Abstract
The late phase of long-term potentiation (LTP) at glutamatergic synapses, which is thought to underlie long-lasting memory, requires gene transcription in the nucleus. However, the mechanism by which signaling initiated at synapses is transmitted into the nucleus to induce transcription has remained elusive. Here, we found that induction of LTP in only a few dendritic spines was sufficient to activate extracellular signal-related kinase (ERK) in the nucleus and regulate downstream transcription factors. Signaling from individual spines was integrated over a wide range of time (>30 min) and space (>80 μm). Spatially dispersed inputs over multiple branches activated nuclear ERK much more efficiently than clustered inputs over one branch. Thus, biochemical signals from individual dendritic spines exert profound effects on nuclear signaling.
Activity-dependent gene transcription is essential for the maintenance of long-term potentiation (LTP) and memory consolidation (1, 2). LTP induction in single dendritic spines activates signaling that can either be restricted to the stimulated spine or spread into the parent dendrite over 5-10 μm (3-5). However, it is unknown whether signaling initiated at single dendritic spines can be transmitted into the nucleus to regulate gene transcription. Extracellular signal-related kinase (ERK) is important both for signaling within the stimulated spine and adjacent dendrites (3, 6, 7) and also for activating transcription factors in the nucleus during LTP (2, 8-11). Thus, ERK signaling may play an important role in relaying signals from the stimulated spines to the nucleus.
To monitor the activity of ERK in the nucleus, we ballistically transfected cultured organotypic hippocampal slices of rats with nuclear-targeted ERK activity reporter (EKARnuc) (12), and imaged CA1 pyramidal neurons with 2-photon fluorescence lifetime imaging microscopy (2pFLIM). The expression of EKARnuc was highly localized to the nucleus (12). Using the weak EKAR expression in the cytosol, fluorescence intensity measurements were used to monitor structural plasticity of dendritic spines on secondary and tertiary apical dendritic branches (Fig. 1).
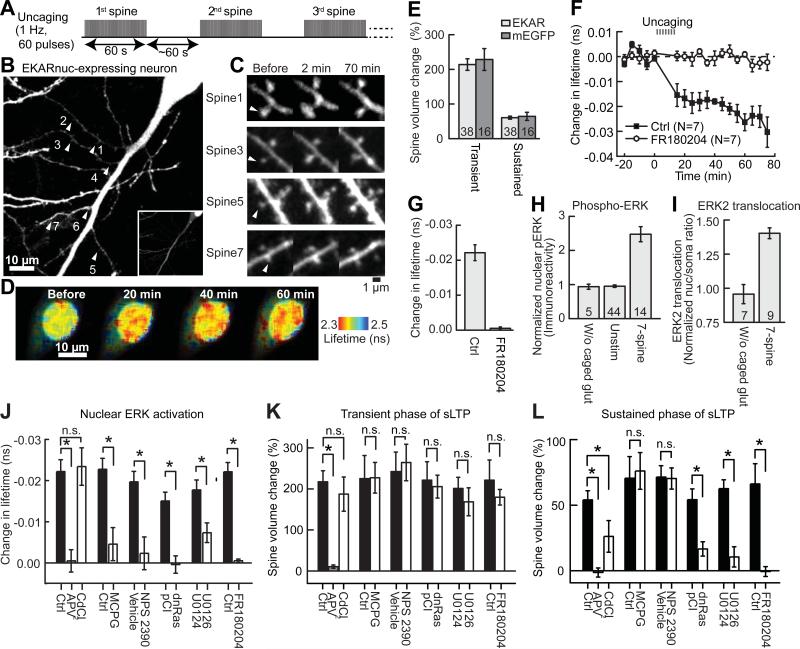
Fig. 1. Stimulation of 7 spines in a sequential fashion triggers activation of ERK in the nucleus.
