The autocatalytic activation of an initiator caspase, exemplified by caspase-9 in mammals or its ortholog, Dronc, in fruit flies, is facilitated by a multimeric adaptor complex known as the apoptosome. Pang et al. report two cryo-EM structures: the complete Dark apoptosome at an overall resolution of 4.0 Å and a complex between the Dark apoptosome and the CARD of Dronc at 4.1 Å resolution. The structural findings, together with structure-guided biochemical analyses, allow delineation of the molecular mechanisms for Dronc activation.
Keywords: programmed cell death, apoptosis, caspase activation, Dark, Dronc, cryo-EM structure
Abstract
Apoptosis is executed by a cascade of caspase activation. The autocatalytic activation of an initiator caspase, exemplified by caspase-9 in mammals or its ortholog, Dronc, in fruit flies, is facilitated by a multimeric adaptor complex known as the apoptosome. The underlying mechanism by which caspase-9 or Dronc is activated by the apoptosome remains unknown. Here we report the electron cryomicroscopic (cryo-EM) structure of the intact apoptosome from Drosophila melanogaster at 4.0 Å resolution. Analysis of the Drosophila apoptosome, which comprises 16 molecules of the Dark protein (Apaf-1 ortholog), reveals molecular determinants that support the assembly of the 2.5-MDa complex. In the absence of dATP or ATP, Dronc zymogen potently induces formation of the Dark apoptosome, within which Dronc is efficiently activated. At 4.1 Å resolution, the cryo-EM structure of the Dark apoptosome bound to the caspase recruitment domain (CARD) of Dronc (Dronc-CARD) reveals two stacked rings of Dronc-CARD that are sandwiched between two octameric rings of the Dark protein. The specific interactions between Dronc-CARD and both the CARD and the WD40 repeats of a nearby Dark protomer are indispensable for Dronc activation. These findings reveal important mechanistic insights into the activation of initiator caspase by the apoptosome.
Apoptosis, also known as programmed cell death, plays a central role in the development and tissue homeostasis of all metazoans (Horvitz 2003; Danial and Korsmeyer 2004; Fuchs and Steller 2011). Apoptosis is executed by a cascade of caspase activation (Yan and Shi 2005). An effector caspase responsible for dismantling the cell is activated by an initiator caspase through an intrachain cleavage (Yan and Shi 2005). The initiator caspase, on the other hand, requires an autocatalytic cleavage for maturation; this process is facilitated by a multimeric protein complex, often known as the apoptosome (Chai and Shi 2014). The initiator caspase is exemplified by caspase-9 in mammals or its functional ortholog, Dronc, in the fruit fly Drosophila melanogaster. Caspase-9 and Dronc are activated by the Apaf-1 apoptosome and the Dark apoptosome, respectively (Chai and Shi 2014).
Research in the past 15 years has led to a clear delineation of the sequential events required for apoptosome formation (Chai and Shi 2014). In mammalian cells, assembly of the Apaf-1 apoptosome is triggered by the release of cytochrome c (Cyt. c) from mitochondria into the cytoplasm (Liu et al. 1996), where Cyt. c binds Apaf-1 in its monomeric, autoinhibited state. Next, the replacement of ADP by dATP or ATP in Apaf-1 causes marked conformational changes, resulting in the formation of a heptameric apoptosome (Kim et al. 2005; Bao et al. 2007). The Apaf-1 apoptosome facilitates the autocatalytic cleavage of the caspase-9 zymogen. The activated caspase-9 remains associated with the Apaf-1 apoptosome as a holoenzyme, which exhibits a catalytic activity two to three orders of magnitude higher than that of free caspase-9 (Rodriguez and Lazebnik 1999). In fruit flies, Dark, the functional ortholog of Apaf-1 (Kanuka et al. 1999; Rodriguez et al. 1999; Zhou et al. 1999), is required for Dronc activation. Formation of the Dark apoptosome can be induced by the presence of dATP (Yu et al. 2006) but not Cyt. c.
Apaf-1 and Dark share an identical domain structure, with a caspase recruitment domain (CARD) at the N terminus followed by a nucleotide-binding domain (NBD), a helical domain (HD1), a winged-helix domain (WHD), a second helical domain (HD2), and 15 WD40 repeats at the C terminus. Of these domains, NBD, HD1, and WHD are collectively referred to as the nucleotide-binding oligomerization domain (NOD).
Despite advances in understanding the sequential events required for apoptosome formation, how the initiator caspase—caspase-9 or Dronc—is activated by the relevant apoptosome remains largely unknown. During the past decade, the electron cryomicroscopic (cryo-EM) structures of the Apaf-1 apoptosome (Acehan et al. 2002; Yu et al. 2005; Yuan et al. 2010, 2011a, 2013) and the Dark apoptosome (Yu et al. 2006; Yuan et al. 2011b) have been elucidated at relatively low resolutions. These structures allowed placement of individual domains within Apaf-1 or Dark but failed to reveal specific determinants that support the function of the apoptosomes. Elucidation of the activation mechanism for an initiator caspase requires structural information of the apoptosome at an atomic resolution and structure-guided biochemical analyses.
In this study, we report two cryo-EM structures: the complete Dark apoptosome at an overall resolution of 4.0 Å and a complex between the Dark apoptosome and the CARD of Dronc at 4.1 Å resolution. The resolution limits are ∼3.0–3.5 Å for the important core regions of the structures, which allow determination of the specific atomic interactions. The structural findings, together with structure-guided biochemical analyses, allow delineation of the molecular mechanisms for Dronc activation.
