Abstract
After a 75-year absence from Florida, substantial local transmission of dengue virus (DENV) occurred in Key West, Monroe County, Florida in 2009 and continued in 2010. The outbreak culminated in 85 reported cases. In 2011 and 2012, only isolated cases of local DENV transmission were reported in Florida, none were reported in Key West. In 2013, a new outbreak occurred, but this time in Martin County about 275 miles North of Key West with 22 reported cases. As the Key West and Martin County outbreaks involved DENV serotype 1 (DENV-1), we wanted to investigate whether the same strain or a different strain of DENV was responsible for the outbreaks. In this study, we report the sequence and phylogenetic analysis of the E generegion from a patient diagnosed with dengue in Martin County. Our results indicate that the 2013 Martin County DENV-1 strain is distinct from the 2009–2010 Key West DENV-1 and that it is most closely related to viruses from a recent expansion of South American DENV-1 strains into the Caribbean. We conclude that the 2013 Martin County outbreak was the result of a new introduction of DENV-1 in Florida.
INTRODUCTION
Currently, about 40% of the world’s population lives in areas at risk of dengue infection (1) and the incidence of dengue is increasing in range and intensity worldwide. A recent meta-analysis estimated 390 million dengue infections per year, more than three times the burden previously estimated by the World Health Organization (1, 2). Dengue is caused by a mosquito-transmitted flavivirus, dengue virus (DENV). DENV infection typically manifests as an acute febrile illness with highly variable outcomes ranging from in apparent symptoms to hemorrhagic fever, shock syndrome or even death. There are four distinct serotypes of DENV (DENV-1, -2, - 3, and -4). Infection with one serotype typically provides lifetime protection against the same serotype, but the resulting immune response can lead to increased disease severity during a secondary infection with a different serotype. There are currently no commercially available vaccines to prevent DENV infection or specific drugs to inhibit viral replication. The primary means of disease prevention and spread is vector control.
DENV is primarily transmitted by peridomestic Aedes aegypti and Ae. albopictus mosquito vectors. Both of these species are typically found in tropical and subtropical regions. However, in recent years, the range of these mosquito vectors has increased, leading to a subsequent expansion of the range of DENV transmission (3, 4). Global travel and commerce have played key roles in range expansion and increasing transmission. Transported mosquito adults and larvae, as well as DENV infected travelers returning from regions where DENV is endemic, can initiate de novo local DENV transmission if the mosquito vectors are present. As a result, sporadic outbreaks of locally transmitted DENV have occurred in numerous temperate regions including France, Croatia, and the United States (U.S.) (5–7). According to the U.S. Geological Survey, in 2013, there were 519 laboratory-confirmed imported DENV cases in 39 states (http://diseasemaps.usgs.gov/dep_us_human.html). Given the variable symptoms and lack of clinical experience with DENV in the U.S., this is almost certainly an underestimate of the true number of imported cases. Ae. aegypti is found in 19 of these states with its range stretching across the southeastern U.S., up the east coast to New York and west to Kentucky and Indiana, and Ae. albopictus is now established on the Atlantic seaboard from Florida to southern New York (8).
