Abstract
Objectives
Aquaporin-1 (AQP1) is a candidate oncogene that is epigenetically modified in adenoid cystic carcinoma (ACC). We sought to (1) assess AQP1 promoter methylation and expression in an ACC cohort, (2) identify correlations between AQP1 and clinical outcomes, and (3) explore the role of AQP1 in tumor progression in vitro.
Study design
Laboratory study, retrospective chart review.
Setting
Academic medical center.
Methods
DNA and RNA were isolated from ACC tumors and control salivary gland tissues. Quantitative methylation-specific polymerase chain reaction (PCR) was performed on bisulfite-treated DNA. Quantitative reverse transcription PCR was performed after cDNA synthesis. Cell lines stably overexpressing an AQP1 plasmid or empty vector were generated. Cell scratch and Matrigel invasion assays were performed. Retrospective chart review was performed for collection of clinical information.
Results
Methylation results from 77 tumors and 30 controls demonstrated that AQP1 was hypomethylated in tumors (P < .0001). Fifty-eight tumors (75.3%) displayed AQP1 hypomethylation compared with controls. AQP1 expression levels assessed in 58 tumors and 23 controls demonstrated a trend toward increased expression in tumors (P = .08). Univariate analysis revealed that AQP1 hypermethylation was associated with increased overall survival. No associations between AQP1 expression level and survival were found. AQP1 overexpression did not affect cell migratory or invasive capacities in vitro.
Conclusion
AQP1 promoter hypomethylation is common in ACC, and AQP1 tends to be overexpressed in these tumors. Increased AQP1 methylation is associated with improved prognosis on univariate analysis, but expression is not associated with outcomes. Further in vitro studies are necessary to clarify the role of AQP1 in ACC.
Keywords: adenoid cystic carcinoma, epigenetics, promoter methylation
Adenoid cystic carcinoma (ACC) of the salivary glands is a rare malignancy, accounting for 10% of all cancers of the head and neck. Clinically, ACC is characterized by slow growth but a propensity for frequent perineural invasion and late distant metastases. The mainstays of treatment remain surgical resection and postoperative radiation therapy, but over 40% of patients may develop metastatic disease, sometimes decades later.1 Few chemotherapeutic options exist for patients with recurrent or metastatic disease, given the relative paucity of knowledge of the underlying molecular mechanisms of ACC tumorigenesis and growth.
Epigenetic regulation of gene expression through DNA methylation is widely accepted as playing a role in the car-cinogenesis of many human cancers.2 Several epigenetic modifications leading to changes in gene expression have been identified in ACC.3–7 Our group previously conducted a genome-wide screen for oncogene candidates that are epigenetically modified in ACC, and aquaporin-1 (AQP1) emerged as the most promising candidate. In our previous study, AQP1 promoter hypomethylation was shown in 2 small independent ACC cohorts, and AQP1 overexpression at both the RNA and protein levels was demonstrated in a small cohort of tumor samples versus controls. We also verified that AQP1 expression was upregulated by CpG island demethylation in an ACC cell line.8
AQP1 is a small transmembrane protein that selectively transports water across cell membranes. It is highly expressed in several tumor types, including lung and colorectal cancers, and has been implicated in tumor cell proliferation, extravasation, migration, and metastasis.9–11 AQP1 is therefore an attractive candidate gene in ACC, given the tumor’s propensity to metastasize.
In the current study, we investigated the role of AQP1 in salivary gland ACC. We found that AQP1 is significantly hypomethylated in ACC tumors compared with normal salivary gland tissues. A trend toward upregulation of AQP1 expression at the mRNA level was also demonstrated. On univariate analysis of retrospective clinical data, promoter hypermethylation of AQP1 was associated with improved overall survival; however, this was not significant on multivariate analysis. No association was found between AQP1 expression levels and survival outcomes. AQP1 overexpression in vitro did not affect cell migratory or invasive capacities in scratch and invasion assays.
