Abstract
Stress granules (SGs) and processing bodies (PBs) are microscopically visible ribonucleoprotein granules that cooperatively regulate the translation and decay of messenger RNA1–3. Using an RNA-mediated interference-based screen, we identify 101 human genes required for SG assembly, 39 genes required for PB assembly, and 31 genes required for coordinate SG and PB assembly. Although 51 genes encode proteins involved in mRNA translation, splicing and transcription, most are not obviously associated with RNA metabolism. We find that several components of the hexosamine biosynthetic pathway, which reversibly modifies proteins with O-linked N-acetylglucosamine (O-GlcNAc) in response to stress, are required for SG and PB assembly. O-GlcNAc-modified proteins are prominent components of SGs but not PBs, and include RACK1 (receptor for activated C kinase 1), prohibitin-2, glyceraldehyde-3-phosphate dehydrogenase and numerous ribosomal proteins. Our results suggest that O-GlcNAc modification of the translational machinery is required for aggregation of untranslated messenger ribonucleoproteins into SGs. The lack of enzymes of the hexosamine biosynthetic pathway in budding yeast may contribute to differences between mammalian SGs and related yeast EGP (eIF4E, 4G and Pab1 containing) bodies.
Stress granules and processing bodies are compositionally related ribonucleoprotein (RNP) granules that cooperatively regulate the translation and decay of mRNA1–5. The signature constituents of SGs are non-canonical, translationally silent 48S preinitiation complexes composed of mRNA bound to small ribosomal subunits and the initiation factors eIF4E, eIF3, eIF4A, eIF4G and poly(A)-binding protein (PABP)6,7. SGs also contain proteins that regulate mRNA translation (for example TIA-1 and TIAR) and decay (for example the 5’–3’ exonuclease XRN1), as well as proteins that regulate various cell signalling pathways (for example TRAF2, ubiquitin and SRC-3)1–3,8. The signature constituents of PBs are components of the decay machinery that removes the m7G (7-methylguanosine) cap and degrades mRNA from the 5’ end (for example XRN1, DCP1/DCP2, Hedls/GE-1 and Lsm1–7)5,9–11. Despite this suggestive enzymatic composition, blocking PB assembly does not inhibit mRNA decay9, suggesting that dispersed PB constituents are fully competent for decay. PBs may therefore harbour untranslated mRNAs awaiting activation of the decay machinery4,12. Thus, PBs, like SGs, may regulate translational silencing as well as mRNA decay.
Although SGs and PBs are coordinately induced by treatment with arsenite, PB assembly does not require phospho-eIF2α13; the stress-induced signalling pathways controlling PB assembly therefore remain unidentified. Conversely, clotrimazole and heat shock trigger SG assembly but not PB assembly13. For the systematic identification of proteins involved in SG and PB assembly, we devised a functional RNA-mediated interference (RNAi) screen for human genes involved in these processes. A human U2OS cell (RDG3) was established that stably expressed fluorescently tagged SG and PB markers (green fluorescent protein (GFP)–G3BP and red fluorescent protein (RFP)–DCP1a)14. Unstressed RDG3 cells contain PBs (in 30–50% of cells) but no SGs, similarly to parental U2OS cells3. Arsenite causes RFP–DCP1a to assemble into PBs and GFP–G3BP to assemble into SGs in about 90% of RDG3 cells (Supplementary Information, Fig. S1a). Time-lapse microscopy reveals synchronous SG and PB assembly on treatment with arsenite (Supplementary Information, Video S1). siRNAs targeting known SG and PB constituents were used to inhibit the assembly of RNA granules (Supplementary Information, Fig. S1b). In accord with previous reports, siRNAs targeting heme regulated inhibitor (HRI) prevent the assembly of SGs but not PBs, whereas siRNAs targeting Rck (p54) strongly inhibit PB assembly, but SG assembly only weakly (Supplementary Information, Fig. S1b, c). As reported previously10,15, decreased expression of DCP2 or the 5’–3’ exonuclease XRN1 promotes PB assembly, a probable consequence of decreased mRNA decay (Supplementary Information, Fig. S1b). Moreover, siRNAs targeting GW182, Lsm1 and Lsm4 significantly inhibit the assembly of PBs (Supplementary Information, Fig. S1b). For the screening assay, we used HRI and Rck as positive controls for SG and PB assembly, respectively.
We used this assay to screen the druggable genome (Dharmacon) targeting 7,317 genes with pools of four different siRNAs per gene (Supplementary Information, Fig. S1f). Pools scoring positive in the initial assay were subjected to a secondary screen with each of the four siRNAs individually. Genes that reduced SG and/or PB assembly when targeted by at least two independent siRNAs (Fig. 1a) are listed in Supplementary Information, Table S1. Not surprisingly, transcription and translation factors make up 26.8% of the genes detected (Fig. 1b). For example, knockdown of RNA polymerases or their regulatory subunits (for example BDP1; Fig. 1c) prevents the assembly of both SGs and PBs.
Figure 1.
RNAi screen identifies candidate genes involved in SG and/or PB assembly. (a) Venn diagram of genes affecting SG and/or PB assembly. (b) Graphic depiction of functional classification of genes revealed with the DAVID bioinformatics tool. (c) Effect of siRNA-mediated knockdown on the arsenite-induced assembly of SGs and PBs in RDG3 cells. Representative siRNA treatments from the primary screen alter the assembly of stress granules (green, GFP–G3BP) and P-bodies (red, RFP–DCP1a). SORT1, BDP1 and RPL10 knockdowns eliminate both SGs and PBs; RAB36 prevents PB assembly but not SG assembly, eIF3S2 knockdown prevents SG assembly but not PB assembly, and PRPF18 eliminates SG assembly but induces huge PBs.