Results
Structure of the Dark apoptosome
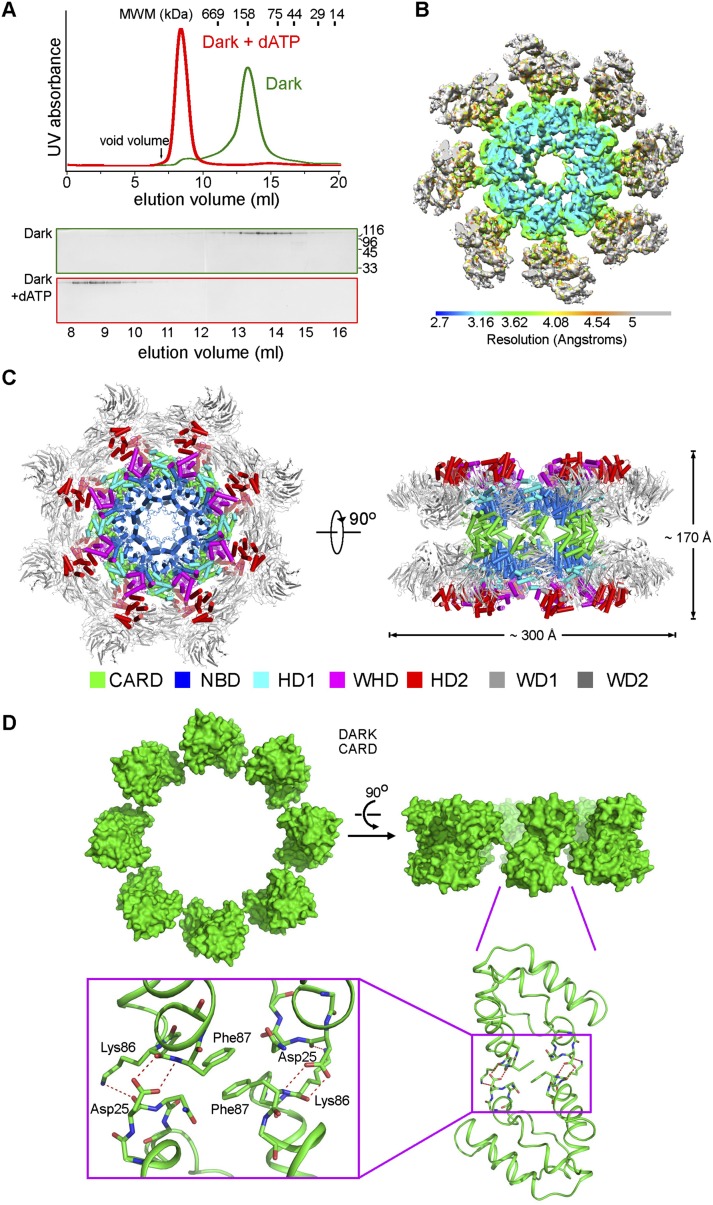
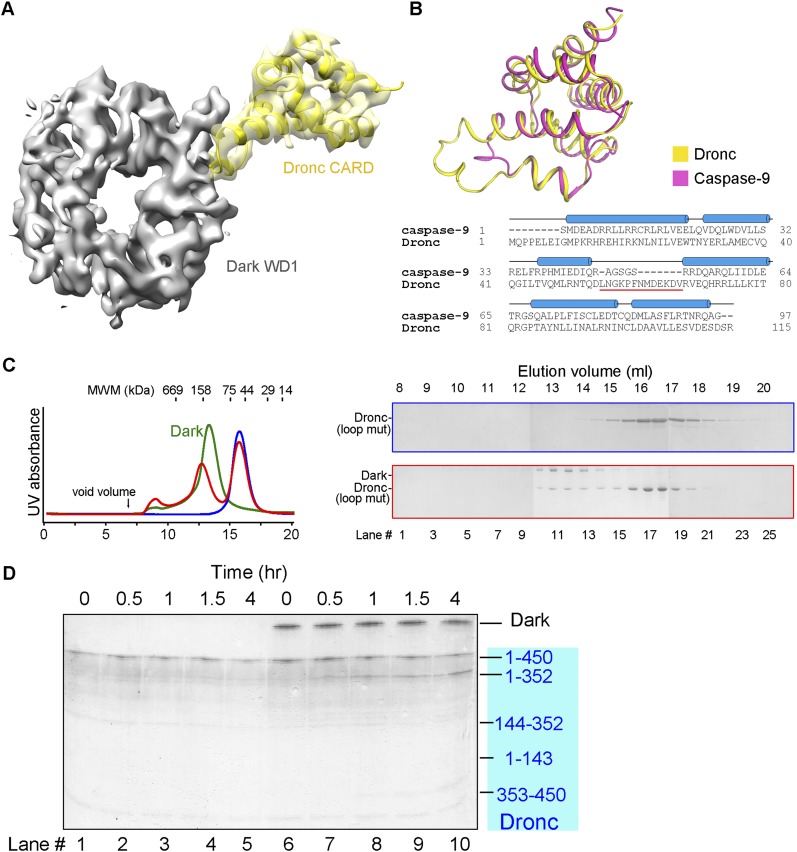
We purified to homogeneity the recombinant, full-length Dark protein (residues 1–1440) from baculovirus-infected insect cells. The purified Dark was eluted from gel filtration with an apparent molecular mass of ∼160 kDa, consistent with that of a Dark monomer (Fig. 1A). This observation suggests that, similar to Apaf-1, Dark may exist in an autoinhibited conformation in the basal state. As previously reported (Yu et al. 2006), incubation of Dark with 5 mM dATP resulted in the formation of a Dark apoptosome, which exhibited an apparent molecular mass of ∼2.5 MDa (Fig. 1A). The Dark apoptosome was stable in solution and displayed clearly identifiable features by cryo-EM single-particle analysis (Supplemental Fig. S1A,B). We selected 9354 particles for reconstruction and determined the structure at an overall resolution of 4.0 Å (Supplemental Figs. S1C,D, S2A,B). The overall correctness of the density map and its handedness were confirmed by the tilt pair test (Supplemental Fig. S2C; Rosenthal and Henderson 2003).
Figure 1.
Structure of the Dark apoptosome at an overall resolution of 4.0 Å. (A) The monomeric Dark protein assembles into a multimeric Dark apoptosome in the presence of 5 mM dATP. The gel filtration chromatograms are shown in the top panel, and SDS-PAGE gels of the peak fractions (stained by Coomassie blue) are shown in the bottom panel. (B) The overall EM density map of the Dark apoptosome is color-coded to indicate a range of resolution. Despite an overall resolution of 4.0 Å, much of the central region of the Dark apoptosome is resolved at resolutions better than 3.5 Å. This and other figures that display the EM density maps were generated in Chimera (Pettersen et al. 2004). (C) Overall structure of the Dark apoptosome is shown in two perpendicular views. The individual domains are color-coded. All cartoon diagrams of the structures were prepared using PyMol (http://www.pymol.org). (D) The 16 molecules of Dark-CARD assemble into a discontinuous ring. The ring contains two layers, one from each Dark octamer. Two Dark-CARDs, each from a different layer, stack closely against each other. Each Asp25 residue mediates three intermolecular H bonds, whereas two Phe87 residues from the two layers interact with each other through van der Waals contacts.
The resolution limit for much of the central region of the Dark apoptosome goes beyond 3.5 Å (Fig. 1B), allowing assignment of specific interactions. This central region, including CARD and the entire NOD (Fig. 1C), contains all of the characterized domains that are essential for Dronc activation. In contrast, the periphery, mostly involving the WD40 repeats, displayed considerably lower resolution. The uneven distribution of the resolution limit likely reflects the inherently stable conformation at the center and the relatively flexible nature at the periphery. On the basis of the EM density, an atomic model for the central region and the bulk of the rest was built using COOT (Emsley and Cowtan 2004) and refined using REFMAC (Murshudov et al. 2011; Amunts et al. 2014).
