Abstract
Neurodevelopmental and neuropsychiatric disorders result from complex interactions between critical genetic factors and as-yet-unknown environmental components. To gain clinical insight, it is critical to develop a comprehensive understanding of these genetic components. RBFOX1, an RNA splicing factor, regulates expression of large genetic networks during early neuronal development and haploinsufficiency causes severe neurodevelopmental phenotypes including autism spectrum disorder (ASD), intellectual disability, and epilepsy. Genomic testing in individuals and large patient cohorts has identified phenotypically similar cases possessing copy number variations in RBFOX1, implicating the gene as an important cause of neurodevelopmental disease. However, a significant proportion of the observed structural variation is inherited from phenotypically normal individuals, raising questions regarding overall pathogenicity of variation at the RBFOX1 locus. In this article, we discuss the molecular, cellular, and clinical evidence supporting the role of RBFOX1 in neurodevelopment and present a comprehensive model for the contribution of structural variation in RBFOX1 to ASD.
Keywords: autism, autism spectrum disorder, RNA splicing, RBFOX1, copy number variation, neurodevelopment, A2BP1, FOX1
Neurodevelopment and the RBFOX1 RNA Splicing Factor
During development, a series of intricate programs of gene regulation must specifically occur within neurons, resulting in both temporal and spatial patterns of distinct gene expression. This results in an organized program of molecular and cellular actions and interactions that translate into the connectivity that underlies the function of the neurotypical human brain. Not surprisingly, disruption of these regulatory programs has been shown to cause a broad range of neurodevelopmental disorders including autism spectrum disorder, schizophrenia, and many others (Bill & Geschwind, 2009; B. L. Fogel et al., 2012; Pescosolido, Yang, Sabbagh, & Morrow, 2012). Over the past decade, we have come to better understand the workings of various aspects of this complex system through the study of key regulatory factors that guide these neurodevelopmental cascades. Many of these factors directly influence gene expression through transcription and/or pre-mRNA alternative splicing, both fundamental processes to the development of tissue-specific genetic programs.
The RBFOX1 gene (also referred to as A2BP1 or FOX1) encodes a splicing regulatory factor, specifically expressed in neurons and muscle, responsible for widespread effects, both enhancing and inhibiting the alternative splicing of many cellular pre-mRNAs (B. L. Fogel, et al., 2012; Underwood, Boutz, Dougherty, Stoilov, & Black, 2005; E. T. Wang et al., 2008). Several lines of evidence indicate that RBFOX1 regulates the alternative splicing of large tissue-specific gene networks including multi-species comparative genomics of splicing regulatory elements (Yeo, Nostrand, & Liang, 2007), bioinformatic analysis of genes alternatively spliced during embryonic cell differentiation (Yeo et al., 2007), genome-wide target site prediction strategies (Zhang et al., 2008), genome-wide transcriptome assessment (E. T. Wang, et al., 2008), multi-species RNA-binding protein motif analysis (Ray et al., 2013), and gene-specific knockdown in differentiated human neural progenitor cell lines (B. L. Fogel, et al., 2012). In addition to mediating alternative splicing, functional roles for RBFOX1 have been identified in the transcriptional regulation of additional gene networks by directly mediating RNA stability (Ray, et al., 2013), influencing transcription (Usha & Shashidhara, 2010), or indirectly through effects on other regulators of gene expression (B. L. Fogel, et al., 2012).
RBFOX1 Genetic Variation and Autism Spectrum Disorder
The available molecular and cellular evidence described above supports RBFOX1 as a high-level regulatory factor in early brain development, so it is not surprising that a growing number of patients with neurodevelopmental phenotypes have been identified with mutations disrupting RBFOX1. These phenotypes, some of which are quite severe, include syndromes of autism spectrum disorder (ASD), intellectual disability, and epilepsy as well as other neuropsychiatric phenotypes.
The RBFOX1 gene is quite large, spanning 2.4 MB on chromosome 16p13.3, making it one of the largest genes in the human genome. It must be noted that the nomenclature used to describe the RBFOX1 genetic architecture over the years has not been consistent across the literature, and it can often be difficult to compare the functional significance of reported variants. This is due, in part, to the fact that the RBFOX1 gene utilizes multiple promoters and undergoes a wide variety of alternative splicing in a tissue-specific fashion, with many of the functional cellular transcripts likely not yet characterized or annotated (B. L. Fogel, et al., 2012; Underwood, et al., 2005) (Figure 1). Furthermore, the major neuronal transcripts initially reported in the literature for humans have been shown to actually be muscle-specific transcripts, the same as originally reported in the mouse (B. L. Fogel, et al., 2012). We advocate the use of the naming scheme first published in Underwood et al. (B. L. Fogel, et al., 2012; Underwood, et al., 2005) for describing variants found in patients (Figure 1A; Supplemental Table 1). Standardized nomenclature is particularly important for determining the pathogenicity of an individual sequence or structural change likely to affect neuron-specific transcripts or isoforms.
