Abstract
Single nucleotide polymorphisms (SNPs) in the PI3K/PTEN/AKT/mTOR signaling pathway may contribute to carcinogenesis. We genotyped five potentially functional PIK3R1 and mTOR SNPs in 1116 esophageal squamous cell cancer (ESCC) patients and 1117 cancer-free controls to assess their associations with ESCC risk. We observed no association with ESCC risk for any of the selected SNPs. However, the combined analysis of these SNPs revealed that subjects with one-to-three risk genotypes had an increased ESCC risk. Stratified analysis by body mass index (BMI) found that ESCC risk was significantly associated with each of three mTOR SNPs among subjects with BMI < 25.0. Specifically, we found that subjects carrying ≥ 1 risk genotypes had significantly increased ESCC risk, particularly for males, ever-smokers, ever-drinkers, and those with age > 60, or BMI < 25.0. Moreover, three mTOR haplotypes were associated with an increase in ESCC risk. Our meta-analysis of mTOR rs2295080 and cancer risk provided further evidence that mTOR SNPs might modulate cancer susceptibility. In this population, such risk effects might be modified by other risk factors, highlighting the importance of gene-environment interaction in esophageal carcinogenesis. Additional, larger studies are warranted to validate our findings.
Esophageal cancer is the sixth most common cause of cancer-related death in the world and also one of the most aggressive malignant tumors. According to GLOBOCAN 2008 estimates, approximately 482,300 new esophageal cancer cases and 406,800 deaths occurred in that year worldwide1. Esophageal cancer occurs as a result of multifaceted gene-environment interactions, as illustrated by its diverse patterns of incidence in different geographic regions of the world. The “esophageal cancer belt” (i.e., the highest risk area), including China, extends from Northern Iran through Central Asia to Northern-Central China. In particular, esophageal squamous cell carcinomas (ESCC) constitutes 90% of the cases in China1,2. Besides well-established risk factors, such as poor nutritional status, low intake of fruits and vegetables, smoking, alcohol intake, and drinking hot beverages, genetic variation in some key genes has been also suggested to modulate ESCC risk3,4. For example, numerous studies have demonstrated the existence of the association between ESCC risk and heritable genetic variants in genes involved in metabolism (e.g., Glutathione S-transferase Mu 1 and Methylenetetrahydrofolate reductase), DNA repair (e.g., X-ray repair cross-complementing protein 1 and Oxoguanine glycosylase), and cell cycle control (p53)3,4,5. Apart from these, genetic variation in genes of other cancer-related pathways may also play a role in ESCC susceptibility.
Phosphatidylinositols (PtdIns) 3-kinases (PI3Ks), a family of lipid kinases, are divided into three different classes (I, II, and III), based on primary structure and biological features. Class I PI3Ks have been extensively studied because they are responsible for the production of PtdIns(3,4,5)P3 (also called PIP3)6. Class I PI3Ks are heterodimeric molecules comprising a regulatory subunit and a catalytic subunit. Among them, PI3K regulatory subunit 1 (alpha) (PIK3R1, alias: p85-α) and PI3K catalytic subunit alpha (PIK3CA, alias: p110-α) have been extensively studied, which are encoded by PIK3R1 and PIK3CA genes, respectively7. PI3Ks debuted in the cancer research field back in the mid-1980s. Since then, the dysregulation of the PI3K/PTEN/AKT/mTOR pathway has been observed in a variety of human cancers, including cancers of the endometrium, stomach, lung, and esophagus8,9,10,11,12,13,14,15. Today, this pathway is well known to regulate important cellular events, including proliferation, adhesion, survival, and motility, which drive malignant transformation of cells and tumor progression16. Growth factors and hormones, such as epidermal growth factor receptor (EGFR) and insulin growth factor-1(IGF1), can stimulate class I PI3K by binding to the receptor tyrosine kinase (RTK). Activated class I PI3Ks convert PtdIns(4,5)P2 (called PIP2) to PIP3 by phosphorylating the hydroxyl group of the inositol ring of the former at the 3-position. The PIP3 then acts as a second messenger to trigger a downstream signaling cascade that is comprised of AKT, mTOR, and other proteins16. Mammalian target of rapamycin/FK506 binding protein 12-rapamycin associated protein 1 (mTOR/FRAP1), a serine/threonine kinase, is a member of the PI3Ks-related kinase protein family and is known as a central effector of cell growth and proliferation through the regulation of protein synthesis17.
Accumulating evidence has shown that mutations in some genes (PIK3CA, RAS, PTEN, and AKT) of the pathway could result in neoplastic transformation in both cellular and animal models15,16,18,19, suggesting a critical role of the pathway in carcinogenesis. Aberrant activation of this pathway has been closely related to various cancers, including ESCC. For example, it was reported that 11.5% of the tumors from ESCC patients harbored PIK3CA mutations20 and that the aberrant activation of mTOR occurred in 69.5% and 25% of ESCC in Japanese patients11 and Caucasian patients in the Netherlands21, respectively. The relatively high incidence of mutations in the PI3K pathway component provides strong evidence that dysregulation of this signaling pathway may contribute to the development of ESCC. Given the profound influence of aberrant activation of PI3K and mTOR on ESCC carcinogenesis, it is plausible that some potentially functional SNPs in genes encoding these proteins are likely to modulate ESCC susceptibility.
In contrast to extensive investigations regarding the mutations in these pathway genes, there are only a few studies exploring cancer risk associated with genetic variation in the same pathway genes. For example, Slattery et al. demonstrated that single nucleotide polymorphisms (SNPs) in PIK3CA and mTOR/FRAP1 genes were significantly associated with risk of colon and rectal cancers, respectively22. Moreover, our group has previously reported associations of mTOR rs1883965 and rs2536 with risk of cancers of the esophagus, stomach, and prostate9,23,24. Two independent studies indicated that the mTOR rs1883965 SNP was significantly associated with an increased risk of gastric cancer24 and ESCC23.
In the present study, we expanded our previous studies by comprehensively analyzing additional potentially functional SNPs in the genes encoding class I PI3Ks and mTOR for their association with ESCC risk in an Eastern Chinese population.
Results
Characteristics of the Study Population
Overall, demographic characteristics of the case and cancer-free controls were comparable (Table 1). No statistically significant difference in the distributions of age and sex was observed between the cases and controls. With respect to drinking and smoking habits, more cases tended to be smokers and drinkers in comparison with controls. Moreover, a significantly higher percentage of cases had BMI (weight in kilograms/height2 in meters) below 25.0, when compared with the controls. To minimize a possible confounding effect, these variables were then adjusted for in the subsequent multivariate logistic regression analyses.
Table 1. Frequency distributions of selected characteristics of ESCC cases and controls.
| Variables | Cases No. (%) | Controls No. (%) | Pa |
|---|---|---|---|
| All subjects | 1,116 (100.0) | 1,117 (100.0) | |
| Age, yr | 0.890 | ||
| Range | 37–88 | 32–86 | |
| Meanb | 60.4 ± 8.3 | 60.3 ± 10.2 | |
| Age group | |||
| ≤50 | 143 (12.8) | 151 (13.5) | |
| 51–60 | 423 (37.9) | 405 (36.3) | |
| 61–70 | 411 (36.8) | 405 (36.3) | |
| >70 | 139 (12.5) | 156 (13.9) | |
| Sex | 0.410 | ||
| Males | 897 (80.4) | 882 (79.0) | |
| Females | 219 (19.6) | 235 (21.0) | |
| Drinking status | <0.0001 | ||
| Ever | 498 (44.6) | 369 (33.0) | |
| Never | 618 (55.4) | 748 (67.0) | |
| Smoking status | 0.0038 | ||
| Ever | 681 (61.0) | 614 (55.0) | |
| Never | 435 (39.0) | 503 (45.0) | |
| Pack-years | <0.0001 | ||
| 0 | 435 (39.0) | 503 (45.0) | |
| ≤16 (mean) | 150 (13.4) | 246 (22.0) | |
| >16 (mean) | 531 (47.6) | 368 (33.0) | |
| Body mass index | <0.0001 | ||
| <25.0 | 721 (64.6) | 485 (43.4) | |
| ≥25.0 | 395 (35.4) | 632 (56.6) |
aTwo-sided χ2 test for distributions between cases and controls.
bData were presented as mean ± SD.