(A) Schematic for 7-spine stimulation. (B) A neuron transfected with EKARnuc. The arrowheads indicate the locations of 7 stimulated spines. (C) Representative images of spine sLTP induced by glutamate uncaging in the neuron shown in (B). (D) Fluorescence lifetime images of EKARnuc before and after 7-spine stimulation in the same neuron as in (B) and (C). (E) Quantification of the transient (1-2 min) and sustained (~70 min) phases of sLTP following 7-spine stimulation in EKARnuc-expressing neurons and in mEGFP-expressing neurons. N is indicated at the bottom of the graph. (F) Time course of change in fluorescence lifetime of EKARnuc following 7-spine stimulation in the absence (Ctrl) or presence of ERK inhibitor FR180204. (G) Quantification of change in fluorescence lifetime averaged over 40-70 min in (F). (H to I) Stimulation of 7 spines leads to increased nuclear phospho-ERK level (pERK; H) and ERK2 nuclear translocation (I). (J to L) Effects of pharmacological agents and genetic manipulation on nuclear ERK activation (J), the transient (1-2 min; K) and sustained (~70 min; L) phases of sLTP. Fluorescence lifetime change averaged over 40-70 min was quantified for experiments shown in fig. S5 and (F). *P < 0.05, n.s., not significant. N = 14 for Ctrl and 7 for APV and CdCl2, 6 for Ctrl and MCPG, 6 for Vehicle and NPS 2390, 7 for pCI and 8 for dnRas, and 7 for U0124 and U0126. Error bars are SEM.
When a single spine was stimulated with a low-frequency train (1Hz, 60 s; Fig. 1A) of two-photon glutamate uncaging pulses in the absence of Mg2+, the spine volume increased rapidly by 275 ± 18% in 1-2 min (transient phase), which decayed to a sustained level 61 ± 4% larger than the original volume that lasted more than 1 hour (sustained phase) (Fig. 1, C and E), as expected (3-5, 13). The volume increase was similar to that induced in neurons expressing monomeric enhanced green fluorescent protein (mEGFP) (Fig. 1E). This structural LTP (sLTP) of spines is known to be associated with electrophysiological LTP (3-5, 13). Repeating this protocol in different spines one by one (~60 s interval; Fig. 1A), we induced sLTP sequentially in 7 spines on 3-5 different dendritic branches (Fig. 1, B and C). Following the 7-spine stimulation, we observed a slow and sustained elevation of ERK activity in the nucleus, as indicated by a gradual (over ~30 min) shortening of the fluorescence lifetime of EKARnuc (~ -0.02 ns) that was maintained for at least the following 40 min (Fig. 1, D and F). Pharmacological inhibition of ERK with ERK inhibitor FR180204 (50 μM) completely prevented the fluorescence lifetime decrease (Fig. 1, F and G). Inhibition of the classical upstream molecules Ras and MEK (14) with dominant negative Ras (dnRas) expression or MEK inhibitor U0126 prevented the fluorescence lifetime decrease (Fig. 1J and fig. S5). Thus, the change in fluorescence lifetime of EKARnuc acted as a reliable reporter of ERK activation in the nucleus.
We further confirmed that our 7-spine stimulation protocol activates nuclear ERK using two methods independent of 2pFLIM imaging of EKARnuc. First, we performed immunostaining of phosphorylated ERK in CA1 neurons expressing mEGFP. Consistent with the EKARnuc results, the level of phosphorylated ERK in the nucleus was persistently elevated following 7-spine stimulation (15) (Fig. 1H and fig. S1). Second, we performed live imaging of mEGFP-tagged ERK2 in organotypic slices. mEGFP-ERK2 was localized predominantly to the cytoplasm under basal condition, but slowly translocated into the nucleus following 7-spine stimulation (Fig. 1I and fig. S2) (8, 15). Thus, nuclear ERK is activated by sequential activation of a few spines.
Next, we determined the source of intracellular Ca2+ elevation that leads to nuclear ERK activation. Uncaging-induced Ca2+ elevation was largely restricted to the stimulated spines, and spreading along the dendrite was limited to 2-3 μm (figs. S3 and S4) (15, 16). This local Ca2+ elevation was dependent mainly on N-methyl-daspartate-type glutamate receptors (NMDARs) and there was negligible contribution from voltage-sensitive Ca2+ channels (VSCCs), metabotropic glutamate receptor (mGluR)-mediated internal Ca2+ release, or Ca2+-permeable α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid receptor (AMPARs) (fig. S4F). Inhibition of NMDARs with 2-amino-5-phosphonopentanoic acid (APV, 50 μM) completely prevented nuclear ERK activation (Fig. 1J and fig. S5A) as well as sLTP (Fig. 1, K-L) (13). In contrast, blockade of VSCCs with CdCl2 (200 μM) did not prevent nuclear ERK activation (Fig. 1J and fig. S5A), although the sustained phase of sLTP was partially inhibited (Fig. 1, K-L). Thus, uncaging-induced nuclear ERK activation was not caused by direct membrane depolarization in close proximity to the nucleus and resultant Ca2+ entry through VSCCs. Indeed, uEPSP recordings showed only small somatic voltage changes during uncaging (1.5–4 mV; fig. S3). Inhibition of mGluRs with α-methyl-4-carboxyphenylglycine (MCPG, 1 mM) or NPS 2390 (20 μM) attenuated the nuclear ERK activation (Fig. 1J and fig. S5, B-C). However, inhibition of mGluRs did not affect sLTP (13) (Fig. 1, K-L). Thus, the requirement for nuclear ERK activation is different from that for Ca2+ elevation or sLTP. Because group I mGluR activation increases the production of diacylglycerol, which leads to activation of protein kinase C (PKC) (17), we further tested if PKC is required for the sustained nuclear ERK activation. Because PKC inhibitor blocks sLTP (3), we applied PKC inhibitor bisindolylmaleimide I (BIM, 0.2 μM) ~60 min after induction of nuclear ERK activation. Surprisingly, this delayed application of BIM caused nuclear ERK activity to return to the basal level gradually (fig. S6), suggesting that sustained PKC activity is required for maintaining nuclear ERK activation.