Materials and Methods
Clinical Samples
Formalin-fixed, paraffin-embedded (FFPE) samples were obtained from 77 patients treated surgically by the Department of Otolaryngology–Head and Neck Surgery of the Johns Hopkins Medical Institutions between the years 1988 and 2011. Tissue blocks with high tumor yield were selected after all blocks had been reviewed by an experienced head and neck pathologist (J.A.B.) to confirm the diagnosis of ACC. Ten sections of 15 μm thickness were cut from each tumor and manually micro-dissected to yield at least 80% tumor purity. In addition, 30 FFPE parotid or submandibular glands that were resected either for benign disease or as part of other surgical procedures were included in this study after histologic confirmation that the tissues to be used were distant from any benign or inflammatory lesion. All tissues were obtained with the approval of the institutional review board (JHH IRB # NA_00001336).
DNA and RNA Extraction from FFPE Tissues
Genomic DNA was extracted from FFPE tissues as previously described.12 Briefly, FFPE tissues first underwent deparaffinization in xylenes. Samples were then digested in 1% sodium dodecyl sulfate (SDS) and 0.02% proteinase K (Roche, Indianapolis, Indiana) at 48°C for 72 hours. DNA was purified by phenol-chloroform extraction and ethanol precipitation. DNA was subsequently resuspended in LoTE (10 mmol/L Tris-HCl and 2.5 mmol/L EDTA), and DNA concentration was quantified using the NanoDrop ND-1000 spectrophotometer (Thermo Scientific, Waltham, Massachusetts). DNA was stored at −20°C until further use.
RNA was extracted from FFPE tissues after undergoing deparaffinization in xylenes. RNA extraction was performed using the RecoverAll Total Nucleic Acid Isolation Kit for FFPE (Ambion, Austin, Texas) according to the manufacturer’s instructions. RNA was stored at −80°C until further use.
One microgram of RNA extracted from FFPE tissues was used for cDNA synthesis using both random hexamers and oligo(dT) primers with the SuperScript First-Strand Synthesis System for reverse transcription polymerase chain reaction (RT-PCR) (Invitrogen, Carlsbad, California) according to the manufacturer’s instructions.
Quantitative Methylation-Specific PCR
Quantitative methylation-specific PCR (qMSP) assays were performed in 384-well reaction plates in a Perkin-Elmer Applied Biosystems 7900HT Fast Real Time PCR System instrument and analyzed using SDS 2.3 (Applied Biosystems, Foster City, California), as previously described.8 The AQP1 primer sequences were forward 5′-GGAGGGTAGTGGTGGTCGA-3′ and reverse 5′-CCTTCACGTTATCCTAAACCG-3′. The AQP1 probe sequence was 6FAM-5′-AAAACCCAAAACAAAACCGATACTAAT-3′-TAMRA.13 Beta-actin was also amplified as a representative housekeeping gene to which AQP1 values were normalized. Beta-actin primers were forward 5′-TGGTGATGGAGGAGGTTTAGTAAGT-3′ and reverse 5′-AACCAATAAAACCTACTCCTCCCTTAA-3′. The beta-actin probe was 6FAM-5′-ACCACCACCCAACACACAATAACAAACACA-3′-TAMRA.14 Each reaction was carried out in a reaction volume of 10 μL, with components as previously described.4 Calibration curves were constructed for each plate with serial dilutions of bisulfite-converted universal methylated human DNA (Zymo Research, Irvine, California). Each plate included samples, water blanks, and calibration curves in triplicate. Reaction conditions were as follows: 95°C for 3 minutes followed by 50 cycles at 95°C for 15 seconds and 60°C for 1 minute. Data were obtained following 50 amplification cycles. All samples were within the range of sensitivity and reproducibility of the assay based on the amplification of the internal reference standard (threshold cycle value for beta-actin of 40). The relative level of methylated DNA in each sample was reported as a ratio between the mean values of AQP1 and of beta-actin ([AQP1/beta-actin] × 100).
Cell Line
The ACC cell line SACC83 was a kind gift from Dr Christopher Moskaluk (University of Virginia). The cells were cultured in RPMI 1640 with 1% penicillin/streptomycin and 10% fetal bovine serum (FBS). The cells were maintained at 37°C with 5% CO2 and propagated using standard cell culture techniques.
RNA Extraction from Cell Lines
Total cellular RNA was isolated from cultured cells by lysing cells in QIAzol Lysis Reagent (Qiagen, Valencia, California). Following phase separation with chloroform, RNA was precipitated with isopropanol and washed with ethanol. RNA was subsequently resuspended in diethylpyrocarbonate (DEPC)-treated water, and concentration was quantified using the NanoDrop ND-1000 spectrophotometer (Thermo Scientific). RNA was stored at −80°C until further use.
