimens (12 fresh tumors and 9 MRT-derived cell lines) using high-density single nucleotide polymorphism genotyping microarrays. Although MRT genomes are characterized by common 22q11.2 deletions, affecting the SMARCB1 locus with a frequency of 95.2% (20/21 specimens), other genetic changes have been less frequent. Of the 20 specimens with deletions of 22q11.2, eight specimens showed uniparental disomy of the SMARCB1 locus with homozygous deletions or gene mutations. High-resolution analysis also disclosed the recurrent hemizygous/homozygous deletions of 7q35–q36.1, involving the CNTNAP2 locus in three specimens. Mutations analysis of CNTNAP2 showed a novel R157C missense mutation in a primary case, and methylation analysis showed recurrent hypermethylation of CNTNAP2 in three of nine cell lines. These results demonstrated that CNTNAP2 is one of the additional gene targets, other than SMARCB1, in MRT.
Keywords: CNTNAP2, malignant rhabdoid tumor, SMARCB1, SNP array
Malignant rhabdoid tumor (MRT) is an extremely rare and highly aggressive neoplasm that typically develops in infancy or early childhood.1 Although initially described as being rhabdomyosarcomatous, an aggressive type of kidney tumor,1 subsequent studies have revealed that MRT occurs in various sites, including the central nervous system (CNS), lung, liver, skin, and soft tissues.2 The most frequent location of the tumor is the kidneys, followed by the CNS, and tumors originating from the latter site are referred to as atypical teratoid/rhabdoid tumors (AT/RT).3 Cytogenetic and molecular analyses of MRT have shown recurrent deletions at 22q11.2, which resulted in identification of SMARCB1 (OMIN 601607) as a characteristic gene abnormality of this tumor.4 Germ-line and somatic mutations/deletions of SMARCB1 have been reported in AT/RT as well as in epithelioid sarcoma, familial schwannomatosis, and renal medullary carcinoma.5–8 The SMARCB1 gene is a member of the ATP-dependent SWI/SNF chromatin-remodeling complex and is recruited to promoters of genes that regulate cell cycle, growth, and differentiation.4 In MRT, SMARCB1 appears to function as a classic tumor suppressor gene, such that germ-line mutations and deletions predispose to the development of these malignancies; somatic loss or mutation of the other allele constitutes the second hit.
In recent years, genome-wide copy number analysis using single nucleotide polymorphism (SNP) arrays (SNP-chip) has been shown to have outstanding power to reveal detailed profiles of genomic abnormalities and identify new genetic targets in various cancers.9 A previous report showed that high-resolution SNP-chip analysis could detect bi-allelic alterations in SMARCB1 in almost all MRT cases, which suggests that SMARCB1 is the primary mutational gene target responsible for the development of MRT and provided further evidence for the clinical utility of molecular diagnostic testing.10 However, some MRT cases retain expression of the protein, and a small number of familial MRTs have been reported to not be associated with SMARCB1 inactivation.11 These findings suggest the possibility of additional relevant genetic loci distinct from SMARCB1. However, the detailed genetic abnormalities in MRT other than chromosome 22 have not been fully understood. Therefore, to clarify the additional genetic lesions involved in the pathogenesis of MRTs, we carried out SNP-chip analysis of 21 MRT samples.
Materials and Methods
Specimens
This study was approved by the ethics board of the University of Tokyo (Tokyo, Japan) (Approval Number 1598). Primary tumor specimens were obtained at the time of the initial surgery or biopsy from patients who were diagnosed as having MRT or AT/RT at collaborating hospitals. In total, 12 primary MRT specimens (four samples of AT/RT) and 9 cell lines derived from patients with MRT (KYM-1, TM87-16, TTC-1240, TTC-549, TTC-642, TTN-45, YAMRT, RTK(J)-4N, and STM-91-01) were analyzed in this study. The original tumor sites of the primary and cell-line specimens are described in Table S1. The TTC series and STM-91-01 were established from MRT patients. KYM-1, YAMRT, TTN-45, TM87-16, and RTK(J)-4N were generous gifts from Dr. Inoue (St. Jude Children's Research Hospital, Memphis, TN, USA), Dr. Kanegane (Toyama University, Toyama, Japan), Dr. Shimada (Children's Hospital Los Angeles), and Dr. Yokomori (Graduate School of Medicine, University of Tokyo). A neuroblastoma cell line, SJNB-1, was used as a control in the methylation analysis. All cell lines were cultured in RPMI-1640 (Gibco, Gaithersburg, MD, USA) supplemented with 9% FBS.12
Microarray analysis
High molecular weight DNA was isolated from tumor specimens and subjected to SNP-chip analysis using Affymetrix GeneChip Mapping 50K and/or 250K arrays (Affymetrix, Santa Clara, CA, USA), according to the manufacturer's protocol (Table S1). After appropriate normalization of mean array intensities, signal ratios between tumor and normal cells were calculated, and allele-specific copy numbers were inferred from the observed signal ratios based on the hidden Markov model using CNAG/AsCNAR software (http://www.genome.umin.jp/CNAG_DLpage/CNAG_top.html).9 Chromosomal losses detected by SNP-chip analysis were confirmed by quantitative genomic PCR (Q-genomic PCR) for CNTNAP2, LRP1B, FHIT, ROBO1, AUTS2, PTPRD, and ATBF1 loci.13 GAPDH was used as control. Primer sets for Q-genomic PCR are listed in Table S2.