Association between selected SNPs and ESCC risk
Three genes (PIK3R1, PIK3CA, and mTOR) were initially searched for potentially functional SNPs. However, we only investigated SNPs in the PIK3R1 and mTOR genes in this study, because no potentially functional SNP in the PIK3CA gene met the SNP selection criteria. The genotype frequency distributions of all the selected SNPs in control subjects were in accordance with the Hardy Weinberg equilibrium (HWE). The minor allele frequencies (MAFs) of the SNP in these controls were similar to those reported in the CHB data from HapMap: 0.19 vs. 0.175 for rs3730089, 0.12 vs. 0.128 for rs3730090, 0.21 vs. 0.239 for rs2295080, 0.19 vs. 0.18 for rs1057079, and 0.078 vs. 0.109 for rs1064261, respectively.
Odds ratios (ORs) were determined by logistic regression analyses with adjustment for the covariates, i.e., age, sex, drinking status, smoking status, and BMI. Results including genotype frequencies, crude OR and 95% confidence interval (CI), and adjusted OR (95% CI) are shown in Table 2. Risk estimates indicated that none of the individual SNPs had a main effect on ESCC risk; that is, there was no significant association identified between any of the selected SNPs and ESCC risk. Next, SNPs under investigation were combined, and the risk estimates revealed that subjects with one, two, or three putative risk genotypes had significantly or borderline significantly increased risk of developing ESCC, compared with those without any such putative risk genotypes. Such an additive effect on risk did not seem to be risk-genotype dose-dependent, since there was no trend of increased ORs for individuals who carried one, two, or three risk genotypes (adjusted OR = 1.34, 95% CI = 1.07–1.68 for one risk genotype; adjusted OR = 1.42, 95% CI = 1.07–1.87 for two risk genotypes; adjusted OR = 1.29, 95% CI = 0.99–1.67 for three risk genotypes; Table 2). Furthermore, all participants were dichotomized according to the presence of risk genotypes. Subjects in the reference group had no risk genotypes, while subjects having one or more risk genotypes were categorized into the other group. Compared with the reference group, those with one or more risk genotypes had statistically, significantly increased ESCC risk (adjusted OR = 1.33, 95% CI = 1.10–1.62).
Table 2. Logistic regression analysis of associations between the genotypes of PIK3R1 & mTOR and ESCC risk.
| Variants | Genotypes | Cases (N = 1,116) | Controls (N = 1,117) | Pa | Crude OR (95% CI) | P | Adjusted OR (95% CI) | Pb |
|---|---|---|---|---|---|---|---|---|
| PIK3R1 rs3730089 | ||||||||
| GG | 736 (66.0) | 729 (65.4) | 0.880 | 1.00 | 1.00 | 0.763 | ||
| AG | 331 (29.7) | 345 (30.9) | 0.95 (0.79–1.14) | 0.594 | 0.98 (0.81–1.19) | 0.857 | ||
| AA | 48 (4.3) | 41 (3.7) | 1.16 (0.76–1.78) | 0.496 | 1.19 (0.76–1.85) | 0.452 | ||
| AG/AA | 379 (34.0) | 386 (34.6) | 0.97 (0.82–1.16) | 0.755 | 1.00 (0.84–1.20) | 0.972 | ||
| PIK3R1 rs3730090 | ||||||||
| CC | 837 (75.3) | 849 (76.4) | 0.463 | 1.00 | 1.00 | 0.475 | ||
| CT | 255 (23.0) | 249 (22.3) | 1.04 (0.86–1.27) | 0.708 | 1.04 (0.85–1.28) | 0.690 | ||
| TT | 19 (1.7) | 14 (1.2) | 1.38 (0.69–2.76) | 0.369 | 1.34 (0.65–2.75) | 0.434 | ||
| CT/TT | 274 (24.7) | 263 (23.6) | 1.06 (0.87–1.28) | 0.578 | 1.06 (0.87–1.30) | 0.570 | ||
| mTOR rs2295080 | ||||||||
| TT | 674 (60.6) | 702 (63.1) | 0.304 | 1.00 | 1.00 | 0.202 | ||
| GT | 390 (35) | 362 (32.5) | 1.12 (0.94–1.34) | 0.199 | 1.13 (0.94–1.36) | 0.185 | ||
| GG | 49 (4.4) | 49 (4.4) | 1.04 (0.69–1.57) | 0.841 | 1.12 (0.73–1.71) | 0.614 | ||
| GT/GG | 439 (39.4) | 411 (36.9) | 1.11 (0.94–1.32) | 0.222 | 1.13 (0.95–1.36) | 0.167 | ||
| mTOR rs1057079 | ||||||||
| TT | 702 (63.0) | 725 (65.0) | 0.321 | 1.00 | 1.00 | 0.248 | ||
| CT | 367 (32.9) | 349(31.3) | 1.09 (0.91–1.30) | 0.368 | 1.10 (0.91–1.32) | 0.322 | ||
| CC | 45 (4.1) | 41 (3.7) | 1.13 (0.73–1.75) | 0.573 | 1.18 (0.75–1.85) | 0.484 | ||
| CT/CC | 412 (37.0) | 390 (35.0) | 1.09 (0.92–1.30) | 0.324 | 1.11 (0.93–1.33) | 0.261 | ||
| mTOR rs1064261 | ||||||||
| AA | 916 (82.2) | 945 (84.8) | 0.153 | 1.00 | 1.00 | 0.134 | ||
| AG | 194 (17.4) | 164 (14.7) | 1.22 (0.97–1.53) | 0.085 | 1.22 (0.96–1.55) | 0.098 | ||
| GG | 4 (0.4) | 6 (0.5) | 0.69 (0.19–2.45) | 0.563 | 0.87 (0.24–3.22) | 0.840 | ||
| AG/GG | 198 (17.8) | 170 (15.2) | 1.20 (0.96–1.50) | 0.109 | 1.21 (0.96–1.53) | 0.108 | ||
| Combined effect of risk genotypesc | ||||||||
| 0 | 273 (24.6) | 322 (29.1) | 1.00 | 1.00 | 0.107 | |||
| 1 | 362 (32.6) | 338 (30.6) | 1.27 (1.03–1.58) | 0.029 | 1.34 (1.07–1.68) | 0.012 | ||
| 2 | 177 (16.0) | 157 (14.2) | 1.34 (1.03–1.75) | 0.032 | 1.42 (1.07–1.87) | 0.014 | ||
| 3 | 209 (18.9) | 200 (18.1) | 1.24 (0.97–1.60) | 0.089 | 1.29 (0.99–1.67) | 0.058 | ||
| 4 | 83(7.48) | 83 (7.50) | 1.19 (0.84–1.68) | 0.322 | 1.21 (0.85–1.73) | 0.293 | ||
| 5 | 5 (0.45) | 6 (0.54) | 0.99 (0.30–3.28) | 0.988 | 1.27 (0.35–4.54) | 0.717 | ||
| P trend = 0.181 | P trend = 0.107 | |||||||
| 0 | 280 (25.1) | 333 (29.8) | 1.00 | 1.00 | ||||
| ≥1 | 836 (74.9) | 784 (70.2) | 1.27 (1.05–1.53) | 0.013 | 1.33 (1.10–1.61) | 0.004 |
aFor additive genetic models. The results were in bold, if the 95% CI excluded 1 or P < 0.05.
bAdjusted for age, sex, smoking and drinking status in logistic regress models.
cRisk genotypes used for the calculation were PIK3R1 rs3730089 AG/AA + PIK3R1 rs3730090 CT/TT + mTOR rs2295080 GT/G G + mTOR rs1057079 CT/CC + mTOR rs1064261 AG/GG.
