Dicentric chromosomes are unstable products of erroneous DNA repair events that can lead to further genome rearrangements and extended gene copy number variations. Lopez et al. find that dicentrics without internal telomere sequences preferentially break at pericentromeric regions. In all cases, cleavage does not occur in anaphase but instead requires cytokinesis. Dicentrics cause the spindle pole bodies and centromeres to relocate to the bud neck during cytokinesis, explaining how cytokinesis can sever dicentrics near centromeres.
Keywords: cell cycle exit, centromere, chromosome, cytokinesis, telomere
Abstract
Dicentric chromosomes are unstable products of erroneous DNA repair events that can lead to further genome rearrangements and extended gene copy number variations. During mitosis, they form anaphase bridges, resulting in chromosome breakage by an unknown mechanism. In budding yeast, dicentrics generated by telomere fusion break at the fusion, a process that restores the parental karyotype and protects cells from rare accidental telomere fusion. Here, we observed that dicentrics lacking telomere fusion preferentially break within a 25- to 30-kb-long region next to the centromeres. In all cases, dicentric breakage requires anaphase exit, ruling out stretching by the elongated mitotic spindle as the cause of breakage. Instead, breakage requires cytokinesis. In the presence of dicentrics, the cytokinetic septa pinch the nucleus, suggesting that dicentrics are severed after actomyosin ring contraction. At this time, centromeres and spindle pole bodies relocate to the bud neck, explaining how cytokinesis can sever dicentrics near centromeres.
Erroneous DNA repair events linking two active centromeres generate dicentric chromosomes, whose instability in mitosis is a major source of genome changes and a key contributor to oncogenesis. Dicentrics stem from double-strand break-induced and template switch-induced rearrangements, replication of hairpin-capped ends, and accidental telomere fusions (McClintock 1941; Bajer 1964; van Steensel et al. 1998; Lobachev et al. 2002; Mizuno et al. 2009, 2013; Stimpson et al. 2010, 2012; Marcand 2014). They can also be created by epigenetic neocentromere formation (Ishii et al. 2008; Stimpson and Sullivan 2010). Rare in normal contexts, double-strand break-induced dicentrics are frequent after exposure to ionizing radiation (Suto et al. 2013). Dicentrics may also become more frequent with age; for instance, due to an increased occurrence of chromosome fusions involving transiently exposed short telomeres in cells approaching replicative senescence. During oncogenesis, a bypass of senescence leads to telomere loss and a state of crisis during which chromosome end fusions generate many dicentrics (Jones et al. 2014).
When dicentrics are formed prior to replication, the two centromeres on the same sister chromatid are pulled toward either the same spindle pole or opposite poles (Fig. 1A). Migration toward the same pole propagates the dicentric chromosome intact to the next generation. Migration toward opposite poles generates an anaphase bridge, which often results in chromosome breakage through an unknown mechanism. Importantly, alternatives to dicentric breakage exist. For instance, a kinetochore can sometimes detach from microtubules, leading to chromosome nondisjunction and aneuploidy. In some cell types, mitotic exit is bypassed in response to anaphase bridges, leading to tetraploidy (Denchi et al. 2006; Pampalona et al. 2012). Two close centromeres can also behave as a single centromere.
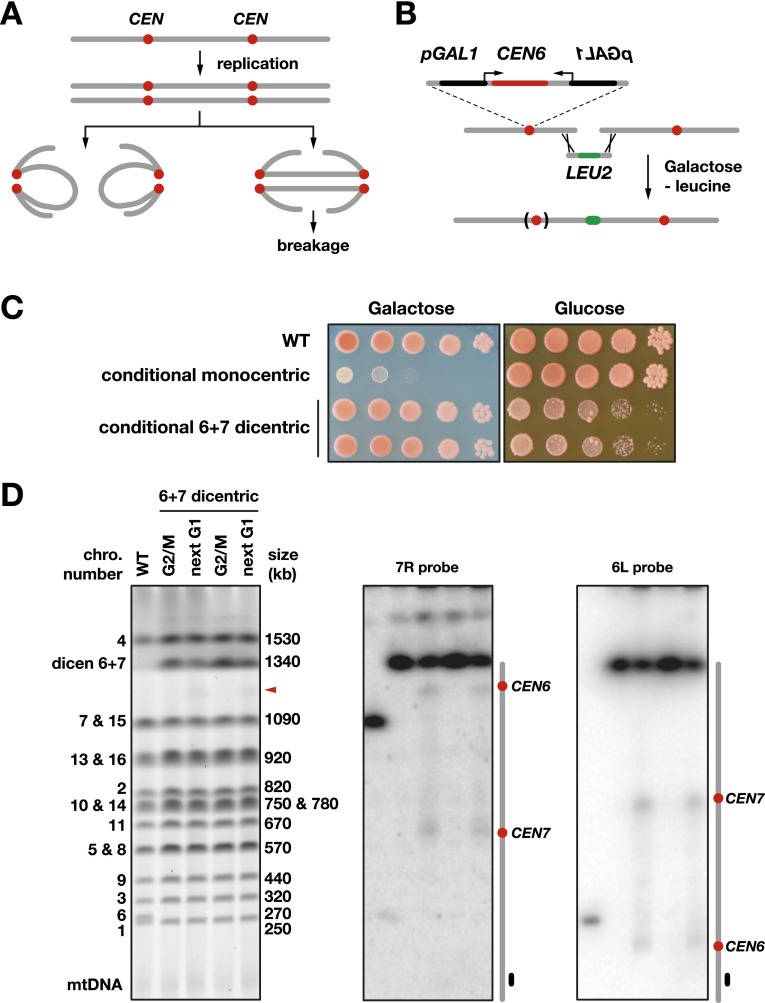
Figure 1.
Breakage of a dicentric formed by chromosome fusion. (A) Segregation during one cell cycle of a dicentric formed in G1. The centromeres are in red. In anaphase, the two centromeres of each sister chromatid can migrate toward the same pole, thus propagating the dicentric intact to the next generation in the two offspring. Alternatively, the centromeres of each sister chromatid can migrate toward opposite poles, forming two anaphase bridges. (B) Fusion of the chromosome 6 right arm to another chromosome end by homologous recombination. (C) Loss of viability on galactose for cells with a conditional monocentric chromosome 6 and on glucose in response to CEN6 reactivation for cells with a conditional 6 + 7 dicentric. Fusion between chromosomes 6 and 7 deletes 1975 bp from the right end of chromosome 6 and 15,271 bp from the left end of chromosome 7. Overnight cultures of strains Lev346 (wild-type [WT]), Lev752 (conditional monocentric), Lev801, and Lev802 (two independent clones with a conditional 6 + 7 dicentric) were diluted by 10-fold serial dilutions, plated on galactose synthetic medium and glucose-rich medium, and incubated for 3 d (galactose) or 2 d (glucose) at 30°C. The ade2-1 mutation present in this background causes the cells to accumulate purine precursors when they start to exhaust the medium. These precursors give a red color to colonies. Colonies with a large fraction of sick or dead cells remain whiter. (D) Breakage of a 6 + 7 dicentric. Cells from strains Lev801 and Lev802 (6 + 7 dicentric) were grown exponentially in galactose-containing medium, synchronized in G1 with α factor, released in glucose-containing medium, and either blocked in G2/M with nocodazole (G2/M) or allowed to proceed through mitosis to be blocked in the next G1 with α factor (next G1). Chromosomes were separated by PFGE, labeled with Gel Red (left panel), and probed with a fragment from the chromosome 7 right arm (middle panel) or chromosome 6 left arm (right panel). The wild-type control was from strain Lev752 grown in glucose-containing medium. The red arrowhead marks the position of the molecules broken near CEN6.
The newly broken ends generated by dicentric breakage cause cell death or are subjected to repair. Fusion of the broken ends can create new dicentrics that break again in the following mitoses, engaging the cell lineage in successive breakage–fusion–bridge cycles (McClintock 1941). Within only a few cell divisions, this can result in large gene copy number aberrations through amplification and deletion of chromosomal regions (Ciullo et al. 2002; Sabatier et al. 2005; Stewénius et al. 2005; Selvarajah et al. 2006; Pennaneach and Kolodner 2009; Marotta et al. 2013). It can also lead to chromothripsis (Sorzano et al. 2013; Li et al. 2014). Dicentric-induced gene copy number alterations are among the pathways allowing malignant cells to emerge and elude the controls preventing cancer (Selvarajah et al. 2006; Martínez-A and van Wely 2011; Gascoigne and Cheeseman 2013). Interestingly, radiotherapy and chemotherapy targeting cancer cells can generate high frequencies of dicentrics, many being purposely lethal, but a few potentially initiating the genetic changes that allow some tumor cells to adapt and escape the therapy. In the germline, rare genome rearrangements created through dicentric breakage contribute directly to karyotype evolution and sometimes rapid adaptation (Croll et al. 2013).
To avoid or exit the recurrent genome instability created by breakage–fusion–bridge cycles, dicentrics need to be converted into monocentric chromosomes. Two phenomena can account for this stabilization. One is centromere inactivation through DNA rearrangements or epigenetic switches at one centromere (Lejeune et al. 1973; Avarello et al. 1992; Kramer et al. 1994; Pennaneach and Kolodner 2009; Mackinnon and Campbell 2011; Sato et al. 2012; Song et al. 2013). The other is telomere addition at the broken ends; for instance, by telomerase or break-induced replication (Murnane and Sabatier 2004; Pennaneach and Kolodner 2009).
We previously observed that dicentric breakage is a more controlled process than initially thought (Pobiega and Marcand 2010). In the budding yeast Saccharomyces cerevisiae, dicentrics formed by telomere fusion often break at the fusion site. This processing restores the parental chromosomes, allowing the cell lineage to recover from the fusion with a normal karyotype. As a rescue pathway able to back up nonhomologous end-joining inhibition at telomeres, it can protect cells from the detrimental consequences of accidental telomere fusions. How telomere fusions are more prone to breakage is unclear. For instance, breakage at the fusion is not correlated to its relative position between the centromeres.
Dicentrics lacking internal telomere sequences usually cause cell death in yeast, presumably because they generate broken ends that are either nonreparable or whose repair leads to inviable outcomes. The survivors that emerge often display a deletion of one centromere (Mann and Davis 1983; Kramer et al. 1994; Pennaneach and Kolodner 2009). These deletions require the nonhomologous end-joining double-strand break repair pathway, indicating that dicentrics are sometimes severed at centromeres (Kramer et al. 1994; Lee et al. 1999). More recently, an indirect mapping of dicentric break sites with a yeast mitotic recombination assay also showed that breakage can occur near one centromere (Song et al. 2013).