Mutation and expression analyses of SMARCB1 and CNTNAP2
Direct sequencing analyses of all coding exons of SMARCB1 and CNTNAP2 were carried out in all samples as previously described.14 The primer sequences and conditions of PCR for mutation analyses of these genes have been described in previous papers.14,15 Total RNA was extracted from the nine cell lines using Isogen reagent (Nippon Gene, Osaka, Japan), according to the manufacturer's instructions and subjected to RT-PCR to synthesize cDNA using the SuperScript Preamplification System for first-strand cDNA synthesis (Life Technologies, Rockville, MD, USA). Semiquantitative RT-PCR analysis for CNTNAP2 expression was carried out as previously described.16
Methylation-specific PCR and 5-aza-2-deoxycytidine treatment
Bisulfate modification of genomic DNA was carried out as previously described.17 For methylation-specific PCR, approximately 10 ng bisulfite-treated DNA was amplified with primers for both the methylated and unmethylated sequences.17 The primer sets for promoter region CNTNAP2 were as described previously,18 and the primer sets for SMARCB1 were as follows: SMARCB1F2, 5′-gtcygtgagaagtcctctac-3′; SMARCB1R2, 5′-gaaatcccaggtcratgagg-3′. Reaction products were separated by electrophoresis on a 2.0% agarose gel. 5-Aza-2-deoxycytidine (Sigma Chemical, Perth, WA, Australia) was dissolved in cold RPMI-1640 immediately before use. Cells were exposed to 0.5 and 1.0 mM 5-aza-2-deoxycytidine for 3 days, with the medium and drug being replaced every 24 h.19 Cells were then harvested and used for RT-PCR analysis.
Histopathology and immunohistochemistry
For light microscopy, tumors from MRT patients were fixed in 10% buffered formalin and embedded in paraffin. Sections from the paraffin-embedded blocks were evaluated by H&E staining and immunoperoxidase techniques using a panel of mAbs, including epithelial membrane antigen, vimentin, a muscle actin-specific monoclonal antibody (HHF35), myoglobin, S100 protein, neuron-specific enolase, and SMARCB1/INI1.14,20
Results
Chromosome 22q11.2 deletions and mutations of SMARCB1
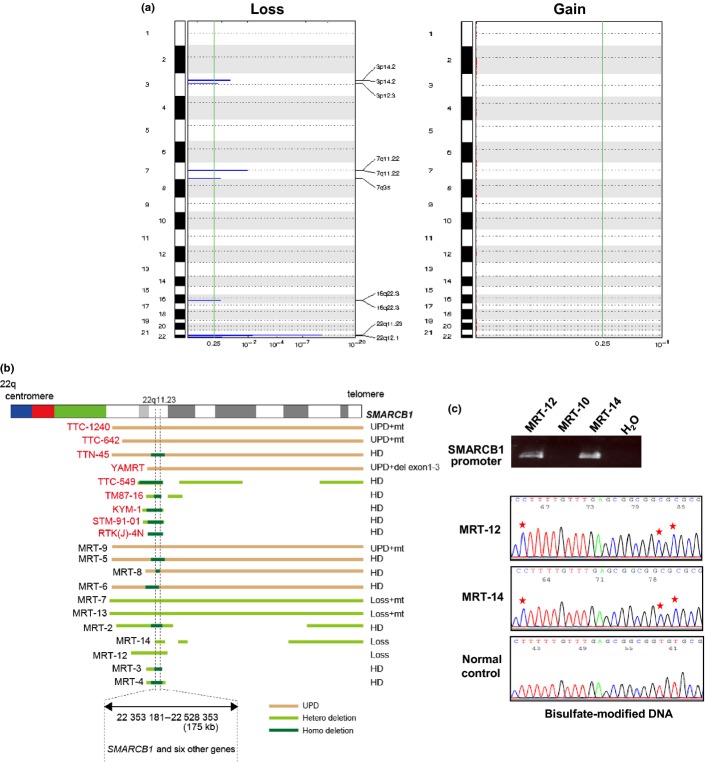
The SNP-chip analysis was carried out for 21 MRT specimens, including 12 fresh tumors and 9 cell lines, using Affymetrix GeneChip 50K XbaI/HindIII and/or 250K NspI/StyI mapping arrays (Table S1). Although many specimens had no matched control DNA and suffered from varying degrees of normal cell contamination, the allelic compositions were accurately determined in most specimens using our CNAG/AsCNAR programs (Fig.1a). In our SNP-chip analysis, the most frequent copy number change detected in MRT was deletion of chromosome 22q11.2 (Fig.1a,b). In total, 20 of 21 specimens (95.2%) had LOH or homozygous deletions at 22q11.2 involving the SMARCB1 locus (Fig.1a,b). In eight samples, uniparental disomy (UPD) of 22q segments caused homozygous mutations/deletions of SMARCB1 (Fig.1b). Ten samples had homozygous focal deletions commonly involving a 175-kb region (ch22:22,353,181-22,528,353), which exclusively included SMARCB1. Subsequent mutation analysis revealed that five samples with heterozygous deletion or UPD at the 22q11.2 locus had mutations in SMARCB1 (Table1). An MRT-derived cell line with 22qUPD (YAMRT) harbored a small deletion involving exons 1–3, which was not detectable by SNP-chip analysis. In our cohort, two specimens showed hemizygous deletion at the SMARCB1 locus, and one case showed no genetic changes within this locus. Immunohistochemical analyses of these three cases showed positive results for vimentin but negative findings for muscle lineage markers