Abstract
MicroRNAs (miRNAs) have emerged as novel post-transcriptional regulators of gene expression. These short non-coding RNAs are involved in diverse biological processes and their dysregulation is often observed under diseased conditions. Therefore, miRNAs hold great potential as clinical biomarkers of physiological and pathological states and extensive efforts are underway to develop efficient approaches for their detection. We review recent advances and discuss the promises and pitfalls of emerging methods of miRNA profiling and detection.
Keywords: biomarkers, clinical applications, detection, microRNAs, miRNAs
MicroRNAs (miRNAs) are small noncoding 19–25 base pair RNAs that act as post-transcriptional regulators of gene expression (Bhardwaj et al. 2010, Janga and Vallabhaneni 2011). A single miRNA can target multiple mRNAs and conversely, a single mRNA can be targeted by several different miRNAs (Pillai 2005, Sethupathy et al. 2006). To date, several hundred miRNAs have been identified in humans and their number continues to increase, which indicates that miRNAs are a major component of the gene regulatory system (Bhardwaj et al. 2010). miRNAs are evolutionarily conserved regulatory molecules and evidence is accumulating that suggests their involvement in a variety of normal and pathophysiological processes (Lewis et al. 2005, O’Driscoll 2006). miRNAs play significant roles in diseases including diabetes (Poy et al. 2004), heart diseases (Cheng et al. 2007, Van et al. 2006), neurological disorders such as fragile X syndrome (Caudy et al. 2002), Alzheimer’s disease (Hebert and De 2009) and many human malignancies (Ventura and Jacks 2009, Visone and Croce 2009). miRNA expression is developmentally and physiologically regulated, and an aberrant expression of miRNAs is often detected under pathological conditions (Cheng et al. 2007, Noh et al. 2009, Pauley et al. 2008, Roldo et al. 2006). Because of the functions of different miRNAs and their differential expression patterns under disease conditions, miRNAs are emerging as a novel class of clinical biomarkers for diagnosis, prognosis, disease classification, and monitoring of progression and response to therapeutic interventions. Therefore, several novel methods have been developed and many are still emerging to achieve better sensitivity, specificity and high throughput under clinical settings. We review here some of the findings that suggest the clinical potential of miRNAs as biomarkers and discuss emerging methods for miRNA profiling.
miRNAs as clinical biomarkers
Biomarkers are specific features that can be used as indicators of biological processes. With recent scientific and technological advancements, newer biomarkers have been identified that are more sensitive, more specific and whose detection is feasible in a clinical setting. There is much in the literature that indicates the potential of miRNAs as clinical biomarkers of disease progression, differential diagnosis, therapy response and molecular typing (Andorfer et al. 2011, Cacchiarelli et al. 2011, Saito et al. 2011, Scapoli et al. 2010, Grizzle et al. in press, Manne et al. 2010) (Table 1). miRNAs have the advantage of greater stability compared to other nucleotide based markers. miRNAs can be detected easily in tissues, body fluids and even formalin fixed, paraffin embedded (FFPE) tissue samples (Cortez and Calin 2009, Nelson et al. 2006a, Bovell et al. 2012). For example, elevated levels of miR-142-3p were detected in the sera of patients with systemic sclerosis and these levels were correlated with the severity of the disease (Makino et al. 2011). Enhanced expression of miRNAs, such as miR-155, miR-146a, miR-132 and miR-16, also has been detected in peripheral blood mononuclear cells of patients suffering from rheumatoid arthritis (Pauley et al. 2008). Nakasa et al. (2008) reported increased expression of miR-146a in different cells and tissues of rheumatoid arthritis patients and suggested that it is associated with the severity of the disease. Kong et al. (2011) investigated the expression of several miRNAs in newly diagnosed type 2 diabetic (nT2D) and pre-diabetic patients and observed that seven miRNAs, i.e., miR-9, miR-29a, miR-30d, miR-34a, miR-124a, miR-146a and miR-375, were elevated significantly in nT2D compared to pre-diabetes. Kapsimali et al. (2007) used miRNA profiling to characterize neural cells, which might facilitate the prediction of likely modes of miRNA function in the central nervous system. Also, a specific miRNA expression was observed during the early phase of acute myocardial infarction (Dong et al. 2009).
Table 1.
List of selected miRNAs and their clinical association
| miRNA | Disease | Clinical association | References |
|---|---|---|---|
| miR-142-3p (↑) | Systemic sclerosis | Severity of disease | Makino et al. 2011 |
| miR-155 (↑), miR-146a (↑), miR-132 (↑), and miR-16(↑) | Rheumatoid arthritis | Pathogenesis of disease | Pauley et al. 2008 |
| miR-9 (↑), miR-29a (↑), miR-30d (↑), miR-34a (↑), miR-124a (↑), miR-146a (↑) and miR-375 (↑) | Newly diagnosed type 2 diabetic | Pathogenesis of disease | Kong et al. 2011 |
| miR-103 (↑), miR-107 (↑), miR-155(↓), miR-21(↑) | Pancreatic cancer | Distinguish tumors with different clinical behavior | Roldo et al. 2006 |
| miR-150 (↓) | Pancreatic cancer | Pathogenesis of disease | Srivastava et al. 2011 |
| miR-141 (↑) | Prostate cancer | Distinguish cancer patients from healthy ones | Mitchell et al. 2008 |
| miR-146a (↓) | Prostate cancer | Development of hormone refractory prostate cancer | Lin et al. 2008 |
| miR-10b (↑) | Prostate cancer | Biochemical relapse of disease | Fendler et al. 2011 |
| miR-10b (↑) | Pancreatic cancer | Aggresive behavior of disease and poor survival of patient | Nakata et al. 2011 |
| miR-451 (↓) | Gastric cancer | Survival of patient | Bandres et al. 2009 |
| miR-34a (↓) | Chronic lymphocytic leukemia | Drug resistance | Zenz et al. 2008 |
| miR-196a (↓) | Barrett’s esophageal metaplasia-dysplasia-invasive adenocarcinoma | Disease progression | Maru et al. 2009 |
| miR-122 (↓) | Hepatocellular carcinoma | Metastatic behavior of disease | Coulouarn et al. 2009 |
| miR-145 (↓), miR-21(↑), miR-129 (↑) | Bladder cancer | Disease progression | Dyrskjot et al. 2009 |
| miR-221 (↓) | Prostate cancer | Aggressive and metastatic behavior and disease | Spahn et al. 2010 |
(↑) Up regulated; (↓) Down regulated
Several groups have exploited the potential of using miRNAs for discriminating cancer from benign diseases and normal conditions (Bloomston et al. 2007, Osaki et al. 2008, Roldo et al. 2006). Correlation of miRNA expression with cancer type, tumor stage and response to drug treatments also has been reported (Lu et al. 2005, Nakajima et al. 2006, Nass et al. 2009, Zenz et al. 2008). Roldo et al. (2006) suggested that miRNA expression profiling may be useful for distinguishing pancreatic tumors with different clinical behavior. These investigators reported that expression of miR-103 and miR-107 associated with lack of expression of miR-155 could discriminate pancreatic tumors from normal tissue, while a set of ten other miRNAs could discriminate endocrine from acinar tumors. Furthermore, they showed that miR-204 was expressed primarily in insulinomas and the overexpression of miR-21 was correlated strongly with high Ki67 proliferation index and occurrence of liver metastasis. We observed recently that miR-150 was expressed differentially and exhibited significant down-regulation in pancreatic cancer tissues compared to normal pancreas (Srivastava et al. 2011). In a study by Bloomston et al. (2007), expression profiling of miRNAs in pancreatic cancer, normal pancreas and chronic pancreatitis demonstrated overexpression of 21 miRNAs and down-regulation of 4 miRNAs in pancreatic cancer, which suggests their utility for distinguishing pancreatic cancer from benign pancreatic tissue. To study the role of miRNAs in the progression of prostate cancer, Lin et al. (2008) profiled miRNA in both prostate cancer cells and human prostate cancerous tissues. They reported that down-regulation of miR-146a was associated with the development of hormone-refractory prostate cancer. Mitchell et al. (2008) determined the level of miR-141 in serum from patients with prostate cancer and reported that the miR-141 level was higher in the serum of cancer patients; thus differential miRNA expression may be useful as potential peripheral blood based biomarkers. Fendler et al. (2011) observed that miR-10b was expressed differentially in primary prostate cancers and normal adjacent tissues obtained by radical prostatectomy, and therefore might have implications in biochemical relapse of prostate cancer.