The physical and catalytic determinants of breakage are unknown. In principle, dicentrics could be cleaved when they are stretched during anaphase. A simple mechanical shearing by spindle forces seems unlikely considering the high elasticity of mitotic chromosomes (Marko 2008; Kruithof et al. 2009), but nucleases could act at this stage (Kramer et al. 1994; Shimizu et al. 2005). Dicentrics could also break later during cytokinesis; for instance, in direct response to constriction or through the action of nucleases activated at this time. In the fission yeast Schizosaccharomyces pombe, entangled chromosomes and nondisjoined chromatids are intersected by the septum at cytokinesis (the “cut” phenotype) (Hirano et al. 1986). In budding yeast, it was suggested that lagging chromatin could postpone cytokinesis completion to prevent breakage (the “NoCut” checkpoint) (Norden et al. 2006). However, this prevention, if it exists, is only partial, since entangled chromosomes, nondisjoined chromatids, and decondensed chromosome arms are cleaved during cytokinesis in budding yeast (McGrew et al. 1992; Baxter and Diffley 2008; Cuylen et al. 2013). Which scenario applies to dicentrics remains unknown.
Here we readdress the behavior of dicentrics beside the ones stemming from telomere fusions by looking at breakage directly. We found that dicentrics without internal telomere sequences preferentially break at pericentromeric regions. In all cases, cleavage does not occur in anaphase but instead requires cytokinesis. Dicentrics cause the spindle pole bodies (SPBs) and centromeres to relocate to the bud neck during cytokinesis, explaining how cytokinesis might sever dicentrics near centromeres.
Results
Conditional dicentrics
To address dicentric breakage, we created and stabilized individual dicentrics produced by fusions between native chromosomes using a conditional centromere. In budding yeast, a centromere is a small 125-base-pair (bp) element connected to a single kinetochore microtubule. This connection can be reversibly disrupted by transcription from a galactose-inducible GAL1 promoter directed toward the centromere (Hill and Bloom 1987; Tanaka et al. 2005). On galactose, the inactivated centromere is detached from the microtubule. When transcription is turned off by glucose, the centromere is rapidly reattached to a microtubule, and its function is restored (Tanaka et al. 2005).
In this study, we chose to inactivate the native centromere of chromosome 6. To maximize its inactivation on galactose, a GAL1 promoter was inserted on each side of the centromere (Fig. 1B; Pobiega and Marcand 2010). CEN6 inactivation will stabilize chromosome fusion between chromosome 6 and another chromosome. In addition, it will create a positive selection for the fusions, since the missegregation of an unfused acentric chromosome 6 has a detrimental effect on cell growth, whereas chromosome fusion will rescue normal segregation and viability (Pobiega and Marcand 2010).
First, we fused the end of the chromosome 6 right arm to the end of the chromosome 7 left arm to generate a conditional dicentric whose centromeres are 600 kb apart. To do so, haploid cells were transformed with a DNA fragment containing a klLEU2 selectable marker flanked by sequences homologous to those near each chromosome end. Clones in which homologous recombination had fused the two chromosome ends were selected on galactose medium lacking leucine (Fig. 1B). Selected clones are sensitive to glucose; i.e., to CEN6 reactivation (Fig. 1C; Supplemental Fig. S1). When grown in galactose medium, they all display the same fusion between chromosome 6 and chromosome 7 (Fig. 1D, left panel).
Dicentrics preferentially break at pericentromeric regions
Next, we asked how the fused chromosomes break when the function of CEN6 is restored. To look at dicentric breakage during a single cell cycle, cells grown exponentially in the presence of galactose were synchronized in G1 with α factor and released from the arrest in glucose-containing medium. Cells were either blocked at the G2/M transition by nocodazole or allowed to progress through mitosis until they were blocked again in the next G1 by α factor that was added back to the medium. Chromosomes were separated by pulse field gel electrophoresis (PFGE) and hybridized with probes from either side of the dicentric (Fig. 1D). During progression through mitosis, full-length dicentric molecules partially fade away (Fig. 1D, left panel). Southern blots of the pulse field gel hybridized with probes specific to the right arm of chromosome 7 or the left arm of chromosome 6 show that break sites are not evenly distributed between the centromeres and occur predominantly in their vicinity (Fig. 1D). The large DNA fragments generated by breakage near CEN6 are also directly detectable after in-gel DNA staining (Fig. 1D, left panel, arrowhead). Contrary to what was previously proposed based on the result of an indirect recombination assay (Song et al. 2013), the pericentromeric regions near the normal centromere and the conditional centromere are equally prone to break. Biases introduced by the selection of a subclass of events in the recombination assay could account for the difference. In our assay, the rare stable survivors of CEN6 reactivation often display a deletion of CEN6 but not CEN7 (Supplemental Fig. S1; see also Kramer et al. 1994). This result highlights the importance of looking at breakage directly and not only by means of the viable and potentially biased repair products of the breaks.
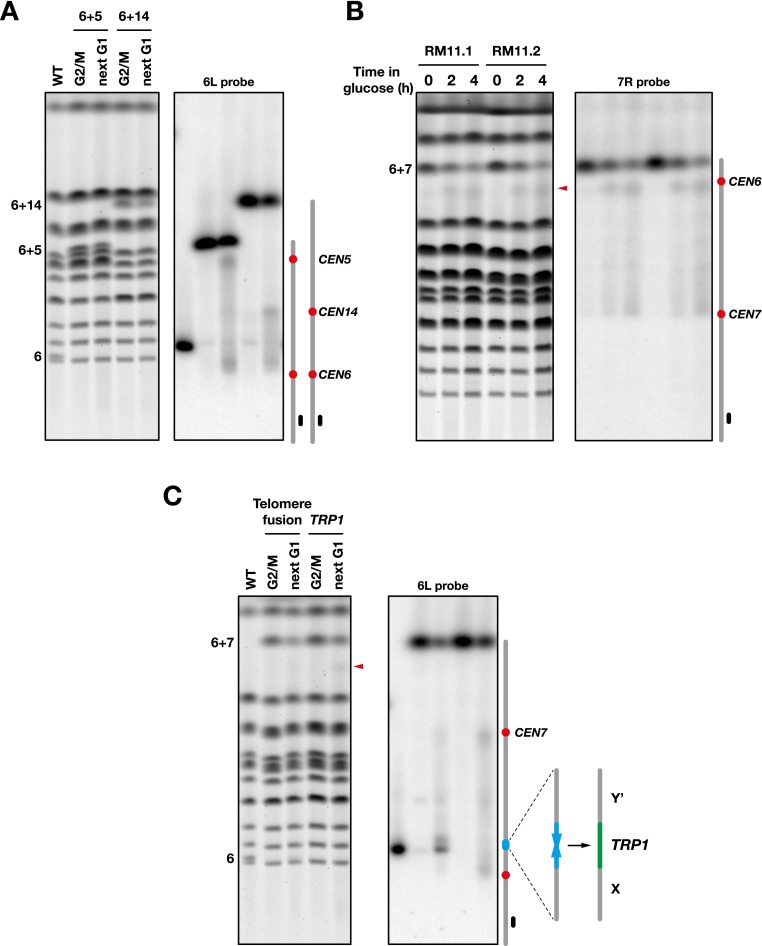
Preferential breakage near the two centromeres is also observed in dicentrics fusing chromosome 6 to other chromosomes (for instance, with chromosomes 5 and 14) (Fig. 2A) as well as in cells from a distant genetic background (the vineyard isolate RM11-1a) (Fig. 2B). Together, these results show that dicentrics in budding yeast do not rupture randomly between the centromeres: The pericentromeric regions behave as hot spots more prone to breakage.
Figure 2.
Breakage of dicentrics with or without telomere fusion. (A) Breakage of 6 + 5 and 6 + 14 dicentrics. Fusion between chromosomes 6 and 5 or 14 deletes 1975 bp from the right end of chromosome 6 and ∼3000 bp from the Y′ subtelomeric element at the right end of chromosome 5 or 14 (strain Lev826 and Lev822, respectively). Chromosomes were separated by PFGE, labeled with Gel Red (left panel) and probed with a fragment from the chromosome 6 left arm (right panel). (B) Breakage of a 6 + 7 dicentric in a genetic background derived from the vineyard isolate RM11-1a (Dimitrov et al. 2009). Cells from strains 134-1a/dic and 134-1d/dic (RM11.1 and RM11.2) were grown exponentially in galactose-containing medium (time 0) and shifted in glucose-containing medium for 2 and 4 h. Chromosomes were separated by PFGE, labeled with Gel Red (left panel), and probed with a fragment from the chromosome 7 right arm (right panel). The red arrowhead marks the position of the molecules broken near CEN6. (C) Breakage at a telomere fusion. Cells from strains Lev1258 and Lev1258/TRP1 were grown exponentially in galactose-containing medium, synchronized in G1, released in glucose-containing medium, and either blocked in G2/M or allowed to proceed to the next G1. Chromosomes were separated by PFGE, labeled with Gel Red (left panel), and probed with a fragment from the chromosome 6 left arm (right panel). In Lev1258/TRP1, the native subtelomeric sequences remain.
Telomere fusions are more prone to breakage than pericentromeric regions
Dicentrics formed by head-to-head telomere fusions between chromosomes often break at the telomere fusion during progression through mitosis (Pobiega and Marcand 2010). In this context, breakage at the telomere fusion predominates, and breakage near centromeres is barely apparent. To test the cis requirement of dicentric breakage at telomere fusions, we took a conditional dicentric with a telomere fusion between the right telomere of chromosome 6 and the right telomere of chromosome 7 and replaced by homologous recombination the TG1–3 telomeric sequence with a klTRP1 marker. As shown in Figure 2C, the loss of the telomeric sequences abolishes breakage at the fusion. It concomitantly reinforces breakage at pericentromeric regions as well as weaker and evenly distributed breakage between the centromeres. Thus, dicentric breakage at telomere fusions depends on telomeric sequences and predominates over breakage at pericentromeric regions.