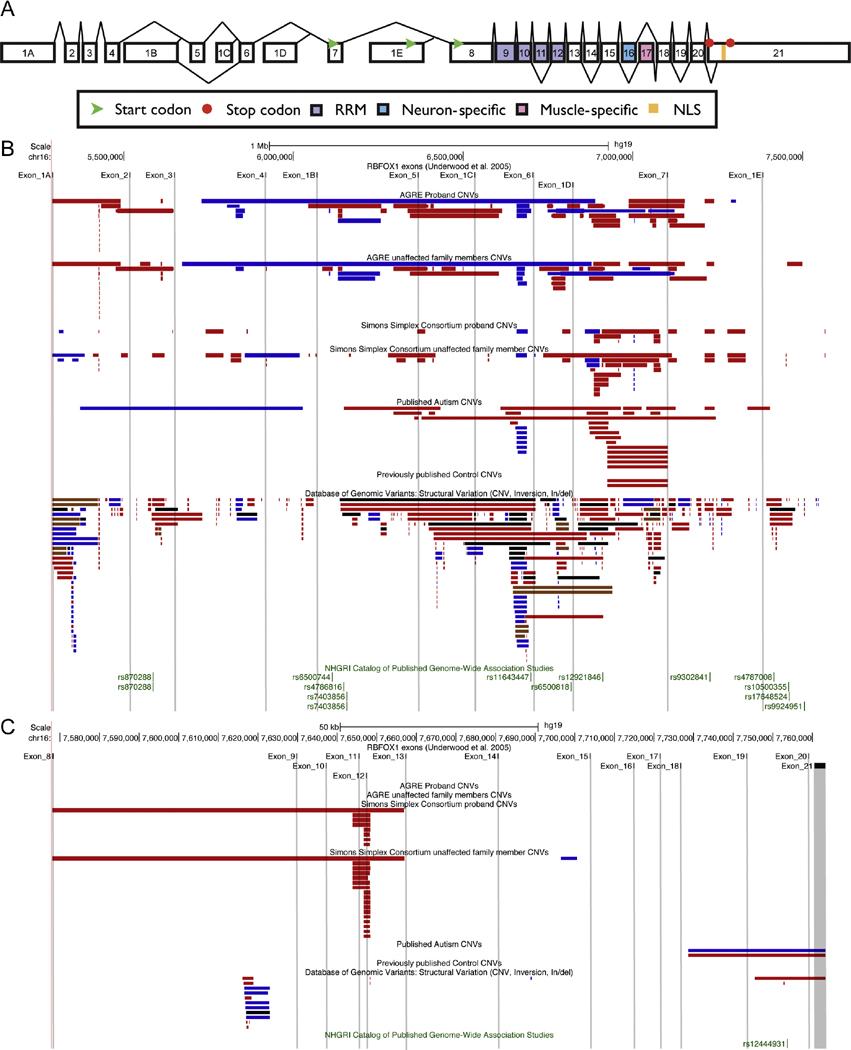
Figure 1. RBFOX1 Genomic Architecture and Copy Number Variation associated with Autism Spectrum Disorder.
A) RBFOX1 gene structure is complex with multiple transcriptional-start sites, translational-start sites, multiple isoforms, and alternate endings. The distribution of CNVs associated with autism spectrum disorder in the RBFOX1 locus [HG19-chr16:5289804–7763340] are shown and compared to the database of genomic variants. B) Exons 1A through 1E. C) Exons 8 through 21. Observed structural variation is shown clustered at the 5′ untranslated region between exons 1 through 7. Single nucleotide polymorphisms related to the locus and showing genome-wide association in ASD also follow a similar distribution pattern (green). Red bars = deletions; Blue bars = duplications; Brown bars = insertions and deletions; Black bars = unknown feature.
One of the earliest clinical descriptions of a neurodevelopmental phenotype associated with RBFOX1 was of two patients with likely ASD, mental retardation, and epilepsy associated with haploinsufficiency caused by de novo translocations involving chromosome 16 (Bhalla et al., 2004). This was followed by the description of another patient with a similar syndrome, also caused by a haploinsufficiency due to a de novo translocation, which incorporated validated rating scales to clinically diagnose the patient with autism (Martin et al., 2007). These two initial case reports provided strong confirmatory evidence of causality between RBFOX1 mutation and human disease, resulting in its classification as an ASD candidate gene (Simons Foundation Autism Research Initiative gene database; https://sfari.org/).
Other structural variations in RBFOX1, most notably copy number variations (CNVs; arbitrarily defined as deletions or duplications greater than 1 kilobase in size), have also been associated with human neurodevelopmental disease. CNVs occur commonly in the population and represent a significant source of human genetic variation (Brent L. Fogel & Geschwind, 2012) (Database of Genomic Variants, DGV; http://dgv.tcag.ca/dgv/app/home) (Iafrate et al., 2004). The link between RBFOX1 CNVs and autism has been suggested by the presence of de novo CNVs within ASD cohorts (Sebat et al., 2007) and in samples from isolated autistic patients (Wintle et al., 2011).
More recently, a number of case reports have highlighted specific RBFOX1 CNVs linked to ASD. Mikhail et al. reported a 3-year-old microcephalic boy with developmental delay, language delay, and an intragenic deletion involving noncoding exons 6 and 1D in RBFOX1 of unknown inheritance status as his parents could not be tested (Mikhail et al., 2011). Davis et al. reported the case of a 12-year-old boy with autism determined through validated rating scales, global hypotonia, a mild developmental left hemiparesis, and deficits in motor planning and coordination with the dominant right hand (Davis et al., 2012). The patient was found to have a deletion involving noncoding exon 5 of RBFOX1 inherited through the father who was not clinically evaluated (Davis, et al., 2012). Most recently, Zhao reported 13 patients with deletions (1 maternally inherited involving noncoding exon 6, 1 de novo involving multiple coding exons between 1D and 10, and the inheritance of the rest could not be determined) and 1 patient with a duplication (of undetermined inheritance) within the RBFOX1 locus (Zhao, 2013). Two patients in this report were excluded from phenotypic analysis due to 1) the finding of an alternate Mendelian genetic diagnosis in addition to a deletion involving RBFOX1 exon 5, and 2) the presence of multiple non-neurological congenital anomalies of unclear etiology in a patient with an RBFOX1 duplication involving exons 1D and 7 (Zhao, 2013). The major clinical findings in the other patients (who all possessed intronic deletions except for one that involved noncoding exon 6) included global developmental delay (58%), epilepsy (50%), macro- or microcephaly (50%), and renal problems (33%) (Zhao, 2013). Of note, 50% of the patients with epilepsy also had developmental delay, one patient with epilepsy but without developmental delay was noted as intellectually disabled, and one patient with developmental delay but without epilepsy was given a clinical diagnosis of autism (Zhao, 2013). The extent of the clinical evaluation for ASD each patient received in this cohort was unfortunately not reported, so it cannot be assumed the other patients lacked such phenotypes.
In general, it has been presumed that the above CNVs lead to reduced RBFOX1 expression. If true, then it would be consistent with the observations that RBFOX1 haploinsufficiency results in a syndrome characterized primarily by neurodevelopmental and neurological phenotypes including ASD, intellectual disability, and epilepsy (Bhalla, et al., 2004; Martin, et al., 2007). Data from knockdown studies in human neural progenitor cell lines modeling haploinsufficiency during neuronal differentiation demonstrate widespread changes in RNA splicing and gene expression (B. L. Fogel, et al., 2012), and studies of the Rbfox1 neural-specific knockout mouse show alterations in synaptic transmission, increased membrane excitability, and a predisposition to seizures (Gehman et al., 2011). Interestingly, whole transcriptome analysis in the brains of autistic patients reveals decreased levels of RBFOX1 and dysregulation of RBFOX1-dependent alternative splicing (Voineagu et al., 2011), similar to the effects seen in haploinsufficient neuronal cell lines (B. L. Fogel, et al., 2012). However, in the majority of cases, the impact of the identified CNV to RBFOX1 expression or function is unclear, as evidenced by the presence of identical CNVs in controls and unaffected family members (Figure 1B and 1C).