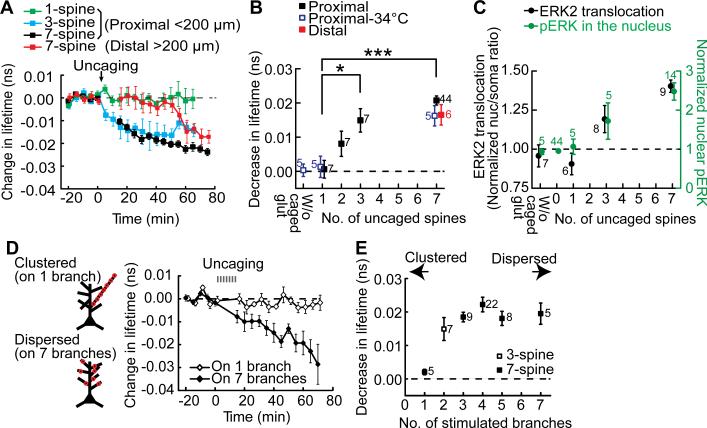
What is the spatial pattern of synaptic activation required for nuclear ERK activation? We first tested how many synapses are required for nuclear ERK activation. We stimulated different numbers (i.e. 1, 3, 7) of spines at a fixed density (i.e. 1-3 spines per dendritic branch, separated by more than 10 μm; Fig. 1, A and B). Glutamate uncaging at a single spine failed to cause any detectable signal in the nucleus, whereas glutamate uncaging at 3 spines led to significantly elevated nuclear ERK activity (Fig. 2, A and B). A similar correlation with the number of stimulated spines was found for EKARnuc signal at a near-physiological temperature (34oC) (Fig. 2B and fig. S7) as well as for the levels of nuclear phospho-ERK and ERK2 translocation into the nucleus (Fig. 2C and figs. S1-S2).
Fig. 2. Spatial stimulation pattern required for nuclear ERK activation.
(A) Time course of fluorescence lifetime change of EKARnuc following stimulation of varied numbers of spines under a fixed density of stimulation. 3-spine stimulation was on 2 separated dendritic branches. Proximal dendrites (<200 μm from the nucleus) were stimulated in most of the experiments, but distal dendrites (<200 μm) were stimulated in some experiments (red). (B) Quantification of the sustained fluorescence lifetime change (averaged over 40-70 min for proximal 7-spine stimulation, 30-60 min for 1- and 3-spine stimulations, 65-75 min for distal 7-spine stimulation). As a negative control, 7-spine stimulation experiments without caged glutamate were performed (W/o caged glut). N for each condition is indicated next to the data points; *P < 0.05, ***P < 0.001. (C) Dependency of the magnitudes of nuclear translocation of mEGFP-ERK2 (black) and pERK in the nucleus (green) on the number of stimulated dendritic spines. The level of pERK was measured at 75 min after stimulation (see fig. S1). The nuclear/somatic ratio was averaged over 55-70 min for 1-spine stimulation and 60-75 min for other groups (see fig. S2). (D) Time course of fluorescence lifetime change of EKARnuc following stimulation of 7 spines that reside on the same branch or on 7 different branches. (E) Quantification of the sustained fluorescence lifetime change following stimulation of varied numbers of dendritic branches. Data are presented as mean ± SEM.
In most of the experiments, we stimulated proximal branches within 200 μm from the soma. However, when we stimulated distal branches at more than 200 μm away from the soma, ERK activation showed a long delay (~40 min) before it started to increase to the level similar to that caused by proximal stimulation (Fig. 2, A and B). This slow process suggests that fast biochemical processes such as Ca2+ waves (18) and electrochemical signaling (2, 19) are unlikely to underlie the nuclear ERK activation induced by activating a few spines.