Pericentromeric breakage hot spots are ∼25–30 kb long
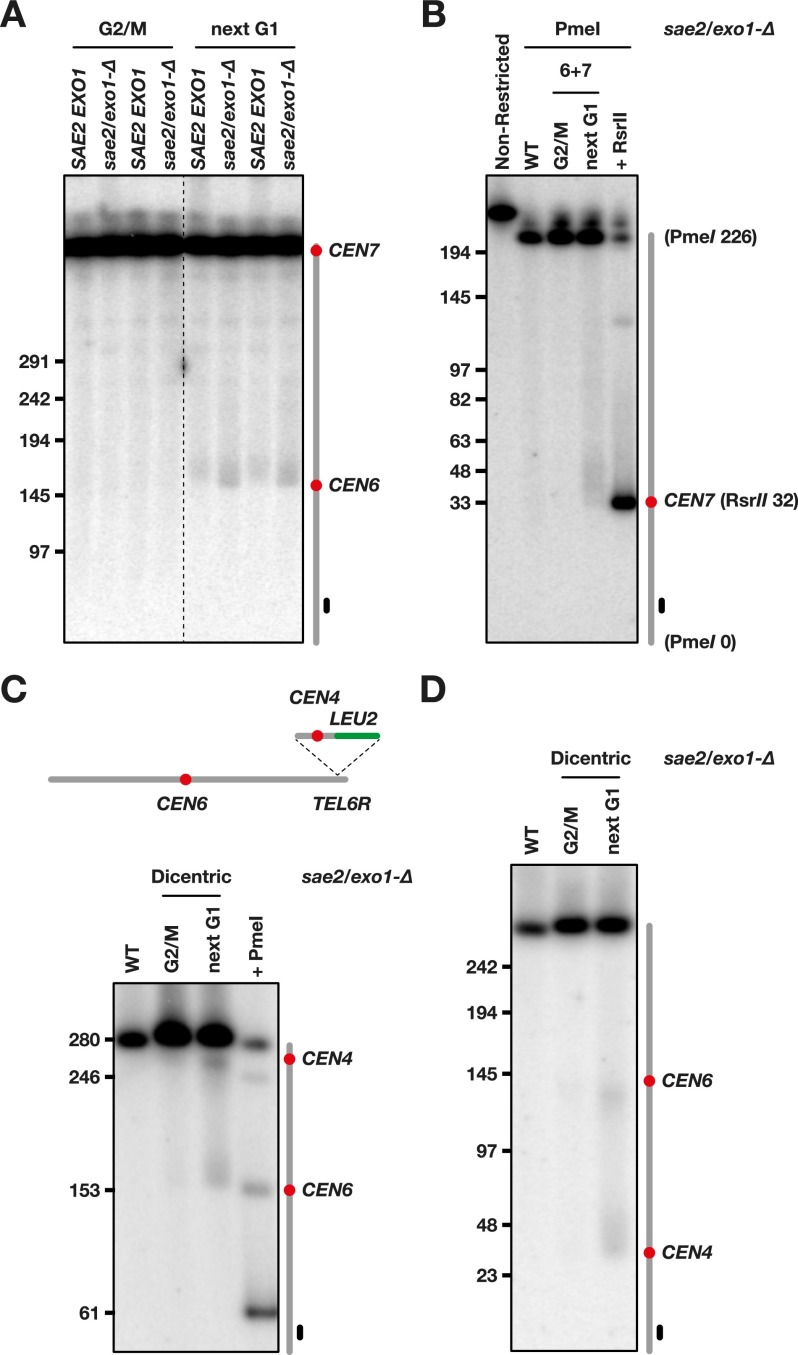
Next, we wanted to estimate more precisely where breakage occurs in the vicinity of the centromeres. We first had a close-up view at CEN6 (Fig. 3A). In cells with a 6 + 7 dicentric, a smear appears after mitosis when DNA is probed from the left arm of chromosome 6. The smearing signal can reflect the break site distribution, but since single-stranded tails alter DNA migration in PFGE, it can also be a consequence of the limited 5′-to-3′ degradation occurring at broken ends in G1 cells (Zierhut and Diffley 2008; Westmoreland et al. 2009). To prevent the formation of single-strand tails after breakage, we looked at the behavior of dicentrics in cells lacking Sae2 and Exo1, the two nucleases required for 5′-to-3′ degradation in G1-arrested cells (Chen et al. 2011; Symington 2014; R Lescasse and S Marcand, unpubl.). As shown in Figure 3A, the loss of the nucleases slightly shifts downward the smear appearing after mitosis. This indicates that the 3′ single-strand tails generated after breakage in wild-type cells partially blur the determination of the break points and that a nuclease-deficient context allows circumvention of this problem. The borders of the smear are at ∼150 and 180 kb. Since the distance between CEN6 and the left telomere is ∼150 kb, breakage occurs in an ∼30-kb region bordered by CEN6 and spreads toward the other centromere.
Figure 3.
Dicentric breakage at pericentromeric regions. (A) Cells from strains Lev801 (SAE2 EXO1) and Lvl103 (sae2/exo1-∆) were grown exponentially in galactose-containing medium, synchronized in G1, released in glucose-containing medium, and either blocked in G2/M or allowed to proceed to the next G1. Chromosomes were separated by PFGE (Supplemental Fig. S3) and probed with a fragment from the chromosome 6 left arm. DNA molecules larger than ∼500 kb comigrate together. (B) DNA from strain Lvl103 (sae2/exo1-∆) was restricted with PmeI in agarose plug prior migration. To mark the position of CEN7, DNA from a wild-type strain lacking dicentrics was further restricted with RsrII (the restriction site is ∼0.2 kb from CEN7). (C) Insertion of an ectopic centromere in the chromosome 6 right arm. Cells from strains Ynb15 (sae2/exo1-∆) were grown exponentially in galactose-containing medium, synchronized in G1, released in glucose-containing medium, and either blocked in G2/M or allowed to proceed to the next G1. Chromosomes were separated by PFGE (Supplemental Fig. S3) and probed with a fragment from the chromosome 6 left arm. To mark the position of CEN6, DNA from a wild-type strain lacking dicentrics was partially restricted with PmeI (the restriction site is ∼2 kb from CEN6). (D) DNA from the same cells was separated by PFGE (Supplemental Fig. S3) and probed with a fragment from the chromosome 6 right end.
Next, we looked at breakage near CEN7 (Fig. 3B). DNA from cells lacking Sae2 and Exo1 was restricted in agarose plugs prior to gel electrophoresis. To map the position of CEN7, we used a native RsrII site at 0.2 kb from the centromere (Fig. 3B, last lane on the right). After mitosis, a smear appears just above the position of CEN7. This indicates that breakage occurs on a region bordered by the centromere and situated toward the other centromere. Its width is ∼25–30 kb. Thus, the two pericentromeric breakage hot spots of this dicentric are similar in length and position relative to the centromere.
Breakage near centromeres is a consequence of centromere structure or function
In yeast, the sequences surrounding the centromeres are unrelated to each other. Since in dicentrics they are more prone to breakage during mitosis, we asked whether this is related to the sequences themselves or is a cis effect from the centromeres. To this purpose, we inserted a 1-kb fragment containing the centromere of chromosome 4 at a distal position of the right arm of chromosome 6 in nuclease-deficient cells. In this conditional dicentric, the two centromeres are 100 kb apart. As shown in Figure 3C, the ectopic centromere establishes a hot spot of breakage. A close-up view from the right end of the chromosome shows that breakage occurs on an ∼25- to 30-kb region bordered by CEN4 and situated toward CEN6 (Fig. 3D). Thus, a 1-kb ectopic centromere sequence is sufficient to create an extended hot spot indistinguishable from the hot spots adjacent to native centromeres. This observation and the fact that unrelated pericentromeric sequences from different centromeres (CEN6, CEN7, CEN5, and CEN14) (Figs. 1D, 2A) all behave as dicentric breakage hot spots indicate that the higher propensity to break is a consequence of centromere structure or function and is unrelated to the surrounding native sequences.
Dicentric breakage requires anaphase exit
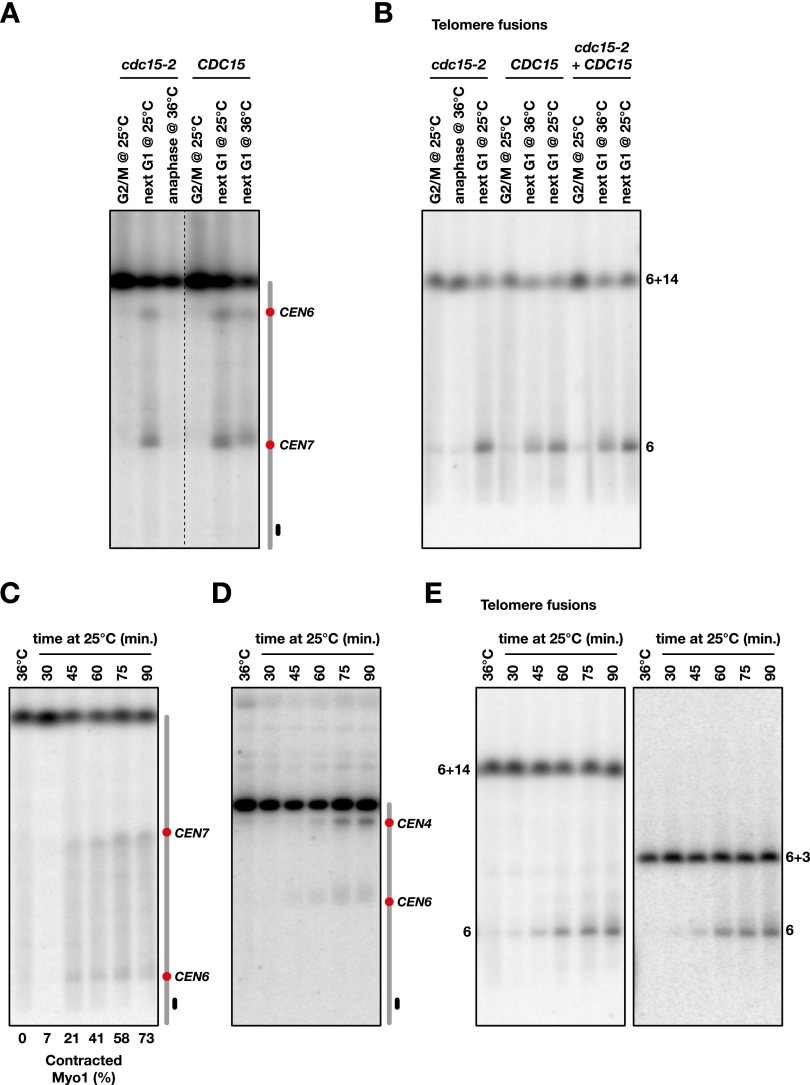
Dicentrics could break in response to the pulling force exerted by the elongated mitotic spindle or the pushing force exerted by cytokinesis. They could also be broken by a nuclease activated during mitotic exit. To distinguish between these scenarios, we first determined the timing of breakage during mitosis. Conditional dicentrics with and without telomere fusion were created in cdc15-2 cells to arrest cell cycle progression in late anaphase at a restrictive temperature. Cells grown exponentially at a permissive temperature (25°C) in the presence of galactose were synchronized in G1 with α factor and released from the arrest in glucose-containing medium. Cells were either blocked by nocodazole at the G2/M transition at permissive temperature, blocked in late anaphase at a restrictive temperature (36°C), or allowed to progress through mitosis at a permissive temperature until they were blocked in the next G1 by α factor. As shown in Figure 4, A and B, dicentrics with or without telomere fusion do not break in anaphase-arrested cells. In wild-type cells (CDC15), dicentric breakage still occurs at 36°C, although the pericentromeric and telomere fusion hot spots are more dispersed, a blurring that might be due in part to increased in vivo 5′ degradation at the broken ends at higher temperatures. These results indicate that the tension established by the elongated mitotic spindle on the anaphase bridges is not sufficient to trigger breakage. This remains true with short intercentromeric distances of 30 and 45 kb (Supplemental Fig. S2).