Strikingly, the individual knockdown of six different eIF3 subunits prevents SG assembly. This robust and SG-specific effect indicates that eIF3 is critical to SG assembly. Knockdown of nine ribosomal proteins prevents the assembly of both SGs and PBs (RLP10; Fig. 1c; Supplementary Information, Table S1). The requirement for large ribosomal subunit proteins may be indirect, because large ribosomal subunits are excluded from SGs and PBs6. It is possible that decreased levels of specific large ribosomal proteins slow elongation or termination, thus impairing polysome disassembly and inhibiting the assembly of both SGs and PBs, as do elongation inhibitors such as emetine or cycloheximide13,16. Genes that independently regulate the assembly of SG or PB include the Ras-related protein RAB36 (PB-specific) and the frizzled homologue FZD2 (SG-specific), whereas the glucose regulator SORT1 (sortilin) is required for the assembly of both SG and PB (Fig. 1c). Knockdown of the cyclophilin-binding protein PRPF18 inhibits SGs and causes the assembly of giant PBs (Fig. 1c), suggesting that it regulates a directional flux of mRNA between SGs and PBs. Thus, our screen implicates proteins involved in RNA metabolism, as well as proteins not linked to RNA translation, processing or decay, to SG and PB assembly. Many mechanisms may affect the assembly of SGs and PBs, few of which may be direct. Possibilities include limiting the pool of untranslating mRNAs that are released during stress, trapping cells in mitosis, where SGs and PBs do not form, blocking polysome disassembly, altering the subcellular localization of SG or PB components, and limiting the aggregation of untranslating messenger RNPs (mRNPs). To explore possible mechanisms, we selected sortilin for further analysis.
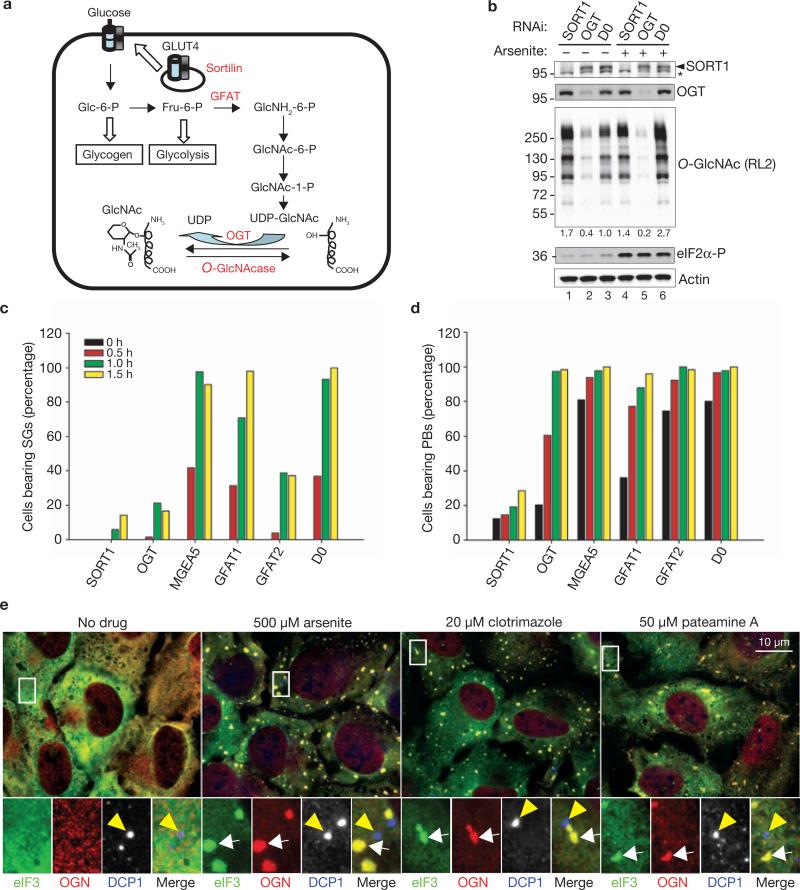
Sortilin is a transmembrane protein regulating the vesicular trafficking of several proteins including the GLUT4 glucose transporter, whose translocation to the plasma membrane controls glucose uptake (Fig. 2a)17. Intracellular glucose can be converted to glycogen for storage, metabolized to pyruvate by glycolysis, or converted to UDP-GlcNAc, a substrate of the hexosamine biosynthetic pathway (HBP)18–21. The reversible O-GlcNAc modification of serine or threonine residues of myriad proteins constitutes an important glucose-sensing signalling pathway in metazoans18–21. Moreover, arsenite promotes the O-GlcNAc modification of cellular proteins22, which confers protection against environmental and physiological stress23–25. O-GlcNAc modification is also activated during nutritional stress26,27. We therefore targeted key HBP enzymes (Fig. 2a, red), to determine whether O-GlcNAc modifications promote arsenite-induced SG or PB assembly. In cells treated with control siRNA (D0), arsenite increases the levels of O-GlcNAc-modified proteins (Fig. 2b, compare lanes 3 and 6). Although knockdown of sortilin modestly increases the levels of constitutive O-GlcNAc-modified proteins, it strongly prevents their arsenite-induced increase (Fig. 2b, compare lanes 1 and 4). Knockdown of O-GlcNAc transferase (OGT) decreases constitutive levels of O-GlcNAc-modified proteins and prevents their stress-induced increase (Fig. 2b, compare lanes 2 and 5). Neither sortilin knockdown nor OGT knockdown affects eIF2α phosphorylation, indicating that O-GlcNAc modifications act independently (Fig. 2b).
Figure 2.