To better understand the contribution of structural variation in RBFOX1 to the development of autism and related disorders, we compiled all published CNVs including those from 2 of the largest ASD cohorts, the Autism Genetic Resource Exchange (AGRE), a primarily multiplex cohort of families with multiple affected siblings, and the Simons Simplex Cohort (SSC), which contains families with only a single affected child (Figure 1B and 1C). In the AGRE cohort, we found that 2.2% of patients carried a CNV in the RBFOX1 locus compared to 0.7% of unaffected siblings (OR=3.19, p=0.006, 95% CI [1.27, 10.28]). In contrast, the SSC did not show a significantly increased odds ratio, with 2.6% of probands and 2.4% of their normal siblings having a CNV (OR=1.11, p=0.77, 95% CI [0.59, 2.12]. While we cannot rule out ascertainment bias or differences in population structure, this data demonstrates a significant enrichment of RBFOX1 CNVs in a multiplex but not in a simplex cohort. This analysis reveals two additional striking features. The first is that, irrespective of cohort, CNVs in RBFOX1 tend to be inherited from an unaffected parent. Second, we observe a locational bias of CNVs toward the 5′ untranslated exons 1–7 (Figure 1B) compared to the constitutively translated exons 8–21 (Figure 1C) with a preponderance of ASD-related deletions occurring in the intron prior to exon 7, which contains a potential translational start site (Figure 1B). Although it is tempting to speculate that these CNVs lead to a correlative alteration of the expression level of RBFOX1, qualitatively, we also observe a large number of CNVs in this intron within unaffected individuals in the DGV (Iafrate, et al., 2004) (Figure 1B and C). Although the CNVs in the DGV appear to be somewhat smaller overall than those in those identified in ASD patients, we unfortunately cannot conclude that larger CNVs have a higher impact on phenotype, primarily due to differences in acquisition between the groups. Therefore, taking all these observations together, we conclude that while RBFOX1 CNVs confer a heritable risk of developing ASD, the majority of RBFOX1 CNVs do not cause haploinsufficiency in isolation and other factors, genetic or environmental, likely contribute.
Further complicating this clinical landscape, the phenotypic spectrum of disease associated with disruption of RBFOX1 regulation and/or function appears to extend into a cacophony of other phenotypes, some commonly found in ASD patients, such as epilepsy and developmental delay. RBFOX1 CNVs have been reported in patients with idiopathic generalized epilepsy, including one patient with epilepsy and ASD that removes exon 7 (Lal et al., 2013). Additionally, the intron prior to exon 7 has a high number of CNVs identified within the International Standard for Cytogenomic Arrays (ISCA) database (https://www.iscaconsortium.org/) that are associated with phenotypes such as global developmental, intellectual, and speech delay. It is possible these reports could reflect populations that are on the ASD spectrum, but incompletely characterized. RBFOX1 CNVs have also been identified in populations of individuals with schizophrenia (International Schizophrenia Consortium, 2008; Melhem et al., 2011; Priebe et al., 2013; Xu et al., 2008), however at a very low rate. The strongest evidence for this association is an increased risk for male-specific schizophrenia (OR=8.2, 95%CI, 0.8–84.4) due to duplications occurring just prior to exon 6, predicted to be inherited from a single founder event (Melhem, et al., 2011). Lastly, various cancer cells also show structural variation in RBFOX1 (Andersen et al., 2011; Bass et al., 2011; Linnebacher et al., 2013) suggesting either a potential post-developmental role in cellular growth and/or differentiation in other tissues or disease consequences of aberrant expression/regulation of RBFOX1 in non-specific tissues.
Finally, although unlikely to be directly causative, RBFOX1 single nucleotide polymorphism (SNP) variants have also be implicated in autism (K. Wang et al., 2009) as well as a diverse array of other human conditions including Alzheimer disease (Kohannim et al., 2012), bipolar disorder (Le-Niculescu et al., 2009), attention-deficit hyperactivity disorder (Elia et al., 2010), schizoaffective disorder (Hamshere et al., 2009), obesity (Ma et al., 2010), and refractive error (Stambolian et al., 2013). It is unclear whether these SNPs may, in some cases, co-segregate with other rare sequence or structural variants directly influencing RBFOX1 expression or regulation.
The Contributions of RBFOX1 Model Systems to Molecular Pathogenesis
The first member of the RBFOX1 family of alternative splicing factors was initially reported in a search for modifiers of sexual differentiation in C. elegans (Hodgkin, Zellan, & Albertson, 1994). Feminization on X (fox-1) was identified as a dominant factor that feminizes XO males, and causes high levels of male lethality due to its ability to splice the xol-1 [XO (male) lethality] gene (Kuroyanagi, 2009). The Drosophila melanogaster homolog (dA2bp1, also known as CG3206) and zebrafish homologs (rbfox1 and rbfox1l) were subsequently identified and shown upon constitutive knockdown to be embryonic lethal, suggesting an early role in embryogenesis (Bajpai, Sambrani, Stadelmayer, & Shashidhara, 2004; Gallagher et al., 2011; Hodgkin, et al., 1994). Although the mechanism underlying this early lethality has not been explained, it provides one potential explanation for the high sequence conservation in RBFOX1 observed from worms to humans and the lack of CNVs found within the coding region of patient and control samples (Figure 1C). Human RBFOX1 was original identified through its interaction with ataxin-2, the protein causing the neurodegenerative disease spinocerebellar ataxia type 2 (SCA2) (Shibata, Huynh, & Pulst, 2000). Although the functional significance of this interaction is not yet fully understood, it likely contributes to ataxin-2’s established role in RNA processing and translation (C. Lim & Allada, 2013). Interestingly, RBFOX1 is present in other protein-protein and gene interaction networks related to cerebellar ataxia (B.L. Fogel & Perlman, 2011; J. Lim et al., 2006), and it affects the splicing and transcription of other ataxia genes in human neural progenitor cells (B. L. Fogel, et al., 2012), an intriguing observation given that cerebellar dysfunction may contribute to the autistic phenotype (Fatemi et al., 2012).