Figure 4.
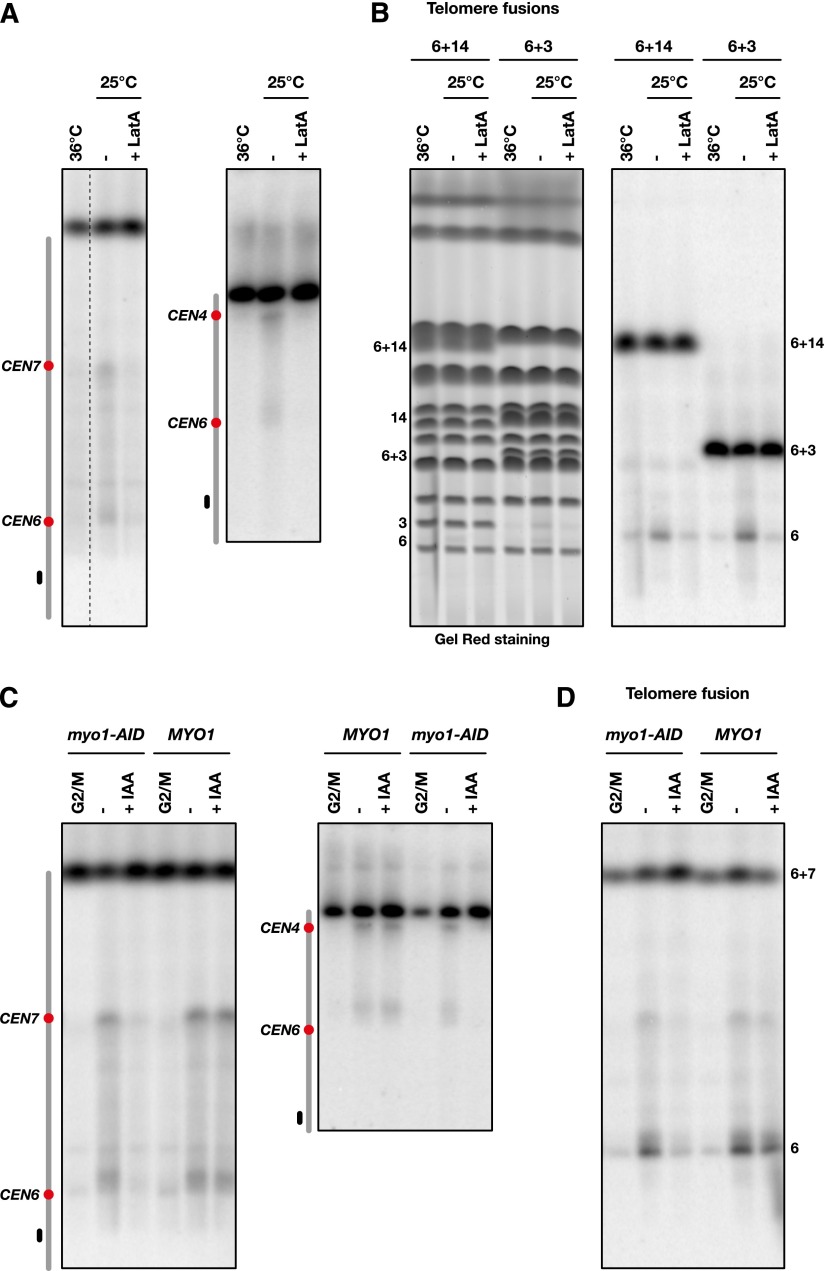
Dicentric breakage requires mitotic exit. (A) Cells from strains Lev803 (cdc15-2) and Lev801 (CDC15) were grown exponentially in galactose-containing medium at 25°C, synchronized in G1, released in glucose-containing medium, and either blocked in G2/M at 25°C or allowed to proceed to the next G1 at 36°C or 25°C. cdc15-2 cells at 36°C remained arrested in anaphase. Chromosomes were separated by PFGE (Supplemental Fig. S3) and probed with a fragment from the chromosome 7 right arm. (B) Cells from strains Lvl200 (cdc15-2), Lev1276 (CDC15), and Lvl200 complemented with a wild-type copy of CDC15 (cdc15-2 + CDC15) were treated as in A. Each contains a telomere fusion between chromosome 6 and chromosome 14. (C) Cells from strain Lvl242 were grown exponentially in galactose-containing medium at 25°C, synchronized in G1, released in glucose-containing medium for 60 min at 36°C, and then released from the anaphase arrest at 25°C. For each time point, the fraction of cells without a Myo1-GFP signal at the bud neck was estimated from the analysis of ∼50 cells. (D) Released time-course experiment with strain Lvl119. CEN4 is inserted at 100 kb from CEN6. (E) Released time-course experiment with strain Lvl200 (telomere fusion with chromosome 14) and strain Lvl198 (telomere fusion with chromosome 3). Chromosomes were separated by PFGE and probed with a fragment from the chromosome 6 left arm.
Next, we looked at dicentric breakage in cells released from anaphase. In yeast, cytokinesis involves a contractile actomyosin ring made of actin filaments assembled by a type II myosin called Myo1 (Wloka and Bi 2012). A Myo1-GFP marker allowed us to follow ring contraction during mitotic exit (Bi et al. 1998). cdc15-2 cells arrested in anaphase at 36°C were shifted to 25°C. Dicentrics started to break ∼45 min after the temperature shift, a time approximately coincident with Myo1 contraction in the released cells observed by microscopy (Fig. 4C–E). This result shows that dicentric breakage requires anaphase exit and coincides with cytokinesis.
Dicentric breakage requires cytokinesis
During cytokinesis, actomyosin ring contraction concurs with cleavage furrow ingression and the growth of the primary septum. The secondary septum then completes the final closure of the cleavage furrow (abscission) (Weiss 2012; Onishi et al. 2013). Actin filaments are essential not only for ring contraction but also to deliver the cargo vesicles essential to septum and membrane growth (Wloka and Bi 2012). To test whether dicentric breakage requires cytokinesis, we looked at breakage in two contexts in which cytokinesis fails.
First, we used the actin-depolymerizing drug latrunculin A (LatA), which destabilizes the actomyosin ring and prevents cargo delivery to the bud neck (Wloka and Bi 2012). When LatA is added to cells released from anaphase, the breakage of dicentrics with or without telomere fusions is prevented (Fig. 5A,B). This indicates that breakage requires actin filaments.
Figure 5.
Dicentric breakage requires cytokinesis. (A) Cells from strains Lev803 (left panel) and Lvl119 (right panel) were grown exponentially in galactose-containing medium at 25°C, synchronized in G1, and released in glucose-containing medium for 60 min at 36°C. LatA (final concentration 5 µM) or its solvent alone (ethanol final concentration 2%) was added prior to release at 25°C. Chromosomes were separated by PFGE (Supplemental Fig. S3) and probed with a fragment from the chromosome 6 left arm. (B) Cells from strains Lvl200 (6 + 14) and Lvl198 (6 + 3) were treated as in A. Each contained a telomere fusion. Chromosomes were separated by PFGE (left panel) and probed with a fragment from the chromosome 6 left arm (right panel). (C) Cells from strains Lvl237 (left panel, myo1-AID), Lvl238 (left panel, MYO1), Lvl233 (right panel, MYO1), and Lvl236 (right panel, myo1-AID) were grown exponentially in galactose-containing medium, synchronized in G1, and released in glucose-containing medium with IAA (final concentration 1 mM) or its solvent alone (ethanol final concentration 0.2%). Chromosomes were separated by PFGE (Supplemental Fig. S3) and probed with a fragment from the chromosome 6 left arm. (D) Cells from strains Lvl246 (myo1-AID) and Lvl252 (MYO1) were treated as in C. Each contained a cloned telomere fusion inserted by homologous recombination next to the klLEU2 marker in a 6 + 7 dicentric.
Second, conditional dicentrics were created in cells carrying a Myo1 auxin-inducible degron (myo1-AID). In the absence of Myo1, the ring is not assembled (Bi et al. 1998), and primary septum formation and cleavage furrow ingression are significantly slowed in the W303 background that we used in our study (Tolliday et al. 2003; Ko et al. 2007). Myo1 was depleted by auxin addition when cells were released from G1 in the presence of glucose and allowed to progress through mitosis until the next G1. As shown in Figure 5, C and D, Myo1 loss results in a strong decrease in dicentric breakage. Together, these results show that dicentric breakage is prevented or significantly delayed when cytokinesis is impaired.
