Harms et al. reveal that PRDM16 binding is highly enriched at a broad set of brown fat-selective genes. PRDM16 physically binds to MED1, a component of the Mediator complex, and recruits it to superenhancers at brown fat-selective genes. PRDM16 deficiency in BAT reduces MED1 binding at PRDM16 target sites and causes a fundamental change in chromatin architecture at key brown fat-selective genes.
Keywords: PRDM16, PRDM3, MED1, Mediator, brown adipose tissue, UCP1
Abstract
PR (PRD1–BF1–RIZ1 homologous) domain-containing 16 (PRDM16) drives a brown fat differentiation program, but the mechanisms by which PRDM16 activates brown fat-selective genes have been unclear. Through chromatin immunoprecipitation (ChIP) followed by deep sequencing (ChIP-seq) analyses in brown adipose tissue (BAT), we reveal that PRDM16 binding is highly enriched at a broad set of brown fat-selective genes. Importantly, we found that PRDM16 physically binds to MED1, a component of the Mediator complex, and recruits it to superenhancers at brown fat-selective genes. PRDM16 deficiency in BAT reduces MED1 binding at PRDM16 target sites and causes a fundamental change in chromatin architecture at key brown fat-selective genes. Together, these data indicate that PRDM16 controls chromatin architecture and superenhancer activity in BAT.
Obesity is a leading cause of preventable death in the United States due to its link to many diseases, including type 2 diabetes, cardiovascular disease, stroke, and certain cancers (Bornfeldt and Tabas 2011). White adipose tissue (WAT), which expands in obesity, is specialized to store energy in the form of lipid, whereas brown and beige adipose tissue expend energy as heat (Harms and Seale 2013). High levels of brown/beige fat activity protect animals against many of the harmful effects of a high-fat diet, including obesity and insulin resistance (Harms and Seale 2013; Stanford et al. 2013; Rosen and Spiegelman 2014). In people, brown/beige fat activity levels also correlate with reduced adiposity, and cold-stimulated brown/beige fat activity promotes weight loss (Saito 2014; Yoneshiro and Saito 2014). Thus, elucidating the molecular pathways that regulate brown/beige fat activity may reveal new approaches to treat obesity and related diseases.
PR (PRD1–BF1–RIZ1 homologous) domain-containing 16 (PRDM16), a zinc finger containing transcription factor, is a critical molecular determinant of brown/beige fat cell fate. Ectopic expression of PRDM16 in fibroblasts or muscle precursors stimulates brown fat cell differentiation (Seale et al. 2007, 2011; Kajimura et al. 2009). PRDM16 is also required for beige fat differentiation and the maintenance of brown adipose tissue (BAT) fate in adult mice (Cohen et al. 2014; Harms et al. 2014). PRDM16 activates genes that are expressed at higher levels in brown relative to white adipocytes (BAT-selective), such as Ucp1, Pparα, and Ppargc1a, while also suppressing the expression of certain WAT-selective genes (Kajimura et al. 2008; Harms et al. 2014). However, the mechanisms by which PRDM16 functions to activate BAT-selective genes were unknown.
We found that PRDM16 binds to chromatin at a broad set of BAT-selective genes in BAT. Deletion of Prdm16 in BAT reduced RNA polymerase II (Pol II) levels at BAT-selective genes and caused a concomitant decrease in the expression levels of these genes. Interestingly, the loss of PRDM16 did not reduce the binding levels of PPARγ or C/EBPβ at PRDM16 target genes and caused only a mild decrease in the levels of H3K27 acetylation (H3K27-Ac), a chromatin mark that is typically associated with active transcription. In contrast, PRDM16 was critically required for the recruitment of MED1, a component of the Mediator complex, to BAT-selective genes. Notably, we found that PRDM16 binds, likely directly, with MED1 through its two zinc finger domains. Knockout of Prdm16 in cultured brown fat cells disrupted higher-order chromatin architecture at Ppargc1a and Pparα, genes that are critical transcriptional targets of PRDM16 in BAT. Finally, we show that PRDM16 marks and regulates the activity of superenhancers (SEs), which are defined as large clusters of transcription factor-binding sites that drive the expression of cell identity genes (Whyte et al. 2013). Taken together, our results reveal that PRDM16 recruits MED1 to SEs in the control of BAT identity.
Results
PRDM16 binding is enriched at BAT-selective genes
PRDM16 induces the expression of brown fat-selective genes (Seale et al. 2007, 2008), but whether PRDM16 acts directly at these genes was unknown. To investigate this, we analyzed the genome-wide binding profile of PRDM16 in BAT using chromatin immunoprecipitation (ChIP) followed by deep sequencing (ChIP-seq) (Supplemental Fig. S1A). The large majority of PRDM16-binding sites identified by ChIP-seq in wild-type BAT were absent in Prdm16 knockout tissue (Supplemental Fig. S1B), which indicated that our ChIP antibody was specific. The genomic distribution of PRDM16 binding was typical for transcription factors with many binding sites located within intergenic and intronic regions (Supplemental Fig. S1C).
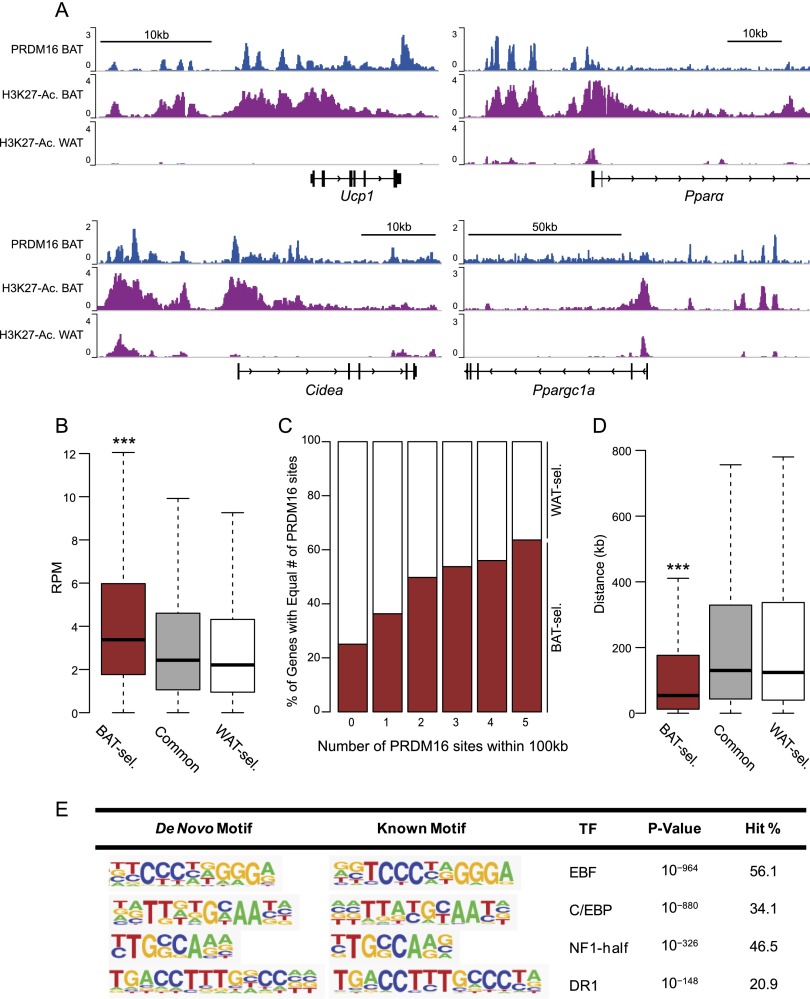
There were numerous PRDM16-binding sites at classical BAT-selective genes, including Ucp1, Pparα, Cidea, and Ppargc1a (Fig. 1A). These PRDM16-bound sites had higher levels of the activating histone mark H3K27-Ac in BAT relative to WAT. As previously reported, PRDM16 was also bound at certain WAT-selective genes, such as Agt and Retn (Supplemental Fig. S1D; Kajimura et al. 2008; Harms et al. 2014). In genome-wide analyses, PRDM16 binding was, in general, highly enriched at BAT-selective genes relative to WAT-selective or common genes (Fig. 1B). Moreover, BAT-selective genes contained more PRDM16-binding sites, and these sites were, on average, located closer to the transcriptional start site (TSS) (Fig. 1C,D). Furthermore, PRDM16-binding levels were highly correlated with the levels of RNA Pol II binding at BAT-selective genes (Supplemental Fig. S1E). Finally, PRDM16 binding was enriched at BAT-selective regions of H3K27-Ac (Supplemental Fig. S1F). Altogether, these results suggest that PRDM16 acts directly in chromatin to promote the transcription of many brown fat-selective genes.