Jin and coworkers were the first to confirm that zebrafish rbfox1l was involved in alternative RNA splicing (Jin et al., 2003). They demonstrated that rbfox1l could bind to an intronic GCAUG pentanucleotide and affect splicing by repressing exon inclusion if the binding site was upstream or enhancing inclusion if the site were downstream (Jin, et al., 2003). The mechanism of repressing exon inclusion involves the hindrance of pre-spliceosomal complexes (Kuroyanagi, 2009), and can be overcome by overexpression of spliceosomal components (Zhou & Lou, 2008). By extension, reduction of RBFOX1 due to haploinsufficiency in patients could allow spliceosome factors to outcompete RBFOX1 for binding, thereby altering splicing for some targets. The mechanism for enhancing exon inclusion is less well defined. Sun and coworkers demonstrated that the c-terminus of RBFOX1 is required for this process, implicating protein-protein interactions as key to enhancing exon usage (Sun, Zhang, Fregoso, & Krainer, 2012). Our recent studies of splicing in human neural progenitor cell lines demonstrated globally that downstream sequences tend to function as enhancers while upstream sequences can mediate both repression and enhancement of specific exons (B. L. Fogel, et al., 2012), suggesting a role for local context in determining the function of RBFOX1. Furthermore, only 56% of the splicing events colocalized with the canonical RBFOX1 binding site, suggesting an interplay of both direct and indirect regulatory mechanisms, or possibly non-canonical binding sites that have yet to be discovered (B. L. Fogel, et al., 2012).
A Model for the Dysregulation of RBFOX1 in Human Neurodevelopmental Disease
As discussed above, a great deal of scientific evidence supports a role for RBFOX1 in the regulation of gene expression during human neurodevelopment. Clinical evidence further supports an association with neurodevelopmental disease in humans when haploinsufficient. The observation of noncoding de novo structural variants in RBFOX1 in patients with ASD would be consistent with the hypothesis that such variation leads to haploinsufficiency. However, the finding that the majority of structural variation in these patients is inherited, including variants similar to those seen de novo (Figure 1), appears to counteract that hypothesis. An imprinting effect seems unlikely as there does not appear to be a sex preference to CNV inheritance and disease association. How then does one reconcile this data with the scientific evidence? One possible conclusion is that much of the noncoding structural variation seen is in fact non-pathogenic and the observation of multiple CNVs in ASD patients is the product of the large size of the RBFOX1 gene, the commonality of structural variation in the human genome (Brent L. Fogel & Geschwind, 2012), and an increased rate in the occurrence of such variation at that locus (Bass, et al., 2011; Yi & Li, 2005). However, if this were the case, it would be expected that CNVs in RBFOX1 would occur at equal frequency in the population as a whole, without enrichment in ASD cohorts. Therefore, the finding of an enrichment in inherited RBFOX1 CNVs using stringent criteria in a large multiplex ASD cohort, but not in a large simplex cohort as described above, points to a different interpretation (see Figure 2). In this model, RBFOX1 CNVs confer an increased risk of developing ASD, dependent upon unknown genetic or environmental factors that influence RBFOX1-regulated cellular programs.
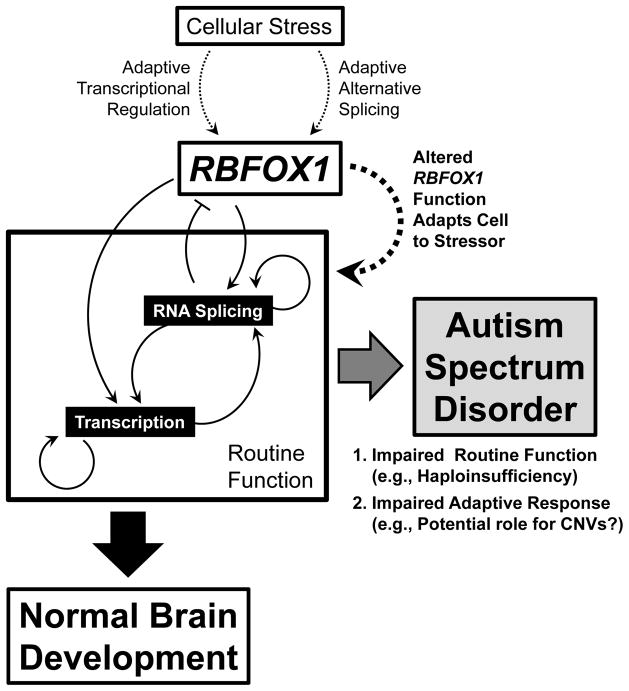
Figure 2. Proposed Model for RBFOX1 Dysregulation in Autism Spectrum Disorder.
During neurodevelopment, RBFOX1 regulates large genetic networks via direct effects on RNA splicing, mRNA stability, and transcriptional regulation as well as indirect effects on gene transcription leading to normal neuronal development. If this function is disrupted (e.g., by haploinsufficiency of RBFOX1), development is sufficiently perturbed resulting in autism spectrum disorder (ASD). As discussed in the text, RBFOX1 may play a further role in the adaptive response to cellular stress by regulating RNA splicing or transcriptional effects in response to environmental stimuli to maintain normal function. If this process is perturbed, this may also result in ASD. Inherited structural variation (CNVs) may damage RBFOX1 RNA processing or transcriptional signals and thus impair this adaptive response, thereby increasing the risk of ASD occurring upon exposure to certain environmental stressors during critical stages in neurodevelopment.