Next, we asked which of clustered or dispersed inputs produce nuclear ERK activation more efficiently, by varying the number of dendritic branches on which the stimulated spines resided. Clustered stimulation of all 7 spines on a single branch failed to induce any nuclear ERK activity increase (Fig. 2, D and E). In contrast, stimulating 3-7 spines distributed over 2-7 branches resulted in marked activation of nuclear ERK (Fig. 2, D and E; all P < 0.05). Thus, signal integration over multiple dendritic branches is required to induce nuclear activation of ERK. We further investigated why dispersed stimulation is more efficient by imaging ERK activation at the branching point in the primary dendritic trunk after stimulating within a dendritic branch (fig. S8). ERK activity at the branching point was saturated when 2 spines within a dendritic branch were stimulated. Thus, additional stimulation to a branch should not cause additional ERK activation in the primary dendrite or in the nucleus.
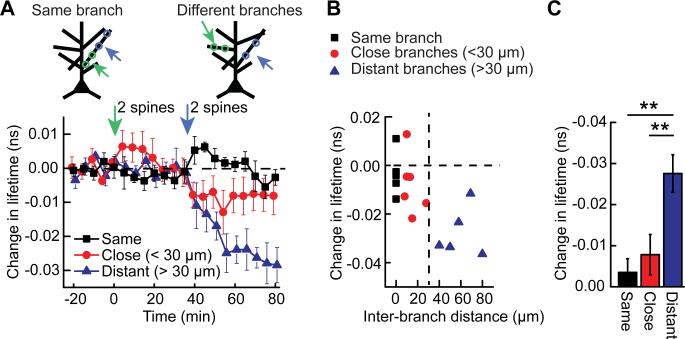
What is the range of the spatiotemporal integration of the nuclear ERK activation? To address this question, we stimulated 2 spines in a dendritic branch first, waited for 30 min, and then stimulated additional 2 spines in the same branch or another dendritic branch 5-80 μm away from the first branch (Fig. 3A). As expected, the first set of stimulation did not activate nuclear ERK because both stimulated spines were on the same branch (see Fig. 2D). However, the second set of stimulation, when applied to another branch separated by more than 30 μm, significantly increased nuclear ERK activity (Fig. 3). Thus, nuclear ERK signaling can integrate synaptic stimulation over more than 30 min. When the second set of stimulation was applied to the same branch or a nearby branch (within 30 μm), we did not observe a significant increase in nuclear ERK activity (Fig. 3). Thus, nuclear ERK is activated more efficiently by spatially distributed pattern of stimulation.
Fig. 3. Wide-range spatiotemporal integration of ERK signaling from different branches.
(A) Time course of EKARnuc fluorescence lifetime in response to sequential stimulation of 2 spines on a branch (green arrow), which does not cause any nuclear ERK activation (as predicted from Fig. 2D), followed by stimulation of 2 additional spines 30 min after the first stimulation (blue arrow). When the second stimulation was on the same branch or a close branch (inter-branch distance <30 μm), ERK was not activated in the nucleus. However, when the second stimulation was applied to a dendritic branch well apart from the first stimulated branch (inter-branch distance >30 μm), significant activation occurred. (N = 6, 6, and 5 neurons for Same, Close and Distant, respectively). (B) Relationship between change in fluorescence lifetime [averaged over 70-80 min from data shown in (A)] and inter-branch distance measured along the primary dendrite. (C) Quantification of ERK signaling shown in (B) (**P < 0.01). Data are presented as mean ± SEM.