The potential of miRNAs for assessing disease aggressiveness and patient survival, and for monitoring therapeutic responses also has been investigated (Hummel et al. 2010, Nakajima et al. 2006, Segura et al. 2010, Yu et al. 2008). Nakata et al. (2011) correlated the expression of miR-10b with severity of pancreatic cancer. They observed that the expression level of miR-10b was significantly greater in pancreatic cancer cells than in normal pancreatic ductal cells and overexpression of miR-10b was significantly associated with poor prognosis of patients with pancreatic cancer. Bandres et al. (2009) demonstrated that down-regulation of miR-451 was linked with prognosis in patients with gastric cancer. In chronic lymphocytic leukemia, reduced expression of miR-34a was correlated with resistance to chemotherapy (Zenz et al. 2008). Maru et al. (2009) identified miR-196a as a potential marker of progression for Barrett’s esophageal metaplasia-dysplasia-invasive adenocarcinoma sequence. In hepatocellular carcinoma, loss of miR-122 expression in tumor cells was correlated with suppression of the hepatic phenotype and development of metastatic properties (Coulouarn et al. 2009). Dyrskjot et al. (2009) identified miR-129 as a potential miRNA for predicting disease progression in bladder cancer. In a large cohort study, miR-221 was determined to be down-regulated progressively in aggressive forms of prostate cancer and also was associated with recurrence of prostate cancer (Spahn et al. 2010). All of these studies strongly suggest that miRNAs can serve as useful biomarkers for multiple clinical applications (Fig. 1).
Fig. 1.
Potential applications of miRNAs as clinical biomarkers. Based on our current knowledge, several clinical applications of miRNAs are proposed as biomarkers of diagnosis, prognosis, therapy response and follow-up, and molecular classification of the disease.
Emerging approaches for miRNA profiling and detection
Conventional methods
Several approaches that have been used for detecting nucleic acids also have been employed for miRNA detection including Northern blotting, reverse-transcription-polymerase chain reaction, and in situ hybridization. We discuss these assays below with the recent modifications that have been made to improve their analytical power and applicability.
Northern blotting
Northern blotting analysis as a technique for miRNA analyses has the advantage that it does not require special equipment and it can be done in normal laboratory settings. Moreover, it can be used to detect both mature and precursor forms of any miRNA. The procedure involves separation of small RNAs by gel electrophoresis, transfer from the gel to a membrane, cross-linking of resolved RNAs by fixation followed by hybridization with labeled probes. One of the major drawbacks of this technique is its low sensitivity. Consequently, the method requires a large quantity of RNA for detection of miRNA, which in most cases, especially for clinical samples, is not possible. Another drawback is that it can detect only one miRNA at a time. Several improvements have been made in traditional Northern blotting protocols to overcome some of these limitations. One of the major improvements is the use of locked nucleic acid (LNA) probes for Northern blot detection, which can increase the sensitivity of the assay by about 10-fold (Valoczi et al. 2004, Varallyay et al. 2007). In addition, Ramkissoon et al. (2006) have developed an improved method that uses digoxigenin labeled RNA probes for miRNA detection. Incorporation of digoxigenin increases the shelf-life of the probes and eliminates the radioactivity hazard. This assay remains time-consuming, however, and requires larger samples, so it is not suitable for clinical settings.
Reverse transcription-polymerase chain reaction (RT-PCR)
Polymerase chain reaction (PCR) based methods have the advantage of being highly sensitive. With technical advancements, it is now possible to detect low copy numbers of both the precursor and the mature forms of miRNAs with great sensitivity and specificity (Chen et al. 2005, Jiang et al. 2005, Schmittgen et al. 2004). Schmittgen et al. (2004) were the first to determine miRNA expression by RT-PCR, but the method could not detect the mature form of the miRNAs owing to their small size. Raymond et al. (2005) subsequently designed a primer for a reverse transcription (RT) primer complementary to the 3′ end of the mature miRNA that also contained a 5′ tail sequence. This created a template that was longer than the mature miRNA for which specific primers could be designed for real time PCR detection (a miRNA specific forward primer and a universal reverse primer recognizing the identical tail sequence). Shi and Chiang (2005) modified this method by adding an artificial poly(A) tail to the miRNAs, which allowed the use of an anchored oligo(dT) RT primer with a 5′ tail. This method exhibited greater sensitivity and could detect miRNA with only 100 pg of total RNA. Chen et al. (2005) developed a somewhat different approach. These investigators observed that using stem loop RT primers with a 3′ adapter specific for each miRNA worked better than the linear RT primer and provided better specificity and sensitivity than linear primers. (Chen et al. 2005). Tang et al. (2006) enhanced the assay further by combining the use of a stem loop RT primer with a single tube reverse transcription followed by a single tube pre-PCR. This technique proved very sensitive and could detect as little as 220 miRNAs from the RNA content of a single embryonic stem cell. Thus, the problems related to the short length of mature miRNAs have been overcome and real time PCR today represents one of the most sensitive and convenient miRNA detection methods.
In situ hybridization
In situ hybridization (ISH) is widely used for miRNA detection. The merit of this technique is that it can be used to determine the subcellular distribution of miRNA (Kloosterman et al. 2006, Nelson et al. 2006b, Wienholds et al. 2005). However, conventional ISH is not efficient for miRNA detection in tissues. The major challenge is fixing small RNAs to prevent their loss during hybridization or washing procedures so that inconsistent or false negative results can be avoided. Furthermore, it is more tedious compared to other miRNA profiling techniques and also is low throughput. Despite these drawbacks, the use of locked nucleic acid (LNA) probes (Kauppinen et al. 2006, Valoczi et al. 2004) enhanced the specificity and sensitivity of miRNA detection by ISH and the technique has been employed in zebra fish, mice, and frog embryos (Kloosterman et al. 2006, Wienholds et al. 2005). Nelson et al. (2006b) modified this method using the miRNA microarray platform RAKE (RNA-primed, array based, Klenow Enzyme) together with LNA based in situ hybridization. This enabled detection of miRNA in FFPE tissues from human brain and oligodendroglia tumors. Deo et al. (2006) improved the specificity further by using fluorescein hapten-linked RNA oligonucleotide probes and modified washing conditions. Another improvement, which combined the use of LNA with tyramide signal amplification, resulted in rapid and efficient detection of miRNAs in frozen tissues (Stenvang et al. 2008). This method obviates the need for microdissecting malignant cells when these are mixed with inflammatory and stromal cells in specific tumors.
Emerging methods
With technological advancement and increasing interest, novel analytical approaches for profiling miRNA expression continue to emerge. Many of these evolving methods are based on nanotechnology principles and address the limitations of conventional methods for clinical applicability by minimizing the sample size required, and increasing sensitivity, precision and dynamic range, while at the same time being fast and less labor-intensive.
Invader assay
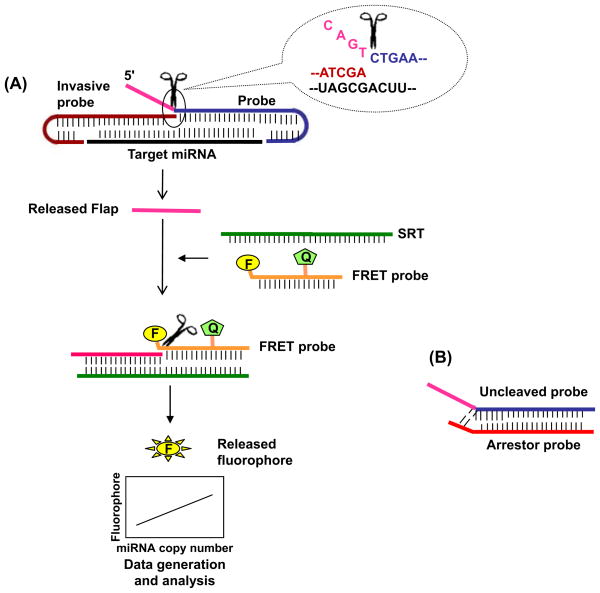
The invader assay developed by Allawi et al. (2004) can discriminate between precursor and mature miRNAs as well as between the target miRNA and its closely related isotypes. The assay combines enzyme based cleavage and fluorescence based reactions in the same tube and is based on a principle similar to that described earlier for the invader mRNA assay (De et al. 2002, Eis et al. 2001). It uses two different oligonucleotide probes: one “probe oligonucleotide” and an “invasive oligonucleotide”; the probes are modified by including 2′-O-methylated stem loops at their 5′ and 3′ ends, respectively, to stabilize hybrids containing the target RNA stacked between the two short base pair duplexes. The invasive oligonucleotide has a 3′ terminal overlapping nucleotide that is not complementary to the miRNA sequence. The probe oligonucleotide has a 5′ flap sequence that is noncomplementary to the miRNA sequence. During assay, both of these probes hybridize to the respective complementary sites of the miRNA, which leaves a 5′ flap in the probe oligonucleotide (Fig. 2A). A structure-specific 5′ nuclease cleaves the 5′ flap sequence from the probe oligonucleotide, which then acts as an invasive oligonucleotide for the 3′ end of a secondary reaction template (SRT). A new probe that contains a fluorophore and quencher hybridizes to the 5′ end of the SRT in a way that slightly overlaps the flap sequence. This promotes a second cleavage in a similar way by 5′ nuclease and separates the fluorophore from its quencher, thus generating a fluorescent signal that can be detected (Fig. 2A). Sensitivity of the invader assay can be increased by adding an arrestor oligonucleotide probe that maintains low background. The arrestor probe hybridizes with the uncleaved probe, thus not allowing it to participate in the secondary reaction (Fig. 2B). This assay is rapid and has high-throughput efficiency. Furthermore, it is extremely sensitive, which allows detection of miRNAs from 50 ng of total RNA (Allawi et al. 2004).