The Hexosamine Biosynthetic Pathway (HBP) is required for SG and PB assembly. (a) Schematic representation of the HBP. (b) Sortilin and OGT regulate stress-induced O-GlcNAc modifications. U2OS cells transfected with the indicated siRNAs (D0, non-targeting control siRNA) were cultured in the absence or presence of arsenite before processing for immunoblots to assess protein expression. Densitometric quantifications of relative O-GlcNAc modification levels are indicated under the O-GlcNAc panel. The asterisk denotes a non-specific band recognized by anti-sortilin antibody. Numbers at the left are molecular masses in kDa. (c) Components of the HBP regulate SG assembly. U2OS cells were treated with the indicated siRNAs, then with arsenite for the indicated times before processing for immunofluorescence microscopy and quantifying of the percentage of cells with SGs. (d) Components of the HBP regulate PB assembly. (e) O-GlcNAc-modified proteins are components of SGs. U2OS cells were treated with the indicated drugs for 40 min, then stained for the stress granule marker eIF3b (eIF3, green), O-GlcNAc (OGN, red), and the P-body marker DCP1a (DCP1, blue). Enlarged views (8.5) of boxed areas show separate channels and merged views. Yellow arrowheads indicate P-bodies; white arrows indicate stress granules. Full scans of blots in b are shown in Supplementary Information, Fig. S7.
We confirmed that sortilin knockdown inhibits arsenite-induced SG assembly (Fig. 2c). OGT knockdown also inhibits SG assembly, whereas knockdown of MGEA5 (O-GlcNAcase, the enzyme that removes O-GlcNAc) had no effect. Knockdown of GFAT2 (glutamine:fructose-6-phosphate amidotransferase 2), but not GFAT1, enzymes that are required for the synthesis of UDP-GlcNAc, also inhibits SG assembly (Fig. 2a, c; Supplementary Information, Fig. S2). Sortilin knockdown inhibits both constitutive and arsenite-induced PB assembly, whereas OGT and GFAT2 inhibit constitutive, but not arsenite-induced, PB assembly (Fig. 2d). We reassayed these genes in U2OS cells stained for endogenous SG markers and examined at different times, to verify that the knockdowns abolished SG assembly rather than merely delaying it (Supplementary Information, Fig. S2). The results confirm that knockdowns of sortilin, OGT or GFAT2 impair SG assembly.
Because high (overexpressed) or low (knockdown) expression of many SG or PB components regulates SG or PB nucleation2, we examined whether sortilin is present in SGs or PBs. We found that endogenous sortilin localizes to the trans-Golgi network and plasma membrane (Supplementary Information, Fig. S3a) but is excluded from arsenite or clotrimazole-induced SGs (Supplementary Information, Fig. S3a, arrows) or PBs (Supplementary Information, Fig. S3b, arrowheads). In addition, we transiently overexpressed Myc-tagged sortilin and detected it with anti-Myc antibody. Overexpression of sortilin potently induces SG assembly without stress (Supplementary Information, Fig. S3b, panels CON-1 and CON-2), yet sortilin remains physically separate from the induced SGs. Different SG markers (TIA-1, eIF3 and eIF4G) and induction conditions (emetine-enforced disassembly; Supplementary Information, Fig. S3b, right panel) confirm that the sortilin-induced foci are genuine SGs. These results indicate that sortilin promotes SG assembly by a non-nucleating mechanism, analogously to overexpression of the S51D phosphomimetic mutant of eIF2α, which induces SGs without physically nucleating their assembly2.
We next examined whether OGT or O-GlcNAc-modified proteins are SG components. A monoclonal antibody against O-GlcNAc-modified proteins strongly detects O-GlcNAc (Fig. 2e, OGN, red) in arsenite-induced SGs (Fig. 2e, eIF3b, green, white arrows) but not in PBs (Fig. 2e, DCP1a, blue, yellow arrowheads). Similar results were obtained with clotrimazole or pateamine A (Fig. 2e) and with a different anti-O-GlcNAc antibody (CTD110.6) in other cell lines (Supplementary Information, Fig. S5c). We conclude that O-GlcNAc-modified proteins are major components of SGs. As reported previously28,29, endogenous OGT is found in nuclei, mitochondria and the cytoplasm of unstressed cells (Supplementary Information, Fig. S3c). In arsenite-treated cells (Supplementary Information, Fig. S3c, middle panel), a small amount of OGT (red) is found in SGs, in contrast to the robust O-GlcNAc signal. SGs induced by clotrimazole contain O-GlcNAc but lack detectable OGT (Supplementary Information, Fig. S3c). These results suggest that OGT-mediated O-GlcNAc modifications may precede the recruitment of modified proteins to SGs, and that sortilin knockdown may act upstream of OGT to inhibit SG assembly. The effects of sortilin (but not OGT) knockdown on arsenite-induced PB assembly may indicate the involvement of additional factors or pathways.