Synthesis of cDNA was performed using qScript cDNA SuperMix (Quanta BioSciences, Gaithersburg, Maryland) according to the manufacturer’s instructions.
Quantitative RT-PCR
Quantitative RT-PCR (qRT-PCR) assays were performed in 384-well reaction plates in a PE Applied Biosystems 7900HT instrument and analyzed using SDS 2.3. AQP1 primers were forward 5′-TCGCCACCGCCATCCTCTCA-3′ and reverse 5′-CCGATGATCTCGATGCCCAGG-3′. Beta-actin was used for normalization. Beta-actin primers were forward 5′-AGTCCTGTGGCATCCACGAAACTA-3′ and reverse 5′-ACTGTGTTGGCGTACAGGTCTTTG-3′.14 Reactions were carried out with SYBR Green PCR Master Mix (Applied Biosystems). Each plate included samples in triplicate and water blanks. Reaction conditions were as follows: 95°C for 5 minutes followed by 50 cycles of 95°C for 15 seconds and 64°C for 30 seconds. Dissociation curves were obtained, and data were analyzed following 50 amplification cycles. The mean threshold cycle numbers of the triplicate samples were calculated for both AQP1 and beta-actin. Relative gene expression was then calculated using the 2−ΔΔCT method.15
Data Analysis
Retrospective chart review was performed for all patients included in this study under a protocol approved by the institutional review board (JHH IRB # NA_00018189). Comparisons between outcomes were performed using chi-square tests for categorical data and t test or nonparametric alternative Wilcoxon rank sum test for continuous data. Multivariate analyses of recurrence-free survival, metastasis-free survival, and overall survival were performed using the Cox proportional hazards model. Multivariate analyses of metastasis outcome were performed using logistic regression. Potential predictors were first investigated by univariate Cox/logistic regression modeling. Variables significant at P < .20 and biomarker variables were then entered into a stepwise Cox/logistic regression model to identify the significance of biomarkers for local recurrence, metastasis, and death.
The level of significance was set at .05 in all analyses. All statistical analyses were performed using SAS version 9.3 (SAS Institute Inc, Cary, North Carolina).
Creation of Stable AQP1-Expressing Clones
An AQP1-expressing plasmid from our laboratory16 and corresponding empty pcDNA3 mammalian expression vectors were transfected into SACC83 using LipoD293 (SignaGen Laboratories, Rockville, Maryland). The pcDNA3 expression vector contains a neomycin resistance gene for selection with G418, and we had previously found that SACC83 was sensitive to G418 (data not shown). Selection of transfected cells and creation of stable polyclonal cell lines was performed using G418 at 500 μg/mL concentration.
Cell Scratch Assay
Stably transfected cells were plated in 6-well plates at ~70% confluency and incubated under standard conditions until cells had adhered completely and 100% confluency had been reached. A linear scarification was made in each well using a p200 pipet tip across the cell monolayer, and debris was removed by gently rinsing with phosphate-buffered saline. Fresh growth medium was then replaced, and images at 3 reference points per well were obtained using a phase-contrast microscope. The cells were then incubated under standard conditions for 12 to 18 hours, and images were taken at the same reference points. The width of the scratch was measured using ImageJ software (http://rsb.info.nih.gov/ij/). All experiments were carried out in triplicate for all cell lines and controls.
Matrigel Invasion Assay
BD Matrigel (BD Biosciences, Franklin Lakes, New Jersey) was diluted using serum-free RPMI, poured into the upper chambers of a 24-well transwell plate, and incubated at 37°C until solidified. Stably transfected cells were then harvested, resuspended in serum-free medium, and added to the chambers. RPMI 1640 containing FBS was added to the lower chambers of the plate. The cells were incubated at 37°C for 24 to 48 hours under standard conditions. The membranes were then fixed with formalin, stained with UV staining dye, washed, and dried overnight. After the membranes had been removed from the inserts, they were mounted on slides and coverslipped. Images were taken using a light microscope, and the number of invaded cells per high power field was counted for 10 total fields per insert. All experiments were carried out in duplicate for all cell lines and controls.