and SMARCB1, supporting the diagnosis of MRT or AT/RT. Thus, to investigate whether abnormal methylation is associated with inactivation of SMARCB1, bisulfate sequencing for the promoter region of SMARCB1 was carried out in these three cases. As shown in Figure1(c), two cases having hemizygous deletions at the SMARCB1 locus displayed complete methylation of the CpG island. However, one case without any genetic abnormality of the SMARCB1 locus lacked PCR products (both methylated and unmethylated) for the promoter region (Fig.1c), suggesting that this case may harbor a small deletion involving the promoter region of SMARCB1, which escaped SNP array detection. In total, 20 of the 21 MRT samples had biallelic aberrations of SMARCB1, indicating genetic homogeneity of MRT. Molecular allelokaryotyping profiles were essentially similar between cell lines and primary tumors, providing some rationale for the combined analysis of both specimens in this study (Fig.1b).
Figure 1.
Copy number changes detected in malignant rhabdoid tumors (MRT). (a) Characteristics of copy number alterations in MRT. Regions showing statistically significant increase or decrease in genomic copy number were detected using the genomic identification of significant targets in cancer (GISTIC) algorithm based on single nucleotide polymorphism array analysis. Because we did not detect any significant chromosomal gains in our cohort, nothing is shown in the right-hand figure. (b) Overall representation of aberrations of chromosome 22q11.2 in MRT. Specimens indicated by red are cell lines. Pink bar indicates uniparental disomy, and yellow and green bars indicate heterozygous deletion and homozygous deletion, respectively. The minimum overlapping deleted region was 175 kb in chromosome 22q11.2, including SMARCB1 and another six genes. SMARCB1 status is indicated at the right. MRT-12 and MRT-14 show heterozygous deletion of the SMARCB1 locus, and the wild-type allele of SMARCB1 was retained in each case. del, deletion; HD, homozygous deletion; mt, mutation; UPD, uniparental disomy. (c) Bisulfate modification- and methylation-specific PCR for SMARCB1 in fresh tumors without biallelic genetic alterations of the SMARCB1 locus. The upper panel shows PCR for the promoter region of SMARCB1. Hypermethylation of the CpG islands in MRT-12 and MRT-14 is shown in the lower panel. CpG islands are marked by asterisks. The bottom panel shows normal control.
Table 1.
Mutations/methylations of SMARCB1 (SNF5/INI1) in malignant rhabdoid tumor (MRT)
| Sample name | Attribute | 22q | Mutation/methylation of SMARCB1 |
|---|---|---|---|
| KYM-1 | Cell line | homoD | homoD |
| TM87-16 | Cell line | homoD | homoD |
| TTC-1240 | Cell line | UPD | G646T (E216 stop) |
| TTC-549 | Cell line | homoD | homoD |
| TTC-642 | Cell line | UPD | C118T (R40 stop) |
| TTN-45 | Cell line | UPD + homoD | homoD |
| YAMRT | Cell line | UPD | del exons 1–3 |
| STM-91-01 | Cell line | homoD | homoD |
| RTK(J)-4N | Cell line | homoD | homoD |
| MRT-2 | Primary | homoD | homoD |
| MRT-3 | Primary | homoD | homoD |
| MRT-4 | Primary | homoD | homoD |
| MRT-5 | Primary | UPD + homoD | homoD |
| MRT-6 | Primary | UPD + homoD | homoD |
| MRT-7 | Primary | heteroD | G646T (E215 stop) |
| MRT-8 | Primary | UPD + homoD | homoD |
| MRT-9 | Primary | UPD | 27 bp to exon 6 553 del |
| MRT-10 | Primary | Normal | |
| MRT-12 | Primary | heteroD | Methylation |
| MRT-13 | Primary | heteroD | ag to aa intron 5, D224 stop |
| MRT-14 | Primary | heteroD | Methylation |
del, deletion; heteroD, heterozygous deletion; homoD, homozygous deletion; UPD, uniparental disomy.