Fig. 2.
Invader assay. A) The assay involves two reactions. In the primary reaction, invasive and probe oligonucleotides with hairpin overhangs for enhancing stability are bound to the target miRNA. Hybridization results in the formation of a 5′ overlap-flap structure from the probe oligonucleotide, which serves as a substrate for the structure-specific 5′ nuclease, which causes release of the 5′ flap. In the secondary reaction, a FRET oligonucleotide labeled with a fluorophore (F) and a quencher (Q) is added to a secondary reaction template (SRT). The released 5′ flap from the primary reaction acts as an invasive probe in the secondary reaction, which leads to the formation of a second fluorophore-conjugated overlap-flap structure. Cleavage of this 5′ flap releases the fluorophore to generate a signal that can be quantified. B) An arrestor oligonucleotide complementary to the probe oligonucleotide is used in the secondary reaction that binds to the uncleaved probe to avoid its interference in the binding of the FRET probe to SRT, which reduces the background noise.
Bioluminescence based methods
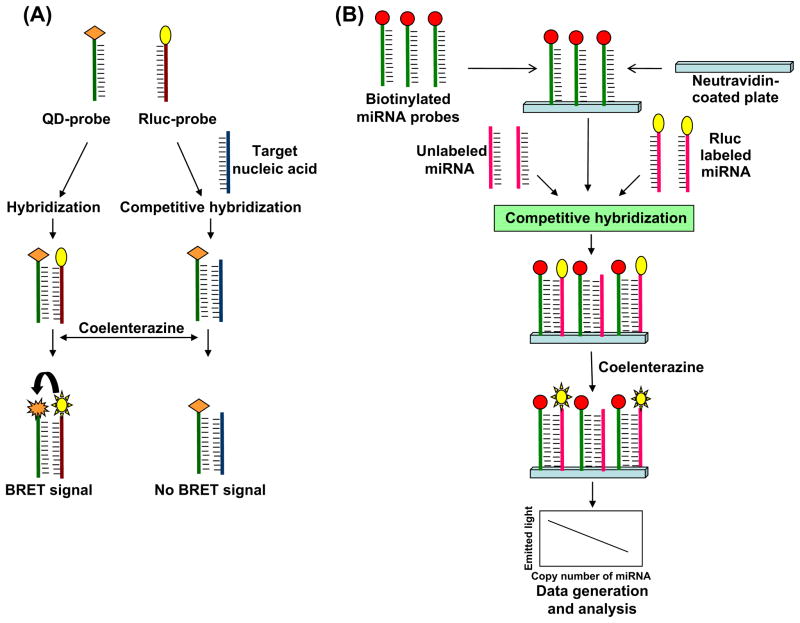
Cissell et al. (2008a) used a bioluminescence resonance energy transfer (BRET) based method for detecting nucleic acids. BRET is a modified form of Förster resonance energy transfer (FRET) in which a bioluminescent molecule is substituted for the donor fluorophore. Although this method was applied first by Xu et al. (1999), these investigators appropriately point out that some of the original BRET observations were made during the isolation of aequorin by Shimomura et al. (1962), because aequorin-GFP compose a BRET pair (Johnson et al. 1962). For nucleic acid assays, Cissell et al. (2008a) used Renilla luciferase (Rluc), a protein that emits bioluminescence in the presence of the substrate, i.e., coelenterazine, as the bioluminescent donor and a red emitting quantum dot (QD705) as an acceptor molecule. Rluc has a bioluminescence emission peak near 480 nm (Xu et al. 1999), while QD705 has the characteristic fluorescent nanocrystal absorption spectrum with high absorption from 300–490 nm (Invitrogen, Life Technologies, Grand Island, NY). QD705 emission occurs at 702 nm. Thus, Rluc-QD705 forms a BRET pair with significant spectral overlap and a very large Stokes’ shift; the former provides high energy transfer efficiency and the latter facilitates spectral separation of Rluc and QD705 emission. Because resonance energy transfer efficiency is dependent to the 6 th power on the donor-emitter intermolecular distance, the BRET assay is highly sensitive to coupling of Rluc to QD705.
The BRET nucleic acid assay uses an amine-modified oligonucleotide probe specific to the target nucleic acid and is labeled with a carboxy-activated QD705. A separate, partially complementary, competitive probe also was created by the addition of a thiol group and labeled with Rluc. The QD705 and Rluc probes hybridize with each other upon mixing and generate a BRET signal at 702 nm caused by the energy transfer from Rluc to QD705 in the presence of coelenterazine. The presence of the nucleic acid target decreases the BRET signal by competitive hybridization with the QD705 probe, which inhibits Rluc-QD705 proximity (Fig. 3A) (Cissell et al. 2008a). One possible limitation of this technique is the reduced emission intensity of bioluminescence assays compared to fluorescence assays. Therefore, a high sensitivity detection system is necessary for this technique and high speed assays may be difficult to implement.
Fig. 3.
Bioluminescence based assay. A) Quantum dot labeled oligonucleotide probe (QD-probe) and Rluc labeled oligonucleotide probe (Rluc-probe) are used in BRET based detection. In the absence of target nucleic acid, QD-probes hybridize with Rluc-probe to generate a signal after addition of coelenterazine. When the target is present, the BRET signal is decreased owing to the competitive inhibition of Rluc hybridization with QD-probe. B) In a modified assay, biotinylated anti-miRNA probes first are immobilized on neutravidin-coated microtiter plate, then Rluc-labeled target miRNAs and unlabeled synthetic target miRNAs are added. Both sample (Rluc labeled) and synthetic (unlabeled) miRNAs compete with each other during hybridization with the immobilized probes. Upon addition of coelenterazine, a decrease in signal intensity is measured as an estimate of target miRNA levels.
Cissell et al. (2008b) also modified the earlier BRET method by developing a competitive oligonucleotide hybridization assay, which uses only Rluc as a direct label for miRNA detection. This method overcomes the need for a dual reporter assay, which simplifies the detection requirements. The method requires a combination of unlabeled synthetic target miRNAs, Rluc-labeled target miRNAs and a biotinylated anti-miRNA probe that is immobilized on a neutravidin-coated microtiter plate (Fig. 3B). The unlabeled miRNA and Rluc-labeled miRNA compete to hybridize with the immobilized biotinylated anti-miRNA probe. After hybridization, unbound miRNAs are removed by washing and Rluc substrate, i.e., coelenterazine, is added. The resulting bioluminescence may be measured by standard radiometric or spectroscopic techniques. The decrease in bioluminescence emission is calculated and related to the concentration of target miRNA using a calibration curve (Fig. 3B). Therefore, this assay permits a simplified detector configuration with very high sensitivity, as little as 1 fmol detection limit, and it can be used for miRNA detection and quantification from crude samples such as blood and saliva. The assay can be performed using a 96- or 384-well plate, which offers potentially high throughput capabilities that could prove extremely useful in clinical settings.
Bead based flow cytometry
Yang et al. (2001) developed a flow cytometry based “beads array for detection of gene expression” (BADGE) method for analysis of expressions of multiple genes. These investigators used fluorescent microspheres conjugated to capture probes that are specific for target cRNA that had been labeled by incorporating biotinylated UTP. The mixture of different beads conjugated with capture probes was allowed to hybridize with the labeled target sample followed by addition of streptavidin-conjugated R-phycoerythrin to allow its binding to the microspheres/capture probe/cRNA complex. Subsequently, flow cytometry was performed using the Luminex 100 system to detect the amount of a specific cRNA in the sample (Yang et al. 2001). This is an inexpensive, rapid and high throughput method that can detect large numbers of targets in a single experiment. Lu et al. (2005) later modified the BADGE method to enable accurate and inexpensive miRNA profiling in which beads conjugated to specific probes were labeled with a mixture of two different fluorescent dyes. These investigators first attached adaptor molecules at both the 5′ and 3′ ends of miRNA and RT was performed using primers complementary to the adaptors. Following RT, miRNA amplification was performed using biotinylated primers, then hybridized to the capture probes complementary to the miRNAs. The carboxylated polystyrene beads permeated with variable mixtures of two different fluorescent dyes can generate up to 100 colors; each color represents a single miRNA. After hybridization, these conjugated beads were stained with streptavidin-phycoerythrin and analyzed using a flow cytometer. The bead color measured by flow cytometry represented a particular miRNA, and intensity of phycoerythrin indicated the abundance of miRNA (Lu et al. 2005).