Stratified Analysis
In an attempt to further scrutinize potential associations between the selected SNPs and ESCC risk, the data were stratified by the dichotomized variables of age, sex, smoking status, drinking status, and BMI, individually, under the dominant genetic model. No significant ESCC risk associated with any of PIK3R1 and mTOR SNPs was detected in the dichotomized subgroups by age, sex, smoking, and drinking status (Tables 3,4,5). However, significant ESCC risk associated with three mTOR SNPs, but not PIK3R1 SNPs, was each individually observed among subjects with BMI < 25.0 under the dominant genetic model (WV/VV vs. WW) (rs2295080: adjusted OR = 1.36, 95% CI = 1.07–1.73; rs1057079: OR = 1.31, 95% CI = 1.03–1.67, rs1014261: OR = 1.39, 95% CI = 1.01–1.92) (Tables 3,4,5).
Table 3. Stratification analysis for associations between variant genotypes of PIK3R1and ESCC risk.
| PIK3R1 3730089 | PIK3R1 3730090 | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| (cases/controls) | (cases/controls) | |||||||||||||
| Variables | GG | AG/AA | Crude OR (95% CI) | P | Adjusted OR (95% CI) | P a | Phom | CC | CT/TT | Crude OR (95% CI) | P | Adjusted ORa (95% CI) | P a | Phom |
| Age | ||||||||||||||
| ≤60 | 373/365 | 192/190 | 0.99 (0.77–1.23) | 0.929 | 0.97 (0.74–1.25) | 0.790 | 0.853 | 415/420 | 150/132 | 1.13 (0.87–1.49) | 0.363 | 1.20 (0.90–1.59) | 0.222 | 0.455 |
| >60 | 363/364 | 187/196 | 0.96 (0.75–1.23) | 0.726 | 1.03 (0.79–1.34) | 0.844 | 422/429 | 128/131 | 0.98 (0.74–1.29) | 0.873 | 0.96 (0.72–1.28) | 0.770 | ||
| Sex | ||||||||||||||
| Females | 153/152 | 65/82 | 0.79(0.53–1.17) | 0.236 | 0.78 (0.51–1.18) | 0.248 | 0.174 | 158/181 | 60/53 | 1.27 (0.83–1.95) | 0.266 | 1.40 (0.88–2.22) | 0.153 | 0.337 |
| Males | 582/576 | 314/305 | 1.02 (0.84–1.24) | 0.825 | 1.08 (0.88–1.33) | 0.450 | 679/668 | 217/213 | 1.01 (0.81–1.25) | 0.948 | 0.98(0.78–1.23) | 0.871 | ||
| Smoking status | ||||||||||||||
| Never | 286/321 | 149/182 | 0.92 (0.70–1.20) | 0.538 | 0.92 (0.70–1.21) | 0.542 | 0.556 | 318/386 | 115/115 | 1.21 (0.90–1.64) | 0.203 | 1.25 (0.92–1.71) | 0.157 | 0.239 |
| Ever | 450/408 | 230/204 | 1.02 (0.81–1.29) | 0.852 | 1.06 (0.83–1.36) | 0.620 | 519/463 | 159/148 | 0.96 (0.74–1.24) | 0.745 | 0.99 (0.76–1.30) | 0.957 | ||
| Drinking status | ||||||||||||||
| Never | 416/494 | 202/252 | 0.95 (0.76–1.19) | 0.670 | 0.97 (0.76–1.22) | 0.776 | 0.257 | 450/565 | 166/178 | 1.17 (0.92–1.50) | 0.207 | 1.16 (0.90–1.50) | 0.256 | 0.271 |
| Ever | 298/235 | 199/134 | 0.97 (0.73–1.28) | 0.832 | 1.09 (0.81–1.47) | 0.574 | 387/284 | 108/85 | 0.93 (0.68–1.29) | 0.670 | 0.90 (0.63–1.25) | 0.507 | ||
| BMI | ||||||||||||||
| <25.0 | 485/320 | 235/164 | 0.95(0.74–1.21) | 0.677 | 0.95(0.74–1.22) | 0.696 | 0.583 | 541/365 | 179/119 | 1.02 (0.78–1.33) | 0.903 | 1.02 (0.78–1.33) | 0.897 | 0.784 |
| ≥25.0 | 251/409 | 143/222 | 1.05(0.81–1.37) | 0.717 | 1.03 (0.79–1.35) | 0.824 | 297/484 | 97/147 | 1.08 (0.80–1.45) | 0.632 | 1.10 (0.82–1.49) | 0.530 | ||
CI, confidence interval; OR, odds ratio.
aObtained in logistic regression models with adjustment for age, sex, smoking status and drinking status.
Phom derived from the homogeneity test.
The results were in bold, if the 95% CI excluded 1 or P < 0.05.
Table 4. Stratification analysis for associations between variant genotypes of mTOR and ESCC risk.
| mTOR rs1057079 | mTOR rs1014261 | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| (cases/controls) | (cases/controls) | ||||||||||||||
| Variables | TT | CT/CC | Crude OR (95% CI) | P | Adjusted OR (95% CI) | P a | Phom | AA | AG/GG | Crude OR (95% CI) | P | Adjusted ORa (95% CI) | P a | Phom | |
| Age | |||||||||||||||
| ≤60 | 364/362 | 201/193 | 1.04 (0.81–1.32) | 0.780 | 1.05 (0.81–1.37) | 0.692 | 0.550 | 464/468 | 101/87 | 1.16 (0.84–1.58) | 0.368 | 1.20 (0.86–1.67) | 0.276 | 0.727 | |
| >60 | 338/363 | 212/197 | 1.15 (0.90–1.47) | 0.261 | 1.15 (0.89–1.48) | 0.284 | 451/477 | 99/83 | 1.25 (0.91–1.72) | 0.172 | 1.27 (0.91–1.77) | 0.167 | |||
| Sex | |||||||||||||||
| Females | 142/150 | 76/84 | 0.96 (0.65–1.41) | 0.818 | 1.01 (0.67–1.53) | 0.956 | 0.453 | 179/201 | 39/33 | 1.33 (0.80–2.20) | 0.273 | 1.24 (0.71–2.15) | 0.451 | 0.665 | |
| Males | 560/575 | 336/306 | 1.13 (0.93–1.37) | 0.225 | 1.13 (0.93–1.39) | 0.226 | 737/744 | 159/137 | 1.17 (0.91–1.50) | 0.215 | 1.17 (0.99–1.52) | 0.243 | |||
| Smoking status | |||||||||||||||
| Never | 267/314 | 166/188 | 1.04 (0.80–1.35) | 0.780 | 1.02 (0.77–1.34) | 0.897 | 0.566 | 352/414 | 81/87 | 1.10 (0.79–1.53) | 0.594 | 1.08 (0.77–1.52) | 0.678 | 0.405 | |
| Ever | 435/411 | 246/202 | 1.15 (0.91–1.45) | 0.231 | 1.21 (0.95–1.54) | 0.132 | 564/531 | 117/83 | 1.33 (0.98–1.80) | 0.069 | 1.31 (0.95–1.80) | 0.104 | |||
| Drinking status | |||||||||||||||
| Never | 388/480 | 228/266 | 1.06 (0.85–1.32) | 0.604 | 1.08 (0.86–1.36) | 0.516 | 0.631 | 506/636 | 110/110 | 1.25 (0.94–1.68) | 0.121 | 1.24 (0.92–1.66) | 0.167 | 0.585 | |
| Ever | 314/245 | 184/124 | 1.16 (0.87–1.54) | 0.309 | 1.16 (0.86–1.57) | 0.322 | 410/309 | 88/60 | 1.11 (0.77–1.58) | 0.585 | 1.10 (0.75–1.61) | 0.632 | |||
| BMI | |||||||||||||||
| <25.0 | 437/323 | 283/161 | 1.30 (1.02–1.65) | 0.033 | 1.31 (1.03–1.67) | 0.031 | 0.022 | 585/415 | 135/69 | 1.39 (1.01–1.91) | 0.042 | 1.39 (1.01–1.92) | 0.043 | 0.167 | |
| ≥25.0 | 265/402 | 129/229 | 0.86 (0.66–1.12) | 0.246 | 0.87 (0.67–1.14) | 0.311 | 331/530 | 63/101 | 1.00 (0.71–1.41) | 0.994 | 0.99 (0.70–1.40) | 0.962 | |||
CI, confidence interval; OR, odds ratio.
aObtained in logistic regression models with adjustment for age, sex, smoking status and drinking status.