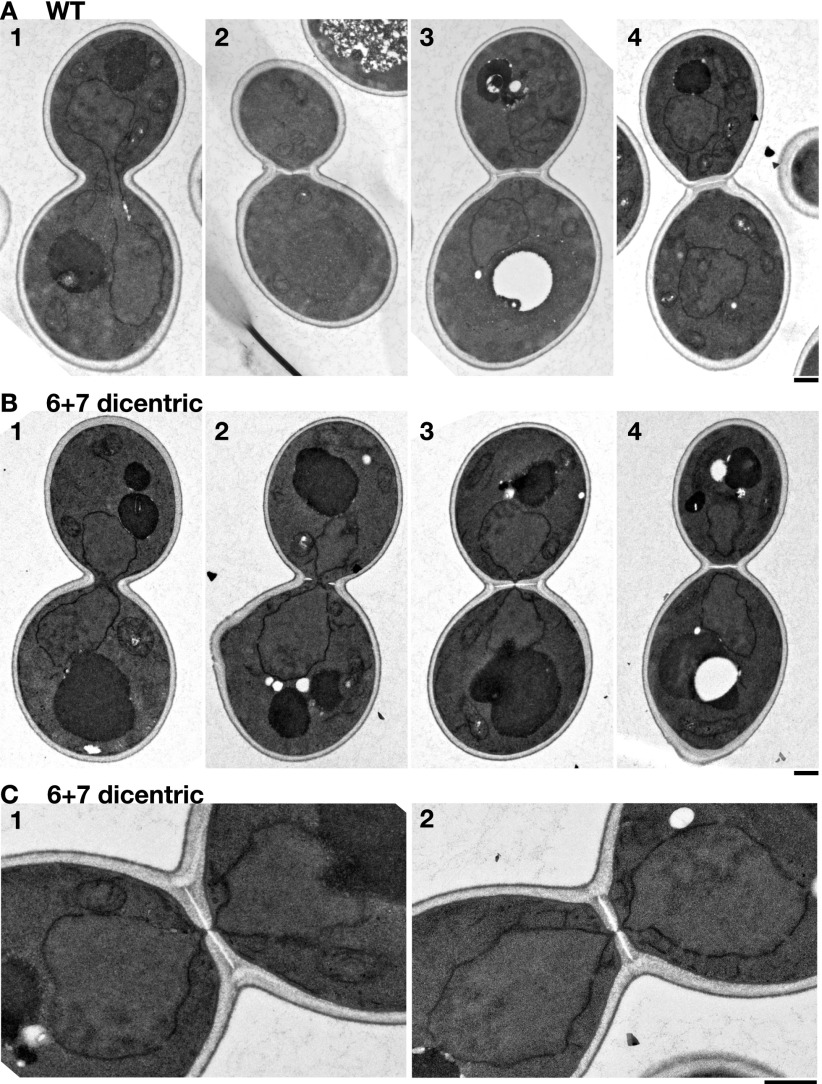
Nuclear separation follows primary septum formation in the presence of dicentrics
In yeast, the nuclear envelope remains assembled during mitosis, and the two segregated nuclei disconnect when the mitotic spindle disassembles during cytokinesis. A coupling of the timings between dicentric breakage and cytokinesis predicts that, when dicentrics form anaphase bridges in mitosis, the two segregated nuclei will remain connected until the cleavage furrow separates them. To test this hypothesis, we used transmission electron microscopy (TEM). Asynchronous cells grown in galactose were switched to glucose for 20–60 min before being fixed and processed for TEM. In control cells without dicentrics, the nuclear envelopes disconnected before initiation of secondary septum formation (Fig. 6A). In contrast, in ∼50% of the cells with a 6 + 7 conditional dicentric lacking internal telomeric sequences, connected nuclear envelopes are located near the bud neck (Fig. 6B). In some cells, the nuclear envelopes are directly pinched by the septum (Fig. 6B, cell 3). Importantly, these cells display a complete primary septum (Fig. 6C, white) accompanied by a secondary septum (Fig. 6C, gray) of nearly final thickness but with a discontinuity through which the nuclear envelop runs (Fig. 6C). In all cells with a closed septum, the nuclei always appeared disconnected (e.g., Fig. 6B, cell 4). These observations support the hypothesis that septum formation triggers dicentric breakage and further suggest that breakage is induced after primary septum formation and actomyosin ring contraction, potentially by secondary septum formation.
Figure 6.
The nuclear envelope is pinched by cytokinesis in the presence of a dicentric. Cells from the control strain Lev752 (A) and the 6 + 7 dicentric strain Lev801 (B,C) were observed as described in Materials and Methods. Cells in A and B are aligned in the inferred sequence of events. (C) Magnified images of the septum region of B, panel 3, and another example, each showing a nuclear envelope pinched by the septum. Time cultured in SC + 2% dextrose was 20 min except for B, panel 3, and C, panels 1 and 2, where it was 60 min. Bars, 0.5 µm.
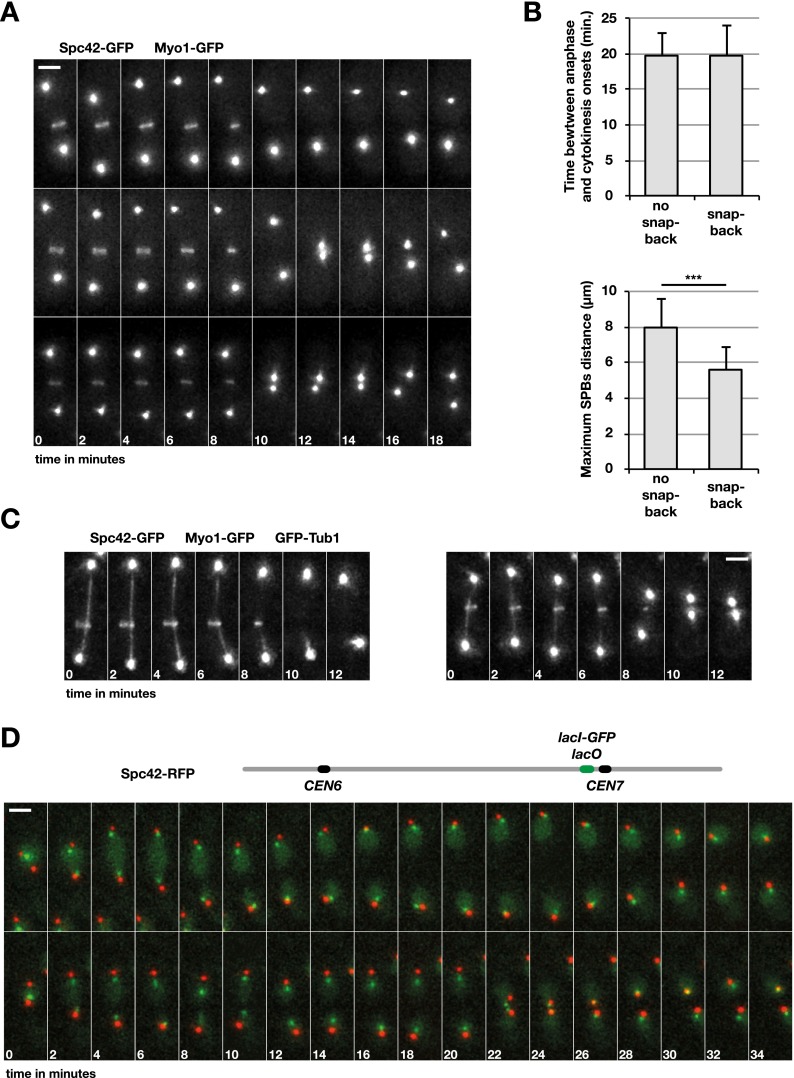
Dicentrics cause the SPBs and the centromeres to relocate toward the bud neck during cytokinesis
The previous data suggest that the closure of the cytokinetic septa triggers dicentric breakage at the bud neck. However, breakage tends to occur near the centromeres. Since the centromeres are segregated to the spindle poles away from the bud neck during mitosis, how can they be preferential sites of cleavage? We addressed this issue using time-lapse microscopy. First, we looked at the segregation of the SPBs relative to actomyosin ring contraction at the onset of cytokinesis. A 6 + 7 conditional dicentric lacking internal telomeric sequences was created in a strain in which Myo1 and the SPB component Spc42 are tagged with GFP. Cells grown in galactose were switched to glucose for ∼60 min prior to image acquisition. In half of the cells (52%, n = 56), the two SPBs behaved normally: They separated from each other and went away from the bud neck during anaphase to reach a maximum distance during ring contraction. Next, they slightly converged back at the end of ring contraction and in the subsequent minutes (e.g., Fig. 7A, top panel). The other half of the cells displayed an unusual behavior: At the end of ring contraction, the two SPBs rapidly snapped back together at the bud neck (average minimum distance between the SPBs: 1 µm ± 0.3 µm) (e.g., Fig. 7A, middle and bottom panels). They remained close to each other for a few minutes and then diverged again. In cells in which this snap back took place, the timing of Myo1 contraction relative to anaphase onset was not different from the one observed in cells in which the snap back did not occur (Fig. 7B, top graph). Interestingly, the maximum distance reached between two SPBs that will snap back is significantly reduced in late anaphase (Figs. 6 [cf. cell 1 in A and B], 7B, bottom graph). This suggests that the mitotic spindle is somewhat constrained and cannot fully elongate.
Figure 7.
A dicentric causes centromeres and the SPBs to relocate to the bud neck at cytokinesis. Experiments were carried out at 25°C. (A) Cells from strains Lev819/dic were grown exponentially in galactose-containing medium and shifted in glucose-containing medium for ∼1 h prior to image acquisition. (B, top panel) Average time between anaphase (SPB distance >2 µm) and Myo1 contraction (the last image with a Myo1-GFP signal). (Bottom panel) Average maximum distance between the two SPBs. (***) P < 0.001 in Student’s t-test. (C) Cells from strains Lev816/dic were grown exponentially in galactose-containing medium and shifted in glucose-containing medium for ∼1 h prior to image acquisition. (D) Cells from strains Lev856/dic were grown exponentially in galactose-containing medium and shifted in glucose-containing medium for ∼1 h prior to image acquisition. Bars, 2 µm.
A simple interpretation of these data is that SPB’s snap back takes place in response to dicentric anaphase bridges that form in half of the cells; i.e., when the centromeres on the same chromatid migrate to opposite poles (Fig. 1A). To address what could be the cause of this unexpected SPB movement, we followed mitotic spindle disassembly in cells expressing the microtubule marker GFP-Tub1 in addition to Spc42-GFP and Myo1-GFP. In the presence of a dicentric, spindle disassembly still takes place with normal timing at the onset of cytokinesis in all cells, including the ones in which the SPBs snap back together (Fig. 7C). Since SPBs converge just after spindle disassembly, the loss of the constraint imposed by the elongated spindle could trigger their movement. Interestingly, a similar post-anaphase convergence had been observed previously in a particular mutant context that allows the spindle to stretch part of the intercentromeric DNA to its full naked length (Thrower and Bloom 2001).