Fluorescence correlation spectroscopy (FCS)
FCS estimates the concentration fluctuations of fluorescent particles in solution by measuring small fluctuations in fluorescence intensity over time (Magde et al. 1974). FCS measurements typically are made within a small interrogation volume that is determined by the spot size of a laser beam in a confocal microscope. Because of the great spatial resolution, FCS measurements can be used to measure subcellular diffusion and reaction rates in living cells (Kim et al. 2007). Therefore, FCS is an accurate and sensitive approach to studying molecular interactions under physiological conditions (Hui et al. 2010). Using this method, the fluorescence emitted from a relatively small number of fluorophores within the interrogation volume can be measured over time. These fluctuations in fluorescence are analyzed using autocorrelation techniques, the results of which subsequently are fitted to a model that describes the diffusion and reaction characteristics of the system (Kim et al. 2007). This technique has been used to quantify properties of single molecules in solution. By using a multicolor confocal system, it is possible to analyze the interactions of many molecules, in which case the technique commonly is referred to as fluorescence cross-correlation spectroscopy (Bacia et al. 2006).
Another flow cytometry based assay for miRNA measurement has been described by Neely et al. (2006). In this approach, single molecules were counted by employing a microfluidics system, laser excitation and avalanche photodiode detection in an optical configuration similar to that employed by many flow cytometers. Two spectrally distinguishable fluorescent probes were used, each complementary to half the target sequence of the miRNA of interest. A quencher containing an oligonucleotide sequence complementary to the unbound labeled probes also was used to decrease the background noise. Two-color laser excitation (532 and 633 nm) was used with laser foci staggered along the length of the flow channel to allow sequential excitation of each fluorophore. In addition, foci from two lasers at identical wavelength were staggered along the length of the flow channel. Triggering was performed post-acquisition based on the cross-correlation of fluorescence emission owing to the excitation of these two identical wavelength lasers. By restricting analysis to events that displayed a high cross-correlation coefficient, the investigators were able to reduce greatly the noise of the detection system to achieve great specificity. Based on this analysis, the investigators were able to detect coincident events between the two fluorescent probes as measured at the two locations corresponding to the foci of the 532 and 633 nm lasers.
One merit of this assay is that it does not require reverse transcription or amplification; rather it is based simply on hybridization of two spectrally distinguishable fluorescent oligonucleotide LNA probes. A second merit is that fluorophore signals from hybridized probes are counted directly on a single molecule level, which yields a detection limit of 500 fmol miRNA. This method has great potential owing to its great sensitivity, speed, and ability to differentiate single base mismatches. Using this technique, Neely et al. (2006) measured the expression profile of 45 different miRNA targets in 16 different human tissues from very small amounts of total RNA.
One potential disadvantage of this assay is that it uses a highly customized combination of micro-fluidics and single photon counting detection that currently is not available as a stand alone system. It may be possible to translate this approach to single FCS systems, but this likely will introduce a great reduction in sensitivity and a great increase in noise, because single molecule counting will not be measurable directly as in the microfluidic flow channel used by Neely et al. (2006). Finally, although this approach did not require the full auto-correlation analysis and fitting to physical models that is employed in traditional FCS techniques (Kim et al. 2007), a basic understanding of molecular transport and reaction models still is encouraged when considering this approach (Kim et al. 2007, Magde et al. 1974).
Surface-enhanced Raman spectroscopy (SERS)
Raman spectroscopy is a highly specific method for molecular characterization, because the inelastic light scatter signal that is generated during Raman scattering depends on the vibrational states of the molecule. Raman spectra typically provide many narrow peaks that can be used to infer much about the structure of a molecule. These narrow peaks also can be used to differentiate similar molecules based on statistical clustering or decomposition techniques, often with great specificity. Unfortunately, traditional Raman scattering produces very weak signals, roughly 10 10 times weaker than fluorescence signals. Because of this, traditional Raman spectroscopy has been limited in terms of sensitivity and signal acquisition times. SERS is a surface-sensitive technique that enhances the Raman scattering signal by performing measurements in the presence of a rough metallic surface. The interaction of laser illumination with the metallic surface creates surface plasmons, which greatly magnify the Raman signal. SERS signal intensities can be on the order of 10 6 times greater than those of unenhanced Raman signals (Kneipp 2007).
SERS has been used successfully as a potential miRNA detection technique by Driskell et al. (2008). These investigators detected enhanced Raman signals from single species of miRNA when blotted onto a silver nanorod array substrate on a glass slide. The silver nanorod substrate, a key element of their detection approach, was created using oblique angle vapor deposition fabrication, which increased the substrate reproducibility (Driskell et al. 2008). After substrate preparation, the investigators spotted different species of miRNA onto the silver nanorod surface. After adsorption of the miRNAs, the SERS spectrum then was obtained for each species. Multiple measurements from each spot and multiple spots for each miRNA species were assessed to test the reproducibility of this approach. Driskell et al. (2008) found that the spectra generated for different miRNAs were similar with regard to the number and location of the scattered bands owing to the presence of similar nucleotides. The relative intensities of the respective bands, however, were different for each miRNA species, because the relative number and composition of nucleotides varied among species. Furthermore, these investigators demonstrated that distinct features of the SERS spectrum changed linearly with changes in the nucleotide content, which indicates that the approach could be applied to gain information from unknown miRNA species. They then used a partial least squares regression approach to demonstrate that miRNA species could be identified accurately based on their SERS spectrum. They also demonstrated that five different miRNAs in a sample set could be discriminated accurately using the SERS spectrum.
The merit of the SERS technique described above lies in its excellent reproducibility and single-nucleotide specificity. Theoretically, this technique could be used to identify and discriminate many different species of miRNAs; however, this approach has yet to be demonstrated effectively with mixtures of miRNAs. Because the peak wavenumbers for the bands are identical and only the relative heights of each peak vary with miRNA species, linear decomposition methods may be limited in their ability to identify correctly the composition of miRNA mixtures. Finally, miRNA detection also may be complicated by the background signals in complex (biological) samples that are generated by SERS. Nevertheless, SERS is an extremely sensitive, accurate, and powerful method for identification and classification of miRNAs and could be employed for routine miRNA expression profiling, after appropriate sample purification (Driskell et al. 2008).
Surface plasmon resonance imaging (SPRI)
SPRI is an imaging technique in which surface plasmons are excited in a thin metallic surface (Pattnaik 2005, Steiner 2004). Although surface plasmons can be excited by other phenomena, such as electron beams, excitation in microscopy systems usually is achieved using a polarized light source, often a laser, combined with an attenuated total reflection coupler, or prism (Steiner et al. 1999). P-polarized light that is incident to the surface excites surface plasmons within a narrow range of incident angles (Steiner 2004). A charge-coupled device camera then is used to detect the reflected light. Because of the polarization coupling requirement, p-polarized light is used to visualize surface absorption, while s-polarized light typically is used as a reference image. Surface plasmon excitation is highly dependent on the thickness and refractive index of the local environment. These parameters can be influenced by subtle changes in the local environment, such as ligand binding or intermolecular interactions, which makes SPRI a highly sensitive molecular detection technique.
SPRI recently has been employed for miRNA profiling by Fang et al. (2006). These investigators developed an amplification approach that enables miRNA detection with very great sensitivity, which was combined with patterning of LNA binding regions on the surface of a thin film to enable detection of a panel of miRNAs. To develop this assay, the investigators immobilized LNA probes that were partially complementary to the target miRNA, leaving a six-nucleotide region, onto an SPRI substrate, presumably gold. Hybridization was effected so that miRNAs were left with a 3′ overhang (non-complementary region to the LNA probes) to which poly(A) tails were added by poly(A) polymerase. Enhancement of the SPRI signal was achieved by subsequent binding of poly(T) coated gold nanoparticles, which bound to the poly(A) tails. This increased the change in the SPRI reflectance image and improved the sensitivity of detection (Fig. 4). This technique promises to be a highly sensitive method for developing panels for miRNA profiling, because the authors demonstrated a detection limit of 10 fmol (Fang et al. 2006). One possible limitation of this technique is that SPRI is highly dependent on changes in temperature (Steiner 2004), which could complicate the development of a portable miRNA profiling tool.