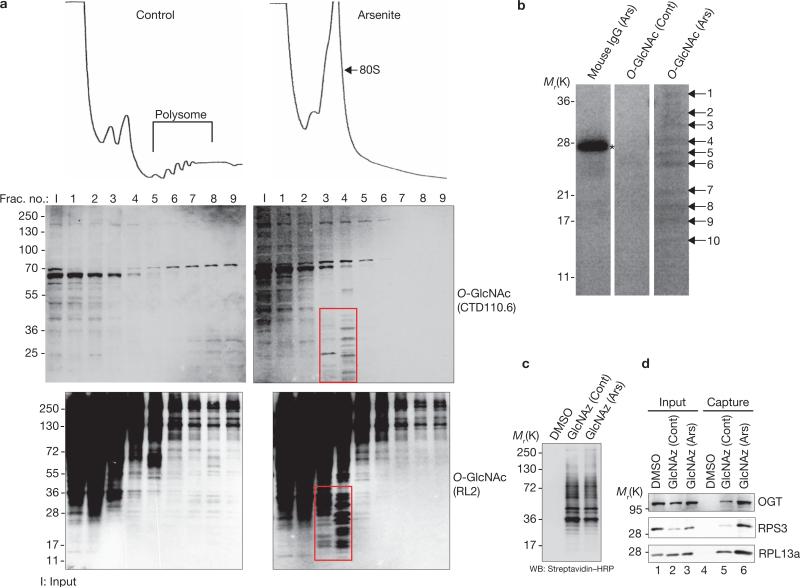
To identify specific O-GlcNAc-modified proteins involved in SG assembly, we analysed sucrose gradient fractions obtained from control versus arsenite-treated U2OS cells (Fig. 3a). Immunoblotting with two different anti-O-GlcNAc antibodies reveals the arsenite-induced O-GlcNAc modification of numerous low-molecular-mass (10–40-kDa) proteins that sediment together with monosomes and untranslated mRNPs (Fig. 3a, boxed regions). These fractions were precipitated with acetone, boiled in 1% SDS to disrupt protein or mRNP complexes completely, then diluted and immunoprecipitated with anti-O-GlcNAc antibodies (see Methods). O-GlcNAc-modified proteins resolved on PAGE were revealed by staining with colloidal blue (Fig. 3b); they were then excised and subjected to mass spectrometry, which identified small and large ribosomal subunit proteins (RPS3, 9, 11 and 24, and RPL6, 13a, 14, 30 and 36a-like), the ribosome-associated protein RACK1, the multifunctional proteins glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and prohibitin-2, and several other proteins (Supplementary Information, Table S2). GAPDH and RPS24 are known O-GlcNAc-modified proteins, thus validating our methodology18,30. Sucrose gradient analysis indicates that RACK1 and prohibitin-2 move from polysome-containing fractions to monosome-containing and ribosomal subunit-containing fractions on treatment with arsenite (Supplementary Information, Fig. S4), suggesting that both proteins associate with the translational machinery. Immunostaining confirms that RACK1 and prohibitin-2 are present in arsenite-induced SGs (Supplementary Information, Fig. S5a, b) and that prohitibitin-2 knockdown inhibits SG assembly in RDG3 cells (Supplementary Information, Fig. S6). These findings suggest that O-GlcNAc modifications of components of the translational machinery promote SG assembly. Other ribosomal protein subunits detected in our screen, including RPL5, RPL10, RPL11 and RPL13a (Supplementary Information, Table S1), are also found to be O-GlcNAc-modified by this approach (Supplementary Information, Table S2). Previous studies indicate that RPL5 depletion does not affect global translation in U2OS cells, suggesting that inhibition of SG or PB assembly by RPL5 ablation is not due to impaired basal translation31.
Figure 3.
Immunopurification and identification of arsenite-induced O-GlcNAc-modified proteins. (a) O-GlcNAc-modified proteins are specifically enriched in fractions containing 80S ribosomes and ribosome subunits in response to oxidative stress. Mock and arsenite-treated U2OS cells were lysed, fractionated over sucrose gradients (10–50%; top panels), and probed by immunoblotting with two different O-GlcNAc antibodies (middle two panels, CTD110.6; bottom two panels, RL2) to quantify O-GlcNAc-modified proteins. The indicated fractions (red boxes) were pooled, precipitated with acetone, and denatured with SDS before reprecipitation with anti-O-GlcNAc antibodies (see Methods). Numbers at the left of the blots are molecular masses in kDa. (b) Reprecipitates were separated by SDS–PAGE and stained with colloidal blue. Ten clearly distinguished bands were excised and subjected to mass spectrometry. A contaminating band is marked with an asterisk in the mouse IgG purified lane as a control. (c) Detection of biotinylated proteins from dimethylsulfoxide-treated (DMSO) U2OS cells (mock) or GlcNAz-treated (Azido-glucosamine) U2OS cells metabolically labelled with or without arsenite treatment (0.5 mM, 45 min) with the use of chemiluminescent substrate after blotting with horseradish peroxidaseconjugated streptavidin. Small aliquots (5%) of resolubilized biotinylated protein pellets were treated with SDS sample buffer and subjected to SDS– PAGE. (d) Biotinylated proteins before (input) or after (capture) pulldown with streptavidin beads were probed by immunoblotting with antibodies against RPS3, RPL13a and OGT (positive control). Full scans of blots in d are shown in Supplementary Information, Fig. S7.
To confirm that these proteins are indeed O-GlcNAc-modified in response to arsenite, we labelled cells metabolically for 2 days with tetraacetylated N-azidoacetylglucosamine (GlcNAz) (Invitrogen Click-iT kit; see Methods). Untreated cells, labelled cells and labelled cells treated with arsenite were harvested in parallel and lysed, and total GlcNAz-labelled proteins were biotinylated with a chemoselective ligation reaction (Fig. 3c). Biotinylated proteins were purified with streptavidin– agarose beads, resolved by SDS–PAGE and blotted for OGT, a known O-GlcNAc-modified protein. As shown in Fig. 3d, OGT is present only in labelled cells (compare lanes 5 and 6 with lane 4). Treatment with arsenite increases the amount of bound OGT (compare lanes 5 and 6), indicating that OGT is O-GlcNAc-modified on treatment with arsenite22. We then probed for candidate O-GlcNAc-modified ribosomal proteins identified by O-GlcNAc immunoprecipitation and mass spectrometric analysis. RPS3 and RPL13a are clearly detected, and both show an arsenite-induced increase in signal, confirming that proteins from both small and large ribosomal subunits are O-GlcNAc-modified in response to arsenite.