Figure 1.
PRDM16 binding is enriched at BAT-selective genes. (A) ChIP-seq profiles in reads per million total reads (RPM) for PRDM16 in BAT and H3K27-Ac in BAT and WAT at Ucp1, Pparα, Cidea, and Ppargc1a. (B) Box plot comparing PRDM16 ChIP signal (RPM) around BAT-selective (BAT-sel.), common, and WAT-selective (WAT-sel.) genes. (***) P < 10−20. (C) Box plot comparing the distance (in kilobases) of the closest PRDM16-binding site from the TSS of BAT-selective, common, or WAT-selective genes. (D) Proportion (percentage) of BAT- and WAT-selective genes grouped by number of PRDM16-binding sites within 100 kb of the TSS. (***) P < 10−25. (E) De novo motif analysis of PRDM16-binding sites.
An important question is whether PRDM16 binds to DNA directly or is recruited to the genome by other factors. De novo motif analyses showed that consensus sequences for EBF, C/EBP, NF1, and PPAR (DR1 motif) were highly enriched at PRDM16-binding sites (Fig. 1E). While there were potentially interesting changes in the rank order of these motifs, the same set of motifs was most highly enriched at both positively and negatively regulated PRDM16 target genes (Supplemental Fig. S2A). Of note, the putative binding motif for PRDM16/PRDM3 (Nishikata et al. 2003) was quite poorly represented in the full set of PRDM16-binding regions in BAT. Among the transcription factors that bind to identified sequence motifs, PRDM16 interacts, likely in a direct manner, with PPARγ and C/EBPβ (Seale et al. 2008; Kajimura et al. 2009). Using sequential ChIP analysis, we found that PRDM16 (along with PPARγ) was colocalized with C/EBPβ at BAT-selective genes (Supplemental Fig. S2B). These results suggest that PRDM16 is recruited to chromatin in BAT through interacting with its DNA-binding partners, including C/EBPβ and PPARγ (Seale et al. 2008; Kajimura et al. 2009).
PRDM16 recruits MED1 to BAT-selective genes
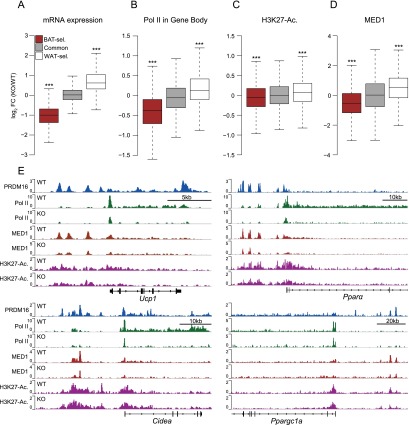
Prdm16 deficiency causes a severe loss in the thermogenic function of BAT in adult mice (Harms et al. 2014). On a global scale, Prdm16 deficiency in adult BAT reduced the expression levels of BAT-selective genes, increased WAT-selective gene levels, and did not affect the expression levels of common genes (Fig. 2A). RNA Pol II binding was significantly reduced within the gene bodies of BAT-selective genes in knockout relative to wild-type BAT, suggesting that PRDM16 regulates the transcription of BAT-selective genes to affect mRNA levels (Fig. 2B). We first postulated that PRDM16 may act to promote or stabilize the association of its direct DNA-binding partners, PPARγ and C/EBPβ, with chromatin. However, ChIP and quantitative PCR (ChIP-qPCR) experiments revealed that there were equivalent levels of PPARγ and C/EBPβ bound to PRDM16 target sites at several BAT-selective genes in wild-type and Prdm16-deficient BAT (Supplemental Fig. S3A). Furthermore, Prdm16 deficiency caused only a modest, albeit significant, reduction in H3K27-Ac levels at BAT-selective genes (Fig. 2C).
Figure 2.
PRDM16 deficiency reduces MED1 levels at BAT-selective genes. (A) Box plot depicting changes in gene expression of BAT-selective (BAT-sel.) genes (red), common genes (gray), and WAT-selective (WAT-sel.) genes (white) in Prdm16 knockout (KO)/wild-type (WT) BAT. (***) P < 10−100. (B) Box plot showing changes in Pol II levels within the gene body of BAT-selective, common, and WAT-selective genes in Prdm16 knockout/wild-type BAT. (***) P < 10−70. (C) Box plot showing H3K27-Ac levels within 100 kb of the TSS of BAT-selective, common, and WAT-selective genes in Prdm16 knockout/wild-type BAT. (D) Box plot depicting MED1 occupancy within 100 kb of the TSS of BAT-selective, common, and WAT-selective genes in Prdm16 knockout/wild-type BAT. (***) P < 10−15. (E) ChIP-seq profiles for PRDM16 (blue), Pol II in wild-type and knockout BAT (green), MED1 in wild-type and knockout BAT (red), and H3K27-Ac in wild-type and knockout BAT (magenta).