The Dark apoptosome consists of two octameric rings with a diameter of 300 Å and a thickness of 170 Å (Fig. 1C). Each ring comprises eight molecules of Dark, with their CARDs protruding above the plane of the ring and forming a discontinuous ring (Fig. 1D). Each CARD stacks against a CARD from the other octameric ring. Specifically, the carboxylate side chain of Asp25 from one CARD makes a pair of intermolecular hydrogen bonds (H bonds): one to the carbonyl oxygen of Lys86 and the other to the amide nitrogen of Phe87, both residues from the other CARD. In addition, the carbonyl oxygen of Asp25 accepts an intermolecular H bond from the side chain of Lys86 (Fig. 1D). Notably, these H bonds are stabilized by the π–π stacking interactions between two Phe87 residues from the two neighboring CARDs.
ATP binding and interdomain interactions
The highest resolutions previously achieved were 6.9 Å for the Dark apoptosome (Yuan et al. 2011b) and 9.5 Å for the Apaf-1 apoptosome (Yuan et al. 2010), neither of which was sufficient for identification of atomic interactions. The current overall resolution of 4 Å, especially the range of 3.0–3.5 Å in the central region of the Dark apoptosome, represents a qualitative advance and allows assignment of specific side chains and atomic interactions both between neighboring domains within the same Dark protein and between two Dark protomers (Supplemental Fig. S1D). These molecular determinants, visualized for the first time, are responsible for the formation of the Dark apoptosome and ultimately control its function of facilitating Dronc activation.
ATP is bound at the interface between the NBD and HD1, surrounded by a number of polar amino acids, including Gly154, Gly156, Lys157, Thr158, Asn246, Arg267, and Tyr304 (Supplemental Fig. S3A). These residues directly contribute to ATP coordination through H bonds. In particular, four residues (Gly154/Gly156/Lys157/Thr158), invariant between Apaf-1 and Dark, constitute the signature P-loop sequence (Gly–Xaa–Gly–Lys–Thr/Ser) for nucleotide binding. Although detailed structural information of the activated Apaf-1 remains to be elucidated, a crystal structure of the CED-4 apoptosome is available (Qi et al. 2010). CED-4 is the Apaf-1 homolog in the nematode worm Caenorhabditis elegans. The ATP-binding mode in the Dark apoptosome is reminiscent of that in the CED-4 apoptosome (Supplemental Fig. S3B; Qi et al. 2010).
Formation of the Dark apoptosome involves elaborate interactions both within the same Dark protein and between Dark protomers (Supplemental Fig. S4A). The intermolecular interactions mainly come from neighboring NBDs, confirming the NBD as the primary element of oligomerization. Additional contacts occur between residues in the HD1 of one protomer and the corresponding amino acids in the NBD of a neighboring protomer, exemplified by potential H bonds between Arg332 and Asp273 (Supplemental Fig. S4B). The intramolecular interactions occur between the CARD and the NBD or among the NBD, HD1, and WHD (Supplemental Fig. S4B).
Dronc activation within a multimeric Dronc–Dark complex
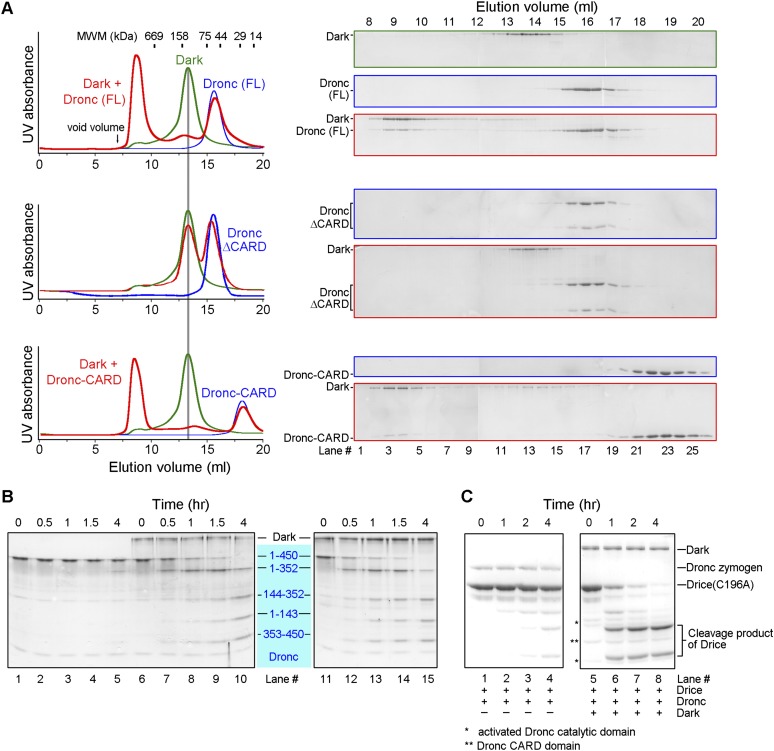
During characterization of the Dark apoptosome, we made a serendipitous discovery: The Dronc zymogen and Dark assemble into a multimeric complex in the absence of any exogenous nucleotide. Upon incubation with the full-length Dronc zymogen (residues 1–450, C318A), the full-length, monomeric Dark was converted to a large oligomer as judged by gel filtration analysis (Fig. 2A, top panels). Reminiscent of the caspase-9–Apaf-1 holoenzyme (Rodriguez and Lazebnik 1999), Dronc was also shifted into this multimeric complex, indicating stable association between Dronc and Dark. A Dronc variant that lacks the CARD (Dronc-ΔCARD) was unable to interact with Dark or induce its oligomerization (Fig. 2A, middle panels). These observations demonstrate that the CARD of Dronc (Dronc-CARD) is required for the interactions between Dronc and Dark as well as for the formation of the multimeric Dark complex. Next, we examined the possibility that Dronc-CARD alone may be sufficient to induce formation of the multimeric Dark complex. Our experimental result unambiguously confirmed this possibility (Fig. 2A, bottom panels). As will be seen later, structure of the multimeric Dark complex in the Dronc-CARD–Dark oligomer is identical to that of the Dark apoptosome.
Figure 2.
The Dronc zymogen and Dark assemble into a multimeric complex that mediates Dronc activation. (A, top panels) The Dronc zymogen (residues 1–450, C318A) and Dark assemble into a multimeric complex. (Middle panels) The caspase domain of Dronc (residues 131–352 and 353–450, C318A) fails to interact with Dark. (Bottom panels) The Dronc-CARD (residues 1–130) also induces formation of the multimeric complex. Gel filtration chromatograms are shown in the left panels, and SDS-PAGE gels of the peak fractions (stained by Coomassie blue) are shown in the right panels. (B) The wild-type (WT) Dronc zymogen undergoes rapid autocatalytic activation in the presence of Dark. The wild-type Dronc zymogen remained intact in the absence of Dark (lanes 1–5) but was fully autoproteolyzed in the presence of Dark (lanes 6–10). (Lanes 11–15) Incubation of the wild-type Dronc zymogen with the preassembled, dATP-induced Dark apoptosome also led to autoproteolysis of the Dronc zymogen. (C) The autocatalytically processed Dronc exhibits robust protease activity. (Lanes 1–4) The wild-type Dronc zymogen had little protease activity toward its physiological substrate, Drice (residues 1–339, C196A). (Lanes 5–8) In contrast, preincubation of the wild-type Dronc zymogen with Dark resulted in robust protease activity.