Results
AQP1 Is Hypomethylated in Primary ACC Tumors Compared with Controls
We determined the methylation status of AQP1 in a cohort of 77 primary ACC tumors and 30 normal salivary gland tissues using qMSP. The demographic and pathologic information of this cohort is summarized in Table 1.
Table 1.
Clinical and Pathologic Characteristics of the Study Cohort.a
| Adenoid Cystic Carcinoma (n = 77) | Controls (n = 30) | |
|---|---|---|
| Age in years, mean (range) | 54.8 (18–86) | 57.6 (32–86) |
| Sex | ||
| Male | 30 (38.9) | 19 (63.3) |
| Female | 47 (61.0) | 11 (36.7) |
| Race | ||
| Caucasian | 57 (74.0) | 25 (83.3) |
| African-American | 11 (14.3) | 3 (10.0) |
| Other | 9 (11.7) | 2 (6.7) |
| Smoking status | ||
| Ever-smoker | 36 (46.8) | 16 (53.3) |
| Never smoker | 30 (39.0) | 13 (43.3) |
| Unknown | 11 (14.3) | 1 (3.3) |
| Primary site | ||
| Major salivary gland | 34 (44.2) | |
| Minor salivary gland | 43 (55.8) | |
| Tumor stage | ||
| T1-T2 | 27 (35.1) | |
| T3-T4 | 38 (49.4) | |
| Unknown | 12 (15.6) | |
| Nodal status | ||
| N0 | 54 (70.1) | |
| N11 | 11 (14.3) | |
| Unknown | 12 (15.6) | |
| Dominant pattern | ||
| Tubular | 12 (15.6) | |
| Cribriform | 55 (71.4) | |
| Solid | 10 (13.0) | |
| Perineural invasion | ||
| Negative | 7 (9.1) | |
| Positive | 38 (49.4) | |
| Unknown | 32 (41.6) | |
| Local recurrence | ||
| Negative | 58 (75.3) | |
| Positive | 19 (24.7) | |
| Regional recurrence | ||
| Negative | 73 (94.8) | |
| Positive | 4 (5.2) | |
| Distant metastasis | ||
| Negative | 58 (75.3) | |
| Positive | 19 (24.7) | |
| Length of follow-up in months, mean (range) | 69.8 (0–299.7) | |
Values are n (%) unless otherwise indicated.
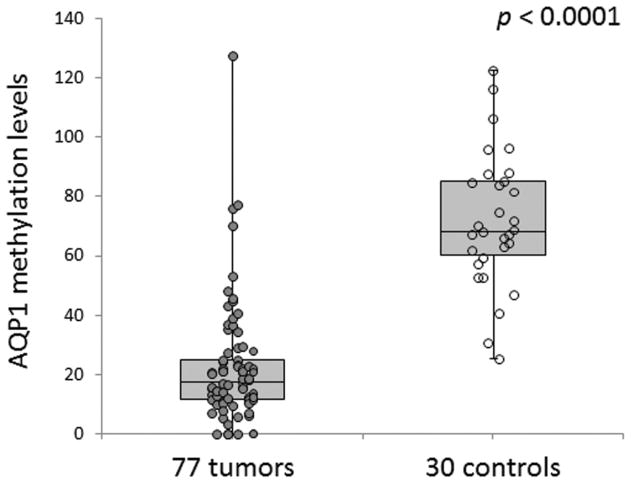
Our qMSP data demonstrated that AQP1 is significantly hypomethylated in ACC tumors compared with control tissues (P < .0001) (Figure 1). AQP1 promoter methylation levels in ACC samples had an average value of 24.0 (range, 0.0–138.7; median 18.4), while normal samples had an average value of 71.9 (range, 25.3–122.6; median 68.3). A total of 58 tumors (75.3%) displayed AQP1 promoter hypomethylation compared with controls.
Figure 1.
Aquaporin-1 is hypomethylated in adenoid cystic carcinoma tumors.
AQP1 Expression Is Increased in Primary ACC Tumors Compared with Controls
We next assessed AQP1 expression at the mRNA level in a subset of 58 tumors and 23 controls using qRT-PCR. There was a trend toward increased expression of AQP1 in tumors compared with controls, although this did not reach statistical significance (P = .08) (Figure 2). The average relative mRNA expression level was 0.06 (range, 0.0–0.8; median 0.0) in ACC samples and 0.03 (range, 0.0–0.1; median 0.0) in control samples.