Fig. 4.
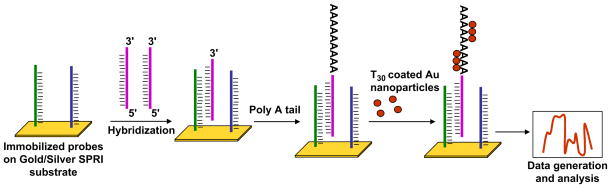
SPRI based method. Probes partially complementary to the target miRNA are immobilized on a metal based surface (gold/silver SPRI surface). Target miRNA hybridizes with the probes leaving a 3′ overhang. Subsequently, a poly(A) tail is added onto the 3′ overhang and later poly(T) coated gold nanoparticles are adsorbed onto the poly(A) tails and detected with SPRI.
Electrical detection based methods
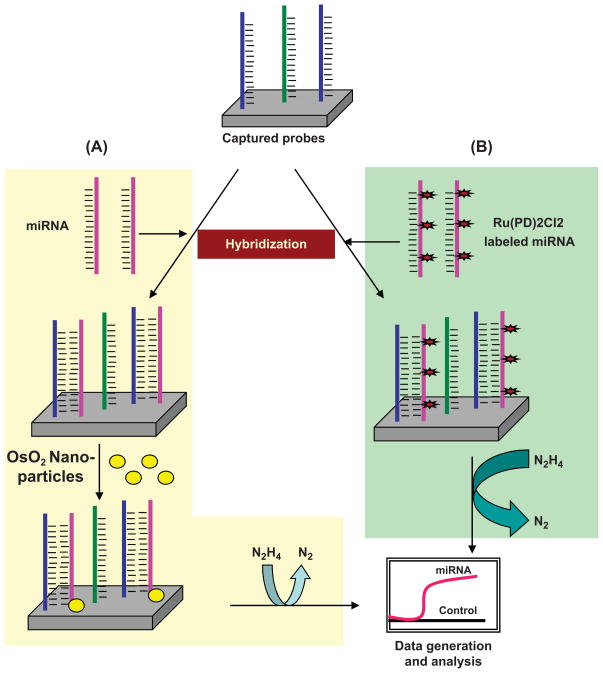
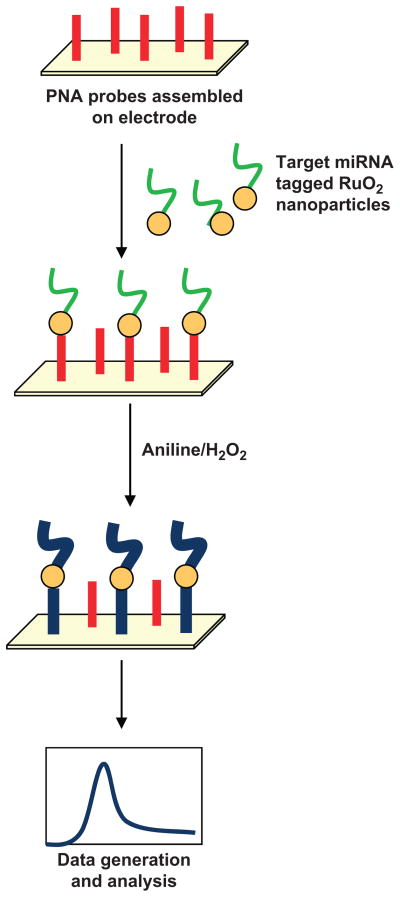
Electrical detection based methods are emerging as highly specific, rapid and high throughput assays for profiling mature miRNAs. These methods are based on changes in circuit properties as a consequence of miRNA hybridization. Gao and Yang (2006) used the electrocatalytic nanoparticle tags method, which is based on the use of isoniazid-capped OsO2 nanoparticle tags. Sodium periodate derivatives were hybridized to oligonucleotide capture probes on the electrode. Thereafter, signals were generated that linked periodate-treated miRNAs with the OsO2 nanoparticles, which oxidized hydrazine and resulted in an electrocatalytic activity at 0.10 V (Fig. 5A). This method proved highly sensitive and could detect miRNA at 80 fmol concentration (Gao and Yang 2006). Later, these investigators modified their strategy to increase the sensitivity further by labeling the target miRNA directly with a redox active and electrocatalytic moiety, Ru(PD)2Cl2 (PD = 1,10-phenanthroline-5, 6-dione), which has excellent electrocatalytic activity toward the oxidation of hydrazine (Fig. 5B); therefore, it permits detection of miRNAs at low concentration, i.e., 20 fmol (Gao and Yu 2007). Recently, Yang et al. (2009) developed another extremely sensitive detection method, which could quantify less than a 10 amol concentration of miRNA. These investigators used a Fe-Ru redox pair as a reporter and used an amplification scheme based on a nano-structured electrode platform. After hybridization of miRNA with peptide nucleic acid capture probes, Ru 3+ was reduced at the nanoelectrode surface and generated signals. These signals were amplified further by the ferricyanide-coupled electrochemical reduction, which regenerated Ru 3+ from Ru 2+. This approach achieved greater sensitivity together with high specificity for mature miRNA detection (Yang et al. 2009). Zhang et al. (2009) developed a label-free, rapid and sensitive direct hybridization assay for miRNA detection using silicon nanowires (SiNW). In this assay, peptide nucleic acids (PNA) were immobilized on the surface of the SiNW device that acted as receptor for miRNA recognition. Upon incubation of PNA functionalized SiNW with complementary miRNA targets, the changes in resistance were measured before and after hybridization and were considered to be a direct estimate of the concentrations of the hybridized target miRNA. This method provided a detection limit of 1 fmol and could discriminate between fully matched and single base mismatched miRNA sequences (Zhang et al. 2009). Peng and Gao (2011) developed a strategy that produced great sensitivity with low background. They first tagged target miRNAs with RuO2 nanoparticles and allowed them to hybridize with PNA probes assembled on a gold electrode. A mixture of aniline/H2O2 then was applied. The RuO2 nanoparticles polymerize aniline to form polyaniline only at hybridized miRNA strands, thus generating a clean background (Fig. 6).
Fig. 5.
Electrical detection based assay. A) Sodium periodate derivatives of target miRNAs are hybridized to probes captured on a metallic electrode followed by addition of OsO2 nanoparticles and incubation to allow their ligation with hybridized miRNAs. Subsequently, electrochemical signals are generated owing to the oxidation of hydrazine. (PD = 1, B) To increase sensitivity, target miRNAs are labeled directly with a electrocatalytic moiety, i.e., Ru(PD)2Cl2 10-phenanthroline-5,6-dione), and hybridized to captured probes. Owing to excellent electrocatalytic activity of Ru(PD)2Cl2 toward the oxidation of hydrazine, low concentrations of miRNA can be detected using this method.
Fig. 6.
Schematic of the RuO2-initiated aniline polymerization based biosensor for miRNA detection. PNA probes are assembled on an electrode and target miRNAs tagged with RuO2 nanoparticles are allowed to hybridize with PNA probes; an aniline/H2O2 mixture then is added. Catalytic activity of RuO2 nanoparticles facilitates polymerization of aniline, which causes deposition of polyaniline (Pan) on the miRNA template and the signal is monitored.
Perspectives
MiRNAs have received significant attention as a new class of gene regulators and accumulating evidence clearly indicates their important roles in diverse biological functions. Data emerging on miRNA functions and dysregulated expression under various pathophysiological conditions have provided strong support for their clinical use as functional biomarkers. Their use can determine the molecular characteristics of diseases. Similarly, miRNAs may be used for early diagnosis of diseases as well as to predict progression and therapeutic outcomes. Interest in miRNAs has increased greatly and the associated literature is expanding rapidly with reports on novel miRNAs, their validated gene targets and their expression profiling under various disease conditions. At a similar pace, novel methods for miRNA profiling also are emerging that facilitate the development of clinically feasible approaches for detecting miRNA. Several new assays have been developed to overcome the size limitation of mature miRNAs, sequence similarity among various members, and their low occurrence in clinical samples. Some of these assays are simple, but not sufficiently sensitive, while others are highly sensitive and support high throughput assays, but may require costly equipment and tedious procedures. Nevertheless, the tremendous progress that has been made in this area owing to increasing collaborations among basic biologists, bioengineers and clinical scientists, as well as enhanced support from funding agencies for such collaborative endeavors, should foster assessment of the clinical utility of miRNAs. Extensive efforts are required to validate these scientific advancements, however, before bringing them to clinical practice.
Acknowledgments
This research was funded by NIH/NCI (CA137513), DOD/US Army (PC0739930, PC093309) and USAMCI.
Footnotes
Conflict of interest statement: The authors have no potential conflict of interest to disclose.