What is the function of the multimeric complex between Dronc and Dark? To answer this question, we purified the wild-type, full-length Dronc to homogeneity and examined its autocatalytic activation under contrasting conditions. In the absence of the monomeric Dark protein, the autocatalytic cleavage of the Dronc zymogen occurred at a basal and barely detectable level (Fig. 2B, lanes 1–5). In sharp contrast, incubation of the Dronc zymogen with Dark resulted in rapid and complete autocatalytic cleavage of Dronc within 4 h (Fig. 2B, lanes 6–10). Presumably, Dronc and Dark formed a multimeric complex within which autocatalytic activation of the Dronc zymogen was greatly accelerated. Importantly, the autocleaved Dronc exhibited a markedly higher protease activity compared with the Dronc zymogen using Drice (residues 1–339, C196A) as the substrate (Fig. 2C). These observations demonstrate that the function of the multimeric complex between Dronc and Dark is to facilitate the autocatalytic activation of the Dronc caspase zymogen. Notably, unlike caspase-9 activation by the Apaf-1 apoptosome, the Dronc zymogen can be efficiently activated by Dark in the absence of any exogenous nucleotide or nucleotide exchange.
The monomeric Dark is known to assemble into an apoptosome in the presence of dATP (Fig. 1A). Incubation of the Dronc zymogen with the dATP-induced Dark apoptosome also led to efficient autocatalytic activation of Dronc (Fig. 2B, lanes 11–15). Once activated, the caspase domain of Dronc was dissociated from the dATP-induced Dark apoptosome, as judged by gel filtration analysis (Supplemental Fig. S5A). Importantly, the free caspase domain of Dronc exhibited robust protease activity toward its physiological substrate, Drice (residues 1–339, C196A) (Supplemental Fig. S5B). As anticipated, however, Dronc-CARD remained associated with the Dark apoptosome (Supplemental Fig. S5A). These observations indicate that formation of the multimeric Dronc–Dark complex can be either induced directly by the Dronc zymogen or assembled by incubating the Dronc zymogen with the dATP-induced, preassembled Dark apoptosome. Thus, in cells, the decisive factor that controls Dronc activation appears to be the availability of the Dronc zymogen to the monomeric Dark protein because the presence of the Dronc zymogen alone is sufficient to trigger formation of the Dark apoptosome regardless of the dATP status.
Structure of the Dark apoptosome bound to Dronc-CARD
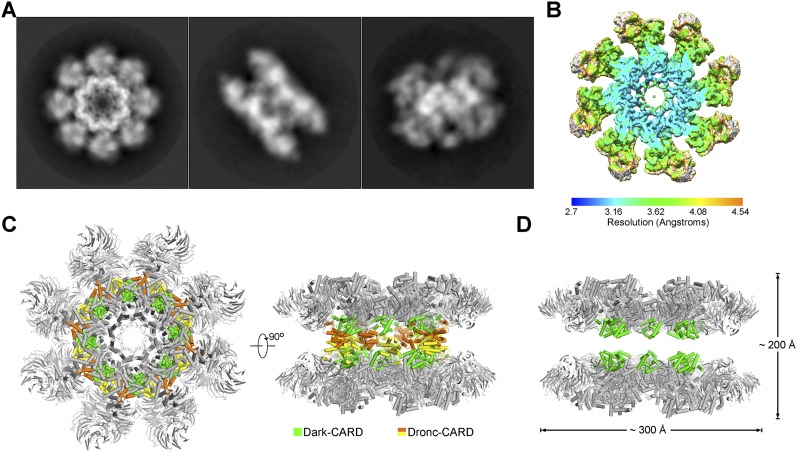
To elucidate the mechanism of Dronc activation, we also performed cryo-EM single-particle analysis on the Dark apoptosome bound to Dronc-CARD. The Dark–Dronc complex was clearly resolved, and the structure was determined at an overall resolution of 4.1 Å (Fig. 3A; Supplemental Fig. S6). Similar to the structure of the dATP-induced Dark apoptosome, the Dark–Dronc complex also displays a double-ring architecture (Fig. 3A). The central region of the Dark–Dronc complex is resolved at a resolution of 3.5 Å or better (Fig. 3B), allowing assignment of side chains and specific interactions. As a consequence of incorporation of Dronc-CARD, the two rings of the Dark apoptosome are separated further apart by ∼30 Å (Fig. 3C). Intriguingly, the Dronc-CARDs are sandwiched between the two rings of Dark apoptosome, and there is no direct interaction between these two rings (Fig. 3D).
Figure 3.
Structure of the multimeric complex between Dark and Dronc-CARD at an overall resolution of 4.1 Å. (A) Three representative two-dimensional classification averages of the complex between Dark and Dronc-CARD. Three contrasting views of the complex are shown. (B) The overall EM density map of the Dark–Dronc-CARD complex is color-coded to indicate a range of resolution. The bulk of the central region of the complex is resolved at resolutions better than 3.5 Å. (C) The overall structure of the complex between Dark and Dronc-CARD is shown in two perpendicular views. Dronc-CARDs are colored yellow and orange, whereas Dark-CARDs are shown in green. (D) The two rings of the Dark octamer make no direct interactions with each other. The 16 molecules of Dronc-CARD are removed to show the separation of the two Dark rings.
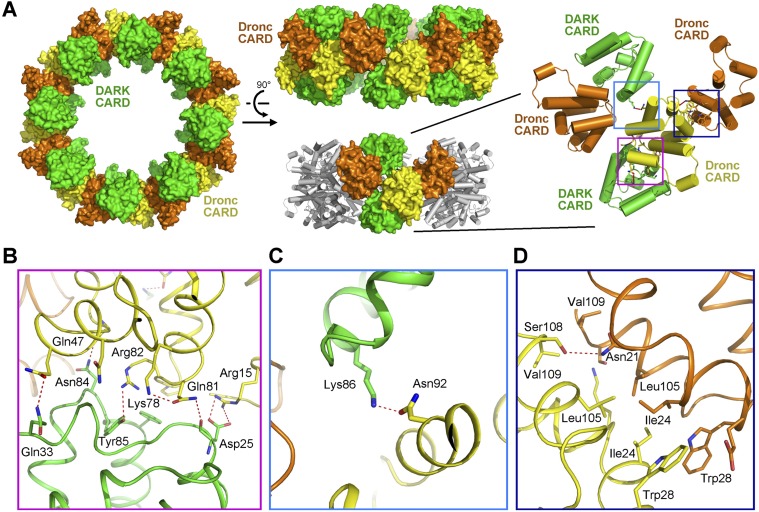
In mammalian cells, the recruitment and activation of caspase-9 zymogen by Apaf-1 strictly depend on the interactions between their respective CARDs (Li et al. 1997; Qin et al. 1999). Our experimental evidence reported here demonstrates the same conclusion for the Dronc zymogen and Dark (Fig. 2A; Supplemental Fig. S5A). In the structure of the Dark–Dronc complex, 16 molecules of Dronc-CARD associate with 16 molecules of Dark-CARD to form a closed ring (Fig. 4A). The 16 Dark-CARDs are arranged into two parallel layers and are separated by the 16 Dronc-CARDs, which are also organized into two layers. Within the CARD ring, each Dronc-CARD interacts with neighboring CARDs through three interfaces: one with a Dronc-CARD from another layer and the other two with Dark-CARDs from two different layers (Fig. 4A).