Other copy number changes detected in MRT
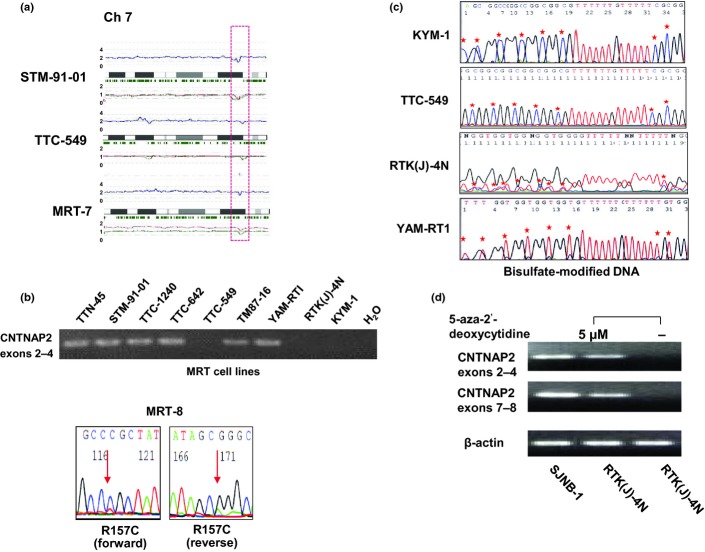
Although recurrent copy number changes other than 22q11.2 deletions were less frequent in MRT (Fig.1a), seven loci of gains and eight loci of losses were commonly detected in multiple samples (Table2). Importantly, some of the regions contained potential gene targets that were known to be associated with tumorigenesis of other cancers, such as CCNL1, POT1, CNTNAP2, and PRPTD (Table2).18,21,22 Detection of homozygous deletions was also of interest because they provide an important clue to pinpoint tumor suppressor loci. In fact, SMARCB1 was identified from homozygously deleted regions at 22q11.2.4 Of note, we found a homozygous deletion of the CNTNAP2 locus at 7q35–q36 in one specimen (STM-91-01), and another two cases (TTC-549 and MRT-7) showed a hemizygous deletion in this region (Fig.2a). The commonly deleted region of the CNTNAP2 locus was expanded in a 534-kb region (ch7:146,183,963-146,718-030) at 7q35–q36, and a homozygous deletion was involved in only exons 9–10 of CNTNAP2. Unfortunately, we were not able to completely exclude the possibility that this may represent copy number variations (CNV) rather than real homozygous/hemizygous deletions, because these homozygous deletions were found in established MRT cell lines. However, in our SNP array database, we did not find any CNV at the CNTNAP2 locus in 100 normal samples, but hemizygous/homozygous deletions were observed in two neuroblastoma cell lines (NB-16 and NB-19) (data not shown), suggesting that this deletions would be somatic events rather than CNV. Complete or incomplete losses of genetic materials at seven loci were confirmed by quantitative genomic PCR (Fig. S1).
Table 2.
Recurrent chromosomal gains and losses in malignant rhabdoid tumor
| Chromosome | Position† |
Length (kb) | No. of affected samples | Gene(s) | |
|---|---|---|---|---|---|
| Start | End | ||||
| Gain | |||||
| 1 | 120 804 640 | 245 519 990 | 124 715 | Four (CL: TM87-16, TTC-549, YAMRT; P: MRT-8) | Many genes |
| 2 | 41 745 732 | 42 067 857 | 322 | One (CL: TTC1240) | None |
| 3 | 158 297 982 | 158 507 647 | 209 | Two (CL: TM87-16; P: MRT-8) | CCNL1, VEPH1 |
| 7 | 123 981 822 | 124 568 564 | 587 | Four (CL: TM87-16, TTC-1240, TTC-549, TTN-45) | POT1 |
| 7 | 125 675 684 | 125 906 795 | 231 | Two (CL: TM87-16, KYM-1) | GRM8 |
| 7 | 130 358 924 | 130 939 457 | 580 | Three (CL: TM87-16, TTC-1240, TTN-45) | None |
| 16 | 76 268 665 | 77 096 360 | 828 | Two (CL: TM87-16, KYM-1) | CLEC3A, WWOX |
| Loss | |||||
| 2 | 140 875 055 | 141 241 576 | 366 | One (CL: STM-91-01) | LRP1B |
| 3 | 60 099 620 | 60 538 672 | 439 | Three (CL: TM87-16, TTC-642, YAMRT) | FHIT |
| 3 | 78 740 688 | 78 995 893 | 255 | Two (CL: TM87-16; P: MRT-8) | ROBO1 |
| 4 | 182 649 464 | 182 977 255 | 328 | Three (CL: TTC-642, KYM-1, RTK(J)-4N) | None |
| 7 | 69 446 093 | 69 593 327 | 147 | Two (CL: TM87-16, TTC-642) | AUTS2 |
| 7 | 146 183 963 | 146 718 030 | 534 | Three (CL: STM-91-01, TTC-549; P: MRT-7) | CNTNAP2 |
| 9 | 9 582 260 | 10 072 420 | 490 | Three (CL: TM87-16, TTC-1240, KYM-1) | PTPRD |
| 16 | 71 464 505 | 71 708 905 | 244 | Two (CL: TTC-642; P: MRT-8) | ATBF1 |
†NCBI build 35. List of lesions detected in more than two cases, and known genes in the regions. CL, cell line; P, primary sample.