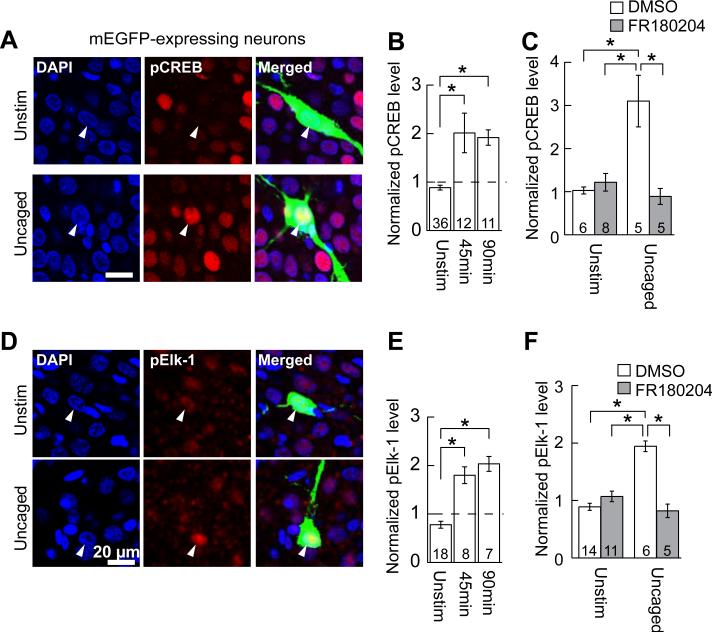
Is nuclear ERK activation induced by stimulation of a few dendritic spines sufficient to regulate gene transcription? To address this question, we used immunostaining to measure the activity of transcription factors cAMP response element-binding protein (CREB) and E26-like transcription factor-1 (Elk-1)(Fig. 4), because these molecules are known to be activated by neuronal activity via ERK activation (14). After glutamate uncaging at 7 spines of mEGFP-expressing CA1 neurons, the slices were immunostained for CREB phosphorylated at Ser-133 (Fig. 4, A and B) or Elk-1 phosphorylated at Ser-383 (Fig. 4, D and E), the phosphorylation sites required for their transcriptional activity (20, 21). The levels of phosphorylated CREB and Elk-1 were higher in uncaged neurons than in surrounding untransfected neurons at 45 and 90 min after uncaging (Fig. 4, B and E). In contrast, mEGFP-positive, unstimulated neurons in the same slices did not show any increase in phosphorylated CREB and Elk-1 (Fig. 4). Furthermore, CREB and Elk-1 phosphorylation was abolished when ERK inhibitor FR180204 was applied before stimulation (Fig. 4, C and F). Thus, stimulation of a few spines regulates activities of transcription factors CREB and Elk-1 through ERK.
Fig. 4. Transcription factors are phosphorylated in response to 7-spine stimulation in an ERK-dependent fashion.
(A) Immunofluorescent images of phosphorylated CREB (pCREB; red). Seven spines of neurons expressing mEGFP (green) were stimulated and the slices were fixed 45 min after the stimulation (“Uncaged”). Unstimulated mEGFP neurons in the same slices are also shown as negative controls (“Unstim”). The nuclei were stained with DAPI (blue). The relatively high basal level of pCREB in a subpopulation of neurons is probably due to spontaneous circuit activity (7). (B) Quantification of the fluorescence intensity of pCREB in the nuclei identified with DAPI. The fluorescence in the nuclei of mEGFP-expressing neurons was normalized to the average fluorescence in nuclei of 5 surrounding untransfected neurons. Uncaged neurons were always paired with unstimulated neurons in the same slices (15). The numbers of neurons are indicated at the bottom of the graph (*P < 0.05). (C) ERK inhibitor FR180204 blocks uncaging-induced CREB phosphorylation. DMSO was used as vehicle control. (D to F) Same as (A) to (C), except phosphorylated Elk-1 (pElk-1) instead of pCREB was analyzed. Data are presented as mean ± SEM.
Here, induction of structural LTP in a few (3-7) spines led to nuclear ERK activation and subsequent regulation of downstream transcription factors CREB and Elk-1. Because each CA1 pyramidal neuron has roughly 10,000 synapses, activation of only a tiny fraction (< 0.1%) of its synapses can activate nuclear signaling that regulates gene transcription. Many studies have demonstrated that somatonuclear Ca2+ transients, caused by somatic depolarization and Ca2+ wave propagation, play an important role in regulation of activity-dependent gene transcription [(19, 22), reviewed by (23)]. However, under our experimental condition, this mechanism is unlikely to play a role. Our sLTP induction protocol produced Ca2+ elevation highly restricted to the vicinity of the stimulated spines and only small somatic voltage changes. In addition, activation of VSCCs was not required for nuclear ERK activation. Moreover, the slow signal transmission strongly suggests that biochemical messengers relay information from activated synapses to the nucleus (10, 11, 24-27). Because Ras activity is known to spread over ~10 μm upon single-spine stimulation (3), downstream ERK may diffuse further and invade the nucleus as a result of multiple-spine stimulation. Consistent with this hypothesis, we observed that ERK2 translocates into the nucleus in response to stimulation of a few spines. Further, the onset of ERK activation in response to distal dendrite stimulation was consistent with the speed of ERK diffusion (15). Considering that the size of the nucleus is ~2,000 times larger than that of an average spine (2) and that the phosphatase-rich cytosol needs to be traversed by phosphorylated ERK, additional mechanisms such as a PKC-ERK positive feedback loop and physical protection would be required to aid the long-distance, persistent synapse-to-nucleus ERK signaling (27, 28).