In yeast, kinetochore microtubules link each centromere to a SPB throughout the cell cycle except during replication, which only transiently disrupts these attachments (Kitamura et al. 2007; Marco et al. 2013). Thus, centromeres and SPBs remain close most of the time in cycling cells. To test whether this remains true for dicentric centromeres during mitotic exit, we tracked the position of a centromere using a lacO/lacI-GFP array. A 6 + 7 conditional dicentric was created in a strain in which CEN7 and the SPBs were tagged with GFP and RFP, respectively. The lacO repeats were ∼16 kb from CEN7. Time-lapse imaging was carried out on cells grown in glucose where both centromeres are active. In cells proceeding normally through mitosis, the lacO array signal remained close to a SPB from anaphase to the next G1 (on average, 0.5 µm ± 0.1 µm) (Fig. 7D, top panel). In cells in which the SPBs snapped back together, the distance between the lacO array and the closest SPB was a bit larger during anaphase (on average, 0.8 µm ± 0.2 µm) but not during the snap back and G1 entry (on average, 0.4 µm ± 0.2 µm) (Fig. 7D, bottom panel). Importantly, CEN7 migrated toward the bud neck along with the SPBs. The preservation of the link between centromeres and SPBs during mitotic exit suggests that recoiling of the stretched chromatin bridges may drive SPB movement toward the bud neck. Irrespective of its cause, centromere relocation reconciles the possibility of breakage near centromeres with a process triggered at the bud neck by cytokinesis.
Discussion
This study provides a coherent framework to understand dicentric breakage at the molecular level by showing its precise context in yeast: the positions on chromosomes, the location in the cell, and the timing during mitotic exit. Our findings reveal that cytokinesis cuts dicentric chromosomes. Furthermore, cleavage takes place preferentially at a few hot spots; namely, at pericentromeric regions and, when present, telomere fusions.
Mechanism of dicentric breakage
Several arguments rule out the idea that the pulling force from the elongated mitotic spindle can rupture dicentrics. First and foremost, breakage requires anaphase exit (Fig. 4; Supplemental Fig. S2). Second, the yeast mitotic spindle at the end of anaphase remains small (<10 µm) relative to naked dsDNA length (10 µm per 30 kb), thus not allowing breakage by stretching only. In addition, the forces needed to melt or overstretch dsDNA (>50 pN) are greater than the estimated forces needed to rupture yeast microtubule–kinetochore interaction (∼9 pN), extend naked DNA (<1 pN), unfold the 30-nm chromatin fiber (∼4 pN), or fully unwrap DNA from histones (∼15–25 pN) (Fisher et al. 2009; Kruithof et al. 2009; Akiyoshi et al. 2010). In other words, during anaphase, the chromatin between centromeres remains very elastic and is more likely to unfold or detach from the spindle than break. Instead, breakage appears to be an option of last resort postponed to cytokinesis, the latest stage of mitosis. This is likely needed to prevent breakage of transiently overstretched chromatin regions that occur during normal chromosome segregation; for instance, due to short-lived entanglements and residual uncleaved cohesins (Renshaw et al. 2010; Biebricher et al. 2013).
What could trigger dicentric breakage during cytokinesis? Simple scenarios can be postulated: The lateral forces applied by cytokinesis might be sufficient to cause DNA cleavage or could trigger unknown nucleases that break DNA. Lateral pressure applied on DNA can partially melt it, generating atypical structures (Lei et al. 2010). The latter might be recognized and cleaved off by structure-specific nucleases. In this context, the simultaneous actions of endonucleases on both strands could cause double-strand breaks. Alternatively, a signal emanating from the contracting cytokinesis apparatus could set off nucleases without any direct pressure on DNA.
The forces that cytokinesis is susceptible to generate are not well defined. At the end of actomyosin ring contraction, the ring internal diameter is estimated to be in the 100- to 200-nm range (Pinto et al. 2012). The ring then disassembles to allow the further growth of the septa to fill the remaining hole. Secondary septum growth is involved in the final steps, including resolution of the plasma membrane furrow (Onishi et al. 2013). Contrary to cytokinesis in animal cells, the ESCRT III complex has no demonstrated role at this stage in yeast (McMurray et al. 2011; Elia et al. 2013). In the presence of anaphase bridges created by dicentrics, the contracted actomyosin ring is probably large enough to accommodate the nuclear envelope and two sister chromatids. SPB snap back toward the bud neck occurs at or shortly after the time of ring disassembly (Fig. 7A), suggesting that breakage has not taken place yet. Once the contracted ring has been disassembled, septum growth can reach a point where it directly presses the nuclear envelope (Fig. 6B,C). This may trigger breakage. If DNA is not broken at this stage, the nuclear envelope is likely to collapse under further pressure from secondary septum growth and closure (Onishi et al. 2013), with chromatin being severed concomitantly to abscission. Once the septum is closed, the nuclei appear to be disconnected (e.g., Fig. 6B, cell 4), indicating that chromosomes have broken by this stage. Breakage itself does not seem to have any impact on viability at the cellular level (such as cell lysis due to membrane damage or incomplete septum formation) because the daughter cells proceed normally into S phase (as estimated by budding) (data not shown) and are viable as long as the genomic contents allow for their survival (Supplemental Fig. S1A; Pobiega and Marcand 2010). Interestingly, cytokinesis is also able to break an overstabilized mitotic spindle, although the details of this process remain unknown too (Woodruff et al. 2010).
Dicentric breakage hot spots
In dicentrics, pericentromeric regions and telomere fusions are hot spots more prone to breakage. A passive recoiling of the stretched anaphase bridges can, in principle, account for the observed centromeres and SPBs relocation in the vicinity of the bud neck after spindle disassembly. In this context, what could explain that specific regions are more likely to be cleaved? Depending on how cytokinesis breaks DNA, we can speculate different possibilities.
If breakage results from a lateral pressure applied by cytokinesis on chromatin, a higher propensity to break should be the consequence of a higher chance of being entrapped at the bud neck when cytokinesis is completed. This may be related to the three-dimensional folding of chromatin domains, larger structures being less likely to translocate freely through the narrow channel left at the end of actomyosin contraction. A positioning at the constriction point might also be an active process involving interactions with the cytokinesis machinery through the nuclear envelope. Finally, breakage at pericentromeric regions could reflect a preferential relocation of the folded intercentromeric chromatin on one side of the bud neck due to thermodynamical properties. For instance, stochastic refolding initiation in one nuclear lobe may favor a full refolding in this lobe. Intercentromeric chromatin may also tend to translocate fully toward one or the other nuclear lobe due to an entropic gain (Panja et al. 2013). The size of the SPBs and their anchoring to centromeres would then likely prevent chromosome translocation through the closing neck behind the pericentromeric regions, favoring breakage there.
If cytokinesis triggers nucleases without directly contacting chromatin, preferential cleavage at telomere fusions and within the pericentromeric regions may result from a preferential recruitment of the nucleases at these loci. Once cleaved, each released chromatin fragment could refold in one nuclear lobe, clearing the space at the bud neck to allow abscission.
What could be the function of dicentric breakage hot spots? Breakage at a telomere fusion restores a normal karyotype, a process offering a straightforward advantage against accidental telomere fusions and whose maintenance could be under positive selection (Pobiega and Marcand 2010). Breakage at pericentromeric regions creates a more ambiguous outcome: Often lethal, it may also allow rearrangements concomitant to double-strand break repair that result in a centromere deletion in some cells, thus converting dicentrics into mitotically stable monocentric chromosomes without gene loss or gain (Supplemental Fig. S1; Kramer et al. 1994). Since these cells survive, preferential breakage near centromeres can offer some selectable fitness advantage. A conservation of the pericentromeric breakage hot spot in other species would argue in favor of a physiological process. However, it can also be a default state governed by thermodynamics, as suggested above, and thus not necessarily under selection.
In multicellular organisms, dicentric breakage hot spots could similarly favor the restoration to a stable karyotype, thus limiting dicentric-induced gene copy number changes. In somatic cells, this would establish an unsuspected and potentially important barrier against the mutagenesis that leads to oncogenesis.
Materials and methods
Strains and plasmids
The strains used in this study are listed in Supplemental Table S1. To fuse the chromosome 6 right end to the chromosome 7 left end at ADH4, cells were transformed with plasmid sp1003 cut with XhoI and NotI. To fuse the chromosome 6 right end to a Y′ subtelomeric element, cells were transformed with plasmid sp1007 cut with XhoI and NotI. Integration is relatively inefficient but specific, probably due to the simultaneous double selection on galactose plates lacking leucine. The KANr-pGAL1-CEN6-pGAL1-skHIS3 allele was backcrossed four times with RM11-1a to create the strains 134-1a and 134-1d (both strains possess a wild-type BUD4 allele, as determined by PCR). Individual telomere fusions were generated with a rap1-(∆) allele, selected on galactose, and complemented with a wild-type copy of RAP1 as previously described (Pobiega and Marcand 2010). The fused telomere repeats in Lev1258 were replaced by the klTRP1 marker through a PCR-mediated transformation as described in Longtine et al. (1998). A 1038-bp fragment encompassing CEN4 (coordinates 449214–450251 of chromosome 4) was cloned into HindIII–BamHI-cut pFA6-klLEU2-MX6. The CEN4-klLEU2 cassette was inserted in the chromosome 6 right arm (coordinate 245700) through a PCR-mediated transformation. The myo1-AID allele was created with plasmid pHyg-AID*-9myc (Morawska and Ulrich 2013). The SPC42-GFP and MYO1-GFP alleles were created with plasmids pYM26 and pYM25, respectively, and the SPC42-RFP allele was created with the plasmid pFA6-mCherry-HPH-MX6. The GFP-TUB1 allele comes from strain AC021 (Woodruff et al. 2009) and was backcrossed four times in the W303 background. The lacI**-GFP allele was integrated at the URA3 locus (Dubarry et al. 2011). The lacO array was integrated in chromosome 7 (coordinate 484500) 16.5 kb from CEN7. LatA was obtained from Cayman Chemical company (reference 10010630), and IAA was obtained from Sigma-Aldrich (reference I2886).