Figure 4.
CARD–CARD interfaces in the multimeric complex between Dark and Dronc-CARD. (A) Sixteen molecules of Dronc-CARD and 16 molecules of Dark-CARD assemble into a closed ring. Two perpendicular views are shown in the left and middle panels. A close-up view on five neighboring CARDs is shown in the right panel. The orange and yellow colors refer to two layers of the Dronc CARD, each containing eight molecules. (B) Each Dronc-CARD interacts with Dark-CARD mainly through an interface that contains multiple interdomain H bonds. Gln81 and Arg82 from Dronc-CARD mediate three H bonds at the center of this interface. (C) Dronc-CARD also interacts with Dark-CARD through a secondary interface. This interface has a single H bond. (D) Two neighboring Dronc-CARDs interact with each other in a pseudosymmetric manner. This interface involves mainly van der Waals contacts.
The two interfaces between Dronc-CARD and the two Dark-CARDs are mediated exclusively by H bonds. In particular, one interface contains seven H bonds (Fig. 4B). At the center of this interface, Gln81 and Arg82 from Dronc-CARD each donates an H bond to Dark-CARD; these interactions are buttressed by an intramolecular H bond within Dronc-CARD between Lys78 and Gln81. The other interface has only one H bond, between Asn92 of Dronc-CARD and Lys86 of Dark-CARD (Fig. 4C). In contrast, the interface between two Dronc-CARDs has an approximate twofold symmetry and is dominated by van der Waals contacts (Fig. 4D). Four hydrophobic residues from Dronc-CARD—Ile24, Trp28, Leu105, and Val109—stack against the corresponding amino acids from a neighboring Dronc-CARD.
To assess the functional importance of these CARD–CARD interactions, we introduced two missense mutations, Q81A and R82A, into the Dronc zymogen (residues 1–450, C318A). These mutations are predicted to abolish the two H bonds at the center of the CARD–CARD interface between Dronc and Dark (Fig. 4B). The resulting Dronc zymogen was examined for its ability to induce formation of the Dark apoptosome. Unlike the Dronc zymogen (Fig. 2A), the variant Q81A/R82A (residues 1–450, C318A) failed to interact with the wild-type Dark protein (Supplemental Fig. S7A). Consequently, the variant Dronc zymogen (residues 1–450, Q81A/R82A) failed to undergo autocatalytic activation upon incubation with Dark (Supplemental Fig. S7B). These results further corroborate the essential role of the observed CARD–CARD interface in the Dronc–Dark complex.
Dronc-CARD directly binds WD40 repeats of Dark
Analysis of the Dronc–Dark complex unexpectedly reveals a direct interaction between Dronc-CARD and the WD40 repeats of Dark (Fig. 5A). The 15 WD40 repeats of Dark are grouped into two β propellers, hereafter referred to as WD1 (residues 597–965) and WD2 (residues 966–1373). WD1 and WD2 consist of seven and eight WD40 repeats, respectively. The intervening loop sequence between the third and fourth α helices of Dronc-CARD, L55NGKPFDMDEKDV67, makes close contact with residues on the side of WD1 (Fig. 5A). The relatively low resolution of EM density in this region does not allow assignment of specific interactions. This interface is likely unique between Dronc and Dark because the corresponding loop in caspase-9 CARD is markedly shorter than that in Dronc-CARD (Fig. 5B).
Figure 5.
The interactions between Dronc-CARD and the WD40 repeats of Dark are essential for Dronc activation. (A) An extended loop sequence between helices 3 and 4 of Dronc makes direct contact to the WD40 repeats in WD1 of Dark. The relatively low resolution at this region of the EM density does not allow definitive assignment of specific side chains. (B) Structural and sequence alignment between the CARDs of Dronc and caspase-9. Unlike Dronc, caspase-9 only contains a short loop between helices 3 and 4. (C) The interactions between Dronc-CARD and the WD40 repeats of Dark are essential for formation of a multimeric complex. The extended loop between helices 3 and 4 of Dronc was replaced by that from caspase-9. The resulting Dronc failed to form a multimeric complex with Dark. Gel filtration chromatograms are shown in the left panel, and peak fractions were visualized on SDS-PAGE by Coomassie staining in the right panel. (D) The loop-replaced Dronc fails to be activated by Dark.
To assess the functional importance of the interactions between Dronc-CARD and WD1 of Dark, we replaced the WD1-interacting loop (L55NGKPFDMDEKDV67) with that from caspase-9 (A46GSGS50) in the Dronc zymogen (residues 1–450, C318A). The resulting Dronc zymogen retained stable association with the wild-type Dark protein but failed to induce formation of the Dark apoptosome (Fig. 5C). This finding demonstrates that the interactions between Dronc-CARD and WD1 of Dark are indispensable for formation of the multimeric Dronc–Dark complex but are not required for association between Dronc and Dark. Consequently, the loop-replaced Dronc zymogen failed to undergo appropriate autocatalytic activation upon incubation with Dark (Fig. 5D). These results identify an essential role for the observed interactions between Dronc-CARD and WD1 of Dark.