The Mediator complex plays a crucial role in regulating transcription, in part through the bridging of transcription factor-bound enhancer regions with the general transcriptional machinery and RNA Pol II at gene promoters (Malik and Roeder 2010). Of particular interest, the MED1 subunit of Mediator regulates the function of important transcription factors in adipocytes, including PPARγ, PPARGC1a, C/EBPβ, and the thyroid receptor (TR) (Yuan et al. 1998; Wallberg et al. 2003; Li et al. 2008; Borggrefe and Yue 2011), which themselves also cooperate with PRDM16. Med1 is expressed at slightly higher levels in BAT relative to epididymal or inguinal WAT, and its expression levels increase during the process of adipogenesis in brown fat cells (Supplemental Fig. S3B,C). In sequential ChIP-qPCR experiments, we observed that MED1 was enriched along with PRDM16 and PPARγ at C/EBPβ-bound regions of BAT-selective genes (Ucp1, Cidea, Pparα, and Ppargc1a) (Supplemental Fig. S2B).
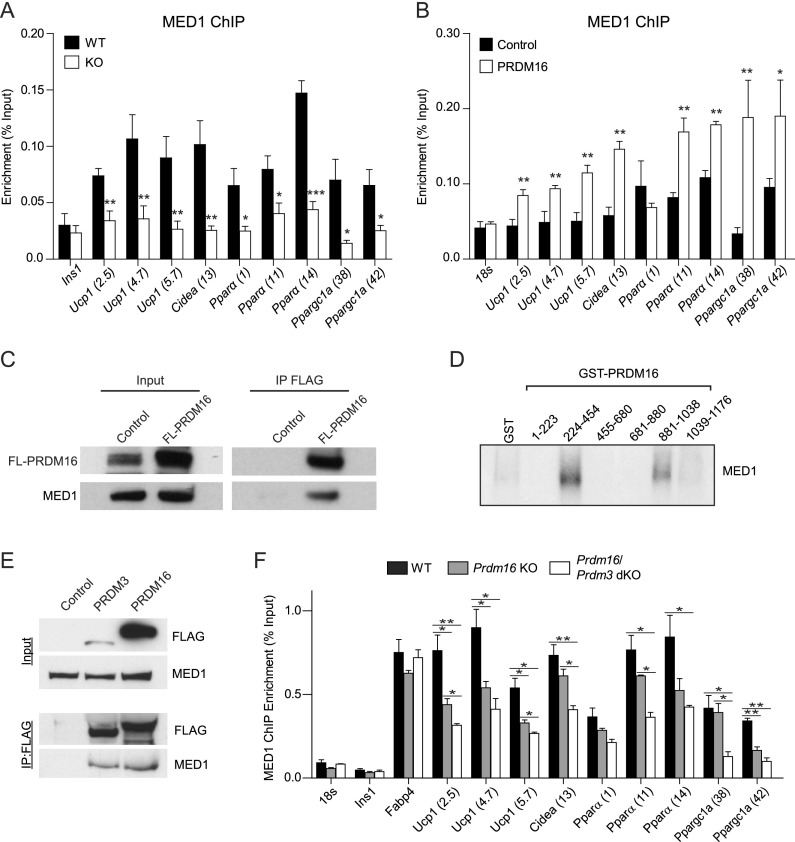
To assess whether there might be an important interaction between PRDM16 and MED1/Mediator in BAT, we profiled the genome-wide chromatin-binding activity of MED1 in wild-type and Prdm16 knockout BAT using ChIP-seq. These analyses showed that PRDM16 deficiency caused a striking and broad decrease in MED1-binding levels at BAT-selective genes (Fig. 2D; Supplemental Fig. S3D). These effects were easily visualized at several genes, including Ucp1, Pparα, Cidea, and Ppargc1a, where Prdm16 knockout samples had greatly diminished MED1 (and RNA Pol II) binding with only modest decreases in H3K27-Ac levels at PRDM16-binding sites (Fig. 2E). ChIP-qPCR analyses on independent BAT samples confirmed that PRDM16 deficiency reduced MED1 binding at PRDM16-binding sites in Ucp1, Pparα, Cidea, and Ppargc1a (Supplemental Fig. S3E).
We next tested whether PRDM16 could increase the binding of MED1 to BAT-selective genes. To do this, we transduced Prdm16 knockout brown adipocytes with Prdm16-expressing or control (empty backbone) retrovirus. Prdm16-deficient adipocytes, which accumulated lipids and expressed normal levels of general adipogenic genes, displayed reduced binding levels of MED1 at BAT-selective genes (Ucp1, Cidea, Pparα, and Ppargc1a) and a corresponding decrease in the mRNA levels of these genes (Fig. 3A; Harms et al. 2014). Importantly, MED1 mRNA or protein levels were not affected by PRDM16 deficiency in brown adipocytes (Supplemental Fig. S4A,B). Ectopic (re-)expression of PRDM16 in knockout cells led to increased binding of MED1 to Ucp1, Cidea, Pparα, and Ppargc1a and activated the expression of these genes without elevating Med1 expression levels (Fig. 3B; Supplemental Fig. S4C).
Figure 3.
PRDM16 binds to MED1 and recruits it to BAT-selective genes. (A) Chip-qPCR analysis of MED1 binding at BAT-selective genes in wild-type (WT) and Prdm16 knockout (KO) adipocytes. Ins1 was used as a nonspecific binding site (mean ± SD). n = 3; (*) P < 0.05; (**) P < 0.01; (***) P < 0.001. (B) ChIP-qPCR analysis of MED1 binding at BAT-selective genes in Prdm16 knockout adipocytes that express PRDM16 or a control (puro) virus. 18s was used as a nonspecific binding site (mean ± SD). n = 3; (*) P < 0.05; (**) P < 0.01. (C) Coimmunoprecipitation of MED1 and PRDM16 in brown adipocytes that express control or Flag-PRDM16. (D) Fluorography of GST fusion proteins containing different regions of PRDM16 that had been incubated with in vitro translated and S35-labeled MED1. (E) Coimmunoprecipitation of MED1, PRDM3, and PRDM16 in brown preadipocytes that express control, Flag-PRDM3, or Flag-PRDM16. (F) Chip-qPCR analysis of MED1 binding at BAT-selective genes in wild-type, Prdm16 knockout, and Prdm16/Prdm3 double-knockout (dKO) tissue. Ins1 and 18s rRNA were used as nonspecific binding sites (mean ± SD). n = 3; (*) P < 0.05; (**) P < 0.01.
To determine whether MED1 was required for the expression of BAT-selective genes, we acutely knocked down MED1 expression in mature brown adipocytes using a siRNA. This caused a sharp decrease in the expression levels of BAT-selective genes while also reducing the expression levels of some common adipocyte genes (Supplemental Fig. S4D). However, MED1 depletion did not affect the expression levels of other adipocyte-related genes, including Agt, Tle3, and Gpr64 (Supplemental Fig. S4D). Importantly, the efficient knockdown of MED1 protein in brown fat cells did not significantly disrupt the association of PRDM16 with chromatin at its target genes (Supplemental Fig. S4E), suggesting that MED1 regulates the transcriptional function but not necessarily the binding activity of PRDM16.