The long-distance spatiotemporal integration in inducing nuclear ERK activation may have significant implications for the functional organization of dendritic inputs. Many studies have shown that synaptic potentiation tends to occur in a spatially clustered fashion (29-31), due to electrical integration and biochemical crosstalk within a short stretch of dendrite (32-35). However, because potentiated synapses would recruit stronger local membrane depolarization and biochemical signaling in the surrounding region in a positive-feedback manner, this mechanism potentially leads to accumulation of potentiated synapses in one dendritic branch (36). The nuclear signaling efficiently induced by spatially dispersed inputs may be important for counter-balancing the tendency to accumulate potentiated spines in one branch and developing balanced spatial distribution of synaptic weights.
Supplementary Material
Acknowledgments
We thank A. West, S. Dudek, S. Soderling, K. Tanaka and G. Augustine for discussion, and N. Hedrick and L. Colgan for comments on the manuscript. We also thank A. Wan for preparing cultured slices and D. Kloetzer for laboratory management. This study was funded by Howard Hughes Medical Institute, National Institute of Mental Health, and National Institute of Neurological Disorders and Stroke. The authors made the following contributions: S.Z. and R.Y. designed experiments. S.Z. collected the majority of the data. E.D.A. performed the GCaMP imaging with pharmacological inhibitors. P.P.-B. performed the patch clamp experiments. S.Z. and R.Y. analyzed the data and wrote the paper. All authors discussed the results and commented on the manuscript.
References and Notes
- 1.Nguyen PV, Abel T, Kandel ER. Requirement of a critical period of transcription for induction of a late phase of LTP. Science. 1994;265:1104. doi: 10.1126/science.8066450. [DOI] [PubMed] [Google Scholar]
- 2.Adams JP, Dudek SM. Late-phase long-term potentiation: getting to the nucleus. Nat Rev Neurosci. 2005;6:737. doi: 10.1038/nrn1749. [DOI] [PubMed] [Google Scholar]
- 3.Harvey CD, Yasuda R, Zhong H, Svoboda K. The Spread of Ras Activity Triggered by Activation of a Single Dendritic Spine. Science. 2008;321:136. doi: 10.1126/science.1159675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee S-JR, Escobedo-Lozoya Y, Szatmari EM, Yasuda R. Activation of CaMKII in single dendritic spines during long-term potentiation. Nature. 2009;458:299. doi: 10.1038/nature07842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Murakoshi H, Wang H, Yasuda R. Local, persistent activation of Rho GTPases during plasticity of single dendritic spines. Nature. 2011;472:100. doi: 10.1038/nature09823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Patterson MA, Szatmari EM, Yasuda R. AMPA receptors are exocytosed in stimulated spines and adjacent dendrites in a Ras-ERK-dependent manner during long-term potentiation. Proc Natl Acad Sci U S A. 2010;107:15951. doi: 10.1073/pnas.0913875107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhu JJ, Qin Y, Zhao M, Van Aelst L, Malinow R. Ras and Rap Control AMPA Receptor Trafficking during Synaptic Plasticity. Cell. 2002;110:443. doi: 10.1016/s0092-8674(02)00897-8. [DOI] [PubMed] [Google Scholar]
- 8.Impey S, et al. Cross Talk between ERK and PKA Is Required for Ca2+ Stimulation of CREB-Dependent Transcription and ERK Nuclear Translocation. Neuron. 1998;21:869. doi: 10.1016/s0896-6273(00)80602-9. [DOI] [PubMed] [Google Scholar]
- 9.West AE, Griffith EC, Greenberg ME. Regulation of transcription factors by neuronal activity. Nat Rev Neurosci. 2002;3:921. doi: 10.1038/nrn987. [DOI] [PubMed] [Google Scholar]
- 10.Martin KC, et al. MAP Kinase Translocates into the Nucleus of the Presynaptic Cell and Is Required for Long-Term Facilitation in Aplysia. Neuron. 1997;18:899. doi: 10.1016/s0896-6273(00)80330-x. [DOI] [PubMed] [Google Scholar]
- 11.Deisseroth K, Mermelstein PG, Xia H, Tsien RW. Signaling from synapse to nucleus: the logic behind the mechanisms. Curr. Opin. Neurobiol. 2003;13:354. doi: 10.1016/s0959-4388(03)00076-x. [DOI] [PubMed] [Google Scholar]
- 12.Harvey CD, et al. A genetically encoded fluorescent sensor of ERK activity. Proc Natl Acad Sci U S A. 2008;105:19264. doi: 10.1073/pnas.0804598105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Matsuzaki M, Honkura N, Ellis-Davies GCR, Kasai H. Structural basis of long-term potentiation in single dendritic spines. Nature. 2004;429:761. doi: 10.1038/nature02617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thomas GM, Huganir RL. MAPK cascade signalling and synaptic plasticity. Nat Rev Neurosci. 2004;5:173. doi: 10.1038/nrn1346. [DOI] [PubMed] [Google Scholar]
- 15.See Supplementary Materials on Science Online.