Figure 2.
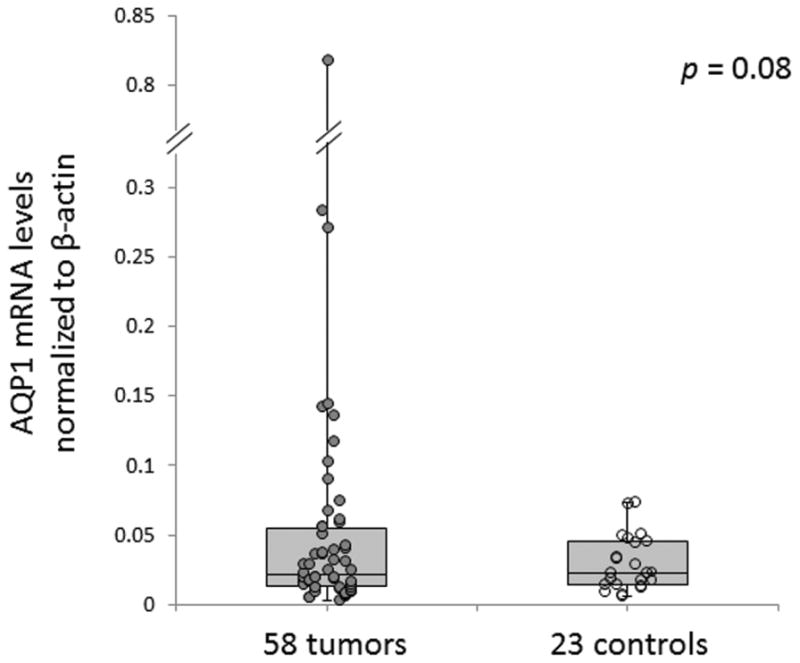
Aquaporin-1 (AQP1) mRNA expression levels tend to be higher in adenoid cystic carcinoma tumors than in controls.
Clinical Correlation of AQP1 Methylation Status
We next sought to evaluate possible associations between AQP1 promoter methylation levels and clinical and pathologic variables. We completed a retrospective chart review of all patients included in our cohort. The average length of follow-up of the cohort was 69.8 months, with a range of 0 to 299.7 months. AQP1 promoter methylation levels were not correlated with gender, age, ethnicity, primary site, smoking history, tumor (T) stage, nodal (N) stage, presence or absence of metastasis (M stage) at diagnosis, dominant histologic pattern, presence or absence of perineural invasion, or treatment with postoperative radiation. On univariate analysis, AQP1 promoter hypermethylation was associated with improved overall survival (P = .036) but not with recurrence- or metastasis-free survival. No associations were found between AQP1 mRNA expression level and clinical outcomes. Furthermore, on multivariate analyses incorporating patient age and tumor stage at diagnosis, AQP1 promoter hypermethylation was no longer significantly associated with survival. Tumor stage was the strongest predictor of overall survival, with hazard ratios of 15.1 (95% confidence interval [CI], 1.86–122.13) and 13.1 (95% CI, 1.68–101.60) for T3 and T4 relative to T1, respectively. The hazard ratio for age was 1.04 (95% CI, 1.01–1.07).
AQP1 Does Not Play a Significant Role in Cell Migratory or Invasive Capability In Vitro
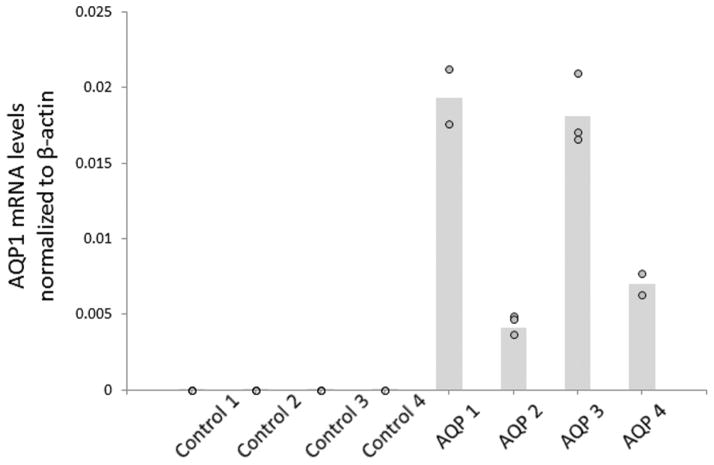
As both metastasis and perineural invasion are characteristic features of salivary gland ACC, we studied the role of AQP1 in cell migration and invasion by the in vitro cell scratch and Matrigel invasion assays. We first generated ACC cell line clones stably overexpressing AQP1. Figure 3 demonstrates the overexpression of AQP1, as confirmed by qRT-PCR, in 4 distinct clones compared with 4 empty vector controls.