Regulation of splicing factors can have profound consequences with regard to neurodevelopmental outcome. There are two examples of Rbfox1 transcriptional regulation. In zebrafish, the homeobox-transcription factor Otp in complex with phospho-Creb binds the rbfox1 promoter in response to physical or osmotic stress (Amir-Zilberstein et al., 2012). Subsequently, Rbfox1 regulates splicing of the Pacap-receptor, pac1, that in turn modulates corticotropin-releasing hormone (Crh) levels, stimulating recovery from stress (Amir-Zilberstein, et al., 2012). Increases in Crh can manifest as increased anxiety or disturbances of sleep (Holsboer & Ising, 2008), both of which are common in ASD (Mazzone, Ruta, & Reale, 2012; Richdale & Schreck, 2009). A second candidate transcriptional regulator is Myt1l, which can bind four distinct binding sites in the Rbfox1 promoter (Hu et al., 2013). In an analysis of human and mouse expression data, consistent coexpression is seen between RBFOX1 and MYT1L in a dataset from patients with frontotemporal dementia (chosen due to an observed 2 fold reduction of MYT1L) and in two large databases that collate expression data; the UCLA Gene expression tool (UGET) and COXPRESdb (Bill, unpublished observation) (Chen-Plotkin et al., 2008) (Day et al., 2009) (Day, Carlson, Dong, O’Connor, & Nelson, 2007; Obayashi & Kinoshita, 2011). Since MYT1L has been shown to be critical for the transdifferentiation of IPSCs (Takahashi & Yamanaka, 2006), and has been associated with ASD (Meyer, Axelsen, Sheffield, Patil, & Wassink, 2012), intellectual delay and obesity (Stevens et al., 2011), and schizophrenia (Y. Lee et al., 2012; Van Den Bossche et al., 2013; Vrijenhoek et al., 2008), this is a finding of particular clinical relevance. As the regulation of RBFOX1 transcription is not yet fully understood, it is possible that CNVs in the 5′ UTR may interfere with this process, either constitutively or in response to certain stressors requiring modifications in RBFOX1 expression (Figure 2).
It is known that many genes, including RBFOX1, undergo extensive alternative splicing (B. L. Fogel, et al., 2012; Underwood, et al., 2005), and that such splicing patterns can be altered by changes to the cellular environment (J. A. Lee, et al., 2009). Rbfox1 RNA has been shown to be regulated via effects on RNA stability (Pistoni et al., 2013) and alternative splicing (Damianov & Black, 2010; Lee, et al., 2009). Rbfox1 can autoregulate the splicing of its own RNA binding domain creating a functional dominant negative, and thus regulate its own activity (Damianov & Black, 2010). Furthermore, Lee et al. have demonstrated that cellular depolarization, known to widely affect RNA splicing, causes alternative splicing of murine Rbfox1 in neurons, resulting in a change in subcellular localization of the protein and subsequent reversal of the effects of depolarization on the splicing of other Rbfox1 targets, which they interpret as a novel adaptive mechanism to chronic stimuli (J. A. Lee, et al., 2009). Regulation of RBFOX1 alternative splicing in response to cellular stressors could therefore play a meaningful role in neurodevelopment, and provide another possible regulatory mechanism CNVs may affect (Figure 2).
Given the extensive networks of genes regulated both directly and indirectly by RBFOX1 (B. L. Fogel, et al., 2012; Ray, et al., 2013), it is likely that various stimuli could occur throughout the course of neuronal development which require an adaptive response from RBFOX1, alone or in conjunction with other factors, to maintain normal expression profiles. Therefore, it may be supposed that mutations in RBFOX1 could exist which impair these adaptive responses while having no, or minimal, effect on routine function. Applying this scenario to human neurodevelopment, noncoding variation that directly disrupts RBFOX1 expression would act to cause haploinsufficiency and therefore disease, whereas variation altering accessory regulatory responses (e.g., various target binding sequences) would only lead to disease under cellular conditions requiring function of those elements. If such conditions are rare, or only transient, then the ability to mount a limited adaptive response may be tolerated, but if the condition is such that the adaptive response is essential for normal brain development, this inability to respond could result in disease. This synergizes well with the suspected contribution of environmental factors to ASD if exposure occurs during critical points in brain development (Herbert, 2010), and suggests a mechanism whereby variation in the RBFOX1 gene could modify ASD risk under certain conditions, but not others. Families tend to stay in the same environment for long periods of time and, therefore, children are often exposed to the same environmental stressors as their siblings. If RBFOX1 CNVs confer ASD risk in a particular environmental context, the pathogenicity of such variants would be most apparent in families with multiple affected children. This model could explain why enrichment of RBFOX1 CNVs is seen in a multiplex, but not a simplex, cohort, as shown above. Further work will be essential to confirm this hypothesis as well as to identify and characterize such regulatory elements and determine what cellular or environmental signals trigger their use. If validated, therapies stimulating such factors or inhibiting the triggering signals could be useful to minimize ASD risk in families with known pathogenic CNVs in RBFOX1 and possibly other genes as well. Until such evidence is available however, we must continue to observe caution in the interpretation of inherited RBFOX1 CNVs as pathogenic when passed on from unaffected individuals.
Supplementary Material
Acknowledgments
The authors wish to thank Stephan Sanders for his invaluable assistance with the assessment of copy number variation in the autism cohorts as well as Daniel H. Geschwind (D.H.G) for his support and helpful suggestions. Data in this manuscript was obtained from the ISCA Consortium database (www.iscaconsortium.org), which generates this information using NCBI’s database of genomic structural variation (dbVar, www.ncbi.nlm.nih.gov/dbvar/), study nstd37. Samples and associated phenotype data were provided by ISCA. We gratefully acknowledge the resources provided by the Autism Genetic Resource Exchange (AGRE) Consortium and the participating AGRE families. The Autism Genetic Resource Exchange is a program of Autism Speaks and is supported, in part, by grant 1U24MH081810 from the National Institute of Mental Health to Clara M. Lajonchere (PI). This work was supported by the National Institutes of Health (9R01MH100027 to D.H.G and K08MH086297 to B.L.F) and Simons SFARI award 206744 to D.H.G.