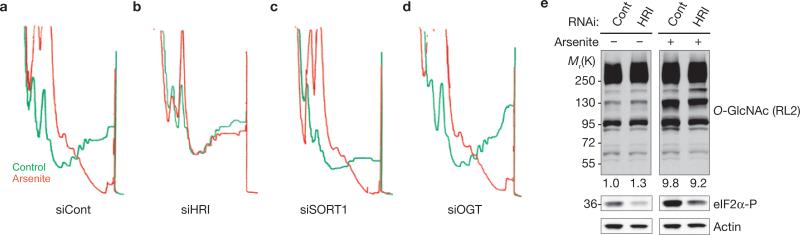
Arsenite-induced eIF2α phosphorylation causes translational arrest, polysome disassembly and SG assembly2. We used sucrose gradient analysis to determine the stage at which sortilin and OGT act. U2OS cells treated with control siRNAs (Fig. 4a) or with siRNAs targeting HRI (Fig. 4b), sortilin (Fig. 4c) or OGT (Fig. 4d) were untreated or arsenite-treated before sucrose gradient analysis. As expected, arsenite efficiently disassembles polysomes in control cells (D0) but not in HRI knockdown cells. In contrast, treatment with arsenite collapses polysomes in cells treated with siRNA targeting sortilin or OGT, indicating that O-GlcNAc modifications do not impair polysome disassembly but are required for 48S complexes to organize into SGs. Sortilin knockdown is significantly toxic, resulting in a lower signal (due to reduced cell number) and partly dismantled polysomes, but treatment with arsenite clearly enhances polysome disassembly (Fig. 4c, compare green and red tracings). No toxicity is seen in OGT knockdowns (Fig. 4d), which behaved identically to the control (Fig. 4a). Because O-GlcNAc-modified ribosome-associated proteins are SG components, and because impaired O-GlcNAc modification prevents SG assembly, it seems that O-GlcNAc modifications of ribosomal subunits contribute to SG assembly downstream of polysome disassembly, probably at the aggregation stage.
Figure 4.
Stress-induced O-GlcNAc modifications are not required for polysome disassembly. (a–d) U2OS cells were treated with the indicated siRNAs (D0 used as a control), then cultured in the absence (green lines) or presence (red lines) of arsenite (0.1 mM, 1 h) before polysome profile analysis. (e) Knockdown of HRI inhibits arsenite-induced eIF2α phosphorylation but not O-GlcNAc modification. U2OS cells treated with the indicated siRNAs were cultured in the absence or presence of arsenite before harvesting and processing for western blotting analysis with the indicated antibodies. Normalized densitometric quantification of O-GlcNAc modification levels is indicated under the respective panels.
Our results underscore the complexity of SG or PB assembly, and suggest a model to describe their interrelationship (Fig. 5). Both the assembly of SGs and that of PBs require polysome disassembly; however, the mode of polysome disassembly determines whether PBs or SGs assemble. Normally (Fig. 5, left pathway), mRNAs targeted for decay are deadenylated, breaking the connection between the 5’ and 3’ ends of the polysomal mRNA. The resulting decreased initiation efficiency causes polysome disassembly, freeing the resulting mRNP for incorporation into a PB. Upon stress (Fig. 5, right pathway), eIF2α phosphorylation causes stalled initiation, the assembly of a 48S mRNP, and polysome disassembly yielding circularized mRNPs, which are assembled into SGs in an eIF3-dependent manner. Polysome disassembly does not require O-GlcNAc addition, but O-GlcNAc modifications of the translation machinery (for example ribosomal protein subunits, RACK1 and prohibitin-2) promote the aggregation of 48S mRNPs into SGs. Because some mRNPs may transit through SGs to PBs2, the inhibition of SG assembly may secondarily affect PB assembly, thus explaining the modest effects of OGT on basal PB levels without stress.
Figure 5.
Model of SG versus PB assembly. Left: normal termination results in an mRNP that is eligible for deadenylation and eventual recruitment to the decapping and decay machinery of a PB. Right: stress-induced polysome disassembly caused by stalled initiation results in a 48S complex that is eligible for O-GlcNAc modification and SG assembly. Some mRNPs from SGs may be translocated to PBs, whereas others may be reinitiated or stored.
Conditions that decrease O-GlcNAc modification decrease the expression of heat-shock protein 70 (Hsp70; ref. 22). Because Hsp70 overexpression enhances SG disassembly32 (or inhibits SG assembly33), Hsp70 should not be a hit in our screen. Indeed, siRNA against Hsp70 was among those screened, but it did not prevent SG or PB assembly in our system.
Finally, we note that glucose deprivation induces budding yeasts to form ‘EGP bodies’ containing some SG markers (eIF4E, eIF4G and Pab1), but lacking 40S subunits and eIF3 (refs 34, 35). EGP bodies often form adjacent to P-bodies, similarly to the pairing of arsenite-induced mammalian SGs and PBs13. Budding yeasts lack OGT, HBP enzymes and some eIF3 subunits; O-GlcNAc modification of the translational machinery may therefore account for differences between yeast EGP bodies and mammalian stress granules. Indeed, O-GlcNAc modification of ribosomal subunits may regulate the assembly of other RNA granules containing ribosomal subunits, such as neuronal granules. The wide variety of genes detected in this screen provides increasing evidence that SG and PB assembly integrate many aspects of cellular metabolism with RNA translation and decay.
METHODS
Cell culture and drug treatment
U2OS (human osteosarcoma), COS7 and RDG3 cells were maintained in DMEM medium (Gibco) supplemented with 10% inactivated fetal bovine serum (Sigma), 1% (v/v) penicillin and streptomycin (Sigma) at 37 °C in 6% CO2. To induce SGs and PBs, cells were treated with sodium arsenite (either 0.1 mM or 0.5 mM) for between 30 min and 1 h. Clotrimazole was used at 20 μM in serum-free medium for 1 h. Pateamine A (Desmethyldesamino-modified; a gift from Jun Lui, Johns Hopkins University) was used at 50 μM for 1 h.
Establishment of RDG3 double stable cell line
Detailed procedures for making RDG3 double stable cells (SG marker, GFP–G3BP; PB marker, RFP–DCP1a) and for their selection and characterization are described elsewhere14.
Antibodies
All antibodies used in this study are listed in Supplementary Information, Table S3.