Figure 3.
Aquaporin-1 (AQP1) is overexpressed in stable SACC83 clones (AQP 1-4) compared with empty vector controls.
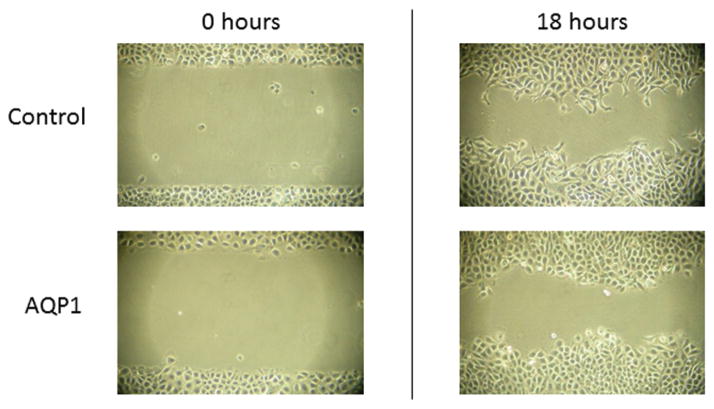
However, we found that introduction of AQP1 into SACC83 did not significantly affect cell migratory ability compared with control cells as assessed by the in vitro scratch test (representative images shown in Figure 4). AQP1 overexpression in ACC83 cells also did not significantly affect invasive capacity in Matrigel cell invasion experiments (representative experiments shown in Figure 5).
Figure 4.
Overexpression of aquaporin-1 (AQP1) does not affect cell migration in SACC83.
Figure 5.
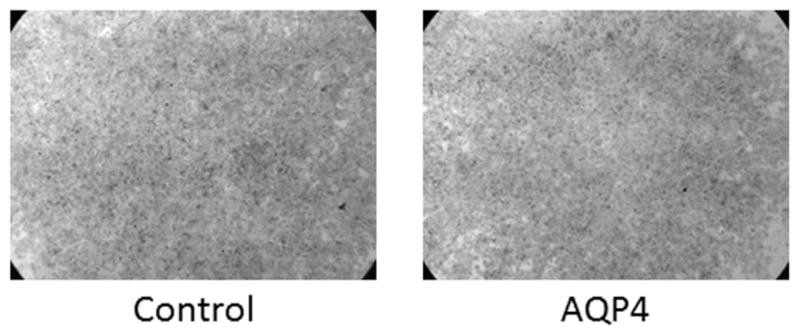
Overexpression of aquaporin-1 (AQP1) does not affect cell invasive capacity in ACC83.
Discussion
Salivary gland ACC is characterized by frequent distant metastasis and perineural invasion. Local control is often achieved with surgery and radiation therapy, but over 40% of patients will develop metastatic disease.1 ACC tumor cells may therefore have a unique potential for cell migration and invasion. Unfortunately, no molecular biomarkers are yet available that can be used to predict prognosis or therapeutic response. In this study, we sought to establish the role of AQP1 in the development of primary and meta-static disease in ACC.
We had previously identified AQP1 as a potential oncogene candidate that is epigenetically altered in ACC by using a genome-wide, array-based approach. AQP1 is a small transmembrane protein that selectively transports water across cell membranes. It has been implicated in cell proliferation, extravasation, migration, and metastasis in a number of tumor models.11 For example, mice injected with AQP1-expressing tumor cells were found to have an increased number of lung metastases and increased tumor cell extravasation across pulmonary vascular endothelium compared with mice injected with AQP1-deficient tumor cells.9 In humans, AQP1 overexpression has been associated with increased rates of metastasis and decreased disease-free survival rates in lung adenocarci-noma.17 AQP1 is therefore an attractive candidate gene in ACC, given the tumor’s propensity to metastasize.