References
- Amir-Zilberstein L, Blechman J, Sztainberg Y, Norton WH, Reuveny A, Borodovsky N, Levkowitz G. Homeodomain protein otp and activity-dependent splicing modulate neuronal adaptation to stress. Neuron. 2012;73(2):279–291. doi: 10.1016/j.neuron.2011.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen CL, Lamy P, Thorsen K, Kjeldsen E, Wikman F, Villesen P, Orntoft TF. Frequent genomic loss at chr16p13.2 is associated with poor prognosis in colorectal cancer. Int J Cancer. 2011;129(8):1848–1858. doi: 10.1002/ijc.25841. [DOI] [PubMed] [Google Scholar]
- Bajpai R, Sambrani N, Stadelmayer B, Shashidhara LS. Identification of a novel target of D/V signaling in Drosophila wing disc: Wg-independent function of the organizer. Gene Expr Patterns. 2004;5(1):113–121. doi: 10.1016/j.modgep.2004.05.005. [DOI] [PubMed] [Google Scholar]
- Bass AJ, Lawrence MS, Brace LE, Ramos AH, Drier Y, Cibulskis K, Meyerson M. Genomic sequencing of colorectal adenocarcinomas identifies a recurrent VTI1A-TCF7L2 fusion. Nat Genet. 2011;43(10):964–968. doi: 10.1038/ng.936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhalla K, Phillips HA, Crawford J, McKenzie OL, Mulley JC, Eyre H, Callen DF. The de novo chromosome 16 translocations of two patients with abnormal phenotypes (mental retardation and epilepsy) disrupt the A2BP1 gene. J Hum Genet. 2004;49(6):308–311. doi: 10.1007/s10038-004-0145-4. [DOI] [PubMed] [Google Scholar]
- Bill BR, Geschwind DH. Genetic advances in autism: heterogeneity and convergence on shared pathways. Curr Opin Genet Dev. 2009;19(3):271–278. doi: 10.1016/j.gde.2009.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen-Plotkin AS, Geser F, Plotkin JB, Clark CM, Kwong LK, Yuan W, Lee VM. Variations in the progranulin gene affect global gene expression in frontotemporal lobar degeneration. Hum Mol Genet. 2008;17(10):1349–1362. doi: 10.1093/hmg/ddn023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damianov A, Black DL. Autoregulation of Fox protein expression to produce dominant negative splicing factors. RNA. 2010;16(2):405–416. doi: 10.1261/rna.1838210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis LK, Maltman N, Mosconi MW, Macmillan C, Schmitt L, Moore K, Cook EH. Rare inherited A2BP1 deletion in a proband with autism and developmental hemiparesis. Am J Med Genet A. 2012;158A(7):1654–1661. doi: 10.1002/ajmg.a.35396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day A, Carlson MR, Dong J, O’Connor BD, Nelson SF. Celsius: a community resource for Affymetrix microarray data. Genome Biol. 2007;8(6):R112. doi: 10.1186/gb-2007-8-6-r112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day A, Dong J, Funari VA, Harry B, Strom SP, Cohn DH, Nelson SF. Disease gene characterization through large-scale co-expression analysis. PLoS ONE. 2009;4(12):e8491. doi: 10.1371/journal.pone.0008491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elia J, Gai X, Xie HM, Perin JC, Geiger E, Glessner JT, White PS. Rare structural variants found in attention-deficit hyperactivity disorder are preferentially associated with neurodevelopmental genes. Mol Psychiatry. 2010;15(6):637–646. doi: 10.1038/mp.2009.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fatemi SH, Aldinger KA, Ashwood P, Bauman ML, Blaha CD, Blatt GJ, Welsh JP. Consensus paper: pathological role of the cerebellum in autism. Cerebellum. 2012;11(3):777–807. doi: 10.1007/s12311-012-0355-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fogel BL, Geschwind DH. Clinical Neurogenetics. In: Daroff R, Fenichel G, Jankovic J, Mazziotta J, editors. Neurology in Clinical Practice. 6. Philadelphia, PA: Elsevier; 2012. pp. 704–734. [Google Scholar]
- Fogel BL, Perlman S. Cerebellar disorders: Balancing the approach to cerebellar ataxia. In: Gálvez-Jiménez N, Tuite PJ, editors. Uncommon Causes of Movement Disorders. 1. Cambridge, NY: Cambridge University Press; 2011. pp. 198–216. [Google Scholar]
- Fogel BL, Wexler E, Wahnich A, Friedrich T, Vijayendran C, Gao F, Geschwind DH. RBFOX1 regulates both splicing and transcriptional networks in human neuronal development. Hum Mol Genet. 2012;21(19):4171–4186. doi: 10.1093/hmg/dds240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallagher TL, Arribere JA, Geurts PA, Exner CR, McDonald KL, Dill KK, Conboy JG. Rbfox-regulated alternative splicing is critical for zebrafish cardiac and skeletal muscle functions. Dev Biol. 2011;359(2):251–261. doi: 10.1016/j.ydbio.2011.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehman LT, Stoilov P, Maguire J, Damianov A, Lin CH, Shiue L, Black DL. The splicing regulator Rbfox1 (A2BP1) controls neuronal excitation in the mammalian brain. Nat Genet. 2011;43(7):706–711. doi: 10.1038/ng.841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamshere ML, Green EK, Jones IR, Jones L, Moskvina V, Kirov G, Craddock N. Genetic utility of broadly defined bipolar schizoaffective disorder as a diagnostic concept. Br J Psychiatry. 2009;195(1):23–29. doi: 10.1192/bjp.bp.108.061424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbert MR. Contributions of the environment and environmentally vulnerable physiology to autism spectrum disorders. Curr Opin Neurol. 2010;23(2):103–110. doi: 10.1097/WCO.0b013e328336a01f. [DOI] [PubMed] [Google Scholar]
- Hodgkin J, Zellan JD, Albertson DG. Identification of a candidate primary sex determination locus, fox-1, on the X chromosome of Caenorhabditis elegans. Development. 1994;120(12):3681–3689. doi: 10.1242/dev.120.12.3681. [DOI] [PubMed] [Google Scholar]
- Holsboer F, Ising M. Central CRH system in depression and anxiety--evidence from clinical studies with CRH1 receptor antagonists. Eur J Pharmacol. 2008;583(2–3):350–357. doi: 10.1016/j.ejphar.2007.12.032. [DOI] [PubMed] [Google Scholar]