- 16.Noguchi J, Matsuzaki M, Ellis-Davies GC, Kasai H. Spine-neck geometry determines NMDA receptor-dependent Ca2+ signaling in dendrites. Neuron. 2005;46:609. doi: 10.1016/j.neuron.2005.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Niswender CM, Conn PJ. Metabotropic Glutamate Receptors: Physiology, Pharmacology, and Disease. Annu. Rev. Pharmacol. Toxicol. 2010;50:295. doi: 10.1146/annurev.pharmtox.011008.145533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nakamura T, Barbara JG, Nakamura K, Ross WN. Synergistic release of Ca2+ from IP3-sensitive stores evoked by synaptic activation of mGluRs paired with backpropagating action potentials. Neuron. 1999;24:727. doi: 10.1016/s0896-6273(00)81125-3. [DOI] [PubMed] [Google Scholar]
- 19.Dudek SM, Fields RD. Somatic action potentials are sufficient for late-phase LTP-related cell signaling. Proc. Natl. Acad. Sci. USA. 2002;99:3962. doi: 10.1073/pnas.062510599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mayr B, Montminy M. Transcriptional regulation by the phosphorylation-dependent factor CREB. Nat Rev Mol Cell Biol. 2001;2:599. doi: 10.1038/35085068. [DOI] [PubMed] [Google Scholar]
- 21.Besnard A, Galan-Rodriguez B, Vanhoutte P, Caboche J. Elk-1 a transcription factor with multiple facets in the brain. Front Neurosci. 2011;5:35. doi: 10.3389/fnins.2011.00035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hardingham GE, Arnold FJ, Bading H. Nuclear calcium signaling controls CREB-mediated gene expression triggered by synaptic activity. Nat. Neurosci. 2001;4:261. doi: 10.1038/85109. [DOI] [PubMed] [Google Scholar]
- 23.Hagenston AM, Bading H. Calcium signaling in synapse-to-nucleus communication. Cold Spring Harb Perspect Biol. 2011;3:a004564. doi: 10.1101/cshperspect.a004564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Worley P, et al. Thresholds for synaptic activation of transcription factors in hippocampus: correlation with long-term enhancement. J. Neurosci. 1993;13:4776. doi: 10.1523/JNEUROSCI.13-11-04776.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Meffert MK, Chang JM, Wiltgen BJ, Fanselow MS, Baltimore D. NF-[kappa]B functions in synaptic signaling and behavior. Nat. Neurosci. 2003;6:1072. doi: 10.1038/nn1110. [DOI] [PubMed] [Google Scholar]
- 26.Thompson KR, et al. Synapse to Nucleus Signaling during Long-Term Synaptic Plasticity: a Role for the Classical Active Nuclear Import Pathway. Neuron. 2004;44:997. doi: 10.1016/j.neuron.2004.11.025. [DOI] [PubMed] [Google Scholar]
- 27.Karpova A, et al. Encoding and transducing the synaptic or extrasynaptic origin of NMDA receptor signals to the nucleus. Cell. 2013;152:1119. doi: 10.1016/j.cell.2013.02.002. [DOI] [PubMed] [Google Scholar]
- 28.Tanaka K, Augustine GJ. A Positive Feedback Signal Transduction Loop Determines Timing of Cerebellar Long-Term Depression. Neuron. 2008;59:608. doi: 10.1016/j.neuron.2008.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Makino H, Malinow R. Compartmentalized versus Global Synaptic Plasticity on Dendrites Controlled by Experience. Neuron. 2011;72:1001. doi: 10.1016/j.neuron.2011.09.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.De Roo M, Klauser P, Muller D. LTP Promotes a Selective Long-Term Stabilization and Clustering of Dendritic Spines. PLoS Biol. 2008;6:e219. doi: 10.1371/journal.pbio.0060219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fu M, Yu X, Lu J, Zuo Y. Repetitive motor learning induces coordinated formation of clustered dendritic spines in vivo. Nature. 2012;483:92. doi: 10.1038/nature10844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Govindarajan A, Israely I, Huang S-Y, Tonegawa S. The Dendritic Branch Is the Preferred Integrative Unit for Protein Synthesis-Dependent LTP. Neuron. 2011;69:132. doi: 10.1016/j.neuron.2010.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Losonczy A, Makara JK, Magee JC. Compartmentalized dendritic plasticity and input feature storage in neurons. Nature. 2008;452:436. doi: 10.1038/nature06725. [DOI] [PubMed] [Google Scholar]