The observed requirement for PRDM16 in the binding of MED1 to BAT-selective genes raised the question of whether PRDM16 physically interacts with MED1. Using coimmunoprecipitation assays, we detected a robust interaction between MED1 and PRDM16 in brown adipocytes (Fig. 3C). Binding experiments using bacterially purified GST-PRDM16 protein fragments and in vitro translated MED1 revealed that MED1 interacts with the two zinc finger domains of PRDM16 (242–454 and 881–1038) (Fig. 3D; Supplemental Fig. S4F), suggesting that the PRDM16–MED1 interaction is direct. Altogether, these results demonstrate that PRDM16 plays a critical role in physically recruiting MED1 to its BAT-selective target genes in adipocytes.
Prdm16 deficiency in the brown fat lineage causes an adult-onset loss of thermogenic gene levels, mitochondria, and function in the interscapular BAT of mice (Harms et al. 2014). This age dependency in the decline of brown fat-specific fate is due in large part to the action of PRDM3 in young BAT, which can compensate for the absence of PRDM16. As such, the concurrent deletion of Prdm16 and Prdm3 causes a more severe and earlier deficit in brown fat development (Harms et al. 2014). These observations raised the question of whether PRDM3 also binds and recruits MED1 to brown fat-specific genes. Using immunoprecipitation assays, we found that PRDM3, like PRDM16, interacts with MED1 in brown preadipose cells (Fig. 3E). Importantly, MED1 binding at BAT-specific genes (Ucp1, Cidea, Pparα, and Pparc1a) was lower in BAT lacking both Prdm16 and Prdm3 (double knockout) relative to wild-type or Prdm16 knockout BAT (Fig. 3F). These results demonstrate that either PRDM16 or PRDM3 can participate in the recruitment of MED1 to brown fat-specific genes in adipocytes.
PRDM16 controls higher-order chromatin structure
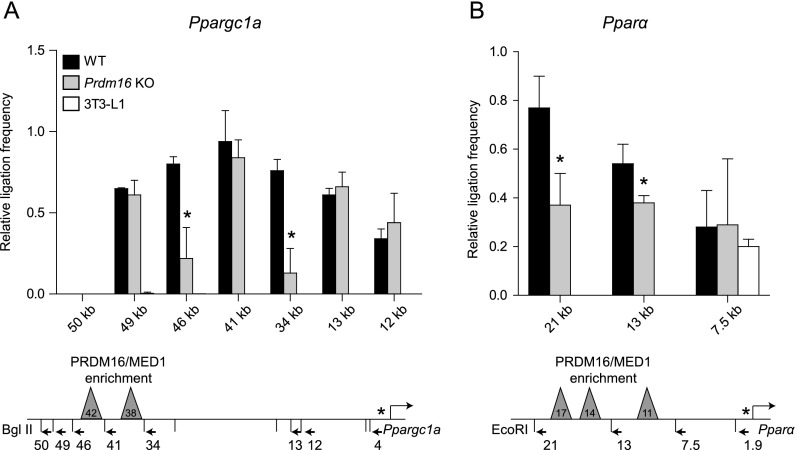
We hypothesized that PRDM16 was essential for promoting long-range chromatin interactions between gene regulatory regions and their associated promoters. To test this, we used chromatin conformation capture (3C) assays and examined whether PRDM16 regulates chromatin architecture at Ppargc1a and Pparα. These genes have distal PRDM16-binding sites that are located >10 kb from the TSS and are thus amenable to 3C analysis. At Ppargc1a, the region surrounding two PRDM16/MED1-binding sites (−34 kb to −46 kb) interacts with the promoter in brown adipocytes but not in 3T3-L1 white adipocytes (Fig. 4A). The interactions between the promoter and the −34-kb and −46-kb regions were significantly decreased in Prdm16 knockout adipocytes, whereas the interaction between the promoter and the −41-kb region was unaffected. At Pparα, there were higher levels of interaction between the promoter and two upstream regions (containing all three sites of PRDM16/MED1 enrichment) in wild-type relative to Prdm16 knockout adipocytes or 3T3-L1 white adipocytes. (Fig. 4B). These results document a requirement for PRDM16 in the assembly of higher-order (and active) chromatin structure at critical BAT-selective genes.
Figure 4.
PRDM16 regulates the chromatin architecture at BAT-selective target genes. (A,B) 3C analysis of the Ppargc1a (A) and Pparα (B) loci in wild-type (WT) and Prdm16 knockout (KO) brown adipocytes and 3T3-L1 white adipocytes. The schematic linear depiction of gene loci shows the locations of the restriction sites and PCR primers used. (*) Anchor primer that resides on a fragment containing the TSS (mean ± SD). n = 3; (*) P < 0.05.
PRDM16 controls BAT-selective SEs
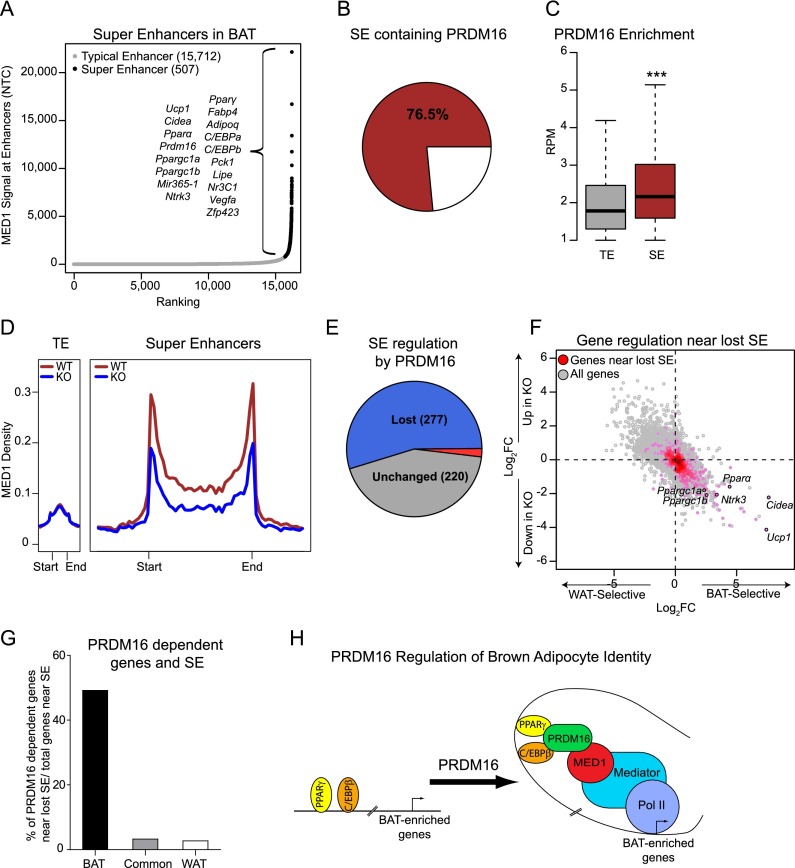
Cell type-specific identity genes are often regulated by large enhancer regions called SEs that are bound by master transcription factors (Whyte et al. 2013). SEs have higher levels of MED1 binding and multiple MED1-binding sites as compared with typical enhancers (TEs) (Whyte et al. 2013). Given the role of PRDM16 in controlling BAT identity and because PRDM16 interacted with MED1, we tested whether PRDM16 was associated with SE activity. We used the genome-wide binding profile of PPARγ, the master transcriptional regulator of white and brown adipocytes, to define a set of constituent enhancers. Next, using the previously described methods (Whyte et al. 2013), we identified 507 SEs and 15,712 TEs in BAT and assigned them to the nearest gene (Fig. 5A). As anticipated, genes having a nearby SE were expressed at higher levels, as reflected by higher levels of RNA Pol II in gene bodies (Supplemental Fig. S5A). Many BAT-selective genes had a nearby SE, including Prdm16, Pparα, Ppargc1a, Ucp1, and Cidea. Reassuringly, gene ontology (GO) analysis identified brown fat differentiation, fat cell differentiation, oxidative reduction, and fatty acid metabolic process as the top scoring gene categories associated with SE-linked genes in BAT (Supplemental Fig. S5B).