- Hu J, Ho AL, Yuan L, Hu B, Hua S, Hwang SS, et al. Neutralization of terminal differentiation in gliomagenesis. Proc Natl Acad Sci U S A. 2013;110(36):14520–14527. doi: 10.1073/pnas.1308610110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iafrate AJ, Feuk L, Rivera MN, Listewnik ML, Donahoe PK, Qi Y, Lee C. Detection of large-scale variation in the human genome. Nat Genet. 2004;36(9):949–951. doi: 10.1038/ng1416. [DOI] [PubMed] [Google Scholar]
- Jin Y, Suzuki H, Maegawa S, Endo H, Sugano S, Hashimoto K, Inoue K. A vertebrate RNA-binding protein Fox-1 regulates tissue-specific splicing via the pentanucleotide GCAUG. Embo J. 2003;22(4):905–912. doi: 10.1093/emboj/cdg089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohannim O, Hibar DP, Jahanshad N, Stein JL, Hua X, Toga AW, Thompson PM. Predicting Temporal Lobe Volume on Mri from Genotypes Using L(1)-L(2) Regularized Regression. Proc IEEE Int Symp Biomed Imaging. 2012:1160–1163. doi: 10.1109/ISBI.2012.6235766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroyanagi H. Fox-1 family of RNA-binding proteins. Cell Mol Life Sci. 2009;66(24):3895–3907. doi: 10.1007/s00018-009-0120-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lal D, Trucks H, Moller RS, Hjalgrim H, Koeleman BP, de Kovel CG, Sander T. Rare exonic deletions of the RBFOX1 gene increase risk of idiopathic generalized epilepsy. Epilepsia. 2013;54(2):265–271. doi: 10.1111/epi.12084. [DOI] [PubMed] [Google Scholar]
- Le-Niculescu H, Patel SD, Bhat M, Kuczenski R, Faraone SV, Tsuang MT, Niculescu AB., 3rd Convergent functional genomics of genome-wide association data for bipolar disorder: comprehensive identification of candidate genes, pathways and mechanisms. Am J Med Genet B Neuropsychiatr Genet. 2009;150B(2):155–181. doi: 10.1002/ajmg.b.30887. [DOI] [PubMed] [Google Scholar]
- Lee JA, Tang ZZ, Black DL. An inducible change in Fox-1/A2BP1 splicing modulates the alternative splicing of downstream neuronal target exons. Genes Dev. 2009;23(19):2284–2293. doi: 10.1101/gad.1837009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y, Mattai A, Long R, Rapoport JL, Gogtay N, Addington AM. Microduplications disrupting the MYT1L gene (2p25.3) are associated with schizophrenia. Psychiatr Genet. 2012;22(4):206–209. doi: 10.1097/YPG.0b013e328353ae3d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim C, Allada R. ATAXIN-2 activates PERIOD translation to sustain circadian rhythms in Drosophila. Science. 2013;340(6134):875–879. doi: 10.1126/science.1234785. [DOI] [PubMed] [Google Scholar]
- Lim J, Hao T, Shaw C, Patel AJ, Szabo G, Rual JF, Zoghbi HY. A protein-protein interaction network for human inherited ataxias and disorders of Purkinje cell degeneration. Cell. 2006;125(4):801–814. doi: 10.1016/j.cell.2006.03.032. [DOI] [PubMed] [Google Scholar]
- Linnebacher M, Ostwald C, Koczan D, Salem T, Schneider B, Krohn M, Prall F. Single nucleotide polymorphism array analysis of microsatellite-stable, diploid/near-diploid colorectal carcinomas without the CpG island methylator phenotype. Oncol Lett. 2013;5(1):173–178. doi: 10.3892/ol.2012.1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L, Hanson RL, Traurig MT, Muller YL, Kaur BP, Perez JM, Baier LJ. Evaluation of A2BP1 as an obesity gene. Diabetes. 2010;59(11):2837–2845. doi: 10.2337/db09-1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin CL, Duvall JA, Ilkin Y, Simon JS, Arreaza MG, Wilkes K, Geschwind DH. Cytogenetic and molecular characterization of A2BP1/FOX1 as a candidate gene for autism. Am J Med Genet B Neuropsychiatr Genet. 2007;144(7):869–876. doi: 10.1002/ajmg.b.30530. [DOI] [PubMed] [Google Scholar]
- Mazzone L, Ruta L, Reale L. Psychiatric comorbidities in asperger syndrome and high functioning autism: diagnostic challenges. Ann Gen Psychiatry. 2012;11(1):16. doi: 10.1186/1744-859X-11-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melhem N, Middleton F, McFadden K, Klei L, Faraone SV, Vinogradov S, Myles-Worsley M. Copy number variants for schizophrenia and related psychotic disorders in Oceanic Palau: risk and transmission in extended pedigrees. Biol Psychiatry. 2011;70(12):1115–1121. doi: 10.1016/j.biopsych.2011.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer KJ, Axelsen MS, Sheffield VC, Patil SR, Wassink TH. Germline mosaic transmission of a novel duplication of PXDN and MYT1L to two male half-siblings with autism. Psychiatr Genet. 2012;22(3):137–140. doi: 10.1097/YPG.0b013e32834dc3f5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikhail FM, Lose EJ, Robin NH, Descartes MD, Rutledge KD, Rutledge SL, Carroll AJ. Clinically relevant single gene or intragenic deletions encompassing critical neurodevelopmental genes in patients with developmental delay, mental retardation, and/or autism spectrum disorders. Am J Med Genet A. 2011;155A(10):2386–2396. doi: 10.1002/ajmg.a.34177. [DOI] [PubMed] [Google Scholar]
- Obayashi T, Kinoshita K. COXPRESdb: a database to compare gene coexpression in seven model animals. Nucleic Acids Res. 2011;39(Database issue):D1016–1022. doi: 10.1093/nar/gkq1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pescosolido MF, Yang U, Sabbagh M, Morrow EM. Lighting a path: genetic studies pinpoint neurodevelopmental mechanisms in autism and related disorders. Dialogues Clin Neurosci. 2012;14(3):239–252. [PMC free article] [PubMed] [Google Scholar]