Phom derived from the homogeneity test.
The results were in bold, if the 95% CI excluded 1 or P < 0.05.
Table 5. Stratification analysis for associations between variant genotypes of mTOR and ESCC risk.
| mTOR rs2295080 | Combined risk genotypes | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| (cases/controls) | (cases/controls) | ||||||||||||||
| Variables | TT | GT/GG | Crude OR (95% CI) | P | Adjusted OR (95% CI) | P a | Phom | 0 | ≥1 | Crude OR (95% CI) | P | Adjusted ORa (95% CI) | P a | Phom | |
| Age | |||||||||||||||
| ≤60 | 342/350 | 223/205 | 1.12 (0.88–1.42) | 0.363 | 1.14 (0.89–1.48) | 0.303 | 0.092 | 325/341 | 240/214 | 1.18 (0.93–1.49) | 0.182 | 1.20 (0.93–1.55) | 0.157 | 0.674 | |
| >60 | 332/345 | 218/205 | 1.11 (0.87–1.41) | 0.414 | 1.12 (0.87–1.44) | 0.381 | 312/350 | 238/210 | 1.27 (1.00–1.61) | 0.055 | 1.28 (1.01–1.65) | 0.049 | |||
| Sex | |||||||||||||||
| Females | 136/146 | 82/88 | 1.00 (0.68–1.46) | 1.000 | 1.05 (0.70–1.57) | 0.813 | 0.542 | 134/146 | 84/88 | 1.04 (0.71–1.52) | 0.837 | 1.11 (0.74–1.66) | 0.631 | 0.362 | |
| Males | 538/556 | 358/325 | 1.14 (0.94–1.38) | 0.174 | 1.14 (0.94–1.38) | 0.181 | 504/546 | 392/335 | 1.27 (1.05–1.53) | 0.014 | 1.28 (1.05–1.56) | 0.014 | |||
| Smoking status | |||||||||||||||
| Never | 249/303 | 184/198 | 1.13 (0.87–1.47) | 0.357 | 1.12 (0.85–1.47) | 0.419 | 0.972 | 237/299 | 198/204 | 1.22 (0.95–1.59) | 0.126 | 1.21 (0.92–1.58) | 0.166 | 0.937 | |
| Ever | 425/399 | 255/213 | 1.12 (0.90–1.41) | 0.315 | 1.18 (0.93–1.51) | 0.172 | 401/393 | 280/221 | 1.24 (0.99–1.55) | 0.059 | 1.31 (1.03–1.67) | 0.028 | |||
| Drinking status | |||||||||||||||
| Never | 376/467 | 240/277 | 1.08 (0.86–1.34) | 0.513 | 1.11 (0.88–1.39) | 0.388 | 0.640 | 357/460 | 261/288 | 1.17 (0.94–1.45) | 0.162 | 1.20 (0.95–1.50) | 0.119 | 0.527 | |
| Ever | 298/235 | 199/134 | 1.17 (0.89–1.55) | 0.265 | 1.20 (0.90–1.62) | 0.216 | 281/232 | 217/137 | 1.31 (0.99–1.72) | 0.056 | 1.34 (1.00–1.79) | 0.051 | |||
| BMI | |||||||||||||||
| <25.0 | 420/315 | 300/169 | 1.34(1.06–1.70) | 0.016 | 1.36 (1.07–1.73) | 0.014 | 0.016 | 392/312 | 328/172 | 1.51 (1.19–1.91) | 0.001 | 1.52 (1.20–1.94) | 0.001 | 0.006 | |
| ≥25.0 | 256/386 | 138/245 | 0.87(0.67–1.13) | 0.285 | 0.88 (0.67–1.14) | 0.323 | 244/380 | 150/251 | 0.92 (0.71–1.29) | 0.544 | 0.95 (0.73–1.24) | 0.711 | |||
CI, confidence interval; OR, odds ratio.
aObtained in logistic regression models with adjustment for age, sex, smoking status and drinking status.
Phom derived from the homogeneity test.
The results were in bold, if the 95% CI excluded 1 or P < 0.05.
Since the three mTOR SNPs were not in complete LD, their individual effects might be additive. We further explored the combined effects of these three SNPs in stratified analyses by age, sex, smoking status, drinking status, and BMI and found that significantly increased ESCC risk was identified for subjects carrying at least one of the three putative risk genotypes (i.e., rs2295080 GT/GG, rs1057079 CT/CC, and rs1014261 AG/GG) among the following subgroups: >60 years of age (adjusted OR = 1.28, 95% CI = 1.01–1.65), males (adjusted OR = 1.28, 95% CI = 1.05–1.56), ever-smokers (adjusted OR = 1.31, 95% CI = 1.03–1.67), ever-drinkers (adjusted OR = 1.34, 95% CI = 1.00–1.79) or BMI < 25.0 (adjusted OR = 1.52, 95% CI = 1.20–1.94) (Table 5). Moreover, while evaluating the strength of associations between mTOR SNPs and ESCC risk among subgroups with BMI < 25.0, the OR (1.52, 95% CI = 1.20–1.94) of combined risk genotypes was larger than the ORs (1.31, 95% CI = 1.03–1.67 for rs1057079; 1.39, 95% CI = 1.01–1.92 for rs1014261; 1.36, 95% CI = 1.07–1.73 for rs2295080) of any individual risk genotype (Table 5), indicating that there was likely a combined effect of these three SNPs. These findings suggested that the effect of each SNP is likely necessary but not sufficient, depending on the presence of other genetic variants.
Association of High-Order Interactions with ESCC Risk by Multiple Dimension Reduction (MDR) Analysis
To further investigate the existence of possible gene-environmental interaction in association with ESCC risk, high-order interactions assessed by using the MDR analysis was performed with inclusion of the five selected SNPs (i.e., rs3730089, rs3730090, rs2295080, rs1057079, and rs1014261) and five known risk factors (i.e., age, sex, smoking status, drinking status, and BMI). In the MDR analysis, BMI was the best one-factor model with the highest cross-validation consistency (CVC) and the lowest prediction error among all ten factors, indicating that BMI was the strongest risk factor for ESCC. Moreover, the ten-factor model had a maximum CVC and a minimum prediction error, with the prediction error being statistically significant (Table 6). Taken together, the ten-factor model showed a better prediction than the one-factor model and represented the best model to predict ESCC risk for this study population.
Table 6. MDR analysis for the prediction of ESCC risk with and without PIK3R1 & mTOR genotypes.
| Best interaction models | Cross-validation | Average prediction error | P a |
|---|---|---|---|
| 1 | 100/100 | 0.394 | < 0.0001 |
| 1, 2 | 100/100 | 0.394 | < 0.0001 |
| 1, 2, 3 | 98/100 | 0.386 | < 0.0001 |
| 1, 2, 3, 4 | 93/100 | 0.379 | < 0.0001 |
| 1, 2, 3, 4, 5 | 100/100 | 0.370 | < 0.0001 |
| 1, 2, 3, 4, 5, 6 | 100/100 | 0.367 | < 0.0001 |
| 1, 2, 3, 4, 5, 6, 7 | 92/100 | 0.358 | < 0.0001 |
| 1, 2, 3, 4, 5, 6, 7, 8 | 94/100 | 0.350 | < 0.0001 |
| 1, 2, 3, 4, 5, 6, 7, 8, 9 | 100/100 | 0.341 | < 0.0001 |
| 1, 2, 3, 4, 5, 6, 7, 8, 9, 10 | 100/100 | 0.340 | < 0.0001 |
MDR, multifactor dimensionality reduction.
aP value for 1000-fold permutation test.