Figure 5.
PRDM16 controls BAT-selective SEs. (A) MED1 ChIP-seq signal (normalized tag count) across 16,219 identified enhancers in BAT. (B) Pie chart showing the percentage of SEs that are bound by PRDM16. (C) Box plot comparing PRDM16-binding strength (RPM) at TEs and SEs. (***) P < 10−25. (D) Metagene representation of MED1 signal at TEs and SEs in wild-type (WT) (red) and Prdm16 knockout (KO) (blue) BAT. The X-axis schematizes the start and end of enhancers. The Y-axis is the average signal in RPM. (E) Pie chart showing BAT SEs that are lost (blue), induced (red), or unchanged (gray) by PRDM16 deficiency. Lost or induced was defined as a twofold or greater change of MED1 signal. (F) Scatter plot showing correlation between SEs lost in Prdm16 knockout BAT and gene expression levels in BAT versus WAT (X-axis) and Prdm16 knockout versus wild-type BAT (Y-axis). (G) Proportion of BAT-selective (BAT-sel.), common, or WAT-selective (WAT-sel.) genes that have reduced gene expression in Prdm16 knockout BAT and are associated with lost SEs. (H) Model for PRDM16-mediated gene activation. PRDM16 interacts with enhancer regions at BAT-selective genes through C/EBPβ and PPARγ. PRDM16 recruits MED1/Mediator to bridge enhancer-bound transcription factors with RNA Pol II/the general transcription machinery.
Remarkably, 78% of BAT SEs contain at least one PRDM16-binding site, and the strength of PRDM16 binding was higher at SEs than at TEs (Fig. 5B,C). SEs were also much more sensitive than TEs to the loss of PRDM16 (Fig. 5D). Out of 507 SEs in wild-type BAT, more than half (277) were “lost” (defined as a reduction in MED1 levels) in Prdm16 knockout BAT, while only 15 SEs were induced (Fig. 5E). The “lost SEs” were linked to genes that had reduced expression levels in Prdm16 knockout relative to wild-type BAT, including Ucp1, Pparα, Cidea, Ntrk3, Ppargc1a, and Ppargc1b (Fig. 5F). Globally, ∼50% of the SE-linked BAT-selective genes had decreased mRNA levels in Prdm16 knockout BAT (Fig. 5G).
SE activity is regulated by BRD4, a bromodomain protein that interacts with Mediator (Loven et al. 2013). To examine whether the transcription of SE-linked, BAT-selective genes is dependent on BRD4 activity, we treated brown adipocytes with the BRD4 inhibitor JQ1, an inactive analog (JQ1−), or vehicle control for 6 h. We then measured the expression levels of the top eight BAT-selective genes that had a linked SE. The levels of seven out of eight of these genes were decreased by JQ1, whereas several highly expressed TE-associated genes were unaffected (Supplemental Fig. S5C). These results suggest that PRDM16-activated genes in BAT depend on BRD4 function and thus are likely controlled by SEs.
Discussion
PRDM16 is a powerful activator of the thermogenic gene program in adipocytes. In this study, we show that PRDM16 interacts with MED1 at brown fat-specific genes to promote gene transcription. The binding of MED1 to PRDM16 appears to be direct, since these factors are able to bind together in vitro as purified proteins. MED1 is a component of the Mediator complex, which plays a key role in regulating gene expression through a variety of mechanisms (Chen and Roeder 2011). Among its various functions, Mediator bridges enhancer regions and their associated transcription factor complexes with RNA Pol II and the transcriptional machinery at gene promoters (Malik and Roeder 2010; Chen and Roeder 2011). Our findings suggest that PRDM16 (or PRDM3) binds to chromatin at enhancers, many of which are SEs, in BAT-selective genes via PPARγ and C/EBPβ (and likely other factors). At these sites, PRDM16 recruits MED1/Mediator and, by doing so, organizes higher-order chromatin structure and promotes preinitiation complex assembly to enhance target gene transcription (Fig. 5H).
Loss of PRDM16 (and MED1) disrupts higher-order chromatin architecture at certain brown fat-specific target genes without impeding the chromatin binding of other transcription factors, including PRDM16-interacting partners PPARγ and C/EBPβ. At Ppargc1a, our results suggest that PRDM16 facilitates the formation of an active chromatin hub (Malik and Roeder 2010) that links at least two enhancer elements with the promoter region. Interestingly, in the absence of PRDM16, only the interaction between the more distal enhancer (−42k) and the promoter is abolished. At Pparα, enhancer regions containing the three sites of PRDM16/MED1 enrichment interact with the promoter in brown but not white adipocytes, and the strength of these interactions is reduced in Prdm16 knockout cells. Future studies will be needed to assess whether PRDM16 affects higher-order chromatin structure at a broader or even full set of target genes. Additionally, it will be important to try to identify mutations in PRDM16 that prevent its association with MED1 in order to more precisely define the role of MED1/Mediator in PRDM16 action.
PRDM16 also physically interacts with the methyltransferase EHMT1 and/or CtBPs in brown adipocytes to repress the transcription of certain white fat- or muscle-selective genes (Kajimura et al. 2008; Ohno et al. 2013; Harms et al. 2014). It is not known whether EHMT1 and/or CtBP are also part of an activating PRDM16 complex at BAT-selective genes. Conversely, it is unclear whether PRDM16 interacts with MED1/Mediator at PRDM16-repressed “white” genes, given that the Mediator complex can also participate in transcriptional repression (Ding et al. 2008). How the activating versus repressive function of PRDM16 is determined at different types of target genes is an important and unresolved question. Presumably, the PRDM16-activated and PRDM16-repressed enhancers/genes are bound by different sets of transcription factors/cofactors that dictate transcriptional output. Unfortunately, our computational analyses did not reveal any obvious candidate DNA motifs that were enriched specifically at either white fat- or brown fat-specific PRDM16 target sites. Given that PRDM16 has DNA-binding activity (Nishikata et al. 2003), one hypothesis is that PRDM16 binds to its recognition site directly at repressed genes but indirectly (via PPARγ and C/EBPβ) at BAT genes. Such a putative PRDM16-binding motif may not emerge easily from global analyses if relatively few genes are directly repressed by PRDM16. Additional ChIP-seq analyses of CtBP and EHMT1 binding in BAT may be informative in identifying genes that are directly repressed by PRDM16. Another possibility is that PRDM16 acts at brown and white genes during different temporal stages of adipogenesis. So far, we only analyzed PRDM16 binding in formed BAT and fully differentiated brown adipocytes. It would be interesting to examine whether PRDM16 has distinct targets and potentially different partner proteins at earlier stages of brown adipocyte differentiation.