References
- Allawi HT, Dahlberg JE, Olson S, Lund E, Olson M, Ma WP, Takova T, Neri BP, Lyamichev VI. Quantitation of microRNAs using a modified Invader assay. RNA. 2004;10:1153–1161. doi: 10.1261/rna.5250604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andorfer CA, Necela BM, Thompson EA, Perez EA. MicroRNA signatures: clinical biomarkers for the diagnosis and treatment of breast cancer. Trends Mol Med. 2011;17:313–319. doi: 10.1016/j.molmed.2011.01.006. [DOI] [PubMed] [Google Scholar]
- Bacia K, Kim SA, Schwille P. Fluorescence cross-correlation spectroscopy in living cells. Nat Methods. 2006;3:83–89. doi: 10.1038/nmeth822. [DOI] [PubMed] [Google Scholar]
- Bandres E, Bitarte N, Arias F, Agorreta J, Fortes P, Agirre X, Zarate R, Diaz-Gonzalez JA, Ramirez N, Sola JJ, Jimenez P, Rodriguez J, Garcia-Foncillas J. MicroRNA-451 regulates macrophage migration inhibitory factor production and proliferation of gastrointestinal cancer cells. Clin Cancer Res. 2009;15:2281–2290. doi: 10.1158/1078-0432.CCR-08-1818. [DOI] [PubMed] [Google Scholar]
- Bhardwaj A, Singh S, Singh AP. MicroRNA based cancer therapeutics: big hope from small RNAs. Mol Cell Pharmacol. 2010;2:213–219. doi: 10.4255/mcpharmacol.10.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bovell L, Shanmugam C, Katkoori VR, Zhang B, Vogtmann E, Grizzle WE, Manne U. miRNAs are stable in colorectal cancer archival tissue blocks. Front Biosci (Elite Ed) 2012;1:1937–1940. doi: 10.2741/514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloomston M, Frankel WL, Petrocca F, Volinia S, Alder H, Hagan JP, Liu CG, Bhatt D, Taccioli C, Croce CM. MicroRNA expression patterns to differentiate pancreatic adenocarcinoma from normal pancreas and chronic pancreatitis. JAMA. 2007;297:1901–1908. doi: 10.1001/jama.297.17.1901. [DOI] [PubMed] [Google Scholar]
- Cacchiarelli D, Legnini I, Martone J, Cazzella V, D’Amico A, Bertini E, Bozzoni I. miRNAs as serum biomarkers for Duchenne muscular dystrophy. EMBO Mol Med. 2011;3:258–265. doi: 10.1002/emmm.201100133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caudy AA, Myers M, Hannon GJ, Hammond SM. Fragile X-related protein and VIG associate with the RNA interference machinery. Genes Dev. 2002;16:2491–2496. doi: 10.1101/gad.1025202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Ridzon DA, Broomer AJ, Zhou Z, Lee DH, Nguyen JT, Barbisin M, Xu NL, Mahuvakar VR, Andersen MR, Lao KQ, Livak KJ, Guegler KJ. Real-time quantification of microRNAs by stem-loop RT-PCR. Nucl Acids Res. 2005;33:e179. doi: 10.1093/nar/gni178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y, Ji R, Yue J, Yang J, Liu X, Chen H, Dean DB, Zhang C. MicroRNAs are aberrantly expressed in hypertrophic heart: do they play a role in cardiac hypertrophy? Am J Pathol. 2007;170:1831–1840. doi: 10.2353/ajpath.2007.061170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cissell KA, Campbell S, Deo SK. Rapid, single-step nucleic acid detection. Anal Bioanal Chem. 2008a;391:2577–2581. doi: 10.1007/s00216-008-2215-5. [DOI] [PubMed] [Google Scholar]
- Cissell KA, Rahimi Y, Shrestha S, Hunt EA, Deo SK. Bioluminescence based detection of microRNA, miR21 in breast cancer cells. Anal Chem. 2008b;80:2319–2325. doi: 10.1021/ac702577a. [DOI] [PubMed] [Google Scholar]
- Cortez MA, Calin GA. MicroRNA identification in plasma and serum: a new tool to diagnose and monitor diseases. Expert Opin Biol Ther. 2009;9:703–711. doi: 10.1517/14712590902932889. [DOI] [PubMed] [Google Scholar]
- Coulouarn C, Factor VM, Andersen JB, Durkin ME, Thorgeirsson SS. Loss of miR-122 expression in liver cancer correlates with suppression of the hepatic phenotype and gain of metastatic properties. Oncogene. 2009;28:3526–3536. doi: 10.1038/onc.2009.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De AM, Lyamichev VI, Eis PS, Iszczyszyn W, Kwiatkowski RW, Law SM, Olson MC, Rasmussen EB. Invader technology for DNA and RNA analysis: principles and applications. Expert Rev Mol Diagn. 2002;2:487–496. doi: 10.1586/14737159.2.5.487. [DOI] [PubMed] [Google Scholar]
- Deo M, Yu JY, Chung KH, Tippens M, Turner DL. Detection of mammalian microRNA expression by in situ hybridization with RNA oligonucleotides. Dev Dyn. 2006;235:2538–2548. doi: 10.1002/dvdy.20847. [DOI] [PubMed] [Google Scholar]
- Dong S, Cheng Y, Yang J, Li J, Liu X, Wang X, Wang D, Krall TJ, Delphin ES, Zhang C. MicroRNA expression signature and the role of microRNA-21 in the early phase of acute myocardial infarction. J Biol Chem. 2009;284:29514–29525. doi: 10.1074/jbc.M109.027896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driskell JD, Seto AG, Jones LP, Jokela S, Dluhy RA, Zhao YP, Tripp RA. Rapid microRNA (miRNA) detection and classification via surface-enhanced Raman spectroscopy (SERS) Biosens Bioelectron. 2008;24:923–928. doi: 10.1016/j.bios.2008.07.060. [DOI] [PubMed] [Google Scholar]
- Dyrskjot L, Ostenfeld MS, Bramsen JB, Silahtaroglu AN, Lamy P, Ramanathan R, Fristrup N, Jensen JL, Andersen CL, Zieger K, Kauppinen S, Ulhoi BP, Kjems J, Borre M, Orntoft TF. Genomic profiling of microRNAs in bladder cancer: miR-129 is associated with poor outcome and promotes cell death in vitro. Cancer Res. 2009;69:4851–4860. doi: 10.1158/0008-5472.CAN-08-4043. [DOI] [PubMed] [Google Scholar]
- Eis PS, Olson MC, Takova T, Curtis ML, Olson SM, Vener TI, Ip HS, Vedvik KL, Bartholomay CT, Allawi HT, Ma WP, Hall JG, Morin MD, Rushmore TH, Lyamichev VI, Kwiatkowski RW. An invasive cleavage assay for direct quantitation of specific RNAs. Nat Biotechnol. 2001;19:673–676. doi: 10.1038/90290. [DOI] [PubMed] [Google Scholar]
- Fang S, Lee HJ, Wark AW, Corn RM. Attomole microarray detection of microRNAs by nanoparticle-amplified SPR imaging measurements of surface polyadenylation reactions. J Am Chem Soc. 2006;128:14044–14046. doi: 10.1021/ja065223p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fendler A, Jung M, Stephan C, Honey RJ, Stewart RJ, Pace KT, Erbersdobler A, Samaan S, Jung K, Yousef GM. miRNAs can predict prostate cancer biochemical relapse and are involved in tumor progression. Int J Oncol. 2011;39:1183–1192. doi: 10.3892/ijo.2011.1128. [DOI] [PubMed] [Google Scholar]
- Gao Z, Yang Z. Detection of microRNAs using electrocatalytic nanoparticle tags. Anal Chem. 2006;78:1470–1477. doi: 10.1021/ac051726m. [DOI] [PubMed] [Google Scholar]
- Gao Z, Yu YH. Direct labeling microRNA with an electrocatalytic moiety and its application in ultra-sensitive microRNA assays. Biosens Bioelectron. 2007;22:933–940. doi: 10.1016/j.bios.2006.04.020. [DOI] [PubMed] [Google Scholar]
- Hebert SS, De SB. Alterations of the microRNA network cause neurodegenerative disease. Trends Neurosci. 2009;32:199–206. doi: 10.1016/j.tins.2008.12.003. [DOI] [PubMed] [Google Scholar]
- Hui YY, Zhang B, Chang YC, Chang CC, Chang HC, Hsu JH, Chang K, Chang FH. Two-photon fluorescence correlation spectroscopy of lipid-encapsulated fluorescent nanodiamonds in living cells. Opt Express. 2010;18:5896–5905. doi: 10.1364/OE.18.005896. [DOI] [PubMed] [Google Scholar]
- Hummel R, Hussey DJ, Haier J. MicroRNAs: predictors and modifiers of chemo- and radiotherapy in different tumour types. Eur J Cancer. 2010;46:298–311. doi: 10.1016/j.ejca.2009.10.027. [DOI] [PubMed] [Google Scholar]
- Invitrogen. [Accessed: 20-Sep-2011];Life Technologies [Online] www.invitrogen.com.