Discussion
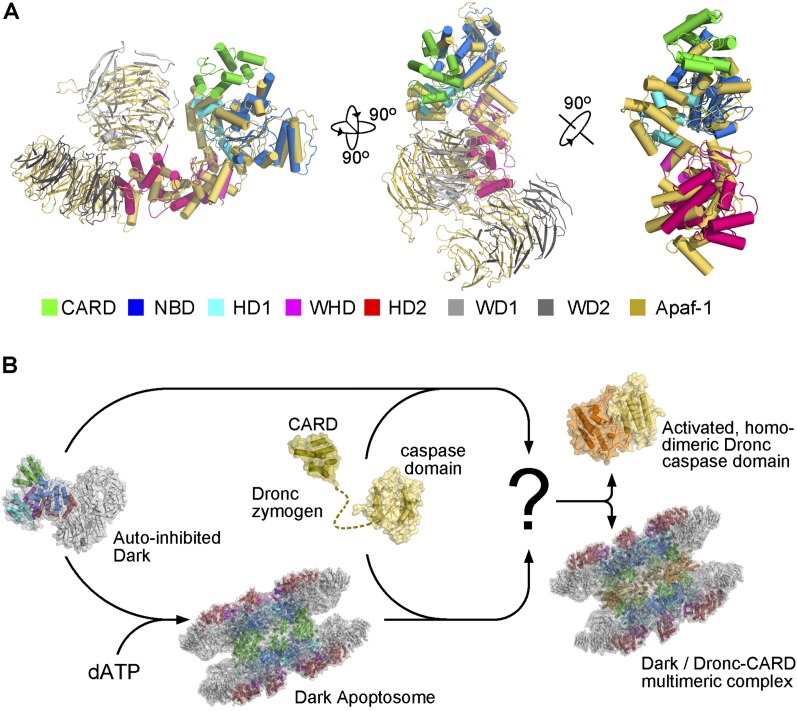
In this study, we present the cryo-EM structures of the Dark apoptosome and the multimeric Dronc–Dark complex at overall resolutions of 4.0 and 4.1 Å, respectively. Notably, the EM density in the central region of the structures exhibits considerably higher resolutions, which allow assignment of specific side chains and atomic interactions. Because the overall domain organization of Dark is identical to that of Apaf-1, the structures reveal for the first time conserved atomic features of an apoptosome from a higher organism. The observed structural features of the Dark apoptosome, most of which are likely preserved in the Apaf-1 apoptosome, reveal the underpinnings of initiator caspase activation. Supporting this analysis, structure of the Dark protomer can be very well aligned with that of the activated Apaf-1 protomer from the Apaf-1 apoptosome (Fig. 6A; Yuan et al. 2013).
Figure 6.
A working model for Dronc activation mediated by the Dark apoptosome. (A) The Dark protomer from the Dark apoptosome is structurally similar to Apaf-1 in its activated conformation. Shown here are three views of the structural superposition of the Dark protomer with Apaf-1 from the Apaf-1 apoptosome (Yuan et al. 2013). Other than the CARD, which is absent in the cryo-EM structure of the Apaf-1 apoptosome (Yuan et al. 2013), all other domains of Dark are very well overlaid with the corresponding domains of Apaf-1. (B) A working model for Dronc activation mediated by the Dark apoptosome. In the basal state, Dark exists as an autoinhibited monomer. Elevated levels of dATP trigger assembly of the Dark apoptosome, which recruits Dronc zymogen to form a multimeric complex (indicated by a question mark). The same multimeric complex can also be formed through incubation of the Dronc zymogen and autoinhibited Dark. Autocatalytic activation of the Dronc zymogen occurs within the multimeric complex, resulting in the release of the free Dronc caspase domain from the multimeric complex between Dronc-CARD and DARK.
Unlike caspase-9, which remains associated with the Apaf-1 apoptosome as a holoenzyme, the activated Dronc caspase domain is dissociated from the Dark apoptosome but exhibits robust protease activity toward its physiological substrate, Drice (Supplemental Fig. S5). For caspase-9, both the uncleaved zymogen and the cleaved two-chain protein exist predominantly as a monomer in solution (Shiozaki et al. 2003) and thus have little protease activity. Only through association with the Apaf-1 apoptosome, the cleaved two-chain caspase-9, but not the uncleaved zymogen, gains robust protease activity (Hu et al. 2013). In sharp contrast, the intrachain cleavage of the Dronc zymogen converts a monomer into a stable homodimer (Yan et al. 2006). Whereas the monomeric Dronc zymogen exhibits little protease activity, the homodimeric, cleaved Dronc displays robust protease activity (Yan et al. 2006). Thus, unlike the Apaf-1 apoptosome, the sole function of the Dark apoptosome is to facilitate the autocatalytic cleavage of the Dronc zymogen.
In mammalian cells, caspase-9 alone cannot induce oligomerization of Apaf-1, and formation of the Apaf-1 apoptosome strictly depends on binding to Cyt. c and exchange of ADP by dATP or ATP. In sharp contrast, the Dronc zymogen alone induces efficient formation of the Dark apoptosome independent of exogenous dATP or ATP (Fig. 2A). This unanticipated finding suggests that, as dATP/ATP is constantly available in cells, formation of the active Dark apoptosome may depend on the availability of the Dronc zymogen to the monomeric Dark protein. Perhaps Dark and Dronc are sequestered in distinct cellular locations prior to cell death stimulation.
Our biochemical and structural investigation reveals important insights into the mechanism by which the Dark apoptosome facilitates the autocatalytic cleavage of the Dronc zymogen. The CARD of Dronc by itself is sufficient to induce the formation of a multimeric Dark apoptosome (Fig. 2A) within which 16 molecules of Dronc-CARD are sandwiched between two layers of Dark-CARD (Fig. 4A). Disruption of key interactions at the interface between Dronc-CARD and Dark-CARD led to the loss of interactions between Dronc and Dark (Supplemental Fig. S7A) and the consequent loss of Dark-mediated activation of Dronc (Supplemental Fig. S7B). Structural analysis also identified an interface between Dronc-CARD and the WD1 of Dark (Fig. 5A). Unlike the interface between Dronc-CARD and Dark-CARD, this interface is dispensable for Dronc–Dark association but is required for formation of the multimeric Dronc–Dark complex (Fig. 5C).
These findings allow us to propose a working model to explain Dronc activation. In the absence of apoptotic stimuli, Dark exists in cells in a monomeric, autoinhibited conformation, which is likely to be similar to that of the autoinhibited Apaf-1 (Fig. 6B). The monomeric Dark can be activated by elevated levels of dATP to form the Dark apoptosome, which further recruits and mediates activation of the Dronc zymogen. Alternatively, the monomeric Dark and the Dronc zymogen can directly assemble into a multimeric complex within which Dronc is activated (Fig. 6B). Within the Dark apoptosome, multiple molecules of the Dronc zymogen are placed in close proximity to each other, which likely facilitates secondary interactions involving their caspase domains and triggers their consequent proteolytic activity. For both routes, the end products are the same: a free, activated homodimer of the Dronc caspase domain and a multimeric complex between Dark and Dronc-CARD. Clearly, further mechanistic understanding of Dronc activation by the Dark apoptosome requires detailed structural information on the multimeric complex between the Dark apoptosome and the full-length Dronc zymogen (Fig. 6B).