In our previous study, AQP1 promoter hypomethylation was confirmed in 2 small independent ACC cohorts, and AQP1 overexpression at both the RNA and protein levels was demonstrated in a small cohort of tumor samples versus controls. The basic biologic functions of AQP1 were also examined in vitro using cells transiently transfected with an AQP1-expressing plasmid or empty control vectors. Overexpression of AQP1 resulted in increased cell proliferation and colony formation. Conversely, silencing of AQP1 in lung cancer cell lines led to decreased cell proliferation and colony formation.8 However, the effects of AQP1 specifically on invasion and metastatic potential in vitro have not previously been assessed. In addition, the small sample size included in our prior study, even with both cohorts combined, limited our ability to draw meaningful correlations between AQP1 methylation status or expression and clinical outcomes.
In the current study, we confirmed in a large cohort of patients that AQP1 was significantly hypomethylated in ACC tumors compared with controls. Furthermore, we demonstrated in a subset of these patients that AQP1 expression at the mRNA level tended to be increased in ACC, although this trend did not reach statistical significance. Regardless, this suggests that promoter hypermethylation of AQP1 plays a role in the regulation of AQP1 gene expression, although epigenetic modifications are not its sole regulatory mechanism. Further studies are ongoing to assess AQP1 expression at the protein level in the same ACC cohort using a tissue microarray.
We next evaluated whether correlations exist between AQP1 methylation or expression levels and clinical variables. Univariate analysis revealed that AQP1 promoter hypermethylation was associated with increased overall survival (P = .036), although not with recurrence- or metastasis-free survival. However, in multivariate modeling that included patient age and tumor stage at diagnosis, this association was no longer statistically significant. In addition, no associations between AQP1 expression level and clinical outcomes were found. However, the number of patients for which we were able to obtain mRNA expression results was small, due to the technical challenges associated with RNA isolation from FFPE tissues. The number of ACC tumors that will be analyzed for AQP1 protein expression on the tissue microarray is considerably larger, which may allow adequate statistical power if associations between AQP1 expression and clinical outcomes do exist.
Last, we evaluated the function of AQP1 overexpression on cell migration and invasion in vitro. We found that stable transfection of AQP1 did not affect cell migratory or invasive capacities in the cell scratch or Matrigel invasion assays. However, there are likely differences in the underlying biological characteristics of stably transfected cells or in cells with profound overexpression of AQP1 compared with native tumor cells or cultured ACC cell lines. Furthermore, the in vitro work presented here used SACC83, the only ACC cell line readily available to us. Further studies are necessary to clarify the role of AQP1 in ACC tumorigenesis and progression.
Conclusions
AQP1 is frequently hypomethylated in ACC tumors compared with control salivary tissues, and AQP1 tends to be overexpressed in these tumors. On univariate analysis, AQP1 promoter hypermethylation is associated with improved overall survival, but AQP1 expression levels are not associated with outcomes. Further in vitro studies are necessary to clarify the role of AQP1 in ACC progression.
Acknowledgments
Sponsorships: None.
Funding source: National Institutes of Health (NIH)/National Institute of Dental and Craniofacial Research (NIDCR) grant (K08-018463); AAO-HNSF Resident Research Grant, awarded by the American Academy of Otolaryngology—Head and Neck Surgery Foundation.
Footnotes
Author Contributions
Marietta Tan, design, data acquisition, analysis, interpretation, drafting, revision, final approval; Chunbo Shao, design, data acquisition, revision, final approval; Justin A. Bishop, data acquisition, analysis, revision, final approval; Zhaoyong Feng, data analysis, interpretation, revision, final approval; Bruce J. Trock, data interpretation, revision, final approval; William H. Westra, data interpretation, revision, final approval; Patrick K. Ha, conception, design, data interpretation, revision, final approval.
Competing interests: None.
This article was presented at the 2013 AAO-HNSF Annual Meeting and OTO EXPO; September 29 to October 3, 2013; Vancouver, British Columbia, Canada.