Cell cycle synchronization
Cells were arrested in G1 with 10−7 M α factor (from a 10−3 M stock solution in ethanol) and released by two washes in rich medium. Cells were arrested in G2/M with 5 µg/mL nocodazole (from 1.5 mg/mL stock solution in DMSO). cdc15-2 cells were arrested in anaphase by gradually increasing the temperature to 36°C (1°C every 4 min) and released by shifting the temperature to 25°C. The efficiency of the arrests and releases was checked by microscopy.
PFGE and Southern blot
Yeast DNA embedded in agarose plugs was prepared as previously described (Pobiega and Marcand 2010). Pulse field electrophoresis was carried out in a 0.9% agarose gel in 0.5× TBE at 14°C with a CHEF DR III from Bio-Rad with a switch time ramping from 60 to 120 sec in 24 h (to resolve the whole karyoptype) or with a switch time ramping from 5 to 30 sec in 22 h or 3 to 20 sec in 20 h (to resolve fragments from 20 to 400 kb). For restriction, DNA plugs were equilibrated twice in 10 mM Tris and 1 mM EDTA (pH 8.0) and twice in restriction buffer (New England Biolabs) prior to addition of the restriction enzyme. The probes for Southern blots were gel-purified PCR products amplified from yeast genomic DNA.
TEM
Cells for TEM analysis were grown in SC + 2% galactose at 30°C to early exponential phase, collected, washed by filtration, and then resuspended in SC + 2% dextrose. At 20 and/or 60 min, portions of cells were fixed and processed as described in Pollard et al. (2012) except that the cells were embedded in 2% low-melting-temperature agarose (SeaPlaque, Lonza) immediately before post-fixation with potassium permanganate.
Time-laspe microscopy
Live-cell images were acquired using a wide-field microscope based on an inverted microscope (Leica DMI-6000B) equipped with adaptive focus control to eliminate Z drift, a 100×/1.4 NA immersion objective with a Prior NanoScanZ Nanopositioning Piezo Z stage system, a CMOS camera (ORCA-Flash4.0, Hamamatsu), and a solid-state light source (SpectraX, Lumencore). The system is piloted by the MetaMorph software (Molecular Device).
Cells were grown exponentially in synthetic medium, and 50 µL of the culture was loaded in microfluidic plates (Y04C plates, ONIX platform, CellASIC) equilibrated at 25°C. GFP and mCherry images were acquired sequentially for each Z section with an exposure time of 20 msec using solid-state 475- and 575-nm diodes and appropriate filters (GFP-mCherry filter, excitation: double BP, 450–490/550–590 nm, and dichroic double BP, 500–550/600–665 nm, Chroma Technology Corp.). For four-dimensional movies, 25 focal steps of 0.25 µm were taken every 2 min for 2 h.
Three-dimensional data sets were deconvolved using the blind deconvolution algorithm of AutoQuant (Media Cybernetics, Inc.) with the point spread function appropriate for our microscope at each emission wavelength. Further processing was done using ImageJ software (National Institutes of Health). All of the images shown are a z projection of z-stack images.
Supplementary Material
Acknowledgments
We thank Julie Cooper for a very welcoming and stimulating sabbatical stay during which this study was initiated, Frank Uhlmann and Thomas Kuilman for the TTye-GFP and cdc15-2 alleles and for suggestions at an early stage of this work, Georjana Barnes for the GFP-TUB1 allele, Angela Taddei for the lacI**-GFP and lacO array plasmids, Dan Gottschling for the RM11-1a strain, Helle Ulrich for the AID toolkit, Armelle Lengronne for advice on AIDs, Rachel Lescasse for unpublished data, Jon Mulholland and John Perrino (Stanford University Cell Sciences Imaging Facility: National Institutes of Health Shared Instrumentation Grant 1S10RR02678001) for assistance with TEM, Mathias Toulouze for help with live imaging, and Greg Ira, Ken Kaplan, Romain Koszul, Jean-Charles Walter, and Tom Petes for fruitful discussions. This work was supported by grants from Association pour la Recherche sur le Cancer (ARC) and Agence Nationale de la Recherche (ANR) (Blanc-SVSE-8-2011-TELO&DICENs). N.B. was supported by a fellowship from the CEA-IRTELIS Ph.D. program and a short-term fellowship from ARC.
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.254664.114.
References
- Akiyoshi B, Sarangapani KK, Powers AF, Nelson CR, Reichow SL, Arellano-Santoyo H, Gonen T, Ranish JA, Asbury CL, Biggins S. 2010. Tension directly stabilizes reconstituted kinetochore-microtubule attachments. Nature 468: 576–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avarello R, Pedicini A, Caiulo A, Zuffardi O, Fraccaro M. 1992. Evidence for an ancestral alphoid domain on the long arm of human chromosome 2. Hum Genet 89: 247–249. [DOI] [PubMed] [Google Scholar]
- Bajer A. 1964. Cine-micrographic studies on dicentric chromosomes. Chromosoma 15: 630–651. [DOI] [PubMed] [Google Scholar]
- Baxter J, Diffley JF. 2008. Topoisomerase II inactivation prevents the completion of DNA replication in budding yeast. Mol Cell 30: 790–802. [DOI] [PubMed] [Google Scholar]
- Bi E, Maddox P, Lew DJ, Salmon ED, McMillan JN, Yeh E, Pringle JR. 1998. Involvement of an actomyosin contractile ring in Saccharomyces cerevisiae cytokinesis. J Cell Biol 142: 1301–1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biebricher A, Hirano S, Enzlin JH, Wiechens N, Streicher WW, Huttner D, Wang LHC, Nigg EA, Owen-Hughes T, Liu Y, et al. . 2013. PICH: a DNA translocase specially adapted for processing anaphase bridge DNA. Mol Cell 51: 691–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Niu H, Chung W-H, Zhu Z, Papusha A, Shim EY, Lee SE, Sung P, Ira G. 2011. Cell cycle regulation of DNA double-strand break end resection by Cdk1-dependent Dna2 phosphorylation. Nat Struct Mol Biol 18: 1015–1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciullo M, Debily M-A, Rozier L, Autiero M, Billault A, Mayau V, Marhomy El S, Guardiola J, Bernheim A, Coullin P, et al. . 2002. Initiation of the breakage-fusion-bridge mechanism through common fragile site activation in human breast cancer cells: the model of PIP gene duplication from a break at FRA7I. Hum Mol Genet 11: 2887–2894. [DOI] [PubMed] [Google Scholar]
- Croll D, Zala M, McDonald BA. 2013. Breakage-fusion-bridge cycles and large insertions contribute to the rapid evolution of accessory chromosomes in a fungal pathogen. PLoS Genet 9: e1003567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuylen S, Metz J, Hruby A, Haering CH. 2013. Entrapment of chromosomes by condensin rings prevents their breakage during cytokinesis. Dev Cell 27: 469–478. [DOI] [PubMed] [Google Scholar]
- Denchi EL, Celli G, de Lange T. 2006. Hepatocytes with extensive telomere deprotection and fusion remain viable and regenerate liver mass through endoreduplication. Genes Dev 20: 2648–2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimitrov LN, Brem RB, Kruglyak L, Gottschling DE. 2009. Polymorphisms in multiple genes contribute to the spontaneous mitochondrial genome instability of Saccharomyces cerevisiae S288C strains. Genetics 183: 365–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubarry M, Loïodice I, Chen CL, Thermes C, Taddei A. 2011. Tight protein–DNA interactions favor gene silencing. Genes Dev 25: 1365–1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elia N, Ott C, Lippincott-Schwartz J. 2013. Incisive imaging and computation for cellular mysteries: lessons from abscission. Cell 155: 1220–1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher JK, Ballenger M, O’Brien ET, Haase J, Superfine R, Bloom K. 2009. DNA relaxation dynamics as a probe for the intracellular environment. Proc Natl Acad Sci 106: 9250–9255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gascoigne KE, Cheeseman IM. 2013. Induced dicentric chromosome formation promotes genomic rearrangements and tumorigenesis. Chromosome Res 21: 407–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill A, Bloom K. 1987. Genetic manipulation of centromere function. Mol Cell Biol 7: 2397–2405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano T, Funahashi S, Uemura T, Yanagida M. 1986. Isolation and characterization of Schizosaccharomyces pombe cutmutants that block nuclear division but not cytokinesis. EMBO J 5: 2973–2979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii K, Ogiyama Y, Chikashige Y, Soejima S, Masuda F, Kakuma T, Hiraoka Y, Takahashi K. 2008. Heterochromatin integrity affects chromosome reorganization after centromere dysfunction. Science 321: 1088–1091. [DOI] [PubMed] [Google Scholar]
- Jones RE, Oh S, Grimstead JW, Zimbric J, Roger L, Heppel NH, Ashelford KE, Liddiard K, Hendrickson EA, Baird DM. 2014. Escape from telomere-driven crisis is DNA ligase III dependent. Cell Reports 8: 1063–1076. [DOI] [PubMed] [Google Scholar]
- Kitamura E, Tanaka K, Kitamura Y, Tanaka TU. 2007. Kinetochore microtubule interaction during S phase in Saccharomyces cerevisiae. Genes Dev 21: 3319–3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko N, Nishihama R, Tully GH, Ostapenko D, Solomon MJ, Morgan DO, Pringle JR. 2007. Identification of yeast IQGAP (Iqg1p) as an anaphase-promoting-complex substrate and its role in actomyosin-ring-independent cytokinesis. Mol Biol Cell 18: 5139–5153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer KM, Brock JA, Bloom K, Moore JK, Haber JE. 1994. Two different types of double-strand breaks in Saccharomyces cerevisiae are repaired by similar RAD52-independent, nonhomologous recombination events. Mol Cell Biol 14: 1293–1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruithof M, Chien F-T, Routh A, Logie C, Rhodes D, van Noort J. 2009. Single-molecule force spectroscopy reveals a highly compliant helical folding for the 30-nm chromatin fiber. Nat Struct Mol Biol 16: 534–540. [DOI] [PubMed] [Google Scholar]
- Lee SE, Pâques F, Sylvan J, Haber JE. 1999. Role of yeast SIR genes and mating type in directing DNA double-strand breaks to homologous and non-homologous repair paths. Curr Biol 9: 767–770. [DOI] [PubMed] [Google Scholar]
- Lei X, Hu J, Fang H. 2010. Radial compression property of ds-DNA molecules studied by a mesoscale model. Europhys Lett 89: 48003. [Google Scholar]
- Lejeune J, Dutrillaux B, Rethore MO, Prieur M. 1973. [Comparison of the structure of chromatids of Homo sapiens and Pan troglodytes (author’s transl)]. Chromosoma 43: 423–444. [DOI] [PubMed] [Google Scholar]