RNAi library screen and hit validation
RDG3 double stable cells were freshly plated 24 h before being screened; 60% confluent RDG3 cells were trypsinized and resuspended in DMEM (15% FBS) at a concentration of 50 cells μl−1. All siRNA transfections were performed in duplicate. In 384-well plates, 9 μl of lipid mix (Lipofectamine 2000 plus Opti-MEM; Invitrogen) and 1.5 μl of siRNA (1 μM, mixture of four different oligonucleotides; Dharmacon SMARTpool) were added and incubated for 30 min. Finally, 1,000 RDG3 cells were plated in each well and incubated for 72 h to induce gene silencing. To induce the assembly of SGs and PBs, sodium arsenite (0.5 mM final concentration) was added to the growth medium and cells were incubated for 30 min. Cells were fixed (4% paraformalde-hyde in PBS, 15 min), permeabilized (ice-cold methanol, 5 min) and stained with Hoechst (Sigma) dye to reveal nuclei. In the secondary screen, the four different siRNAs targeting genes identified in the primary screen were randomly localized in 384-well plates, and the assay was repeated. Only genes that were detected when targeted by at least two independent siRNAs were judged as robust positives; these are summarized in Supplementary Information, Table S1.
RNA interference and quantification of knockdown effect on SGs and PBs
U2OS or RDG3 cells were transfected with siRNAs at 40 nM final concentration using Lipofectamine 2000 (Invitrogen). After 40 h, cells were trypsinized, reseeded and transfected again for a further 40–44 h. For single transfection, cells were treated for 48–72 h and processed for the next step. Knockdown efficiencies were verified by either immunoblotting or real-time quantitative PCR with reverse transcription (RT–qPCR). siRNAs were purchased from Ambion or Dharmacon. Control siRNAs, D0 and U0, were described previously36. siRNA target sequences to deplete each protein used in this study are listed in Supplementary Information, Table S3. Knockdown effects on SGs and PBs were assessed by quantifying the number of cells out of more than 100 cells from different fields.
Immunofluorescence microscopy
General immunofluorescence procedures were performed as described previously3. Cells grown on coverslips were rinsed with PBS (pH 7.4), fixed with 4% paraformaldehyde for 15 min, permeabilized with cold methanol for 10 min or with 0.5% Triton X-100 in PBS in some cases (as specified below) and blocked by incubation in 5% normal horse serum in PBS containing 0.02% sodium azide (NHS/PBS-A) for at least 1 h. Primary antibodies diluted in blocking solution were added and incubated for about 1 h at 22-24 °C. Cells were then washed twice with PBS. Finally, cells were incubated with secondary antibodies (Jackson Immunoresearch ML grade) for 1 h, washed with PBS (three times, 5–10 min each) and mounted in polyvinyl-based mounting medium. Wide-field fluorescence microscopy was performed with an Eclipse E800 (Nikon) equipped with epifluorescence optics and a digital camera (CCD-SPOT RT; Diagnostic Instruments). The images were compiled with Adobe Photoshop (v. 10).
Immunoblot analysis
Cell extracts were prepared with RIPA buffer (25 mM Tris-HCl pH 7.6, 150 mM NaCl, 1% Nonidet P40, 1% sodium deoxycholate, 0.1% SDS) or direct lysis buffer (5 mM MES, pH 6.5, 2% SDS) and proteins were quantified, where indicated, with bicinchoninic acid assay reagent (Pierce). Total proteins (5–10 μg) were subjected to 4–20% gradient PAGE in SDS, transferred to nitrocellulose membranes and probed with the indicated antibodies with PBS containing 5% normal horse serum as a blocking reagent.
Polysome profiling analysis
U2OS cells (4 × 106) cultured at 80–90% confluence were incubated with or without 0.1 mM sodium arsenite for 1 h before the addition of 10 μg ml−1 cycloheximide, washed with cold PBS, then scrape-harvested into 1 ml of polysome lysis buffer (15 mM Tris-HCl pH 7.4, 15 mM MgCl2, 0.3 M NaCl, 1% Triton X-100) supplemented with 0.1% (v/v) 2-mercaptoethanol, 100 μg ml−1 cycloheximide, 0.1 mg ml−1 heparin, protease inhibitor cocktail (EDTA-free; Pierce) and RNAsin (Ambion). After 15 min of tumbling in the cold room, nuclei were pelleted (10,000g for 10 min) and supernatants were fractionated in 10–50% linear sucrose gradients by ultracentrifugation (125,000g. for 3 h 10 min) in a Beckman ultracentrifuge and an SW40 Ti rotor. Gradients were eluted with a gradient fractionator (Brandel) and monitored with a UA-5 detector (ISCO). Fractions were acetone-precipitated, and processed for further analyses.
Immunopurification of O-GlcNAc-modified proteins
Extracts from cells treated for 45 min with 0.5 mM sodium arsenite were prepared from two 90% confluent 150-mm dishes and applied to sucrose gradients as described above. For immunopurification of low-molecular-mass (10–40 kDa) O-GlcNAc-modified proteins induced by arsenite treatment, fractions containing most of the O-GlcNAc-modified proteins from the polysome gradient (18 fractions in total) were pooled, precipitated with acetone (60% final concentration) overnight at −20 °C and centrifuged at 14,000g for 15 min. Air-dried pellets were resuspended in 0.12 ml of denaturing buffer (50 mM Tris-HCl pH 7.5, 2 mM EDTA, 1% SDS), boiled for 5 min, diluted with 1.08 ml of immunoprecipitation (IP) buffer (50 mM Tris-HCl pH 7.5, 2 mM EDTA, 150 mM NaCl, 0.1% Nonidet P40) containing protease inhibitor cocktail (Pierce). Denatured total proteins were incubated with anti-O-GlcNAc antibody (10 μg; ABR) for 1 h at 4 °C followed by incubation overnight with 20 μl of pre-equilibrated protein A/G beads (Pierce) at 4 °C. The bead pellet was washed five times with 1 ml of IP buffer, treated with SDS sample buffer (without reducing agent), and subjected to 14% SDS–PAGE. Colloidal blue stain (Invitrogen) was used to reveal proteins; these were excised from the gel and identified by mass spectrometric analysis (Taplin Biological Mass Spectrometry Facility at Harvard Medical School).