Materials and methods
Protein expression and purification
The full-length D. melanogaster Dark complementary DNA was subcloned into vector pFastBac1 (Invitrogen) with a C-terminal 10xHis tag. The recombinant Dark was expressed using the Bac-to-Bac baculovirus system (Invitrogen). Bacmid DNAs were generated in DH10Bac cells, and the resulting baculoviruses were generated and amplified in Sf9 insect cells (Invitrogen). Dark protein was overexpressed in Hi-5 insect cells (Invitrogen) grown in the SIM HF medium (Sino Biological, Inc.). Forty-eight hours after viral infection, the cells were collected and homogenized in a buffer containing 25 mM Tris (pH 8.0) and 150 mM NaCl. The cells were disrupted by 60 strokes on ice using a Dounce homogenizer. After high-speed centrifugation at 14,000 rpm for 60 min, the supernatant was harvested. The Dark protein was purified to homogeneity by nickel affinity chromatography (Ni-NTA, Qiagen) and anion exchange chromatography (Source-15Q, GE Healthcare).
The full-length Dronc (residues 1–450), Dronc-CARD (residues 1–130), and relevant mutants were subcloned into pET-21b with a C-terminal 8xHis tag. All mutants were generated using a standard PCR-based strategy and verified by plasmid sequencing. All Dronc proteins and mutants were expressed in Escherichia coli strain BL21(DE3) and purified to homogeneity as described (Yan et al. 2006).
Dark apoptosome assembly
Purified recombinant Dark protein was incubated with 5 mM dATP or an excess amount of Dronc-CARD for >4 h at 4°C and further purified by gel filtration chromatography (Superdex-200, GE Healthcare). The final buffer for the Dark apoptosome contained 25 mM Tris (pH 8.0), 150 mM NaCl, and 5 mM dithiothreitol. The peak fractions were collected for cryo-EM studies.
Electron microscopy
Aliquots of 3 μL of assembled Dark apoptosome or the complex between Dark and Dronc-CARD at a concentration of ∼60 nM were placed on glow-discharged holey carbon grids (CuR2/2, Quantifoil) on which a homemade continuous carbon film (estimated to be ∼30 Å thick) had previously been deposited. Grids were blotted for 2 sec and flash-frozen in liquid ethane using an FEI Vitrobot. Grids were transferred to an FEI Tecnai Polara electron microscope that was operating at 300 kV. Images were recorded manually using an FEI Falcon-II detector at a calibrated magnification of 104,478, yielding a pixel size of 1.34 Å. A dose rate of 28 electrons per square angstrom per second and an exposure time of 1 sec were used on the Falcon.
Image processing
The swarm tool in the EMAN2 was used for semiautomated selection of 65,342 particles from 693 micrographs for the Dark apoptosome and 111,689 particles from 1234 micrographs for the complex sample (Tang et al. 2007). Contrast transfer function parameters were estimated using CTFFIND3 (Mindell and Grigorieff 2003). All two- and three-dimensional classifications and refinements were performed using RELION (Scheres 2012). We used reference-free two-dimensional class averaging and three-dimensional classification to discard bad particles and selected 24,460 and 41,476 particles for the Dark apoptosome and the Dark–Dronc-CARD complex, respectively, for a first three-dimensional refinement. A 60 Å low-pass filtered cryo-EM reconstruction of Dark (EMDB-5235) (Yuan et al. 2011b) was used as an initial model for the three-dimensional refinement. For both structures, the resulting maps showed high resolution for the central domain, but the density for the HD2 and the two WD domains was relatively poor. In a subsequent three-dimensional classification run with four classes, an angular sampling of 1.8° was combined with local angular searches around the refined orientations, and the refined model from the first refinement was used as a starting model. In both cases, the largest class showed improved density and was selected for the final reconstruction, yielding a subset of 9354 particles for the Dark apoptosome and 11,359 particles for the complex.
To correct for beam-induced movements, the 16 video frames for each micrograph were first aligned using whole-image motion correction (Bai et al. 2013). Second, particle-based beam-induced movement correction was performed using statistical movie processing in RELION. For these calculations, we used running averages of seven movie frames and a standard deviation of one pixel for the translational alignment. To increase further the accuracy of the per-particle movement correction, we used the particle polishing procedure in RELION-1.3 to fit linear tracks through the optimal translations for all running averages and employ a resolution and dose-dependent model for the radiation damage, where each frame was weighted with a different B factor (temperature factor) as estimated from single-frame reconstructions (Scheres 2014). Reported resolutions were based on the gold standard FSC 0.143 criterion (Chen et al. 2013), and FSC curves were corrected for the effects of a soft mask on the FSC curve using high-resolution noise substitution (Henderson and McMullan 2013). Prior to visualization, all density maps were corrected for the modulation transfer function (MTF) of the detector and then sharpened by applying a negative B factor that was estimated using automated procedures (Rosenthal and Henderson 2003). Local resolution variations were estimated using ResMap (Kucukelbir et al. 2014).
Model building and refinement
The atomic model was manually built in COOT (Emsley et al. 2010) and refined with stereochemical and homology restraints in REFMAC (Murshudov et al. 2011; Amunts et al. 2014) using the modified versions of these programs for cryo-EM maps. Throughout refinement, secondary structure restraints as determined by ProSMART (Nicholls et al. 2012) were applied.
Supplementary Material
Acknowledgments
This work was funded by the National Natural Science Foundation of China (projects 31430020, 31130002, and 31321062 to Y.S.), a European Union Marie Curie Fellowship (to X.-C.B.), and the UK Medical Research Council (MC_UP_A025_1013 to S.H.W.S.).
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.255877.114.