The best model with maximum cross-validation consistency and minimum prediction error rate was in bold.
Labels: 1, BMI; 2, smoking status; 3, sex; 4, age; 5, drink status; 6, PIK3R1 rs3730089; 7, mTOR rs2295080; 8, PIK3R1 rs3730090; 9, mTOR rs1057079; 10, mTOR rs1064061.
mTOR Haplotypes and ESCC Risk
Since PIK3R1 and mTOR are located in different chromosomes, we only explored whether the haplotypes of three mTOR SNPs would influence ESCC risk. As presented in Table 7, seven mTOR haplotypes were identified. When the most frequent haplotype T-T-A was used as the reference group, three haplotypes, T-C-A, T-C-G, and G-T-A, were significantly associated with increased ESCC risk.
Table 7. Haplotype analysis for genotypes of mTOR and ESCC risk.
| Haplotype frequencies | ||||||||
|---|---|---|---|---|---|---|---|---|
| Cases (N = 2232) | Controls (N = 2234) | |||||||
| Haplotypes a | n | % | n | % | Crude OR (95% CI) | P | Adjusted OR (95% CI) | P a |
| T-T-A | 1700 | 76.23 | 1746 | 78.72 | 1.00 | 0.009 | 1.00 | |
| T-C-A | 23 | 1.03 | 9 | 0.41 | 2.63 (1.21–5.69) | 0.015 | 2.96 (1.32–6.67 | 0.023 |
| T-C-G | 19 | 0.85 | 6 | 0.27 | 3.25 (1.30–8.16) | 0.012 | 2.97 (1.16–7.60) | 0.024 |
| G-T-A | 40 | 1.79 | 18 | 0.81 | 2.28 (1.30–4.00) | 0.004 | 2.41 (1.34–4.37) | 0.003 |
| G-T-G | 33 | 1.48 | 27 | 1.22 | 1.26 (0.75–2.10) | 0.385 | 1.38 (0.81–2.35) | 0.234 |
| T-C-G | 265 | 11.88 | 271 | 12.22 | 1.01 (0.84–1.21) | 0.963 | 1.03 (0.85–1.24) | 0.795 |
| G-T-A | 150 | 6.73 | 141 | 6.36 | 1.09 (0.86–1.39) | 0.468 | 1.12 (0.87–1.43) | 0.380 |
aObtained in logistic regression models with adjustment for age, sex, smoking status and drinking status.
Gene-Gene and Gene-Environment Interactions
As presented in Table 8, logistic regression analyses identified significant gene-environment interactions of BMI with either mTOR rs1057079 or rs2295080 SNPs. Moreover, our results further showed significant gene-gene interactions of mTOR rs2295080 with either mTOR rs1057079 or rs1064261. Interestingly, the interaction between smoking status and drinking status were also noticeable (Table 8).
Table 8. Gene-gene and gene-environment interactions (logistic regression).
| P value | ||||||||
|---|---|---|---|---|---|---|---|---|
| rs3730089 | rs3730090 | rs2295080 | rs1057079 | rs1064261 | Drinking | Smoking | BMI | |
| rs3730089 | ||||||||
| rs3730090 | 0.8746 | |||||||
| rs2295080 | 0.3692 | 0.1886 | ||||||
| rs1057079 | 0.6180 | 0.1129 | <0.001 | |||||
| rs1064261 | 0.7348 | 0.5803 | 0.0246 | 0.7329 | ||||
| Drinking | 0.9182 | 0.2707 | 0.6398 | 0.6315 | 0.5853 | |||
| Smoking | 0.5558 | 0.2390 | 0.9725 | 0.5663 | 0.4052 | 0.0016 | ||
| BMI | 0.8435 | 0.1294 | 0.0046 | 0.001 | 0.0797 | 0.9281 | 0.3237 |
Meta-analysis for the Association between mTOR rs2295080 and Cancer Risk
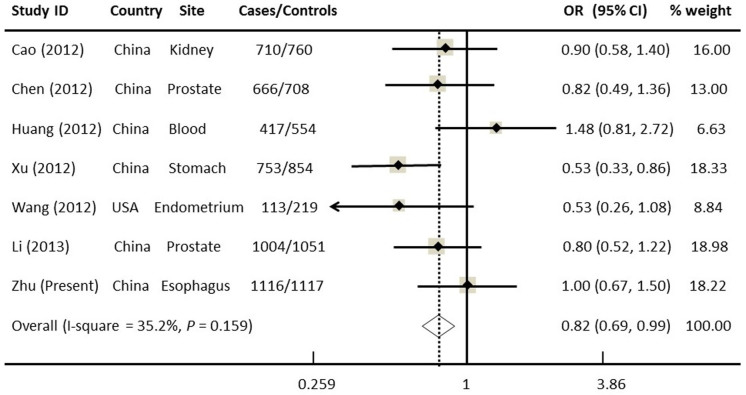
To date, six published studies have explored the association of mTOR rs2295080 with the risk of various cancers but yielded conflicting results8,9,10,13,25,26, whereas fewer studies on other mTOR SNPs have been published. To better evaluate such an association, we performed a meta-analysis with all published studies and our new data, leading to a total of 4772 cases and 5264 controls. When all the data were combined, the mTOR rs2295080 SNP appeared to be modestly protective and significantly associated with a decreased cancer risk under most of the genetic models tested without obvious among-study heterogeneity (homozygous: OR = 0.79, 95% CI = 0.66–0.95; heterozygous: OR = 0.88, 95% CI = 0.78–1.02, dominant: OR = 0.87, 95% CI = 0.80–0.94, recessive: OR = 0.82, 95% CI = 0.69–0.90) (Figure 1). Moreover, the shape of funnel plots seemed symmetrical and Egger's test showed no significance (data not shown), suggesting no publication bias.
Figure 1. Forest plot of overall cancer risk associated with mTOR rs2295080 (a genetic recessive model).
This was derived from a meta-analysis of seven relevant case-control studies. The OR and 95% CI of each study are plotted with a box and a horizontal line. Quadrangles represent pooled ORs and 95% CIs; Chi2, chi-square; df, degrees of freedom; I2, index of heterogeneity.
Discussion
Knowledge on the genetics of cancer can help health care professionals in providing high-risk individuals with better decisions on prevention and intervention strategies, such as cancer screening, early detection, and targeted therapy. Numerous studies have indicated that potentially functional SNPs in the important genes may confer host genetic susceptibility to cancer. Aberrant activation of the PI3K/PTEN/AKT/mTOR signaling pathway is a common event in a wide range of tumor types, suggesting a role for this pathway in carcinogenesis. In the present study, however, none of the studied SNPs in the PIKR1 and mTOR genes exhibited an association with ESCC risk. However, the combined analysis of these SNPs revealed significant risk associations with one, two, and three risk genotypes, compared with zero risk genotype. Actually, lack of the main effect of individual SNPs on cancer risk does not necessarily rule out these SNPs as etiologic factors, because these SNPs may have low penetrance in cancer susceptibility, compared with environmental and life style factors contributing to the risk. It is likely, however, that the relevant exposure, such as smoking, alcohol intake, and hormonal disorder, may interact with genetic factors27. This was the case in the present study. For example, our stratified analysis by BMI suggested a significantly increased ESCC risk associated with mTOR rs2295080, rs1057079, and rs1014261, individually, among subjects with BMI<25.0. When risk genotypes of these three mTOR SNPs were combined, subjects with ≥1 risk genotype exhibited an increased ESCC risk in the older participants (age>60), males, smokers, drinkers, or those with BMI<25.0, supporting gene-environment interactions on ESCC susceptibility.