Figure 2.
Recurrent deletions of chromosome 7q35–q36 and CNTNAP2 alterations in malignant rhabdoid tumor (MRT). (a) Deletions of chromosome 7q35–q36 in three specimens detected by single nucleotide polymorphism (SNP) array. For each panel, total copy numbers (tCNs; red dots), moving averages of tCNs for five consecutive SNPs (blue line), an ideogram of the relevant chromosome, location of heterozygous SNP calls (green bars), and allele-specific copy numbers (AsCNs) averaged for five consecutive SNPs (red and green lines for larger and smaller alleles, respectively) are plotted. (b) Expression and mutation analyses of MRT. Upper panel shows RT-PCR analysis of CNTNAP2 in nine cell lines. Sequence chromatogram of R157C missense mutation detected in a fresh tumor, MRT-8, is shown in the lower panel. (c) Bisulfate modification- and methylation-specific PCR for CNTNAP2 in cell lines. Hypermethylation of CpG islands in KYM-1, TTC-549, and RTK(J)-4N cell lines is shown in the upper panel. The lower panel shows control. CpG islands are marked by asterisks. (d) Representative results of re-expression of transcriptionally silenced CNTNAP2 after treatment with 5-aza-deoxycytidine in MRT cell lines. Reverse transcription-PCR analysis of KYM-1 cell line harvested following 72 h of incubation with control media (−) and 5 μM 5-aza-2-deoxycytidine (+). SJNB-1 neuroblastoma cell line, which expressed abundant CNTNAP2, was used as a positive control.
CNTNAP2 aberrations found in MRT
CNTNAP2 was a single gene found in a novel recurrent homozygous deletion at 7q35–q36. Thus, to assess the involvement of CNTNAP2 in MRT pathogenesis, mutation, expression, and methylation analyses were carried out in our series. Through mutation analysis of the coding region of CNTNAP2, we found a novel R157C missense mutation in a fresh tumor of MRT, which was not found in 60 healthy volunteers and not registered in the dbSNP 137 (http://www.ncbi.nlm.nih.gov/projects/SNP/snp_summary.cgi?view+summary=view+summary&build_id=137) or 1000 Genomes (http://www.1000genomes.org/) databases (Fig.2b). This single nucleotide change was scored as “probably damaging” or “damaging” by two computational prediction software packages, SIFT (http://sift.jcvi.org/) and PolyPhen-2 (http://genetics.bwh.harvard.edu/pph2/). CNTNAP2 expression was not detected in three of nine cell lines by RT-PCR (Fig.2b). Because a homozygous deletion in CNTNAP2 detected in STM-91-01 cells was partial and limited within exons 9–10, the transcript of exons 2–4 of STM-91-01 was detected, although this specimen had a homozygous deletion (Fig.1b).
In addition, these three cell lines lacking CNTNAP2 expression displayed complete or partial methylation of the CpG island of CNTNAP2 (Fig.2b,c). Methylation analysis was also carried out in 20 neuroblastoma cell lines, but we did not detect any methylation of CpG islands of CNTNAP2 (data not shown). To elucidate whether hypermethylation of CNTNAP2 blocked gene expression in MRT cells, we subjected all three cell lines to 3 days' exposure to 5-aza-deoxycytidine. As shown in Figure2(d), CNTNAP2 expression was induced by 5-aza-deoxycytidine treatment in the three cell lines, indicating that methylation of the CpG island is a direct mechanism for CNTNAP2 silencing in MRT.