- Li Y, Schwab C, Ryan SL, Papaemmanuil E, Robinson HM, Jacobs P, Moorman AV, Dyer S, Borrow J, Griffiths M, et al. . 2014. Constitutional and somatic rearrangement of chromosome 21 in acute lymphoblastic leukaemia. Nature 508: 98–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobachev KS, Gordenin DA, Resnick MA. 2002. The Mre11 complex is required for repair of hairpin-capped double-strand breaks and prevention of chromosome rearrangements. Cell 108: 183–193. [DOI] [PubMed] [Google Scholar]
- Longtine MS, McKenzie A 3rd, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14: 953–961. [DOI] [PubMed] [Google Scholar]
- Mackinnon RN, Campbell LJ. 2011. The role of dicentric chromosome formation and secondary centromere deletion in the evolution of myeloid malignancy. Genet Res Int 2011: 643628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann C, Davis RW. 1983. Instability of dicentric plasmids in yeast. Proc Natl Acad Sci 80: 228–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcand S. 2014. How do telomeres and NHEJ coexist? Mol Cell Oncol. doi: 10.4161/23723548.963438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marco E, Dorn JF, Hsu P-H, Jaqaman K, Sorger PK, Danuser G. 2013. S. cerevisiae chromosomes biorient via gradual resolution of syntely between S phase and anaphase. Cell 154: 1127–1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marko JF. 2008. Micromechanical studies of mitotic chromosomes. Chromosome Res 16: 469–497. [DOI] [PubMed] [Google Scholar]
- Marotta M, Chen X, Watanabe T, Faber PW, Diede SJ, Tapscott S, Tubbs R, Kondratova A, Stephens R, Tanaka H. 2013. Homology-mediated end-capping as a primary step of sister chromatid fusion in the breakage–fusion–bridge cycles. Nucleic Acids Res 41: 9732–9740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-A C, van Wely KHM. 2011. Centromere fission, not telomere erosion, triggers chromosomal instability in human carcinomas. Carcinogenesis 32: 796–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClintock B. 1941. The stability of broken ends of chromosomes in Zea Mays. Genetics 26: 234–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGrew JT, Goetsch L, Byers B, Baum P. 1992. Requirement for ESP1 in the nuclear division of Saccharomyces cerevisiae. Mol Biol Cell 3: 1443–1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMurray MA, Stefan X, Wemmer M, Odorizzi G, Emr SD, Thomer J. 2011. Genetic interactions with mutations affecting septin assembly reveal ESeRT functions in budding yeast cytokinesis. Biol Chem 392: 699–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuno K, Lambert S, Baldacci G, Murray JM, Carr AM. 2009. Nearby inverted repeats fuse to generate acentric and dicentric palindromic chromosomes by a replication template exchange mechanism. Genes Dev 23: 2876–2886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuno K, Miyabe I, Schalbetter SA, Carr AM, Murray JM. 2013. Recombination-restarted replication makes inverted chromosome fusions at inverted repeats. Nature 493: 246–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morawska M, Ulrich HD. 2013. An expanded tool kit for the auxin-inducible degron system in budding yeast. Yeast 30: 341–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murnane JP, Sabatier L. 2004. Chromosome rearrangements resulting from telomere dysfunction and their role in cancer. BioEssays 26: 1164–1174. [DOI] [PubMed] [Google Scholar]
- Norden C, Mendoza M, Dobbelaere J, Kotwaliwale CV, Biggins S, Barral Y. 2006. The nocut pathway links completion of cytokinesis to spindle midzone function to prevent chromosome breakage. Cell 125: 85–98. [DOI] [PubMed] [Google Scholar]
- Onishi M, Ko N, Nishihama R, Pringle JR. 2013. Distinct roles of Rho1, Cdc42, and Cyk3 in septum formation and abscission during yeast cytokinesis. J Cell Biol 202: 311–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pampalona J, Frías C, Genescà A, Tusell L. 2012. Progressive telomere dysfunction causes cytokinesis failure and leads to the accumulation of polyploid cells. PLoS Genet 8: e1002679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panja D, Barkema GT, Kolomeisky AB. 2013. Through the eye of the needle: recent advances in understanding biopolymer translocation. J Phys Condens Matter 25: 413101. [DOI] [PubMed] [Google Scholar]
- Pennaneach V, Kolodner RD. 2009. Stabilization of dicentric translocations through secondary rearrangements mediated by multiple mechanisms in S. cerevisiae. PLoS ONE 4: e6389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto IM, Rubinstein B, Kucharavy A, Unruh JR, Li R. 2012. Actin depolymerization drives actomyosin ring contraction during budding yeast cytokinesis. Dev Cell 22: 1247–1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pobiega S, Marcand S. 2010. Dicentric breakage at telomere fusions. Genes Dev 24: 720–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard LW, Onishi M, Pringle JR, Lord M. 2012. Fission yeast Cyk3p is a transglutaminase-like protein that participates in cytokinesis and cell morphogenesis. Mol Biol Cell 23: 2433–2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renshaw MJ, Ward JJ, Kanemaki M, Natsume K, Nédélec FJ, Tanaka TU. 2010. Condensins promote chromosome recoiling during early anaphase to complete sister chromatid separation. Dev Cell 19: 232–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabatier L, Ricoul M, Pottier G, Murnane JP. 2005. The loss of a single telomere can result in instability of multiple chromosomes in a human tumor cell line. Mol Cancer Res 3: 139–150. [DOI] [PubMed] [Google Scholar]
- Sato H, Masuda F, Takayama Y, Takahashi K, Saitoh S. 2012. Epigenetic inactivation and subsequent heterochromatinization of a centromere stabilize dicentric chromosomes. Curr Biol 22: 658–667. [DOI] [PubMed] [Google Scholar]
- Selvarajah S, Yoshimoto M, Park PC, Maire G, Paderova J, Bayani J, Lim G, Al-Romaih K, Squire JA, Zielenska M. 2006. The breakage-fusion-bridge (BFB) cycle as a mechanism for generating genetic heterogeneity in osteosarcoma. Chromosoma 115: 459–467. [DOI] [PubMed] [Google Scholar]
- Shimizu N, Shingaki K, Kaneko-Sasaguri Y, Hashizume T, Kanda T. 2005. When, where and how the bridge breaks: anaphase bridge breakage plays a crucial role in gene amplification and HSR generation. Exp Cell Res 302: 233–243. [DOI] [PubMed] [Google Scholar]
- Song W, Gawel M, Dominska M, Greenwell PW, Hazkani-Covo E, Bloom K, Petes TD. 2013. Nonrandom distribution of interhomolog recombination events induced by breakage of a dicentric chromosome in Saccharomyces cerevisiae. Genetics 194: 69–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorzano COS, Pascual-Montano A, Sánchez de Diego A, Martínez-A C, van Wely KHM. 2013. Chromothripsis: breakage–fusion–bridge over and over again. Cell Cycle 12: 2016–2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewénius Y, Gorunova L, Jonson T, Larsson N, Höglund M, Mandahl N, Mertens F, Mitelman F, Gisselsson D. 2005. Structural and numerical chromosome changes in colon cancer develop through telomere-mediated anaphase bridges, not through mitotic multipolarity. Proc Natl Acad Sci 102: 5541–5546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stimpson KM, Sullivan BA. 2010. Epigenomics of centromere assembly and function. Curr Opin Cell Biol 22: 772–780. [DOI] [PubMed] [Google Scholar]
- Stimpson KM, Song IY, Jauch A, Holtgreve-Grez H, Hayden KE, Bridger JM, Sullivan BA. 2010. Telomere disruption results in non-random formation of de novo dicentric chromosomes involving acrocentric human chromosomes. PLoS Genet 6: e1001061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stimpson KM, Matheny JE, Sullivan BA. 2012. Dicentric chromosomes: unique models to study centromere function and inactivation. Chromosome Res 20: 595–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suto Y, Hirai M, Akiyama M, Kobashi G, Itokawa M, Akashi M, Sugiura N. 2013. Biodosimetry of restoration workers for the Tokyo Electric Power Company (TEPCO) Fukushima Daiichi Nuclear Power Station aAccident. Health Phys 105: 366–373. [DOI] [PubMed] [Google Scholar]
- Symington LS. 2014. End resection at double-strand breaks: mechanism and regulation. Cold Spring Harb Perspect Biol 6: a016436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka K, Mukae N, Dewar H, van Breugel M, James EK, Prescott AR, Antony C, Tanaka TU. 2005. Molecular mechanisms of kinetochore capture by spindle microtubules. Nature 434: 987–994. [DOI] [PubMed] [Google Scholar]
- Thrower DA, Bloom K. 2001. Dicentric chromosome stretching during anaphase reveals roles of Sir2/Ku in chromatin compaction in budding yeast. Mol Biol Cell 12: 2800–2812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolliday N, Pitcher M, Li R. 2003. Direct evidence for a critical role of myosin II in budding yeast cytokinesis and the evolvability of new cytokinetic mechanisms in the absence of myosin II. Mol Biol Cell 14: 798–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Steensel B, Smogorzewska A, de Lange T. 1998. TRF2 protects human telomeres from end-to-end fusions. Cell 92: 401–413. [DOI] [PubMed] [Google Scholar]
- Weiss EL. 2012. Mitotic exit and separation of mother and daughter cells. Genetics 192: 1165–1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westmoreland J, Ma W, Yan Y, Van Hulle K, Malkova A, Resnick MA. 2009. RAD50 is required for efficient initiation of resection and recombinational repair at random, γ-induced double-strand break ends. PLoS Genet 5: e1000656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wloka C, Bi E. 2012. Mechanisms of cytokinesis in budding yeast. Cytoskeleton 69: 710–726. [DOI] [PubMed] [Google Scholar]
- Woodruff JB, Drubin DG, Barnes G. 2009. Dynein-driven mitotic spindle positioning restricted to anaphase by She1p inhibition of dynactin recruitment. Mol Biol Cell 20: 3003–3011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodruff JB, Drubin DG, Barnes G. 2010. Mitotic spindle disassembly occurs via distinct subprocesses driven by the anaphase-promoting complex, Aurora B kinase, and kinesin-8. J Cell Biol 191: 795–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zierhut C, Diffley JFX. 2008. Break dosage, cell cycle stage and DNA replication influence DNA double strand break response. EMBO J 27: 1875–1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.