Although few studies have investigated cancer risk associated with the SNPs we studied, some reported findings that are in line with ours. For example, one study observed an association between mTOR rs2295080 and a reduced risk of renal cancer in 710 cases and 760 controls25. In addition, mTOR rs2295080 was also shown to protect against gastric cancer risk in a Chinese population10. Functional analysis demonstrated that the rs2295080 variant G allele reduced transcriptional activity in both normal gastric mucosa epithelial cell lines (GES-1) and three different gastric cancer cell lines, compared with the wide-type T allele10. Moreover, mTOR mRNA expression levels in gastric cancer tissues with GT/GG genotypes were significantly lower than those with the TT genotype10, indicating that mTOR rs2295080 may decrease gastric cancer risk by affecting mTOR transcription.
Similarly, our meta-analysis of seven studies with 4772 cases and 5264 controls found that rs2295080 was significantly associated with a reduced cancer risk under homogenous (GG vs.TT) and recessive (GG vs.TT/TG) genetic models. However, due to the relatively small sample size in the current meta-analysis, large single studies with different cancer types and ethnic groups are needed to validate our findings. Moreover, additional meta-analyses with stratified analyses by cancer type are warranted to further determine the effect of this SNP on the risk of each specific cancer. For example, in contrast with other cancers, our study indicated that mTOR rs2295080 variant genotypes (GT/GG) were associated with an increased ESCC risk, and the association became significant among subjects with BMI<25.0. One explanation for the discrepancy in the association studies is that the function of rs2295080 SNP may be tissue-specific, but the biological function of this SNP should be further examined in different types of cancer in future studies. It was not possible to perform additional meta-analyses for the remaining studied SNPs, because very few published studies (<3) have investigated the associations between each of those SNPs and cancer risk.
One earlier U.S. study of 1574 colon cancer cases and 1940 healthy controls revealed a significant association between mTOR rs1057079 variant genotypes and increased risk of colon and rectal cancers, while this SNP was also associated with tumor harboring TP53 mutations22. The same study also found an association of PIK3CA rs7651265 SNP with rectal cancer risk22. However, in the present study, significant associations were only confined to certain subgroups exposed to smoking or alcohol, indicating the importance of considering other risk factors when analyzing genetic impact on cancer predisposition. As mentioned above, age and sex also appeared to modify the effect of genetic variants on ESCC risk. Although mechanisms remain unclear, ESCC is far more prevalent among males than females globally1. For example, a large-scale epidemiological study in North China observed that ESCC was more prevalent in males than females and in older populations rather than younger populations28. In the case that mTOR SNPs of interest only have some moderate effects, it is not unreasonable that the associations between the combination of these SNPs and ESCC risk were only detected in males and older groups who may have been exposed to environmental risk factors for a longer period. Therefore, it is plausible that our stratified analyses revealed an association between a combination of three mTOR SNPs and an increased ESCC risk among smokers and drinkers, since smoking and alcohol drinking are well-recognized strong risk factors for ESCC29. We also found that there was significant interaction between smoking and drinking in this study population. These finding were consistent with those reported by others, in which a meta-analysis suggested that tobacco and alcohol consumption synergistically increased the risk of developing ESCC30.
In the present study, significant mTOR SNP-related increases in ERCC risk were detected among subjects with BMI<25, but not among those with BMI ≥ 25.0, suggesting that BMI was a significant effect modifier of ERCC risk associated with mTOR SNPs. To support this finding, further MDR testing of the high-order interaction analysis consistently recognized BMI as a main risk factor for ESCC, which is consistent with the fact that body weight is reversely associated with risk of ESCC as well as with smoking, drinking, and nutrition. For example, previous studies conducted in both Western countries31,32 and China found that both poor nutrition and low BMI were associated with an increased ESCC risk. In the present study, we found a significant interaction between BMI and either mTOR rs1057079 or rs2295080 SNPs. Our results also showed that mTOR rs2295080 significantly interacted with either mTOR rs1057079 or rs1064261, suggesting that these SNPs of interest might collectively confer and modulate ESCC susceptibility.
There are some limitations in the present study. First, although age, sex, smoking, drinking, and BMI were considered, there were also a number of uncollected factors contributing to ESCC risk, including nutrition status; intake of hot beverages, fruits, vegetables; other genetic variations; and socioeconomic status. Failure to adequately control for these factors limited our ability to analyze gene-gene and gene-environment interactions. Second, the number of SNPs genotyped in the manuscript was also very limited, and some potentially functional SNPs in these two genes might be missed. Third, we performed multiple comparisons (single SNPs, SNPs combined, haplotypes, stratified by age, sex, BMI, etc.) in the present study, which may have led to chance findings (e.g., false positive findings). Therefore, these results should be interpreted with caution. Larger, more stringently designed studies are needed to validate our findings. Moreover, PIK3CA is a well-known oncogene, the activation of which has been implicated in various cancers. Failure to investigate the association of PIK3CA polymorphisms with ESCC risk is also a potential limitation.
In summary, we found that rs2295080, rs1057079, and rs1064261 SNPs in the mTOR gene may modify the host's genetic susceptibility to ESCC risk; however, these effects were largely dependent on other risk factors, i.e., BMI, age, sex, smoking and drinking status. Our results emphasize the importance of gene-environment interactions in determining the ESCC susceptibility, supporting the idea that the low-penetrant genetic effects of common SNPs on cancer predisposition may be fundamentally governed by the interplaying of SNPs and specific environmental exposures during the process of carcinogenesis.
Methods
Study Population
This case-control study was conducted at Fudan University, Shanghai Cancer Center. Briefly, the cases (n = 1116) were patients with newly-diagnosed and histopathologically confirmed ESCC from March 2009 to September 2011, who were all genetically unrelated Han Chinese and residents in Eastern China. The patients who had one or more of the following features were excluded: other types of cancer, primary tumors outside the esophagus, and cancers with unknown primary sites. Age and sex-matched, cancer-free controls (n = 1117) were selected from the Taizhou cohort33 following a procedure of matching with cases on age (± 5 years) and sex. A structured questionnaire was used to obtain the following information from each of the participants during personal interviews: demographic data and environmental exposure history such as age, sex, ethnicity, BMI, and tobacco and alcohol consumption. We defined individuals who smoked <100 cigarettes in their lifetime as “non-smokers”, while others were considered “smokers”. Moreover, subjects with alcohol consumption at least once a week for ≥1 year were defined as “drinkers”, and others were “non-drinkers”34. BMI was calculated from self-reported height and weight. We used a BMI value of 25 to divide subjects into two groups with BMI <25 and ≥2534; the World Health Organization (WHO) recommends body mass index (BMI, in kg/m2) ≥25 as a cutoff for categorization of overweightness35. Ninety percent of interviewed subjects consented to participate in this study by signing a written informed consent form. This research protocol was approved by the Institutional Review Board of FUSCC and the experiment on humans was performed in accordance with relevant guidelines and regulations.