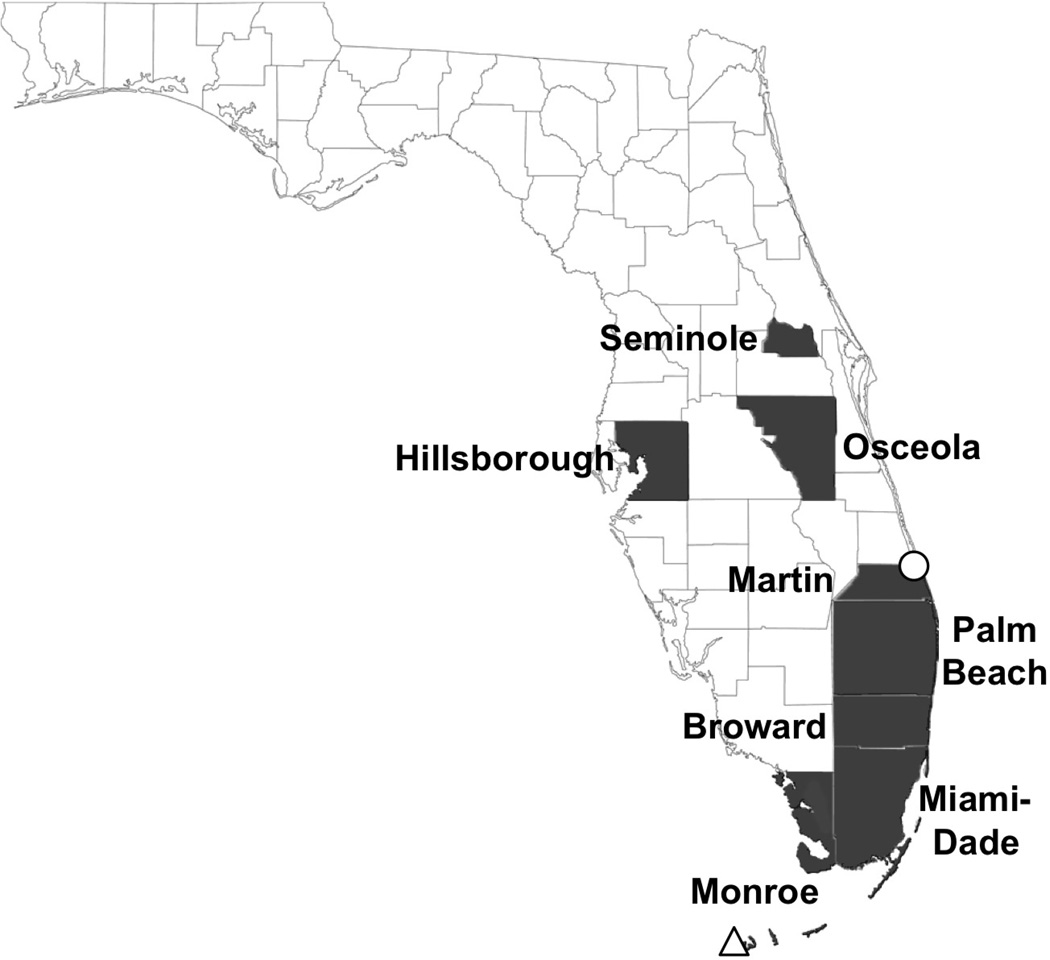
In 2009, after a 75-year absence from Florida, a substantial outbreak of locally transmitted DENV occurred in Key West, Monroe County, Florida. According to the Florida Department of Health, twenty-two cases of locally transmitted DENV were confirmed that year (http://www.floridahealth.gov/diseases-and-conditions/mosquito-borne-diseases/surveillance.html). In 2010, an additional sixty-three cases of locally acquired DENV were reported in Monroe County and one case each in Miami-Dade and Broward Counties. The same strain of DENV serotype 1 (DENV-1)was isolated from both mosquitoes and patients in Monroe County, confirming local transmission (9, 10). No further cases of locally acquired DENV have been reported in Monroe County since 2010, suggesting that DENV had been extirpated from the local vector population in that location. However, small numbers of sporadic cases with no travel history have continued in Florida. In 2011, seven additional cases of locally acquired DENV were reported: three cases in Miami-Dade, two in Palm Beach, and one each in Martin and Hillsborough Counties. In 2012, four more cases were reported: two in Miami-Dade and one each in Osceola and Seminole Counties. Most recently, in 2013, another substantial outbreak occurred where twenty-three cases of locally acquired DENV were reported: twenty-two in Martin and one in Miami-Dade Counties. The Florida counties with reported cases of locally acquired DENV are shown in Figure 1.
Figure 1.
Map of Florida showing counties with reported locally transmitted DENV cases 2009–2013. Old Town Key West (open triangle) in Monroe County, the site of the 2009–2010 DENV-1 outbreak, is approximated 275 miles from the sites of the 2013 DENV-1 outbreak, Jensen Beach and Rio (open circle) in Martin County. Data was derived from the Florida Department of Health Mosquito-borne Diseases Surveillance reports (http://www.floridahealth.gov/diseases-and-conditions/mosquito-borne-diseases/surveillance.html).
In this study, we set out to determine whether the locally transmitted DENV strain from Martin County in 2013 is the same or different as the locally transmitted DENV from Key West in 2009–2010. The answer to this question has major implications for control efforts and epidemiological surveillance. If the two viruses are similar, then that would suggest that a single introduction had spread to multiple areas in Florida due to movement of people and/or mosquitoes within the state. The distance between the neighborhoods of Old Town Key West and Jensen Beach and Rio in Martin County is about 275 miles (Figure 1), linked much of the way by direct interstate highways. If the two viruses are distinct, that would suggest a new introduction of DENV in Florida from outside the U.S. Control and surveillance measures to address these two distinct scenarios would differ in focusing on local versus international transport.