On a related note, deletion of Prdm16 in mature brown adipocytes using the AdipoQCre driver did not cause any reduction in the levels of brown fat-selective genes even in 3-mo-old mice (Cohen et al. 2014), a time point at which deletion of Prdm16 at earlier developmental stages reduces brown fat gene levels (Harms et al. 2014). PRDM16 may thus cooperate with MED1/Mediator to establish chromatin hubs and would then be dispensable for hub stability. In this model, PRDM16 facilitates promoter–enhancer interactions, but once these long-range interactions are established, they are stabilized through other mechanisms. One appealing hypothesis is that structural cohesin proteins, which are documented to interact with MED1 and the mediator complex (Kagey et al. 2010), are recruited to chromatin hubs to stabilize assembled chromatin loops.
In summary, PRDM16 is a crucial component of the transcriptional network that drives the expression of brown fat cell-specific genes in adipocytes. Developing methods to increase the expression and/or activity of PRDM16 in adipocytes may translate into effective therapies for obesity and type 2 diabetes. It has become increasingly clear over recent years that PRDM16 is a transcriptional coregulator that participates in many different complexes to control various gene programs. It may thus be beneficial to activate a PRDM16-driven subprogram in fat cells. A more detailed mechanistic and structural understanding of how PRDM16 interacts and cooperates with MED1/Mediator may uncover novel drug targets aimed at promoting thermogenesis in fat tissue.
Materials and methods
Animals
Myf5Cre mice (stock no. 010529) were obtained from the Jackson Laboratory. Prdm16flox and Prdm3flox were previously described (Goyama et al. 2008; Harms et al. 2014). Adult BAT-selective Prdm16 knockout (Myf5Cre/+; Prdm16flox/flox) mice displayed a near complete loss of PRDM16 mRNA and protein in interscapular BAT (Harms et al. 2014) and were studied here at ≥9 mo of age. All animal experiments were approved by the University of Pennsylvania’s Institutional Animal Care and Use Committee.
Cell culture
Wild-type and Prdm16 knockout brown adipocytes were cultured and differentiated as described before (Harms et al. 2014). siMed1 (Dharmacon L-040964-01) was electroporated into primary brown adipocytes at 6 d of differentiation using the Lonza Amaxa Nucleofector II (program A-033) and nucleofector kit V (VCA-1003). Brown adipocytes were treated with 500 nM JQ1 and its inert analog, JQ1−, for 6 h.
Real-time qPCR
Total RNA was extracted by TRIzol (Invitrogen) followed by purification using PureLink RNA columns (Invitrogen). Isolated mRNA was reverse-transcribed using the high-capacity cDNA synthesis kit (Applied Biosystems) and used in real-time qPCR reactions with SYBR Green master mix (Applied Biosystems) on a 7900 HT (Applied Biosystems). Tata-binding protein (Tbp) was used as an internal normalization control.
ChIP
BAT from wild-type and Prdm16 knockout mice was dissected, washed with PBS, minced, fixed in 1% formaldehyde for 15 min at 4°C, and quenched with 125 mM glycine for 5 min at 4°C. MED1 samples were additionally cross-linked using EGS (Thermo, 21565) as previously described (Zeng et al. 2006). Samples were then washed twice with cold PBS and placed in a lysis buffer containing 20 mM Tris, 0.1% SDS, 1% Triton-100, 150 mM NaCl, 1 mM EDTA, and protease inhibitors and sonicated until a mean chromatin size of 500–1 kb was achieved. Chromatin was spun at 13,000 rpm for 10 min, and antibodies were added for overnight incubation at 4°C on a rotating platform. Protein-A Sepharose (GE 17-0780-01) was added to samples for 4 h at 4°C. Samples were washed twice with 0.1% SDS, 0.1% NaDOC, 1% Triton X-100, 0.15 M NaCl, 1 mM EDTA, and 20 mM Tris (pH 8); once with 0.1% SDS, 0.1% NaDOC, 1% Triton X-100, 0.5 M NaCl, 1 mM EDTA, and 20 mM Tris (pH 8); once with 0.25 M LiCl, 0.5% NaDOC, 0.5% NP-40, 1 mM EDTA, and 20 mM Tris (pH 8); and twice with 1 mM EDTA and 20 mM Tris (pH 8). Samples were eluted overnight at 65°C in a solution containing 20 mM Tris (pH 8), 1 mM EDTA, and 1% SDS. Samples were subsequently treated with RNase A and Proteinase K and column-purified (Qiagen, 28104). Target enrichment was calculated as percent of input. Primer sequences are in Supplemental Table S1. The PRDM16 antibody used for ChIP has been previously described (Harms et al. 2014). The commercial antibodies used were H3K27-Ac (Abcam, ab4729), Pol II (Santa Cruz Biotechnology, sc-899 or sc-9001), PPARγ (Santa Cruz Biotechnology, sc-7196), C/EBPβ (Santa Cruz Biotechnology, sc-150), and MED1 (Bethyl Laboratories, A300-793A). All ChIP-seq data have been deposited in Gene Expression Omnibus (GEO) with the following accession numbers: GSE63964 (H3K27-Ac and RNA Pol II in BAT and WAT) and GSE63965 (PRDM16, RNA Pol II, MED1, H3K27-Ac in wild-type, and PRDM16 knockout BAT).
Sequential ChIP
Chromatin was prepared from BAT as described above. Chromatin was immunoprecipitated overnight using an anti-C/EBPβ antibody and washed as described for standard ChIP. However, elution was conducted in buffer containing 10 mM DTT and 1% SDS and kept for 15 min at 65°C. Eluted chromatin was diluted by 20-fold in ChIP dilution buffer and incubated with IgG or a second antibody of interest. The second elution was conducted identically to a standard ChIP. The isolated chromatin was reverse-cross-linked, purified, and analyzed via qPCR. Sequential ChIP for PRDM16 was performed identically, except that the tissue was additionally cross-linked with EGS (Zeng et al. 2006).