Discussion
Our SNP-chip analysis revealed a high frequency of deletion at 22q11.2 in the MRT genome, indicating the unique patterns in the genome imbalance map that characterizes MRT.10 In our analysis, most MRT specimens showed biallelic genetic aberrations of SMARCB1, but three cases did not show biallelic genetic abnormalities of SMARCB1. Among them, two cases with kidney MRT showed heterozygous deletions of the 22q11.2 locus and complete methylation of the promoter region of SMARCB1, suggesting that not only genetic events, but also epigenetic changes contributed to biallelic inactivation of SMARCB1 in MRT. Although epigenetic silencing of SMARCB1 in MRT has never been reported, epigenetic alterations of promoter regions of SMARCB1 should be considered for the possible alterations for inactivation of SMARCB1 in MRT.
In previous reports, heterozygous knockout mice developed tumors consistent with MRT,23 beginning as early as 5 weeks of age, but mice crossed with SMARCB1+/− and CCND1−/− mice did not develop any spontaneous tumor.23 Thus, these findings suggest that CCND1, located at 11q13.3, may be a key mediator involved in the genesis of MRT. Of interest, our analysis identified recurrent gains at CCNL1, which functions in association with cyclin-dependent kinases, including CCND1, suggesting that CCNL1 acts as one of the mediators involved in the development of MRT. We also found recurrent losses at the PTPRD locus on 9p23 and gains at the POT1 locus on 7q31. PTPRD and POT1 have been shown to be mutated or downregulated in several human cancers, including neuroblastoma, lung cancer, and chronic myeloid leukemia;22,24,25 thus, further analysis of these gene alterations in a large number of MRT samples would be necessary to assess their involvement in the pathogenesis of MRT.
The CNTNAP2 gene encodes a single-pass transmembrane protein, mediating cell–cell interactions in the CNS and/or peripheral nervous system.26 This gene is located in a common fragile site that is inactivated in different types of cancers, including brain tumor, ovarian cancer, and breast cancer.27 Recently, CNTNAP2 was shown to be translocated and methylated in a subset of glioma and demonstrated functional characteristics of a tumor suppressor gene.28 In our analysis, recurrent homozygous/hemizygous deletions involving the CNTNAP2 locus were newly identified, and expression of CNTNAP2 was substantially reduced in 33% of MRT cell lines, supporting the fact that CNTNAP2 is a putative second tumor suppressor gene for MRT. Furthermore, methylation of the promoter region of CNTNAP2 has been reported in not only glioma, but also in pancreatic adenocarcinoma.18 In accordance with this, two cell lines, KYM-1 and TTC-549, showed complete methylation of CNTNAP2 resulting in loss of CNTNAP2 expression. However, one cell line, RTK(J)-4N, displayed partial methylation of the CpG island, although CNTNAP2 expression was absent in this cell line. Therefore, additional mechanisms, such as mutations of the promoter region of CNTNAP2 in the unmethylated allele, would also be associated with silencing of CNTNAP2 in RTK(J)-4N. Because a corresponding normal sample of the MRT-8 case with CNTNAP2 R157C mutation was not available, we could not exclude the possibility that this mutation would be a rare SNP. However, the mutant allele was relatively low compared to the wild-type allele, suggesting that the single nucleotide change detected at R157 in MRT-8 was most likely to be a somatic mutation rather than non-functional SNPs. Taken together, these findings suggested that CNTNAP2 is a candidate gene target in a subset of MRT.
In conclusion, consistent with other reports, our results illustrate that SMARCB1 is the primary gene implicated in the pathogenesis of MRT.10 Although frequencies of recurrent genetic changes other than the SMARCB1 locus were low, our findings suggest that CNTNAP2 is one of the potential second gene targets for MRT. To our knowledge, this is the first report to describe aberrations of CNTNAP2 in MRT. Further studies are necessary to unravel the oncogenic effects of CNTNAP2 in MRT.
Acknowledgments
We are grateful to Ms. Matsumura, Ms. Hoshino, Ms. Yin, Ms. Saito, Ms. Mori, and Ms. Ogino for their excellent technical assistance. We also express our appreciation to Drs. Shimada, Yokomori, Kanegane, and Inoue for their generous gifts of MRT cell lines. This work was supported by: Research on Measures for Intractable Diseases, Health, and Labor Sciences Research Grants, Ministry of Health, Labor and Welfare of Japan; Research on Health Sciences focusing on Drug Innovation; Japan Health Sciences Foundation; Core Research for Evolutional Science and Technology, Japan Science and Technology Agency; and the Project for Development of Innovative Research on Cancer Therapeutics.
Disclosure Statement
The authors have no conflicts of interest.
Funding Information
The Ministry of Education, Culture, Sports, Science and Technology (Project for Development of Innovative Research on Cancer Therapeutics, 23390269), Japan. Japan Foundation for Pediatric Research fund (1171-945).