Here we report the sequence and phylogenetic analyses of the E protein region of DENV amplified from a single patient diagnosed in Martin County in 2013. Our results indicate that the Martin County and Key West DENV are both DENV-1, but that the two strains are distinct. While the Key West DENV-1 was most closely related to viruses from Nicaragua, the Martin County DENV-1 is most closely related to viruses from a recent expansion of South American DENV-1 viruses into the Caribbean.
RESULTS
Semi-nested reverse transcriptase PCR (RT-PCR) was used to identify viremic samples and distinguish the infecting serotype in13 serum samples collected between August 2 and September 26, 2013 from patients with dengue symptoms and no travel history in Martin County, Florida (11). A single patient sample from August 29, 2013 was positive for DENV by first round RT-PCR. No other RT-PCR reaction from any of the other 12 patient samples showed a product consistent with amplification of DENV sequences. The second round, semi-nested PCR from the positive sample identified the serotype as DENV-1. Additional RT-PCR was performed to amplify the complete E protein coding region and adjacent regions. The sequence of this amplification product has been deposited in GenBank (MartinFL_USA2013, accession number KJ415284). The Martin County DENV-1 E sequence was aligned with a set of 188 additional non-redundant American DENV-1 E sequences from the National Center for Biotechnology Information Virus Variation database(12). This set included the majority of deposited Western hemisphere DENV-1 sequences, in addition to several isolates from Hawaii(USA) and Easter Island (Chile) that belong to Asian DENV-1 clades and served as an out group.
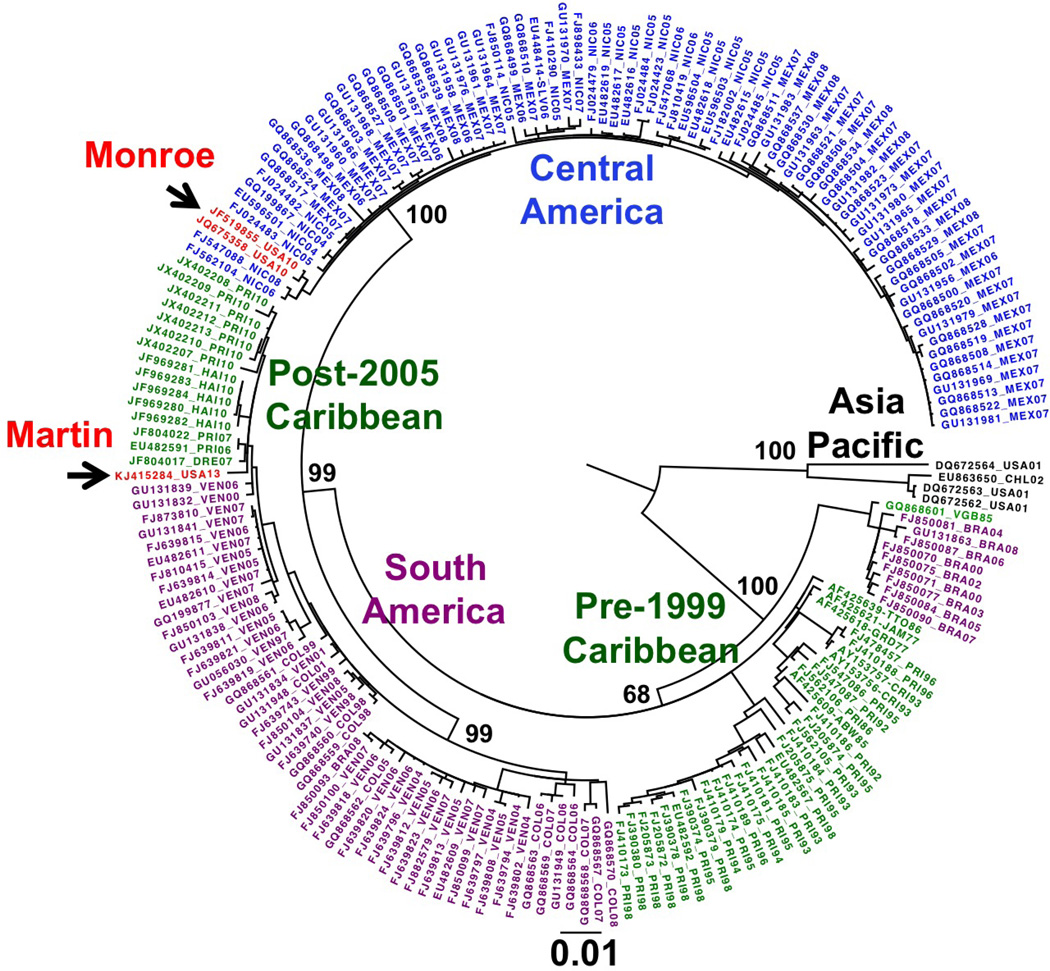
Maximum likelihood, maximum parsimony and distance methods using the 1,480-nt E gene region yielded phylogenetic trees with very similar topologies. The maximum likelihood tree is shown in Figure 2. Western hemisphere DENV-1 strains group in geographically bounded clades. Evidence for clade replacement was seen most strikingly with a group of recent sequences from the Caribbean that are related to earlier South American DENV-1 clades rather than Central American clades (13). The clade containing Central American strains was separated from the clade containing South American and recent Caribbean strains with strong bootstrap support. The Key West 2010 strains grouped with Central American strains, as previously reported (9, 10). The Martin County DENV-1 strain grouped most closely with viruses that have recently appeared in the Caribbean from South American origins. In total, 12 nucleotide changes were observed in the 1,480 nt aligned E gene region between the Martin County DENV-1 and the most closely related Caribbean strain, a DENV-1 found in the Dominican Republic in 2007 (GenBank accession number JF804017). Comparison of the predicted translation products of this Dominican Republic strain and the Martin County strain showed 3 amino acid changes in E. Using DENV-1 E protein numbering, the three altered amino acids were valine at E protein position 55 changed to isoleucine in the Martin County sequence (V55I), threonine at position 160 changed to isoleucine (T160I) and lysine at position 394 changed to arginine (K394R).