ChIP-seq data processing
ChIP-seq reads for PRDM16, H3K27-Ac, Pol II, and MED1 were aligned to mouse genome (mm9) using Bowtie with options -k 1 -m 1-best-strata (Langmead et al. 2009). All redundant reads were discarded except one per genomic position. Peak calling for PRDM16 and MED1 was performed using Homer (Heinz et al. 2010). After initial calling, all peaks were resized to 200 base pairs (bp), and a cutoff of one read per million total reads (RPM) was applied. PRDM16-binding sites were more rigorously defined as follows: After initial peak calling, reproducibility of replicates was assessed by scatter plot before pooling. Final peak calling was performed for pooled wild type using pooled knockout samples as a background control. For H3K27-Ac analysis, we performed differential peak calling for BAT and WAT samples using one depot as a ChIP sample, the other as a control, and vice versa. In all cases, any genomic regions that overlapped with ENCODE blacklist regions (Bernstein et al. 2012) were eliminated. De novo motif search was done within a 200-bp region around peak centers using Homer. In addition to the default motif search for all PRDM16 sites, we also performed a motif search using two subgroups of PRDM16 sites within 100 kb from the TSS of (1) genes up-regulated by Prdm16 knockout and (2) genes down-regulated by Prdm16 knockout. After confirming that all four key motifs are present in both groups, we compared fold enrichment of the key motifs in each group relative to all PRDM16 sites using the common set of motif position weight matrices (PWMs) for fair comparison, and P-value was calculated using a hypergeometric test.
Genome-wide analysis comparing depot-selective genes
BEDTools (Quinlan and Hall 2010) was used for genomic region handling. When comparing PRDM16 signal around depot-selective genes, we used fat pad DNase-hypersensitive sites (DHSs) downloaded from ENCODE (Bernstein et al. 2012) as an unbiased set of regulatory elements, and PRDM16 ChIP-seq signals at DHSs within 100 kb of the TSS were summed for each gene. H3K27-Ac levels were also compared between depot-selective genes anchoring on DHSs, where H3K27-Ac ChIP-seq signal was measured in a 2-kb window around DHSs. Gene transcriptional changes were measured as Pol II ChIP-seq signal in gene bodies, excluding the first 500 bp from the TSS. To compare MED1 occupancy changes, two sets of MED1 peaks from wild type and knockout were pooled, and any overlapping peaks whose center-to-center distance was <100 bp were merged into a single peak.
SE analysis
Previously published PPARγ ChIP-seq data in BAT was downloaded from GEO (Rajakumari et al. 2013). ROSE (Hnisz et al. 2013; Loven et al. 2013) was used for SE calling, where enhancers were ranked by MED1 signal. A gene was associated with enhancers (typical or super) within a 100-kb region around the TSS, where multiple associations were allowed. GO analysis was done using DAVID (Huang et al. 2009).
3C
The 3C procedure was performed as described (Ho et al. 2013) using wild-type and Prdm16 knockout brown adipocytes or 3T3-L1 adipocytes at day 8 of differentiation. Briefly, 20 µg of fixed nuclei was digested with BglII (Ppargc1a) or EcoR1 (Pparα). Half (10 µg) of the digested chromatin was used for the ligation, while the other half was set up identically but without T4 DNA ligase. The PCR products were separated on 1.5% agarose gels, stained with SYBR Gold, imaged with the Typhoon PhosphorImager system (GE Healthcare), and quantified using ImagerQuant TL (GE Healthcare). The ligation efficiency was calculated as the ratio of ligated products to the corresponding random ligation of a mouse BAC clone; this was normalized to 3C analysis at a housekeeping locus (Ercc3) (Ho et al. 2008).
Protein interaction analyses
Protein interactions were assayed as previously described (Kajimura et al. 2008; Seale et al. 2008). Protein extracts were prepared in a buffer containing 20 mM Tris, 10% glycerol, 200 mM NaCl, 2 mM EDTA, 0.1% NP-40, 10 mM NaF, and protease inhibitors. Flag-M2 beads (Sigma, A2220) were added to the lysate and subsequently washed five times. Proteins were separated in 4%–12% Bis-Tris NuPAGE gels (Invitrogen) and transferred to PVDF membranes. Primary antibodies used were anti-PRDM16 (Seale et al. 2007), MED1 (Bethyl A300-793A), and M2-HRP (Sigma, A8592). For in vitro binding assays, GST-PRDM16 fragments were prepared as described (Kajimura et al. 2008). S35-labeled MED1 was prepared using the TNT reticulocyte lysate kit (Promega, L5020). Equal amounts of GST-PRDM16 proteins were incubated overnight at 4°C with in vitro translated MED1 in a buffer containing 20 mM HEPES (pH 7.6), 150 mM KCl, 2.5 mM MgCl2, 0.05% NP40, and 10% glycerol. GST beads were washed three times. Bound proteins were separated in 4%–12% Bis-Tris NuPAGE gels (Invitrogen) and soaked in a solution containing 4% glycerol, 46% MeOH, and 10% acetic acid for 30 min followed by soaking in Amplify (GE/Amersham, NAMP 100) for 30 min. The gel was dried and analyzed via fluorography.
Statistical analysis
All ChIP-qPCR data derived from tissue were reported as mean ± SEM. Data from cell lines and primary cells were reported as mean ± SD. Student’s t-test was used to calculate significance in ChIP-qPCR and tissue culture experiments (P < 0.05 [*], P < 0.01[**], and P < 0.001 [***]) using Excel.
Supplementary Material
Acknowledgments
We thank Samantha Falk for technical help. We are also grateful to the Functional Genomics Core of the Penn Diabetes and Endocrinology Research Center (DK19525) for high-throughput sequencing. This work was supported by a National Institute of General Medical Sciences/National Institutes of Health award (DP2OD007288) and a Searle Scholars Award to P.S., a National Institutes of Health award (R21DK098769) to K.-J.W., and a National Institute of Diabetes and Digestive and Kidney Diseases award (R01 DK49780) and the JPB Foundation to M.A.L. M.J.H., Y.H., S.N.S., J.I., S.R., and D.J.S. performed experiments and analyzed data. H.-W.L. and K.-J.W. performed computational analyses of ChIP-seq and mRNA expression data sets. D.J.S. and M.A.L. provided ChIP-seq data sets for integrated analysis. M.J.H., H.-W.L., K.-J.W., and P.S. designed experiments. M.J.H. and P.S. wrote the manuscript.
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.252734.114.