Supporting Information
Additional supporting information may be found in the online version of this article:
Quantitative-genomic PCR for chromosomal deleted regions detected by single nucleotide polymorphism-chip analysis in malignant rhabdoid tumor.
Analyzed samples of malignant rhabdoid tumor and single nucleotide polymorphism array platform.
Table S2. Primer sets for quantitative-genomic PCR in malignant rhabdoid tumor.
References
- 1.Beckwith JB, Palmer NF. Histopathology and prognosis of Wilms tumors: results from the First National Wilms' Tumor Study. Cancer. 1978;41:1937–48. doi: 10.1002/1097-0142(197805)41:5<1937::aid-cncr2820410538>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
- 2.DeYoung BR, Swanson PE, Argenyi ZB, et al. CD31 immunoreactivity in mesenchymal neoplasms of the skin and subcutis: report of 145 cases and review of putative immunohistologic markers of endothelial differentiation. J Cutan Pathol. 1995;22:215–22. doi: 10.1111/j.1600-0560.1995.tb00741.x. [DOI] [PubMed] [Google Scholar]
- 3.Bhattacharjee M, Hicks J, Langford L, et al. Central nervous system atypical teratoid/rhabdoid tumors of infancy and childhood. Ultrastruct Pathol. 1997;21:369–78. doi: 10.3109/01913129709021935. [DOI] [PubMed] [Google Scholar]
- 4.Versteege I, Sevenet N, Lange J, et al. Truncating mutations of hSNF5/INI1 in aggressive paediatric cancer. Nature. 1998;394:203–6. doi: 10.1038/28212. [DOI] [PubMed] [Google Scholar]
- 5.Biegel JA, Fogelgren B, Wainwright LM, Zhou JY, Bevan H, Rorke LB. Germline INI1 mutation in a patient with a central nervous system atypical teratoid tumor and renal rhabdoid tumor. Genes Chromosom Cancer. 2000;28:31–7. doi: 10.1002/(sici)1098-2264(200005)28:1<31::aid-gcc4>3.0.co;2-y. [DOI] [PubMed] [Google Scholar]
- 6.Carter JM, O'Hara C, Dundas G, et al. Epithelioid malignant peripheral nerve sheath tumor arising in a schwannoma, in a patient with “neuroblastoma-like” schwannomatosis and a novel germline SMARCB1 mutation. Am J Surg Pathol. 2012;36:154–60. doi: 10.1097/PAS.0b013e3182380802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hulsebos TJ, Plomp AS, Wolterman RA, Robanus-Maandag EC, Baas F, Wesseling P. Germline mutation of INI1/SMARCB1 in familial schwannomatosis. Am J Hum Genet. 2007;80:805–10. doi: 10.1086/513207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Calderaro J, Moroch J, Pierron G, et al. SMARCB1/INI1 inactivation in renal medullary carcinoma. Histopathology. 2012;61:428–35. doi: 10.1111/j.1365-2559.2012.04228.x. [DOI] [PubMed] [Google Scholar]
- 9.Chen Y, Takita J, Choi YL, et al. Oncogenic mutations of ALK kinase in neuroblastoma. Nature. 2008;455:971–4. doi: 10.1038/nature07399. [DOI] [PubMed] [Google Scholar]
- 10.Jackson EM, Sievert AJ, Gai X, et al. Genomic analysis using high-density single nucleotide polymorphism-based oligonucleotide arrays and multiplex ligation-dependent probe amplification provides a comprehensive analysis of INI1/SMARCB1 in malignant rhabdoid tumors. Clin Cancer Res. 2009;15:1923–30. doi: 10.1158/1078-0432.CCR-08-2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fruhwald MC, Hasselblatt M, Wirth S, et al. Non-linkage of familial rhabdoid tumors to SMARCB1 implies a second locus for the rhabdoid tumor predisposition syndrome. Pediatr Blood Cancer. 2006;47:273–8. doi: 10.1002/pbc.20526. [DOI] [PubMed] [Google Scholar]
- 12.Takita J, Chen Y, Okubo J, et al. Aberrations of NEGR1 on 1p31 and MYEOV on 11q13 in neuroblastoma. Cancer Sci. 2011;102:1645–50. doi: 10.1111/j.1349-7006.2011.01995.x. [DOI] [PubMed] [Google Scholar]
- 13.Nannya Y, Sanada M, Nakazaki K, et al. A robust algorithm for copy number detection using high-density oligonucleotide single nucleotide polymorphism genotyping arrays. Cancer Res. 2005;65:6071–9. doi: 10.1158/0008-5472.CAN-05-0465. [DOI] [PubMed] [Google Scholar]