- Pistoni M, Shiue L, Cline MS, Bortolanza S, Neguembor MV, Xynos A, Gabellini D. Rbfox1 downregulation and altered calpain 3 splicing by FRG1 in a mouse model of Facioscapulohumeral muscular dystrophy (FSHD) PLoS Genet. 2013;9(1):e1003186. doi: 10.1371/journal.pgen.1003186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Priebe L, Degenhardt F, Strohmaier J, Breuer R, Herms S, Witt SH, Cichon S. Copy number variants in German patients with schizophrenia. PLoS ONE. 2013;8(7):e64035. doi: 10.1371/journal.pone.0064035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rare chromosomal deletions and duplications increase risk of schizophrenia. Nature. 2008;455(7210):237–241. doi: 10.1038/nature07239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray D, Kazan H, Cook KB, Weirauch MT, Najafabadi HS, Li X, Hughes TR. A compendium of RNA-binding motifs for decoding gene regulation. Nature. 2013;499(7457):172–177. doi: 10.1038/nature12311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richdale AL, Schreck KA. Sleep problems in autism spectrum disorders: prevalence, nature, & possible biopsychosocial aetiologies. Sleep Med Rev. 2009;13(6):403–411. doi: 10.1016/j.smrv.2009.02.003. [DOI] [PubMed] [Google Scholar]
- Sebat J, Lakshmi B, Malhotra D, Troge J, Lese-Martin C, Walsh T, Wigler M. Strong association of de novo copy number mutations with autism. Science. 2007;316(5823):445–449. doi: 10.1126/science.1138659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata H, Huynh DP, Pulst SM. A novel protein with RNA-binding motifs interacts with ataxin-2. Hum Mol Genet. 2000;9(9):1303–1313. doi: 10.1093/hmg/9.9.1303. [DOI] [PubMed] [Google Scholar]
- Stambolian D, Wojciechowski R, Oexle K, Pirastu M, Li X, Raffel LJ, Bailey-Wilson JE. Meta-analysis of genome-wide association studies in five cohorts reveals common variants in RBFOX1, a regulator of tissue-specific splicing, associated with refractive error. Hum Mol Genet. 2013;22(13):2754–2764. doi: 10.1093/hmg/ddt116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens SJ, van Ravenswaaij-Arts CM, Janssen JW, Klein Wassink-Ruiter JS, van Essen AJ, Dijkhuizen T, Engelen JJ. MYT1L is a candidate gene for intellectual disability in patients with 2p25.3 (2pter) deletions. Am J Med Genet A. 2011;155A(11):2739–2745. doi: 10.1002/ajmg.a.34274. [DOI] [PubMed] [Google Scholar]
- Sun S, Zhang Z, Fregoso O, Krainer AR. Mechanisms of activation and repression by the alternative splicing factors RBFOX1/2. RNA. 2012;18(2):274–283. doi: 10.1261/rna.030486.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126(4):663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Underwood JG, Boutz PL, Dougherty JD, Stoilov P, Black DL. Homologues of the Caenorhabditis elegans Fox-1 protein are neuronal splicing regulators in mammals. Mol Cell Biol. 2005;25(22):10005–10016. doi: 10.1128/MCB.25.22.10005-10016.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usha N, Shashidhara LS. Interaction between Ataxin-2 Binding Protein 1 and Cubitus-interruptus during wing development in Drosophila. Dev Biol. 2010;341(2):389–399. doi: 10.1016/j.ydbio.2010.02.039. [DOI] [PubMed] [Google Scholar]
- Van Den Bossche MJ, Strazisar M, Cammaerts S, Liekens AM, Vandeweyer G, Depreeuw V, Del-Favero J. Identification of rare copy number variants in high burden schizophrenia families. Am J Med Genet B Neuropsychiatr Genet. 2013;162B(3):273–282. doi: 10.1002/ajmg.b.32146. [DOI] [PubMed] [Google Scholar]
- Voineagu I, Wang X, Johnston P, Lowe JK, Tian Y, Horvath S, Geschwind DH. Transcriptomic analysis of autistic brain reveals convergent molecular pathology. Nature. 2011;474(7351):380–384. doi: 10.1038/nature10110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vrijenhoek T, Buizer-Voskamp JE, van der Stelt I, Strengman E, Sabatti C, Geurts van Kessel A, Veltman JA. Recurrent CNVs disrupt three candidate genes in schizophrenia patients. Am J Hum Genet. 2008;83(4):504–510. doi: 10.1016/j.ajhg.2008.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, Burge CB. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456(7221):470–476. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Zhang H, Ma D, Bucan M, Glessner JT, Abrahams BS, Hakonarson H. Common genetic variants on 5p14.1 associate with autism spectrum disorders. Nature. 2009;459(7246):528–533. doi: 10.1038/nature07999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wintle RF, Lionel AC, Hu P, Ginsberg SD, Pinto D, Thiruvahindrapduram B, Scherer SW. A genotype resource for postmortem brain samples from the Autism Tissue Program. Autism Res. 2011;4(2):89–97. doi: 10.1002/aur.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu B, Roos JL, Levy S, van Rensburg EJ, Gogos JA, Karayiorgou M. Strong association of de novo copy number mutations with sporadic schizophrenia. Nat Genet. 2008;40(7):880–885. doi: 10.1038/ng.162. [DOI] [PubMed] [Google Scholar]
- Yeo GW, Nostrand EL, Liang TY. Discovery and analysis of evolutionarily conserved intronic splicing regulatory elements. PLoS Genet. 2007;3(5):e85. doi: 10.1371/journal.pgen.0030085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo GW, Xu X, Liang TY, Muotri AR, Carson CT, Coufal NG, Gage FH. Alternative splicing events identified in human embryonic stem cells and neural progenitors. PLoS Comput Biol. 2007;3(10):1951–1967. doi: 10.1371/journal.pcbi.0030196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi S, Li WH. Molecular evolution of recombination hotspots and highly recombining pseudoautosomal regions in hominoids. Mol Biol Evol. 2005;22(5):1223–1230. doi: 10.1093/molbev/msi106. [DOI] [PubMed] [Google Scholar]
- Zhang C, Zhang Z, Castle J, Sun S, Johnson J, Krainer AR, Zhang MQ. Defining the regulatory network of the tissue-specific splicing factors Fox-1 and Fox-2. Genes Dev. 2008;22(18):2550–2563. doi: 10.1101/gad.1703108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao WW. Intragenic deletion of RBFOX1 associated with neurodevelopmental/neuropsychiatric disorders and possibly other clinical presentations. Mol Cytogenet. 2013;6(1):26. doi: 10.1186/1755-8166-6-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou HL, Lou H. Repression of prespliceosome complex formation at two distinct steps by Fox-1/Fox-2 proteins. Mol Cell Biol. 2008;28(17):5507–5516. doi: 10.1128/MCB.00530-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.