Figure 2.
Maximum-likelihood phylogenetic tree of the 1,480-nt envelope gene region from Western hemisphereDENV-1 isolates. Sequences are labeled with GenBank accession numbers, country, and year. Countries are given as standard 3-letter codes. Scale bar indicates number of substitutions per site. The Martin County and Key West sequences are shown in red; Central American sequences, in blue; Caribbean sequences, in green; South American sequences, in purple, Asian/Pacific sequences, in black. Isolates from Hawaii (USA) and Easter Island (CHI) are of Asian origin and form an out group. Viruses group by geography and by year. The bootstrap values of the important nodes separating Central American from South American and recent Caribbean strains are shown. The tree was drawn by using FigTree software (http://tree.bio.ed.ac.uk/software/figtree).
DISCUSSION
The 2010 Key West DENV-1 strain grouped most closely with Central American viruses originating in Nicaragua (9) whereas the 2013 Martin County DENV-1 strain originated from a clade of DENV-1 that recently appeared in the Caribbean from South American origins. These two viruses are clearly distinct and are not a result of a single DENV-1 introduction in Florida and subsequent local spread of the virus by humans and/or mosquitoes. The bootstrap values supporting the separation of the clades that distinguish these viruses are high, indicating strong statistical support.
While the Martin County DENV-1 strain groups most closely with Caribbean DENV-1 strains, the branch length between the Martin County DENV-1 strain and all of the related Caribbean DENV-1 strains is relatively long, indicating that the Martin County strain is not very closely related to any specific Caribbean strain that has been sequenced. One possible interpretation for this relatively long branch length is that the Martin County DENV-1 strain had been isolated for some time from its closest relatives, either in the Caribbean, or possibly in Florida. An alternate, and perhaps more likely, scenario is that recent surveillance and sequencing from the Caribbean islands is incomplete and the closest relatives of the Martin County DENV-1 virus have not been sampled to date. As a result, it is difficult to determine with certainty which Caribbean island may have been the source of the 2013 Martin County DENV-1 strain. The Martin County DENV-1 strain has three amino acid changes in the E protein compared to the closest relative from the Dominican Republic. Two of these changes (V55I and K394R) are relatively conservative and are also found in a number of other related Caribbean DENV-1 strains. The T160I change on the other hand, is non-conservative and is not found in any other related DENV-1 strain. This change is structurally located on the exposed surface of the E protein domain I near the domain I/II hinge region, and may play a role in viral entry or immune escape.