- 14.Uno K, Takita J, Yokomori K, et al. Aberrations of the hSNF5/INI1 gene are restricted to malignant rhabdoid tumors or atypical teratoid/rhabdoid tumors in pediatric solid tumors. Genes Chromosom Cancer. 2002;34:33–41. doi: 10.1002/gcc.10052. [DOI] [PubMed] [Google Scholar]
- 15.Zweier C, de Jong EK, Zweier M, et al. CNTNAP2 and NRXN1 are mutated in autosomal-recessive Pitt-Hopkins-like mental retardation and determine the level of a common synaptic protein in Drosophila. Am J Hum Genet. 2009;85:655–66. doi: 10.1016/j.ajhg.2009.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Takita J, Ishii M, Tsutsumi S, et al. Gene expression profiling and identification of novel prognostic marker genes in neuroblastoma. Genes Chromosom Cancer. 2004;40:120–32. doi: 10.1002/gcc.20021. [DOI] [PubMed] [Google Scholar]
- 17.Takita J, Yang HW, Chen YY, et al. Allelic imbalance on chromosome 2q and alterations of the caspase 8 gene in neuroblastoma. Oncogene. 2001;20:4424–32. doi: 10.1038/sj.onc.1204521. [DOI] [PubMed] [Google Scholar]
- 18.Omura N, Li CP, Li A, et al. Genome-wide profiling of methylated promoters in pancreatic adenocarcinoma. Cancer Biol Ther. 2008;7:1146–56. doi: 10.4161/cbt.7.7.6208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Takita J, Hayashi Y, Nakajima T, et al. The p16 (CDKN2A) gene is involved in the growth of neuroblastoma cells and its expression is associated with prognosis of neuroblastoma patients. Oncogene. 1998;17:3137–43. doi: 10.1038/sj.onc.1202232. [DOI] [PubMed] [Google Scholar]
- 20.Hoot AC, Russo P, Judkins AR, Perlman EJ, Biegel JA. Immunohistochemical analysis of hSNF5/INI1 distinguishes renal and extra-renal malignant rhabdoid tumors from other pediatric soft tissue tumors. Am J Surg Pathol. 2004;28:1485–91. doi: 10.1097/01.pas.0000141390.14548.34. [DOI] [PubMed] [Google Scholar]
- 21.Muller D, Millon R, Theobald S, et al. Cyclin L1 (CCNL1) gene alterations in human head and neck squamous cell carcinoma. Br J Cancer. 2006;94:1041–4. doi: 10.1038/sj.bjc.6603036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ramsay AJ, Quesada V, Foronda M, et al. POT1 mutations cause telomere dysfunction in chronic lymphocytic leukemia. Nat Genet. 2013;45:526–30. doi: 10.1038/ng.2584. [DOI] [PubMed] [Google Scholar]
- 23.Tsikitis M, Zhang Z, Edelman W, Zagzag D, Kalpana GV. Genetic ablation of Cyclin D1 abrogates genesis of rhabdoid tumors resulting from Ini1 loss. Proc Natl Acad Sci USA. 2005;102:12129–34. doi: 10.1073/pnas.0505300102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nair P, De Preter K, Vandesompele J, Speleman F, Stallings RL. Aberrant splicing of the PTPRD gene mimics microdeletions identified at this locus in neuroblastomas. Genes Chromosom Cancer. 2008;47:197–202. doi: 10.1002/gcc.20521. [DOI] [PubMed] [Google Scholar]
- 25.Solomon DA, Kim JS, Cronin JC, et al. Mutational inactivation of PTPRD in glioblastoma multiforme and malignant melanoma. Cancer Res. 2008;68:10300–6. doi: 10.1158/0008-5472.CAN-08-3272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Poliak S, Gollan L, Martinez R, et al. Caspr2, a new member of the neurexin superfamily, is localized at the juxtaparanodes of myelinated axons and associates with K+ channels. Neuron. 1999;24:1037–47. doi: 10.1016/s0896-6273(00)81049-1. [DOI] [PubMed] [Google Scholar]
- 27.McAvoy S, Ganapathiraju SC, Ducharme-Smith AL, et al. Non-random inactivation of large common fragile site genes in different cancers. Cytogenet Genome Res. 2007;118:260–9. doi: 10.1159/000108309. [DOI] [PubMed] [Google Scholar]
- 28.Bralten LB, Gravendeel AM, Kloosterhof NK, et al. The CASPR2 cell adhesion molecule functions as a tumor suppressor gene in glioma. Oncogene. 2010;29:6138–48. doi: 10.1038/onc.2010.342. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Quantitative-genomic PCR for chromosomal deleted regions detected by single nucleotide polymorphism-chip analysis in malignant rhabdoid tumor.
Analyzed samples of malignant rhabdoid tumor and single nucleotide polymorphism array platform.
Table S2. Primer sets for quantitative-genomic PCR in malignant rhabdoid tumor.