- Janga SC, Vallabhaneni S. MicroRNAs as post-transcriptional machines and their interplay with cellular networks. Adv Exp Med Biol. 2011;722:59–74. doi: 10.1007/978-1-4614-0332-6_4. [DOI] [PubMed] [Google Scholar]
- Jiang J, Lee EJ, Gusev Y, Schmittgen TD. Real-time expression profiling of microRNA precursors in human cancer cell lines. Nucl Acids Res. 2005;33:5394–5403. doi: 10.1093/nar/gki863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson FH, Shimomura O, Saiga Y. Action of cyanide on Cypridina luciferin. J Cell Comp Physiol. 1962;59:265–272. doi: 10.1002/jcp.1030590306. [DOI] [PubMed] [Google Scholar]
- Kapsimali M, Kloosterman WP, de BE, Rosa F, Plasterk RH, Wilson SW. MicroRNAs show a wide diversity of expression profiles in the developing and mature central nervous system. Genome Biol. 2007;8:R173. doi: 10.1186/gb-2007-8-8-r173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kauppinen S, Vester B, Wengel J. Locked nucleic acid: high-affinity targeting of complementary RNA for RNomics. Handbook Exp Pharmacol. 2006:405–422. doi: 10.1007/3-540-27262-3_21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SA, Heinze KG, Schwille P. Fluorescence correlation spectroscopy in living cells. Nat Methods. 2007;4:963–973. doi: 10.1038/nmeth1104. [DOI] [PubMed] [Google Scholar]
- Kloosterman WP, Wienholds E, de BE, Kauppinen S, Plasterk RH. In situ detection of miRNAs in animal embryos using LNA-modified oligonucleotide probes. Nat Methods. 2006;3:27–29. doi: 10.1038/nmeth843. [DOI] [PubMed] [Google Scholar]
- Kneipp K. Surface-enhanced raman scattering. Phys Today. 2007;60:40–46. [Google Scholar]
- Kong L, Zhu J, Han W, Jiang X, Xu M, Zhao Y, Dong Q, Pang Z, Guan Q, Gao L, Zhao J, Zhao L. Significance of serum microRNAs in pre-diabetes and newly diagnosed type 2 diabetes: a clinical study. Acta Diabetol. 2011;48:61–69. doi: 10.1007/s00592-010-0226-0. [DOI] [PubMed] [Google Scholar]
- Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- Lin SL, Chiang A, Chang D, Ying SY. Loss of mir-146a function in hormone-refractory prostate cancer. RNA. 2008;4:417–424. doi: 10.1261/rna.874808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA, Downing JR, Jacks T, Horvitz HR, Golub TR. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- Magde D, Elson EL, Webb WW. Fluorescence correlation spectroscopy. II. An experimental realization. Biopolymers. 1974;13:29–61. doi: 10.1002/bip.1974.360130103. [DOI] [PubMed] [Google Scholar]
- Makino K, Jinnin M, Kajihara I, Honda N, Sakai K, Masuguchi S, Fukushima S, Inoue Y, Ihn H. Circulating miR-142-3p levels in patients with systemic sclerosis. Clin Exp Dermatol. 2012;37:34–39. doi: 10.1111/j.1365-2230.2011.04158.x. [DOI] [PubMed] [Google Scholar]
- Manne U, Shanmugam C, Bovell L, Katkoori VR, Bumpers HL. miRNAs as biomarkers for management of patients with colorectal cancer. Biomark Med. 2010;4:761–770. doi: 10.2217/bmm.10.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maru DM, Singh RR, Hannah C, Albarracin CT, Li YX, Abraham R, Romans AM, Yao H, Luthra MG, Anandasabapathy S, Swisher SG, Hofstetter WL, Rashid A, Luthra R. MicroRNA-196a is a potential marker of progression during Barrett’s metaplasia-dysplasia-invasive adenocarcinoma sequence in esophagus. Am J Pathol. 2009;174:1940–1948. doi: 10.2353/ajpath.2009.080718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL, Peterson A, Noteboom J, O’Briant KC, Allen A, Lin DW, Urban N, Drescher CW, Knudsen BS, Stirewalt DL, Gentleman R, Vessella RL, Nelson PS, Martin DB, Tewari M. Circulating microRNAs as stable blood based markers for cancer detection. Proc Natl Acad Sci USA. 2008;105:10513–10518. doi: 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakajima G, Hayashi K, Xi Y, Kudo K, Uchida K, Takasaki K, Yamamoto M, Ju J. Non-coding microRNAs hsa-let-7g and hsa-miR-181b are associated with chemoresponse to S-1 in colon cancer. Cancer Genom Proteom. 2006;3:317–324. [PMC free article] [PubMed] [Google Scholar]
- Nakasa T, Miyaki S, Okubo A, Hashimoto M, Nishida K, Ochi M, Asahara H. Expression of microRNA-146 in rheumatoid arthritis synovial tissue. Arthr Rheum. 2008;58:1284–1292. doi: 10.1002/art.23429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakata K, Ohuchida K, Mizumoto K, Kayashima T, Ikenaga N, Sakai H, Lin C, Fujita H, Otsuka T, Aishima S, Nagai E, Oda Y, Tanaka M. MicroR-NA-10b is overexpressed in pancreatic cancer, promotes its invasiveness, and correlates with a poor prognosis. Surgery. 2011;150:916–922. doi: 10.1016/j.surg.2011.06.017. [DOI] [PubMed] [Google Scholar]
- Nass D, Rosenwald S, Meiri E, Gilad S, Tabibian-Keissar H, Schlosberg A, Kuker H, Sion-Vardy N, Tobar A, Kharenko O, Sitbon E, Lithwick YG, Elyakim E, Cholakh H, Gibori H, Spector Y, Bentwich Z, Barshack I, Rosenfeld N. MiR-92b and miR-9/9* are specifically expressed in brain primary tumors and can be used to differentiate primary from metastatic brain tumors. Brain Pathol. 2009;19:375–383. doi: 10.1111/j.1750-3639.2008.00184.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neely LA, Patel S, Garver J, Gallo M, Hackett M, McLaughlin S, Nadel M, Harris J, Gullans S, Rooke J. A single-molecule method for the quantitation of microRNA gene expression. Nat Methods. 2006;3:41–46. doi: 10.1038/nmeth825. [DOI] [PubMed] [Google Scholar]
- Nelson DM, Sadovsky Y, Robinson JM, Croy BA, Rice G, Kniss DA. Advanced techniques in placental biology–workshop report. Placenta. 2006a;27:S87–S90. doi: 10.1016/j.placenta.2006.01.012. [DOI] [PubMed] [Google Scholar]
- Nelson PT, Baldwin DA, Kloosterman WP, Kauppinen S, Plasterk RH, Mourelatos Z. RAKE and LNA-ISH reveal microRNA expression and localization in archival human brain. RNA. 2006b;12:187–191. doi: 10.1261/rna.2258506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noh SJ, Miller SH, Lee YT, Goh SH, Marincola FM, Stroncek DF, Reed C, Wang E, Miller JL. Let-7 microRNAs are developmentally regulated in circulating human erythroid cells. J Transl Med. 2009;7:98. doi: 10.1186/1479-5876-7-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Driscoll L. The emerging world of microRNAs. Anticancer Res. 2006;26:4271–4278. [PubMed] [Google Scholar]
- Osaki M, Takeshita F, Ochiya T. MicroRNAs as biomarkers and therapeutic drugs in human cancer. Biomarkers. 2008;13:658–670. doi: 10.1080/13547500802646572. [DOI] [PubMed] [Google Scholar]
- Pattnaik P. Surface plasmon resonance: applications in understanding receptor-ligand interaction. Appl Biochem Biotechnol. 2005;126:79–92. doi: 10.1385/abab:126:2:079. [DOI] [PubMed] [Google Scholar]
- Pauley KM, Satoh M, Chan AL, Bubb MR, Reeves WH, Chan EK. Upregulated miR-146a expression in peripheral blood mononuclear cells from rheumatoid arthritis patients. Arthr Res Ther. 2008;10:R101. doi: 10.1186/ar2493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Y, Gao Z. Amplified detection of microRNA based on ruthenium oxide nanoparticle-initiated deposition of an insulating film. Anal Chem. 2011;83:820–827. doi: 10.1021/ac102370s. [DOI] [PubMed] [Google Scholar]