There are many similarities between Old Town, Key West and the Jensen Beach and Rio neighborhoods where DENV cases were reported in Martin County in 2013. One that stands out is that neither location is a major port of entry either for aviation or shipping. The number of locally acquired cases was large compared to other Florida counties, indicating that factors existed in both Key West and Martin County that favored local transmission that may not have existed in other sites of local transmission. Locally acquired DENV has been reported in recent years in a number of metropolitan areas in Florida, but transmission was limited to a few cases at most. Therefore, a port of entry with large traffic from DENV endemic countries (for example, Miami-Dade County)may not be sufficient to initiate local DENV transmission and spread. Additional factors need to be considered. As in Key West, many households and business in Jensen Beach and Rio maintain their windows open more than 50% of the time, have vegetation in more than 50% of their property, or keep open-air containers such as birdbaths or other receptacles ((14) and G. Lemire, presented at the 85th Annual Meeting of the Florida Mosquito Control Association, Cape Coral, FL, 17 to 20 November 2013). These factors could contribute to increased human exposure to mosquito bites.
The study of DENV introduction and local transmission in Florida can inform dengue control efforts and epidemiological surveillance. In this instance, we were only able to obtain sequence data from an individual patient sample and it is possible that other strains were also involved. However, to date, there is no evidence to support spread of DENV from one county to another within the state of Florida. The 2009–2010 Key West DENV-1 strain has not resurfaced in Martin County or appeared elsewhere in Florida. The end of the Key West outbreak could be attributed to effective vector control, climatic conditions, or human behaviors that limited exposure to mosquito bites. It remains to be seen whether DENV transmission will resume next rainy season in Martin County.
Frequent travel to dengue endemic countries, transport of goods, the presence of mosquito vectors, and behaviors that promote exposure to mosquito bites contribute to new introductions of DENV in the U.S. Consistent with the northernmost range of Ae. aegypti and Ae. albopictus, Suffolk County, New York was the site of a locally acquired case of DENV in 2013(15). Thus, recent sporatic outbreaks of local DENV transmission in the continental U.S. are not limited to the Texas-Mexico border as they had been in the past. Public awareness about DENV and its symptoms will increase the likelihood that an infected traveler will seek medical care. Effective surveillance of imported dengue cases including rapid diagnosis of infected travelers, quarantine measures (avoidance of mosquitoes), and increasing vector control measures in the immediate vicinity of index cases will play key roles in preventing the spread of DENV in currently non-endemic regions. The key to coordinating and mobilizing these efforts will be effective communication between diagnosing physicians, state health departments, mosquito control districts, local government officials, university research laboratories, and the public at large.
MATERIALS AND METHODS
Source of blood samples
Patient blood samples were collected for diagnostic purposes. The use of de-identified, unused patient samples for DENV analysis was approved by the Institutional Review Boards of Martin Health System and Florida Gulf Coast University (protocol number: 2013-61).
Viral RNA was extracted from serum samples using a Qiagen RNeasy mini kit (Valencia, CA, USA) following the manufacturer’s instructions. DENV RNA was amplified using pan3 dengue virus consensus primers by RT-PCR with a Qiagen One-step kit (Qiagen, Valencia, CA, USA)(11). A second semi-nested round of PCR was performed with serotype-specific primers using Invitrogen Platinum PCR Supermix High Fidelity (Invitrogen, Carlsbad, CA, USA)(11). The pan-DENV and DENV serotype specific amplification products were visualized by agarose gel electrophoresis. For phylogenetic analysis, a larger, 2,594 bp fragment containing the entire E gene was amplified by RT-PCR using the following primers:
Forward 1KB-3F 5’-GCACATGCCATAGGAACATCCATCAC-3’
Reverse 3KBR 5’-ACTTCTCCTGACCCTGCAGAGACCAT-3’
Three separate RT-PCR amplifications were performed and the products were each directly sequenced using terminal and internal primers by Sanger sequencing at double or triple coverage (Functional Biosciences, Madison, WI, USA). Sequence run data were assembled and edited using DNASTAR Laser Gene software. A total of 188 American DENV-1sequences from 1980 to 2013 were obtained through NCBI Virus Variation database to build a dataset for phylogenetic analysis (http://www.ncbi.nlm.nih.gov/genomes/VirusVariation/). Sequences were aligned using the MUSCLE algorithm and phylogenetic trees were generated using the Sea view software package (16).