- 34.Gasparini S, Magee JC. State-Dependent Dendritic Computation in Hippocampal CA1 Pyramidal Neurons. J. Neurosci. 2006;26:2088. doi: 10.1523/JNEUROSCI.4428-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Harvey CD, Svoboda K. Locally dynamic synaptic learning rules in pyramidal neuron dendrites. Nature. 2007;450:1195. doi: 10.1038/nature06416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Goldberg J, Holthoff K, Yuste R. A problem with Hebb and local spikes. Trends Neurosci. 2002;25:433. doi: 10.1016/s0166-2236(02)02200-2. [DOI] [PubMed] [Google Scholar]
- 37.Stoppini L, Buchs PA, Muller DA. A simple method for organotypic cultures of nervous tissue. J. Neurosci. Methods. 1991;37:173. doi: 10.1016/0165-0270(91)90128-m. [DOI] [PubMed] [Google Scholar]
- 38.McAllister AK. Biolistic transfection of neurons. Sci. STKE. 2000;2000:PL1. doi: 10.1126/stke.2000.51.pl1. [DOI] [PubMed] [Google Scholar]
- 39.Yasuda R, et al. Supersensitive Ras activation in dendrites and spines revealed by two-photon fluorescence lifetime imaging. Nat. Neurosci. 2006;9:283. doi: 10.1038/nn1635. [DOI] [PubMed] [Google Scholar]
- 40.Murakoshi H, Lee S-J, Yasuda R. Highly sensitive and quantitative FRET-FLIM imaging in single dendritic spines using improved non-radiative YFP. Brain Cell Biol. 2008;36:31. doi: 10.1007/s11068-008-9024-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pologruto TA, Sabatini BL, Svoboda K. ScanImage: Flexible software for operating laser-scanning microscopes. Biomed. Eng. Online. 2003;2:13. doi: 10.1186/1475-925X-2-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yasuda R, et al. Imaging calcium concentration dynamics in small neuronal compartments. Sci STKE. 2004;2004:pl5. doi: 10.1126/stke.2192004pl5. [DOI] [PubMed] [Google Scholar]
- 43.Tian L, et al. Imaging neural activity in worms, flies and mice with improved GCaMP calcium indicators. Nat Methods. 2009;6:875. doi: 10.1038/nmeth.1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Canagarajah BJ, Khokhlatchev A, Cobb MH, Goldsmith EJ. Activation mechanism of the MAP kinase ERK2 by dual phosphorylation. Cell. 1997;90:859. doi: 10.1016/s0092-8674(00)80351-7. [DOI] [PubMed] [Google Scholar]
- 45.Wiegert JS, Bengtson CP, Bading H. Diffusion and Not Active Transport Underlies and Limits ERK1/2 Synapse-to- Nucleus Signaling in Hippocampal Neurons. J. Biol. Chem. 2007;282:29621. doi: 10.1074/jbc.M701448200. [DOI] [PubMed] [Google Scholar]
- 46.Burack WR, Shaw AS. Live Cell Imaging of ERK and MEK. J. Biol. Chem. 2005;280:3832. doi: 10.1074/jbc.M410031200. [DOI] [PubMed] [Google Scholar]
- 47.Fukuda M, Gotoh Y, Nishida E. Interaction of MAP kinase with MAP kinase kinase: its possible role in the control of nucleocytoplasmic transport of MAP kinase. EMBO J. 1997;16:1901. doi: 10.1093/emboj/16.8.1901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Horgan AM, Stork PJS. Examining the mechanism of Erk nuclear translocation using green fluorescent protein. Exp. Cell Res. 2003;285:208. doi: 10.1016/s0014-4827(03)00037-5. [DOI] [PubMed] [Google Scholar]
- 49.Lidke DS, et al. ERK nuclear translocation is dimerization-independent but controlled by the rate of phosphorylation. J. Biol. Chem. 2010;285:3092. doi: 10.1074/jbc.M109.064972. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.