SNP Selection and Genotyping
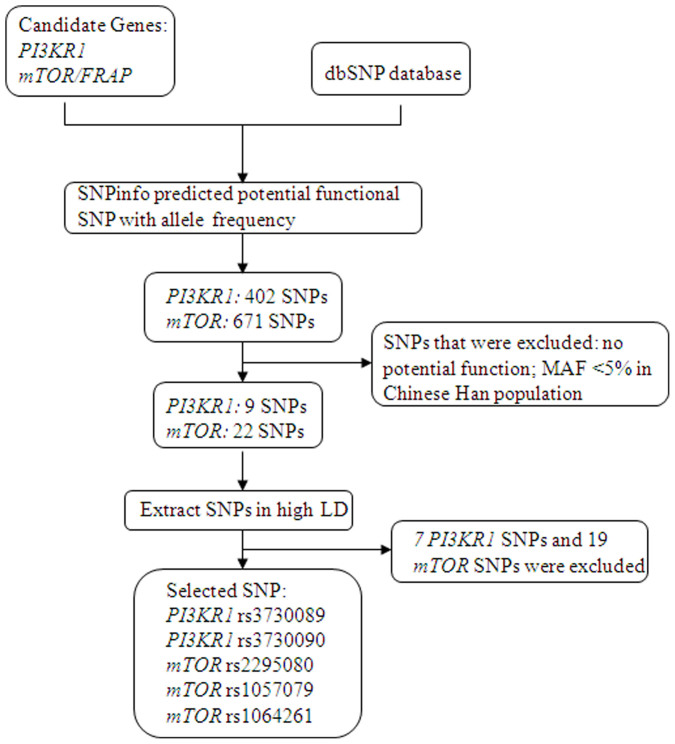
We searched the National Center for Biotechnology Information dbSNP database (http://www.ncbi.nlm.nih.gov/projects/SNP) for common, potentially functional SNPs in the PIK3CA, PIK3R1 and mTOR genes based on the following criteria: (1) located at exons, the 5′ near gene, 5′ untranslated regions (UTR), 3′ UTR, 3′ near gene and splice sites; (2) the minor allele frequency (MAF) ≥ 5% in Chinese Han population; (3) potentially functional SNPs as predicted by SNPinfo software (http://snpinfo.niehs.nih.gov/snpinfo/snpfunc.htm); (4) having low linkage disequilibrium (LD) with each other using an r2 threshold of <0.8; and (5) not investigated in the published genome-wide association studies (GWAS) of ESCC. No SNP in the PIK3CA gene met the criteria. Ultimately, we chose five SNPs (PIK3R1: rs3730089 and rs3730090; mTOR: rs1057079, rs1064261, and rs2295080) for the study. The SNP selection process is indicated in Figure 2.
Figure 2. The SNP selection flow chart.
We isolated genomic DNA from blood samples by using the Qiagen Blood DNA Mini Kit (Qiagen Inc., Valencia, CA) and performed the TaqMan assay for genotyping as reported previously36. Briefly, we labeled allele-specific probes for SNPs of interest with the fluorescent dyes VIC and FAM. During extension, the 5′- exonuclease activity of the Taq polymerase cleaves the fluorophore from the nonfluorescent quencher. By using the ABI 7900 HT Sequence Detection System (Applied Biosystems, Foster City, CA), we used a post-amplification allelic discrimination run on the machine to determine genotype based on the relative amount of fluorescence of VIC and FAM. Finally, we performed PCR reactions in a total reaction volume of 5 μl in 384-well plates. Individuals involved in genotyping were blind to patient status.
Statistical Methods
We used the χ2 test to assess differences in the frequency distributions of the selected demographic variables, risk factors, and genotypes of the selected SNPs between the cases and controls. We tested the Hardy–Weinberg equilibrium (HWE) for genotype distribution in controls by a goodness-of-fit χ2 test. Crude and adjusted ORs and their 95% CIs for the association of ESCC risk with selected SNPs were calculated using both univariate and multivariate logistic regression analyses with adjustment for co-variates including age, sex, smoking, drinking, and BMI, respectively. These co-variates were selected because of their importance in possible interaction with genetic factors, and were entered into the model at the same time as a group of categorical variables defined in Table 3. The P-value for multiplicative interaction between these selected SNPs and co-variates (age, sex, BMI, etc) was calculated by adding the product terms to the logistic regression model. A two-tailed P < 0.05 was used as the criterion of statistical significance. We also evaluated the associations in stratified analyses by age, sex, BMI, and smoking and drinking status. Four genetic models, 1) homozygous (WW vs. VV), 2) heterozygous (WW vs. WV), 3) dominant (WW vs. WV/VV), and 4) recessive (WW/WV vs. VV), were adopted for these analyses, with W and V representing wild and variant alleles of each SNP, respectively. In the present study, we defined the haplotype as a combination of rs2295080, rs1014261, and rs1057079 SNPs in the mTOR gene. The unphased genotype data were used to determine haplotype frequencies and individual haplotypes. Logistic regression analysis was performed to calculate ORs for the association of haplotypes with ESCC risk, while the haplotype of the highest frequency was considered as the reference group. Moreover, genotypes with one or two variant alleles of a SNP were referred to as risk genotype. Risk genotypes used for the calculation were PIK3R1 rs3730089 AG/AA, PIK3R1 rs3730090 CT/TT, mTOR rs2295080 GT/GG, mTOR rs1057079 CT/CC, mTOR rs1064261 AG/GG. All tests were two-sided, and a P< 0.05 was considered statistically significant. All statistical analyses were performed with SAS software (version 9.1; SAS Institute, Cary, NC). Furthermore, we used MDR software (V2.0 beta 8.2) to determine the possible high-order gene-gene or gene-environment interactions in the association37. Briefly, we tested 100-fold cross-validation and 1000-fold permutation under the assumption of no association. The best candidate interaction model was supposed to have the minimum average prediction error and the maximum CVC.
Finally, a meta-analysis was conducted to evaluate the association between mTOR rs2295080 SNP and cancer risk. Briefly, the search terms, inclusion, and exclusion criteria were defined in previous publications38,39. Relevant studies were searched from the MEDLINE, EMBASE and Scopus databases (Last Updated: July 25, 2014). The pooled ORs and 95% CIs were calculated under four different genetic models. Chi-square-based Q-test was used to check heterogeneity assumption. Either the fixed-effects model (the Mantel-Haenszel method) or the random-effects model (the DerSimonian and Laird method) was applied to calculate the pooled OR estimate, depending on the heterogeneity of study populations included in this meta-analysis36,37. Potential publication bias was estimated by the funnel plot and Egger's linear regression test. Sensitivity analysis was performed to assess the stability of the meta-analysis. All statistical tests were performed with STATA (version 11.0; Stata Corporation, College Station, TX). All P values were two-sided, and a P<0.05 was considered statistically significant.
Author Contributions
Conceived and designed the experiments: Q.Y.W., J.Q.X. Performed the experiments: J.H.Z., M.Y.W. Analyzed the data: J.H.Z., M.Y.W. Contributed reagents/materials/analysis tools: M.L.Z., J.H., J.C.W., J.L., X.F.W., Q.Y.W. Wrote the paper: J.H.Z., M.Y.W., J.Q.X., Q.Y.W.