ACKNOWLEDGEMENTS
Research reported in this publication was supported by the National Institute of Allergy and Infectious Diseases of the National Institutes of Health under Award Number R01AI099210. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.World Heath Organization. Geneva: 2009. Dengue: Guidelines for Diagnosis, Treatment, Prevention and Control: New Edition. [PubMed] [Google Scholar]
- 2.Bhatt S, Gething PW, Brady OJ, Messina JP, Farlow AW, Moyes CL, Drake JM, Brownstein JS, Hoen AG, Sankoh O, Myers MF, George DB, Jaenisch T, Wint GR, Simmons CP, Scott TW, Farrar JJ, Hay SI. The global distribution and burden of dengue. Nature. 2013;496:504–507. doi: 10.1038/nature12060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Benedict MQ, Levine RS, Hawley WA, Lounibos LP. Spread of the tiger: global risk of invasion by the mosquito Aedes albopictus. Vector Borne Zoonotic Dis. 2007;7:76–85. doi: 10.1089/vbz.2006.0562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Enserink M. Entomology. A mosquito goes global. Science. 2008;320:864–866. doi: 10.1126/science.320.5878.864. [DOI] [PubMed] [Google Scholar]
- 5.La Ruche G, Souares Y, Armengaud A, Peloux-Petiot F, Delaunay P, Despres P, Lenglet A, Jourdain F, Leparc-Goffart I, Charlet F, Ollier L, Mantey K, Mollet T, Fournier JP, Torrents R, Leitmeyer K, Hilairet P, Zeller H, Van Bortel W, Dejour-Salamanca D, Grandadam M, Gastellu-Etchegorry M. First two autochthonous dengue virus infections in metropolitan France, September 2010. Euro Surveill. 2010;15:19676. [PubMed] [Google Scholar]
- 6.Schmidt-Chanasit J, Haditsch M, Schoneberg I, Gunther S, Stark K, Frank C. Dengue virus infection in a traveller returning from Croatia to Germany. Euro Surveill. 2010;15:19677. doi: 10.2807/ese.15.40.19677-en. [DOI] [PubMed] [Google Scholar]
- 7.Centers for Disease Control and Prevention. Locally acquired Dengue--Key West, Florida, 2009–2010. MMWR Morb Mortal Wkly Rep. 2010;59:577–581. [PubMed] [Google Scholar]
- 8.Darsie RF, Ward RA. Identification and Geographical Distribution of the Mosquitoes of North America, North of Mexico. Gainesville, FL: University of Florida Press; 2005. [Google Scholar]
- 9.Graham AS, Pruszynski CA, Hribar LJ, DeMay DJ, Tambasco AN, Hartley AE, Fussell EM, Michael SF, Isern S. Mosquito-associated dengue virus, Key West, Florida, USA, 2010. Emerg Infect Dis. 2011;17:2074–2075. doi: 10.3201/eid1711.110419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Munoz-Jordan JL, Santiago GA, Margolis H, Stark L. Genetic relatedness of dengue viruses in Key West, Florida, USA, 2009–2010. Emerg Infect Dis. 2013;19:652–654. doi: 10.3201/eid1904.121295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lanciotti RS, Calisher CH, Gubler DJ, Chang GJ, Vorndam AV. Rapid detection and typing of dengue viruses from clinical samples by using reverse transcriptase-polymerase chain reaction. J Clin Microbiol. 1992;30:545–551. doi: 10.1128/jcm.30.3.545-551.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Resch W, Zaslavsky L, Kiryutin B, Rozanov M, Bao Y, Tatusova TA. Virus variation resources at the National Center for Biotechnology Information: dengue virus. BMC Microbiol. 2009;9:65. doi: 10.1186/1471-2180-9-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang C, Mammen MP, Jr, Chinnawirotpisan P, Klungthong C, Rodpradit P, Monkongdee P, Nimmannitya S, Kalayanarooj S, Holmes EC. Clade replacements in dengue virus serotypes 1 and 3 are associated with changing serotype prevalence. J Virol. 2005;79:15123–15130. doi: 10.1128/JVI.79.24.15123-15130.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Radke EG, Gregory CJ, Kintziger KW, Sauber-Schatz EK, Hunsperger EA, Gallagher GR, Barber JM, Biggerstaff BJ, Stanek DR, Tomashek KM, Blackmore CG. Dengue outbreak in Key West, Florida, USA, 2009. Emerg Infect Dis. 2012;18:135–137. doi: 10.3201/eid1801.110130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rochlin I, Ninivaggi DV, Hutchinson ML, Farajollahi A. Climate change and range expansion of the Asian tiger mosquito (Aedes albopictus) in Northeastern USA: implications for public health practitioners. PLoS One. 2013;8:e60874. doi: 10.1371/journal.pone.0060874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gouy M, Guindon S, Gascuel O. Sea View version 4: A multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol Biol Evol. 2010;27:221–224. doi: 10.1093/molbev/msp259. [DOI] [PubMed] [Google Scholar]