References
- Acehan D, Jiang X, Morgan DG, Heuser JE, Wang X, Akey CW. 2002. Three-dimensional structure of the apoptosome: implications for assembly, procaspase-9 binding and activation. Mol Cell 9: 423–432. [DOI] [PubMed] [Google Scholar]
- Amunts A, Brown A, Bai XC, Llacer JL, Hussain T, Emsley P, Long F, Murshudov G, Scheres SH, Ramakrishnan V. 2014. Structure of the yeast mitochondrial large ribosomal subunit. Science 343: 1485–1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai XC, Fernandez IS, McMullan G, Scheres SH. 2013. Ribosome structures to near-atomic resolution from thirty thousand cryo-EM particles. eLife 2: e00461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao Q, Lu W, Rabinowitz JD, Shi Y. 2007. Calcium blocks formation of apoptosome by preventing nucleotide exchange in Apaf-1. Mol Cell 25: 181–192. [DOI] [PubMed] [Google Scholar]
- Chai J, Shi Y. 2014. Apoptosome and inflammasome: conserved machineries for caspase activation. National Science Review 1: 101–118. [Google Scholar]
- Chen S, McMullan G, Faruqi AR, Murshudov GN, Short JM, Scheres SH, Henderson R. 2013. High-resolution noise substitution to measure overfitting and validate resolution in 3D structure determination by single particle electron cryomicroscopy. Ultramicroscopy 135: 24–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danial NN, Korsmeyer SJ. 2004. Cell death: critical control points. Cell 116: 205–219. [DOI] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60: 2126–2132. [DOI] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, Cowtan K. 2010. Features and development of Coot. Acta Crystallogr D Biol Crystallogr 66: 486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs Y, Steller H. 2011. Programmed cell death in animal development and disease. Cell 147: 742–758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson R, McMullan G. 2013. Problems in obtaining perfect images by single-particle electron cryomicroscopy of biological structures in amorphous ice. Microscopy (Oxf) 62: 43–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvitz HR. 2003. Worms, life, and death (Nobel lecture). ChemBioChem 4: 697–711. [DOI] [PubMed] [Google Scholar]
- Hu Q, Wu D, Chen W, Yan Z, Shi Y. 2013. Proteolytic processing of the caspase-9 zymogen is required for apoptosome-mediated activation of caspase-9. J Biol Chem 288: 15142–15147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanuka H, Sawamoto K, Inohara N, Matsuno K, Okano H, Miura M. 1999. Control of the cell death pathway by Dapaf-1, a Drosophila Apaf-1/CED-4-related caspase activator. Mol Cell 4: 757–769. [DOI] [PubMed] [Google Scholar]
- Kim HE, Du F, Fang M, Wang X. 2005. Formation of apoptosome is initiated by cytochrome c-induced dATP hydrolysis and subsequent nucleotide exchange on Apaf-1. Proc Natl Acad Sci 102: 17545–17550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kucukelbir A, Sigworth FJ, Tagare HD. 2014. Quantifying the local resolution of cryo-EM density maps. Nat Methods 11: 63–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P, Nijhawan D, Budihardjo I, Srinivasula SM, Ahmad M, Alnemri ES, Wang X. 1997. Cytochrome c and dATP-dependent formation of Apaf-1/Caspase-9 complex initiates an apoptotic protease cascade. Cell 91: 479–489. [DOI] [PubMed] [Google Scholar]
- Liu X, Kim CN, Yang J, Jemmerson R, Wang X. 1996. Induction of apoptosis program in cell-free extracts: requirement for dATP and cytochrome c. Cell 86: 147–157. [DOI] [PubMed] [Google Scholar]
- Mindell JA, Grigorieff N. 2003. Accurate determination of local defocus and specimen tilt in electron microscopy. J Struct Biol 142: 334–347. [DOI] [PubMed] [Google Scholar]
- Murshudov GN, Skubak P, Lebedev AA, Pannu NS, Steiner RA, Nicholls RA, Winn MD, Long F, Vagin AA. 2011. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr D Biol Crystallogr 67: 355–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholls RA, Long F, Murshudov GN. 2012. Low-resolution refinement tools in REFMAC5. Acta Crystallogr D Biol Crystallogr 68: 404–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. 2004. UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem 25: 1605–1612. [DOI] [PubMed] [Google Scholar]
- Qi S, Pang Y, Hu Q, Liu Q, Li H, Zhou Y, He T, Liang Q, Liu Y, Yuan X, et al. . 2010. Crystal structure of the Caenorhabditis elegans apoptosome reveals an octameric assembly of CED-4. Cell 141: 446–457. [DOI] [PubMed] [Google Scholar]
- Qin H, Srinivasula SM, Wu G, Fernandes-Alnemri T, Alnemri ES, Shi Y. 1999. Structural basis of procaspase-9 recruitment by the apoptotic protease-activating factor 1. Nature 399: 547–555. [DOI] [PubMed] [Google Scholar]
- Rodriguez J, Lazebnik Y. 1999. Caspase-9 and Apaf-1 form an active holoenzyme. Genes & Dev 13: 3179–3184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez A, Oliver H, Zou H, Chen P, Wang X, Abrams JM. 1999. Dark is a Drosophila homologue of Apaf-1/CED-4 and functions in an evolutionarily conserved death pathway. Nat Cell Biol 1: 272–279. [DOI] [PubMed] [Google Scholar]
- Rosenthal PB, Henderson R. 2003. Optimal determination of particle orientation, absolute hand, and contrast loss in single-particle electron cryomicroscopy. J Mol Biol 333: 721–745. [DOI] [PubMed] [Google Scholar]
- Scheres SH. 2012. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J Struct Biol 180: 519–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheres SH. 2014. Beam-induced motion correction for sub-megadalton cryo-EM particles. eLife 3: e03665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiozaki EN, Chai J, Rigotti DJ, Riedl SJ, Li P, Srinivasula SM, Alnemri ES, Fairman R, Shi Y. 2003. Mechanism of XIAP-mediated inhibition of caspase-9. Mol Cell 11: 519–527. [DOI] [PubMed] [Google Scholar]
- Tang G, Peng L, Baldwin PR, Mann DS, Jiang W, Rees I, Ludtke SJ. 2007. EMAN2: an extensible image processing suite for electron microscopy. J Struct Biol 157: 38–46. [DOI] [PubMed] [Google Scholar]
- Yan N, Shi Y. 2005. Mechanisms of apoptosis through structural biology. Annu Rev Cell Dev Biol 21: 35–56. [DOI] [PubMed] [Google Scholar]
- Yan N, Huh JR, Schirf V, Demeler B, Hay BA, Shi Y. 2006. Structure and activation mechanism of the Drosophila initiator caspase Dronc. J Biol Chem 281: 8667–8674. [DOI] [PubMed] [Google Scholar]
- Yu X, Acehan D, Menetret JF, Booth CR, Ludtke SJ, Riedl SJ, Shi Y, Wang X, Akey CW. 2005. A structure of the human apoptosome at 12.8 Å resolution provides insights into this cell death platform. Structure 13: 1725–1735. [DOI] [PubMed] [Google Scholar]
- Yu X, Wang L, Acehan D, Wang X, Akey C. 2006. Three-dimensional structure of a double apoptosome formed by the Drosophila Apaf-1 related killer. J Mol Biol 355: 577–589. [DOI] [PubMed] [Google Scholar]
- Yuan S, Yu X, Topf M, Ludtke SJ, Wang X, Akey CW. 2010. Structure of an apoptosome-procaspase-9 CARD complex. Structure 18: 571–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan S, Yu X, Asara JM, Heuser JE, Ludtke SJ, Akey CW. 2011a. The holo-apoptosome: activation of procaspase-9 and interactions with caspase-3. Structure 19: 1084–1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan S, Yu X, Topf M, Dorstyn L, Kumar S, Ludtke SJ, Akey CW. 2011b. Structure of the Drosophila apoptosome at 6.9 Å resolution. Structure 19: 128–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan S, Topf M, Reubold TF, Eschenburg S, Akey CW. 2013. Changes in Apaf-1 conformation that drive apoptosome assembly. Biochemistry 52: 2319–2327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L, Song Z, Tittel J, Steller H. 1999. HAC-1, a Drosophila homolog of Apaf-1 and CED-4, functions in developmental and radiation-induced apoptosis. Mol Cell 4: 745–755. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.