Metabolic labelling and detection of O-GlcNAc-modified proteins
Metabolic labelling of O-GlcNAc-modified proteins was performed with Click-iT GlcNAz metabolic glycoprotein labelling reagent (Invitrogen). Labelled proteins were biotinylated (Click-iT protein analysis detection kit) and then purified by using streptavidin– agarose beads (Invitrogen); final products were subjected to immunoblot analyses.
Quantitative real-time PCR
Total RNA samples were prepared with TRIzol (Invitrogen) and quantified by spectrophotometry. RNA (5 μg) was treated with DNAase followed by cDNA synthesis with i-script cDNA synthesis kit (Bio-Rad). RT–qPCR was performed based on the SYBRgreen method with SYBR mix (Bio-Rad) in a cycler (MX3000p; Stratagene).
Live-cell imaging
Live-cell videos were taken on a Nikon TE2000-U inverted microscope equipped with a Nikon C1 Plus confocal system (BWH Confocal Core). Cells were maintained at 37 °C with 10% CO2 in an incubated microscope enclosure. Z stacks were acquired with a large (100-μm) pinhole at 0.8 μm steps through the entire cells every minute, and were rendered into two-dimensional images.
Supplementary Material
ACKNOWLEDGEMENTS
We thank C. Shamu and colleagues in the ICCB-L RNAi screening facilities at Harvard Medical School for their assistance. We also thank N. Ramadan at the Drosophila RNAi Screening Centre at Harvard Medical School for assistance with the automated microscope. We thank Jens Lykke-Andersen for the DCP1a antibody, and Carlos Morales for the Myc-tagged sortilin plasmid. We thank Keith Blackwell, Pavel Ivanov and Dan Schoenberg for critical review of the manuscript. This work was supported by NIH grants AI065858, AI033600 and AR0514732.
Footnotes
AUTHOR CONTRIBUTIONS
T.O. was responsible for experimental design, data acquisition, data analysis and the preparation of figures; N.K. contributed to experimental design, data acquisition, data analysis and the preparation of figures; S.T. and T.H. contributed to data acquisition and provided technical assistance; P.A. conceived the project and supervised all experimental activities. The manuscript was written by P.A., N.K. and T.O.
Supplementary Information is available on the Nature Cell Biology website.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
Published online at http://www.nature.com/naturecellbiology/
Reprints and permissions information is available online at http://npg.nature.com/reprintsandpermissions/
References
- 1.Anderson P, Kedersha N. RNA granules. J. Cell Biol. 2006;172:803–808. doi: 10.1083/jcb.200512082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anderson P, Kedersha N. Stress granules: the Tao of RNA triage. Trends. Biochem. Sci. 2008;33:141–150. doi: 10.1016/j.tibs.2007.12.003. [DOI] [PubMed] [Google Scholar]
- 3.Kedersha N, Anderson P. Mammalian stress granules and processing bodies. Methods Enzymol. 2007;431:61–81. doi: 10.1016/S0076-6879(07)31005-7. [DOI] [PubMed] [Google Scholar]
- 4.Parker R, Sheth U. P bodies and the control of mRNA translation and degradation. Mol. Cell. 2007;25:635–646. doi: 10.1016/j.molcel.2007.02.011. [DOI] [PubMed] [Google Scholar]
- 5.Eulalio A, Behm-Ansmant I, Izaurralde E. P bodies: at the crossroads of post-transcriptional pathways. Nature Rev. Mol. Cell Biol. 2007;8:9–22. doi: 10.1038/nrm2080. [DOI] [PubMed] [Google Scholar]
- 6.Kedersha N, et al. Evidence that ternary complex (eIF2-GTP-tRNA(i)(Met))-deficient preinitiation complexes are core constituents of mammalian stress granules. Mol. Biol. Cell. 2002;13:195–210. doi: 10.1091/mbc.01-05-0221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kimball SR, Horetsky RL, Ron D, Jefferson LS, Harding HP. Mammalian stress granules represent sites of accumulation of stalled translation initiation complexes. Am. J. Physiol. Cell Physiol. 2003;284:C273–C284. doi: 10.1152/ajpcell.00314.2002. [DOI] [PubMed] [Google Scholar]
- 8.Kwon S, Zhang Y, Matthias P. The deacetylase HDAC6 is a novel critical component of stress granules involved in the stress response. Genes Dev. 2007;21:3381–3394. doi: 10.1101/gad.461107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eulalio A, Behm-Ansmant I, Schweizer D, Izaurralde E. P-body formation is a consequence, not the cause, of RNA-mediated gene silencing. Mol. Cell. Biol. 2007;27:3970–3981. doi: 10.1128/MCB.00128-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sheth U, Parker R. Decapping and decay of messenger RNA occur in cytoplasmic processing bodies. Science. 2003;300:805–808. doi: 10.1126/science.1082320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lykke-Andersen J, Wagner E. Recruitment and activation of mRNA decay enzymes by two ARE-mediated decay activation domains in the proteins TTP and BRF-1. Genes Dev. 2005;19:351–361. doi: 10.1101/gad.1282305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Franks TM, Lykke-Andersen J. TTP and BRF proteins nucleate processing body formation to silence mRNAs with AU-rich elements. Genes Dev. 2007;21:719–735. doi: 10.1101/gad.1494707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kedersha N, et al. Stress granules and processing bodies are dynamically liked sites of mRNP remodeling. J. Cell Biol. 2005;169:871–884. doi: 10.1083/jcb.200502088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kedersha N, Tisdale S, Hickman T, Anderson P. Methods Enzymol. doi: 10.1016/S0076-6879(08)02626-8. in the press. [DOI] [PubMed] [Google Scholar]