References
- 1.Khan AJ, DiGiovanna MP, Ross DA, et al. Adenoid cystic carcinoma: a retrospective clinical review. Int J Cancer. 2001;96:149–158. doi: 10.1002/ijc.1013. [DOI] [PubMed] [Google Scholar]
- 2.Herman JG, Baylin SB. Gene silencing in cancer in association with promoter hypermethylation. N Engl J Med. 2003;349:2042–2054. doi: 10.1056/NEJMra023075. [DOI] [PubMed] [Google Scholar]
- 3.Li J, El-Naggar A, Mao L. Promoter methylation of p16INK4a, RASSF1A, and DAPK is frequent in salivary adenoid cystic carcinoma. Cancer. 2005;104:771–776. doi: 10.1002/cncr.21215. [DOI] [PubMed] [Google Scholar]
- 4.Durr ML, Mydlarz WK, Shao C, et al. Quantitative methylation profiles for multiple tumor suppressor gene promoters in salivary gland tumors. PLoS One. 2010;5:e10828. doi: 10.1371/journal.pone.0010828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu J, Shao C, Tan M, et al. Molecular biology of adenoid cystic carcinoma. Head Neck. 2012;34:1665–1677. doi: 10.1002/hed.21849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bell A, Bell D, Weber RS, et al. CpG island methylation profiling in human salivary gland adenoid cystic carcinoma. Cancer. 2011;117:2898–2909. doi: 10.1002/cncr.25818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang CY, Mao L, Li L, et al. Promoter methylation as a common mechanism for inactivating E-cadherin in human salivary gland adenoid cystic carcinoma. Cancer. 2007;110:87–95. doi: 10.1002/cncr.22758. [DOI] [PubMed] [Google Scholar]
- 8.Shao C, Sun W, Tan M, et al. Integrated, genome-wide screening for hypomethylated oncogenes in salivary gland adenoid cystic carcinoma. Clin Cancer Res. 2011;17:4320–4330. doi: 10.1158/1078-0432.CCR-10-2992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hu J, Verkman AS. Increased migration and metastatic potential of tumor cells expressing aquaporin water channels. Faseb J. 2006;20:1892–1894. doi: 10.1096/fj.06-5930fje. [DOI] [PubMed] [Google Scholar]
- 10.Papadopoulos MC, Saadoun S, Verkman AS. Aquaporins and cell migration. Pflugers Arch. 2008;456:693–700. doi: 10.1007/s00424-007-0357-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Verkman AS, Hara-Chikuma M, Papadopoulos MC. Aquaporins—new players in cancer biology. J Mol Med (Berl) 2008;86:523–529. doi: 10.1007/s00109-008-0303-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shao C, Bai W, Junn JC, et al. Evaluation of MYB promoter methylation in salivary adenoid cystic carcinoma. Oral Oncol. 2011;47:251–255. doi: 10.1016/j.oraloncology.2011.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Laurie SA, Ho AL, Fury MG, et al. Systemic therapy in the management of metastatic or locally recurrent adenoid cystic carcinoma of the salivary glands: a systematic review. Lancet Oncol. 2011;12:815–824. doi: 10.1016/S1470-2045(10)70245-X. [DOI] [PubMed] [Google Scholar]
- 14.Kim MS, Yamashita K, Baek JH, et al. N-methyl-D-aspartate receptor type 2B is epigenetically inactivated and exhibits tumor-suppressive activity in human esophageal cancer. Cancer Res. 2006;66:3409–3418. doi: 10.1158/0008-5472.CAN-05-1608. [DOI] [PubMed] [Google Scholar]
- 15.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 16.Hoque MO, Soria JC, Woo J, et al. Aquaporin 1 is overexpressed in lung cancer and stimulates NIH-3T3 cell proliferation and anchorage-independent growth. Am J Pathol. 2006;168:1345–1353. doi: 10.2353/ajpath.2006.050596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Machida Y, Ueda Y, Shimasaki M, et al. Relationship of aquaporin 1, 3, and 5 expression in lung cancer cells to cellular differentiation, invasive growth, and metastasis potential. Hum Pathol. 2011;42:669–678. doi: 10.1016/j.humpath.2010.07.022. [DOI] [PubMed] [Google Scholar]