Acknowledgments
This study was supported by the funds from the China Recruitment Program of Global Experts at Fudan University, the National Natural Science Foundation of China (81302101), and Ministry of Health (201002007). We would like to thank Ms. Katherine Zeph for her editing of the manuscript. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- Jemal A. et al. Global cancer statistics. CA Cancer J Clin 61, 69–90, 10.3322/caac.20107 (2011). [DOI] [PubMed] [Google Scholar]
- Tran G. D. et al. Prospective study of risk factors for esophageal and gastric cancers in the Linxian general population trial cohort in China. Int J Cancer 113, 456–463, 10.1002/ijc.20616 (2005). [DOI] [PubMed] [Google Scholar]
- Hiyama T., Yoshihara M., Tanaka S. & Chayama K. Genetic polymorphisms and esophageal cancer risk. Int J Cancer 121, 1643–1658, 10.1002/ijc.23044 (2007). [DOI] [PubMed] [Google Scholar]
- Xing D., Tan W. & Lin D. Genetic polymorphisms and susceptibility to esophageal cancer among Chinese population (review). Oncol Rep 10, 1615–1623 (2003). [PubMed] [Google Scholar]
- Talukdar F. R., Ghosh S. K., Laskar R. S. & Mondal R. Epigenetic, genetic and environmental interactions in esophageal squamous cell carcinoma from northeast India. PloS one 8, e60996, 10.1371/journal.pone.0060996 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okkenhaug K. Signaling by the phosphoinositide 3-kinase family in immune cells. Annu Rev Immunol 31, 675–704, 10.1146/annurev-immunol-032712-095946 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter C. L. et al. Purification and characterization of phosphoinositide 3-kinase from rat liver. J Biol Chem 265, 19704–19711 (1990). [PubMed] [Google Scholar]
- Chen J. et al. Genetic variations in a PTEN/AKT/mTOR axis and prostate cancer risk in a Chinese population. PloS one 7, e40817, 10.1371/journal.pone.0040817 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q. et al. Polymorphisms in the mTOR gene and risk of sporadic prostate cancer in an Eastern Chinese population. PloS one 8, e71968, 10.1371/journal.pone.0071968 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu M. et al. A polymorphism (rs2295080) in mTOR promoter region and its association with gastric cancer in a Chinese population. PloS one 8, e60080, 10.1371/journal.pone.0060080 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirashima K. et al. Aberrant activation of the mTOR pathway and anti-tumour effect of everolimus on oesophageal squamous cell carcinoma. Br J Cancer 106, 876–882, 10.1038/bjc.2012.36 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shigaki H. et al. PIK3CA mutation is associated with a favorable prognosis among patients with curatively resected esophageal squamous cell carcinoma. Clin Cancer Res 19, 2451–2459, 10.1158/1078-0432.CCR-12-3559 (2013). [DOI] [PubMed] [Google Scholar]
- Wang L. E. et al. Roles of genetic variants in the PI3K and RAS/RAF pathways in susceptibility to endometrial cancer and clinical outcomes. J Cancer Res Clin Oncol 138, 377–385, 10.1007/s00432-011-1103-0 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heist R. S. & Engelman J. A. SnapShot: non-small cell lung cancer. Cancer Cell 21, 448 e442 10.1016/j.ccr.2012.03.007 (2012). [DOI] [PubMed] [Google Scholar]
- Chappell W. H. et al. Ras/Raf/MEK/ERK and PI3K/PTEN/Akt/mTOR inhibitors: rationale and importance to inhibiting these pathways in human health. Oncotarget 2, 135–164 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vivanco I. & Sawyers C. L. The phosphatidylinositol 3-Kinase AKT pathway in human cancer. Nat Rev Cancer 2, 489–501, 10.1038/nrc839 (2002). [DOI] [PubMed] [Google Scholar]
- Fingar D. C., Salama S., Tsou C., Harlow E. & Blenis J. Mammalian cell size is controlled by mTOR and its downstream targets S6K1 and 4EBP1/eIF4E. Genes Dev 16, 1472–1487, 10.1101/gad.995802 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z. et al. A murine lung cancer co-clinical trial identifies genetic modifiers of therapeutic response. Nature 483, 613–617, 10.1038/nature10937 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garnett M. J. et al. Systematic identification of genomic markers of drug sensitivity in cancer cells. Nature 483, 570–575, 10.1038/nature11005 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeng C. H. et al. High-throughput genotyping in metastatic esophageal squamous cell carcinoma identifies phosphoinositide-3-kinase and BRAF mutations. PloS one 7, e41655, 10.1371/journal.pone.0041655 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boone J. et al. mTOR in squamous cell carcinoma of the oesophagus: a potential target for molecular therapy? J Clin Pathol 61, 909–913, 10.1136/jcp.2008.055772 (2008). [DOI] [PubMed] [Google Scholar]
- Slattery M. L. et al. Genetic variation in a metabolic signaling pathway and colon and rectal cancer risk: mTOR, PTEN, STK11, RPKAA1, PRKAG2, TSC1, TSC2, PI3K and Akt1. Carcinogenesis 31, 1604–1611, 10.1093/carcin/bgq142 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu M. L. et al. Polymorphisms in mTORC1 genes modulate risk of esophageal squamous cell carcinoma in eastern Chinese populations. J Thorac Oncol 8, 788–795, 10.1097/JTO.0b013e31828916c6 (2013). [DOI] [PubMed] [Google Scholar]
- He J. et al. Genetic variations of mTORC1 genes and risk of gastric cancer in an eastern chinese population. Mol Carcinogenesis 52 Suppl 1, 70–79, 10.1002/mc.22013 (2013). [DOI] [PubMed] [Google Scholar]
- Cao Q. et al. A functional variant in the MTOR promoter modulates its expression and is associated with renal cell cancer risk. PloS one 7, e50302, 10.1371/journal.pone.0050302 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L. et al. Association of genetic variations in mTOR with risk of childhood acute lymphoblastic leukemia in a Chinese population. Leuk Lymphoma 53, 947–951, 10.3109/10428194.2011.628062 (2012). [DOI] [PubMed] [Google Scholar]
- Weiss J. M., Goode E. L., Ladiges W. C. & Ulrich C. M. Polymorphic variation in hOGG1 and risk of cancer: a review of the functional and epidemiologic literature. Mol Carcinogenesis 42, 127–141, 10.1002/mc.20067 (2005). [DOI] [PubMed] [Google Scholar]
- Feng X. S. et al. Prevalence and age, gender and geographical area distribution of esophageal squamous cell carcinomas in North China from 1985 to 2006. Asian Pac J Cancer Prev 15, 1981–1987 (2014). [DOI] [PubMed] [Google Scholar]
- Holmes R. S. & Vaughan T. L. Epidemiology and pathogenesis of esophageal cancer. Seminars in radiation oncology 17, 2–9, 10.1016/j.semradonc.2006.09.003 (2007). [DOI] [PubMed] [Google Scholar]
- Prabhu A., Obi K. O. & Rubenstein J. H. The synergistic effects of alcohol and tobacco consumption on the risk of esophageal squamous cell carcinoma: a meta-analysis. Am J Gastroenterol 109, 822–827, 10.1038/ajg.2014.71 (2014). [DOI] [PubMed] [Google Scholar]
- Lahmann P. H. et al. Body mass index, long-term weight change, and esophageal squamous cell carcinoma: is the inverse association modified by smoking status? Cancer 118, 1901–1909, 10.1002/cncr.26455 (2012). [DOI] [PubMed] [Google Scholar]
- Chow W. H. et al. Body mass index and risk of adenocarcinomas of the esophagus and gastric cardia. J Natl Cancer Inst 90, 150–155 (1998). [DOI] [PubMed] [Google Scholar]
- Wang X. et al. Rationales, design and recruitment of the Taizhou Longitudinal Study. BMC public health 9, 223, 10.1186/1471-2458-9-223 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu M. L. et al. Polymorphisms in the ERCC5 gene and risk of esophageal squamous cell carcinoma (ESCC) in Eastern Chinese populations. PloS one 7, e41500, 10.1371/journal.pone.0041500 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jih J. et al. Using appropriate body mass index cut points for overweight and obesity among Asian Americans. Prev Med 65,1–6 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- He J. et al. Polymorphisms in the XPG gene and risk of gastric cancer in Chinese populations. Hum Genet 131, 1235–1244, 10.1007/s00439-012-1152-8 (2012). [DOI] [PubMed] [Google Scholar]
- Hahn L. W., Ritchie M. D. & Moore J. H. Multifactor dimensionality reduction software for detecting gene-gene and gene-environment interactions. Bioinformatics 19, 376–382 (2003). [DOI] [PubMed] [Google Scholar]
- Zhu J. et al. Association studies of ERCC1 polymorphisms with lung cancer susceptibility: a systematic review and meta-analysis. PloS one 9, e97616, 10.1371/journal.pone.0097616 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y. Q. et al. The association between the polymorphisms of TNF-alpha and non-Hodgkin lymphoma: a meta-analysis. Tumour Biol 35, 12509–12517 10.1007/s13277-014-2569-6 (2014). [DOI] [PubMed] [Google Scholar]