- 15.Cougot N, Babajko S, Seraphin B. Cytoplasmic foci are sites of mRNA decay in human cells. J. Cell Biol. 2004;165:31–40. doi: 10.1083/jcb.200309008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kedersha N, et al. Dynamic shuttling of TIA-1 accompanies the recruitment of mRNA to mammalian stress granules. J. Cell Biol. 2000;151:1257–1268. doi: 10.1083/jcb.151.6.1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hou JC, Pessin JE. Ins (endocytosis) and outs (exocytosis) of GLUT4 trafficking. Curr. Opin. Cell Biol. 2007;19:466–473. doi: 10.1016/j.ceb.2007.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Love DC, Hanover JA. The hexosamine signaling pathway: deciphering the “O-GlcNAc code”. Sci. STKE. 2005;2005:re13. doi: 10.1126/stke.3122005re13. [DOI] [PubMed] [Google Scholar]
- 19.Marshall S. Role of insulin, adipocyte hormones, and nutrient-sensing pathways in regulating fuel metabolism and energy homeostasis: a nutritional perspective of diabetes, obesity, and cancer. Sci. STKE. 2006;2006:re7. doi: 10.1126/stke.3462006re7. [DOI] [PubMed] [Google Scholar]
- 20.Slawson C, Housley MP, Hart GW. O-GlcNAc cycling: how a single sugar post-translational modification is changing the way we think about signaling networks. J. Cell. Biochem. 2006;97:71–83. doi: 10.1002/jcb.20676. [DOI] [PubMed] [Google Scholar]
- 21.Zachara NE, Hart GW. Cell signaling, the essential role of O-GlcNAc. Biochim. Biophys. Acta. 2006;1761:599–617. doi: 10.1016/j.bbalip.2006.04.007. [DOI] [PubMed] [Google Scholar]
- 22.Zachara NE, et al. Dynamic O-GlcNAc modification of nucleocytoplasmic proteins in response to stress. A survival response of mammalian cells. J. Biol. Chem. 2004;279:30133–30142. doi: 10.1074/jbc.M403773200. [DOI] [PubMed] [Google Scholar]
- 23.Jones SP, et al. Cardioprotection by N-acetylglucosamine linkage to cellular proteins. Circulation. 2008;117:1172–1182. doi: 10.1161/CIRCULATIONAHA.107.730515. [DOI] [PubMed] [Google Scholar]
- 24.Zachara NE. The sweet nature of cardioprotection. Am. J. Physiol. Heart Circ. Physiol. 2007;293:H1324–H1326. doi: 10.1152/ajpheart.00697.2007. [DOI] [PubMed] [Google Scholar]
- 25.Zachara NE, Hart GW. O-GlcNAc a sensor of cellular state: the role of nucleocytoplasmic glycosylation in modulating cellular function in response to nutrition and stress. Biochim. Biophys. Acta. 2004;1673:13–28. doi: 10.1016/j.bbagen.2004.03.016. [DOI] [PubMed] [Google Scholar]
- 26.Cheung WD, Hart GW. AMP-activated protein kinase and p38 MAPK activate O-GlcNAcylation of neuronal proteins during glucose deprivation. J. Biol. Chem. 2008;283:13009–13020. doi: 10.1074/jbc.M801222200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Taylor RP, et al. Glucose deprivation stimulates O-GlcNAc modification of proteins through up-regulation of O-linked N-acetylglucosaminyltransferase. J. Biol. Chem. 2008;283:6050–6057. doi: 10.1074/jbc.M707328200. [DOI] [PubMed] [Google Scholar]
- 28.Morris NJ, et al. Sortilin is the major 110-kDa protein in GLUT4 vesicles from adipocytes. J. Biol. Chem. 1998;273:3582–3587. doi: 10.1074/jbc.273.6.3582. [DOI] [PubMed] [Google Scholar]
- 29.Nielsen MS, et al. The sortilin cytoplasmic tail conveys Golgi–endosome transport and binds the VHS domain of the GGA2 sorting protein. EMBO J. 2001;20:2180–2190. doi: 10.1093/emboj/20.9.2180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wells L, et al. Mapping sites of O-GlcNAc modification using affinity tags for serine and threonine post-translational modifications. Mol. Cell Proteomics. 2002;1:791–804. doi: 10.1074/mcp.m200048-mcp200. [DOI] [PubMed] [Google Scholar]
- 31.Dai MS, Lu H. Inhibition of MDM2-mediated p53 ubiquitination and degradation by ribosomal protein L5. J. Biol. Chem. 2004;279:44475–44482. doi: 10.1074/jbc.M403722200. [DOI] [PubMed] [Google Scholar]
- 32.Mazroui R, Di Marco S, Kaufman RJ, Gallouzi IE. Inhibition of the ubiquitin– proteasome system induces stress granule formation. Mol. Biol. Cell. 2007;18:2603–2618. doi: 10.1091/mbc.E06-12-1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gilks N, et al. Stress granule assembly is mediated by prion-like aggregation of TIA-1. Mol. Biol. Cell. 2004;15:5383–5398. doi: 10.1091/mbc.E04-08-0715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brengues M, Parker R. Accumulation of polyadenylated mRNA, Pab1p, eIF4E, and eIF4G with P-bodies in Saccharomyces cerevisiae. Mol. Biol. Cell. 2007;18:2592–2602. doi: 10.1091/mbc.E06-12-1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hoyle NP, Castelli LM, Campbell SG, Holmes LE, Ashe MP. Stress-dependent relocalization of translationally primed mRNPs to cytoplasmic granules that are kinetically and spatially distinct from P-bodies. J. Cell Biol. 2007;179:65–74. doi: 10.1083/jcb.200707010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stoecklin G, Mayo T, Anderson P. ARE-mRNA degradation requires the 5′-3′ decay pathway. EMBO Rep. 2006;7:72–77. doi: 10.1038/sj.embor.7400572. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.