References
- Bernstein BE, Birney E, Dunham I, Green ED, Gunter C, Snyder M, Consortium EP. 2012. An integrated encyclopedia of DNA elements in the human genome. Nature 489: 57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borggrefe T, Yue X. 2011. Interactions between subunits of the Mediator complex with gene-specific transcription factors. Semin Cell Dev Biol 22: 759–768. [DOI] [PubMed] [Google Scholar]
- Bornfeldt KE, Tabas I. 2011. Insulin resistance, hyperglycemia, and atherosclerosis. Cell Metab 14: 575–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W, Roeder RG. 2011. Mediator-dependent nuclear receptor function. Semin Cell Dev Biol 22: 749–758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen P, Levy JD, Zhang Y, Frontini A, Kolodin DP, Svensson KJ, Lo JC, Zeng X, Ye L, Khandekar MJ, et al. . 2014. Ablation of PRDM16 and beige adipose causes metabolic dysfunction and a subcutaneous to visceral fat switch. Cell 156: 304–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding N, Zhou H, Esteve PO, Chin HG, Kim S, Xu X, Joseph SM, Friez MJ, Schwartz CE, Pradhan S, et al. . 2008. Mediator links epigenetic silencing of neuronal gene expression with x-linked mental retardation. Mol Cell 31: 347–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goyama S, Yamamoto G, Shimabe M, Sato T, Ichikawa M, Ogawa S, Chiba S, Kurokawa M. 2008. Evi-1 is a critical regulator for hematopoietic stem cells and transformed leukemic cells. Cell Stem Cell 3: 207–220. [DOI] [PubMed] [Google Scholar]
- Harms M, Seale P. 2013. Brown and beige fat: development, function and therapeutic potential. Nat Med 19: 1252–1263. [DOI] [PubMed] [Google Scholar]
- Harms MJ, Ishibashi J, Wang W, Lim HW, Goyama S, Sato T, Kurokawa M, Won KJ, Seale P. 2014. Prdm16 is required for the maintenance of brown adipocyte identity and function in adult mice. Cell Metab 19: 593–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK. 2010. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell 38: 576–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hnisz D, Abraham BJ, Lee TI, Lau A, Saint-André V, Sigova AA, Hoke HA, Young RA. 2013. Super-enhancers in the control of cell identity and disease. Cell 155: 934–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho Y, Tadevosyan A, Liebhaber SA, Cooke NE. 2008. The juxtaposition of a promoter with a locus control region transcriptional domain activates gene expression. EMBO Rep 9: 891–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho Y, Shewchuk BM, Liebhaber SA, Cooke NE. 2013. Distinct chromatin configurations regulate the initiation and the maintenance of hGH gene expression. Mol Cell Biol 33: 1723–1734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang D, Sherman B, Lempicki R. 2009. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4: 44–57. [DOI] [PubMed] [Google Scholar]
- Kagey MH, Newman JJ, Bilodeau S, Zhan Y, Orlando DA, van Berkum NL, Ebmeier CC, Goossens J, Rahl PB, Levine SS, et al. . 2010. Mediator and cohesin connect gene expression and chromatin architecture. Nature 467: 430–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajimura S, Seale P, Tomaru T, Erdjument-Bromage H, Cooper MP, Ruas JL, Chin S, Tempst P, Lazar MA, Spiegelman BM. 2008. Regulation of the brown and white fat gene programs through a PRDM16/CtBP transcriptional complex. Genes Dev 22: 1397–1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajimura S, Seale P, Kubota K, Lunsford E, Frangioni JV, Gygi SP, Spiegelman BM. 2009. Initiation of myoblast to brown fat switch by a PRDM16–C/EBP-β transcriptional complex. Nature 460: 1154–1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Gade P, Nallar SC, Raha A, Roy SK, Karra S, Reddy JK, Reddy SP, Kalvakolanu DV. 2008. The Med1 subunit of transcriptional mediator plays a central role in regulating CCAAT/enhancer-binding protein-β-driven transcription in response to interferon-γ. J Biol Chem 283: 13077–13086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loven J, Hoke HA, Lin CY, Lau A, Orlando DA, Vakoc CR, Bradner JE, Lee TI, Young RA. 2013. Selective inhibition of tumor oncogenes by disruption of super-enhancers. Cell 153: 320–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik S, Roeder RG. 2010. The metazoan Mediator co-activator complex as an integrative hub for transcriptional regulation. Nat Rev Genet 11: 761–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishikata I, Sasaki H, Iga M, Tateno Y, Imayoshi S, Asou N, Nakamura T, Morishita K. 2003. A novel EVI1 gene family, MEL1, lacking a PR domain (MEL1S) is expressed mainly in t(1;3)(p36;q21)-positive AML and blocks G-CSF-induced myeloid differentiation. Blood 102: 3323–3332. [DOI] [PubMed] [Google Scholar]
- Ohno H, Shinoda K, Ohyama K, Sharp LZ, Kajimura S. 2013. EHMT1 controls brown adipose cell fate and thermogenesis through the PRDM16 complex. Nature 504: 163–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM. 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26: 841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajakumari S, Wu J, Ishibashi J, Lim HW, Giang AH, Won KJ, Reed RR, Seale P. 2013. EBF2 determines and maintains brown adipocyte identity. Cell Metab 17: 562–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosen ED, Spiegelman BM. 2014. What we talk about when we talk about fat. Cell 156: 20–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito M. 2014. Human brown adipose tissue: regulation and anti-obesity potential. Endocr J 61: 409–416. [DOI] [PubMed] [Google Scholar]
- Seale P, Kajimura S, Yang W, Chin S, Rohas LM, Uldry M, Tavernier G, Langin D, Spiegelman BM. 2007. Transcriptional control of brown fat determination by PRDM16. Cell Metab 6: 38–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seale P, Bjork B, Yang W, Kajimura S, Chin S, Kuang S, Scime A, Devarakonda S, Conroe HM, Erdjument-Bromage H, et al. . 2008. PRDM16 controls a brown fat/skeletal muscle switch. Nature 454: 961–967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seale P, Conroe HM, Estall J, Kajimura S, Frontini A, Ishibashi J, Cohen P, Cinti S, Spiegelman BM. 2011. Prdm16 determines the thermogenic program of subcutaneous white adipose tissue in mice. J Clin Invest 121: 96–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanford KI, Middelbeek RJ, Townsend KL, An D, Nygaard EB, Hitchcox KM, Markan KR, Nakano K, Hirshman MF, Tseng YH, et al. . 2013. Brown adipose tissue regulates glucose homeostasis and insulin sensitivity. J Clin Invest 123: 215–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallberg AE, Yamamura S, Malik S, Spiegelman BM, Roeder RG. 2003. Coordination of p300-mediated chromatin remodeling and TRAP/mediator function through coactivator PGC-1α. Mol Cell 12: 1137–1149. [DOI] [PubMed] [Google Scholar]
- Whyte WA, Orlando DA, Hnisz D, Abraham BJ, Lin CY, Kagey MH, Rahl PB, Lee TI, Young RA. 2013. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell 153: 307–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoneshiro T, Saito M. 2014. Activation and recruitment of brown adipose tissue as anti-obesity regimens in humans. Ann Med doi: 10.3109/07853890.2014.911595. [DOI] [PubMed] [Google Scholar]
- Yuan CX, Ito M, Fondell JD, Fu ZY, Roeder RG. 1998. The TRAP220 component of a thyroid hormone receptor-associated protein (TRAP) coactivator complex interacts directly with nuclear receptors in a ligand-dependent fashion. Proc Natl Acad Sci 95: 7939–7944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng PY, Vakoc CR, Chen ZC, Blobel GA, Berger SL. 2006. In vivo dual cross-linking for identification of indirect DNA-associated proteins by chromatin immunoprecipitation. BioTechniques 41: 694–698. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.