- Pillai RS. MicroRNA function: multiple mechanisms for a tiny RNA? RNA. 2005;11:1753–1761. doi: 10.1261/rna.2248605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poy MN, Eliasson L, Krutzfeldt J, Kuwajima S, Ma X, Macdonald PE, Pfeffer S, Tuschl T, Rajewsky N, Rorsman P, Stoffel M. A pancreatic islet-specific microRNA regulates insulin secretion. Nature. 2004;432:226–230. doi: 10.1038/nature03076. [DOI] [PubMed] [Google Scholar]
- Ramkissoon SH, Mainwaring LA, Sloand EM, Young NS, Kajigaya S. Nonisotopic detection of microRNA using digoxigenin labeled RNA probes. Mol Cell Probes. 2006;20:1–4. doi: 10.1016/j.mcp.2005.07.004. [DOI] [PubMed] [Google Scholar]
- Raymond CK, Roberts BS, Garrett-Engele P, Lim LP, Johnson JM. Simple, quantitative primer-extension PCR assay for direct monitoring of microRNAs and short-interfering RNAs. RNA. 2005;11:1737–1744. doi: 10.1261/rna.2148705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roldo C, Missiaglia E, Hagan JP, Falconi M, Capelli P, Bersani S, Calin GA, Volinia S, Liu CG, Scarpa A, Croce CM. MicroRNA expression abnormalities in pancreatic endocrine and acinar tumors are associated with distinctive pathologic features and clinical behavior. J Clin Oncol. 2006;24:4677–4684. doi: 10.1200/JCO.2005.05.5194. [DOI] [PubMed] [Google Scholar]
- Saito M, Schetter AJ, Mollerup S, Kohno T, Skaug V, Bowman ED, Mathe EA, Takenoshita S, Yokota J, Haugen A, Harris CC. The association of microRNA expression with prognosis and progression in early-stage, non-small cell lung adenocarcinoma: a retrospective analysis of three cohorts. Clin Cancer Res. 2011;17:1875–1882. doi: 10.1158/1078-0432.CCR-10-2961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scapoli L, Palmieri A, Lo ML, Pezzetti F, Rubini C, Girardi A, Farinella F, Mazzotta M, Carinci F. MicroRNA expression profiling of oral carcinoma identifies new markers of tumor progression. Int J Immunopathol Pharmacol. 2010;23:1229–1234. doi: 10.1177/039463201002300427. [DOI] [PubMed] [Google Scholar]
- Schmittgen TD, Jiang J, Liu Q, Yang L. A high-throughput method to monitor the expression of microRNA precursors. Nucl Acids Res. 2004;32:e43. doi: 10.1093/nar/gnh040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segura MF, Belitskaya-Levy I, Rose AE, Zakrzewski J, Gaziel A, Hanniford D, Darvishian F, Berman RS, Shapiro RL, Pavlick AC, Osman I, Hernando E. Melanoma microRNA signature predicts post-recurrence survival. Clin Cancer Res. 2010;16:1577–1586. doi: 10.1158/1078-0432.CCR-09-2721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sethupathy P, Megraw M, Hatzigeorgiou AG. A guide through present computational approaches for the identification of mammalian microRNA targets. Nat Methods. 2006;3:881–886. doi: 10.1038/nmeth954. [DOI] [PubMed] [Google Scholar]
- Shi R, Chiang VL. Facile means for quantifying microRNA expression by real-time PCR. Biotechniques. 2005;39:519–525. doi: 10.2144/000112010. [DOI] [PubMed] [Google Scholar]
- Spahn M, Kneitz S, Scholz CJ, Stenger N, Rudiger T, Strobel P, Reedville H, Kneitz B. Expression of microRNA-221 is progressively reduced in aggressive prostate cancer and metastasis and predicts clinical recurrence. Int J Cancer. 2010;127:394–403. doi: 10.1002/ijc.24715. [DOI] [PubMed] [Google Scholar]
- Srivastava SK, Bhardwaj A, Singh S, Aurora S, Wang B, Grizzle WE, Singh AP. MicroRNA-150 directly targets MUC4 and suppresses growth and malignant behavior of pancreatic cancer cells. Carcinogen sis. 2011;32:1832–1839. doi: 10.1093/carcin/bgr223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiner G. Surface plasmon resonance imaging. Anal Bioanal Chem. 2004;379:328–331. doi: 10.1007/s00216-004-2636-8. [DOI] [PubMed] [Google Scholar]
- Steiner G, Sablinskas V, Hubner A, Kuhne C, Salzer R. Surface plasmon resonance imaging of microstructured monolayers 1. J Molec Struct. 1999;509:265–273. [Google Scholar]
- Stenvang J, Silahtaroglu AN, Lindow M, Elmen J, Kauppinen S. The utility of LNA in microRNA based cancer diagnostics and therapeutics. Semin Cancer Biol. 2008;18:89–102. doi: 10.1016/j.semcancer.2008.01.004. [DOI] [PubMed] [Google Scholar]
- Tang F, Hajkova P, Barton SC, O’Carroll D, Lee C, Lao K, Surani MA. 220-Plex microRNA expression profile of a single cell. Nat Protoc. 2006;1:1154–1159. doi: 10.1038/nprot.2006.161. [DOI] [PubMed] [Google Scholar]
- Valoczi A, Hornyik C, Varga N, Burgyan J, Kauppinen S, Havelda Z. Sensitive and specific detection of microRNAs by northern blot analysis using LNA-modified oligonucleotide probes. Nucl Acids Res. 2004;32:e175. doi: 10.1093/nar/gnh171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van RE, Sutherland LB, Liu N, Williams AH, McAnally J, Gerard RD, Richardson JA, Olson EN. A signature pattern of stress-responsive microRNAs that can evoke cardiac hypertrophy and heart failure. Proc Natl Acad Sci USA. 2006;103:18255–18260. doi: 10.1073/pnas.0608791103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varallyay E, Burgyan J, Havelda Z. Detection of microRNAs by Northern blot analyses using LNA probes. Methods. 2007;43:140–145. doi: 10.1016/j.ymeth.2007.04.004. [DOI] [PubMed] [Google Scholar]
- Ventura A, Jacks T. MicroRNAs and cancer: short RNAs go a long way. Cell. 2009;136:586–591. doi: 10.1016/j.cell.2009.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visone R, Croce CM. MiRNAs and cancer. Am J Pathol. 2009;174:1131–1138. doi: 10.2353/ajpath.2009.080794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wienholds E, Kloosterman WP, Miska E, Alvarez-Saavedra E, Berezikov E, de BE, Horvitz HR, Kauppinen S, Plasterk RH. MicroRNA expression in zebrafish embryonic development. Science. 2005;309:310–311. doi: 10.1126/science.1114519. [DOI] [PubMed] [Google Scholar]
- Xu Y, Piston DW, Johnson CH. A bioluminescence resonance energy transfer (BRET) system: application to interacting circadian clock proteins. Proc Natl Acad Sci USA. 1999;96:151–156. doi: 10.1073/pnas.96.1.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Hui A, Pampalakis G, Soleymani L, Liu FF, Sargent EH, Kelley SO. Direct, electronic microRNA detection for the rapid determination of differential expression profiles. Angew Chem Int Ed Engl. 2009;48:8461–8464. doi: 10.1002/anie.200902577. [DOI] [PubMed] [Google Scholar]
- Yang L, Diem K, Wang X. BADGE, beads array for the detection of gene expression, a high-throughput diagnostic bioassay. Genome Res. 2001;11:1888–1898. doi: 10.1101/gr.190901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu SL, Chen HY, Chang GC, Chen CY, Chen HW, Singh S, Cheng CL, Yu CJ, Lee YC, Chen HS, Su TJ, Chiang CC, Li HN, Hong QS, Su HY, Chen CC, Chen WJ, Liu CC, Chan WK, Chen WJ, Li KC, Chen JJ, Yang PC. MicroRNA signature predicts survival and relapse in lung cancer. Cancer Cell. 2008;13:48–57. doi: 10.1016/j.ccr.2007.12.008. [DOI] [PubMed] [Google Scholar]
- Zenz T, Benner A, Dohner H, Stilgenbauer S. Chronic lymphocytic leukemia and treatment resistance in cancer: the role of the p53 pathway. Cell Cycle. 2008;7:3810–3814. doi: 10.4161/cc.7.24.7245. [DOI] [PubMed] [Google Scholar]
- Zhang GJ, Chua JH, Chee RE, Agarwal A, Wong SM. Label-free direct detection of MiRNAs with silicon nanowire biosensors. Biosens Bioelectron. 2009;24:2504–2508. doi: 10.1016/j.bios.2008.12.035. [DOI] [PubMed] [Google Scholar]