Abstract
Resistance of breast cancers to targeted hormone receptor (HR) or human epidermal growth factor receptor 2 (HER2) inhibitors often occurs through dysregulation of the phosphoinositide 3-kinase, protein kinase B/AKT/mammalian target of rapamycin (PI3K/AKT/mTOR) pathway. Presently, no targeted therapies exist for breast cancers lacking HR and HER2 overexpression, many of which also exhibit PI3K/AKT/mTOR hyper-activation. Resistance of breast cancers to current therapeutics also results, in part, from aberrant epigenetic modifications including protein acetylation regulated by histone deacetylases (HDACs). We show that the investigational drug MLN0128, which inhibits both complexes of mTOR (mTORC 1 and mTORC2), and the hydroxamic acid pan-HDAC inhibitor TSA synergistically inhibit the viability of a phenotypically diverse panel of five breast cancer cell lines (HR−/+, HER2−/+). The combination of MLN0128 and TSA induces apoptosis in most breast cancer cell lines tested, but not in the non-malignant MCF-10A mammary epithelial cells. In parallel, the MLN0128/TSA combination reduces phosphorylation of AKT at S473 more than single agents alone and more so in the 5 malignant breast cancer cell lines than in the nonmalignant mammary epithelial cells. Examining polysome profiles from one of the most sensitive breast cancer cell lines (SKBR3), we demonstrate that this MLN0128/TSA treatment combination synergistically impairs polysome assembly in conjunction with enhanced inhibition of 4eBP1 phosphorylation at S65. Taken together, these data indicate that the synergistic growth inhibiting consequence of combining a mTORC1/C2 inhibitor like MLN0128 with a pan-HDAC inhibitor like TSA results from their mechanistic convergence onto the PI3K/AKT/mTOR pathway, profoundly inhibiting both AKT S473 and 4eBP1 S65 phosphorylation, reducing polysome formation and cancer cell viability.
Keywords: mTOR, HDAC, MLN0128, INK128, Trichostatin A, TSA, Breast cancer, AKT, Polysomes, Ribosomes
Introduction
Breast cancer is the second leading cause of cancer-related mortality in women in the United States [1]. Over 80 % of breast cancers are driven by human epidermal growth factor receptor 2 (HER2) and/or estrogen and progesterone receptor (ER/PR) overexpression. Anti-receptor monoclonal antibodies and anti-estrogenic agents (tamoxifen, aromatase inhibitors) target HER2-positive (15–20 %) and ER/PR-positive (60–80 %) breast cancers, respectively. Resistance frequently emerges, often caused by activation of downstream signaling pathways, such as the phosphoinositide 3-kinase, protein kinase B/AKT/mammalian target of rapamycin (PI3K/AKT/mTOR) pathway [1-4].
The PI3K/AKT/mTOR pathway is dysregulated in breast cancers regardless of ER/PR and HER2 status. This can occur through activating mutations in PIK3CA, which codes for the catalytic subunit of PI3K, or through loss of PTEN, a phosphatase that removes phosphate groups from phosphoinositide-3, negatively regulating PI3K. The PI3K/AKT/mTOR pathway promotes proliferation partly by promoting phosphorylation of the small (40S) ribosomal subunit protein S6 (S6) and the eukaryotic initiation factor 4e binding protein 1 (4eBP1), increasing polysome formation and translation of transcripts regulating tumor cell survival [5-8].
Rapalogs are first generation mTOR inhibitors. They allosterically inhibit the target of rapamycin complex 1 (mTORC1), one of two complexes formed by mTOR. The rapalog, everolimus (RAD001, Novartis), is FDA approved in combination with aromatase inhibitors for patients with ER/PR+ breast cancers unresponsive to first line aromatase inhibitors [9, 10]. However, many patients do not respond to this combination. Furthermore, in endocrine-treated patients, increased AKT phosphorylation at S473 in tumors is correlated with poor clinical outcome [11]. Since everolimus inhibits mTORC1 but not mTORC2, it paradoxically increases AKT phosphorylation by relieving a negative feedback loop that otherwise restricts PI3K signaling when S6K is activated [12]. Consistently, AKT S473 phosphorylation is increased in patients treated with everolimus [13].
Next generation mTOR-targeting drugs are ATP-competitive inhibitors, which target both mTORC1 and mTORC2. An example is MLN0128 (formerly INK128), which increases survival in mouse models of BCR-ABL-driven B cell acute lymphoblastic leukemia and MYC-driven Burkitt’s lymphoma and prevents the translation of mRNA’s coding for pro-invasive genes in prostate cancer cells [8, 14, 15]. MLN0128 reduces the size of HER2-transfected MCF7 (ER/PR+, HER2+) xenografts [16], illustrating its potential against breast cancer. MLN0128 is completing phase I clinical trials against breast (HER2−, HER2+) and other epithelial cancers (NCT01058707, NCT01351350).
Furthermore, epigenetic alterations, including dysregulated protein acetylation, affect gene expression and signaling pathways. These alterations contribute to tumorigenesis and drug resistance [17]. Histone deacetylases (HDACs) remove acetyl groups from histone and non-histone proteins, increasing expression of p21 and other tumor suppressors [18]. Classic HDACs are grouped into classes I and II [19]. Pan-HDAC inhibitors, which inhibit both classes, show preclinical and clinical efficacy against breast cancers. For example, the pan-HDAC inhibitor panobinostat (LBH589, Novartis) reduces proliferation and survival of triple negative breast cancer cells, and is in phase II clinical trials against breast cancer [20]. Furthermore, trichostatin A (TSA), another pan-HDAC inhibitor, reduces proliferation of breast cancer cells of various subtypes (ER/PR−/+, HER2−/+) with IC50’s in the nanomolar range; TSA also reduces the growth of mutagen-induced mammary tumors in rats [21, 22]. In addition to epigenetic mechanisms, our lab has shown that HDAC inhibition reduces cancer cell proliferation at the post-translational level by promoting the decay of oncogenic transcripts, such as HER2 mRNA [23, 24].
Previous studies suggest that combining mTOR and HDAC inhibitors may be more effective than single agents at growth inhibition of specific malignant cell lines. Examples of effective combinations include everolimus with panobinostat against prostate cancer cells [25], everolimus with the class I HDAC inhibitor valproic acid against prostate cancer and Burkitt’s lymphoma cells [26, 27], and everolimus with the pan-HDAC inhibitor entinostat (MS-275, Syndax) against acute myelogenous leukemia cells [28]. In other models, ATP-competitive mTOR inhibitors and dual PI3K/mTOR inhibitors demonstrate efficacy in combination with HDAC inhibitors. The dual pan-PI3K/mTORC1/2 inhibitors BEZ235 and BGT226 (Novartis), in combination with panobinostat, inhibit viability of head and neck squamous carcinoma cells to a greater extent than each drug alone. This reduced cell viability is accompanied by a reduction in AKT S473 phosphorylation [29]. Combining the mTORC1/2 inhibitor PP242 and the pan-HDAC inhibitor vorinostat (SAHA, Merck) reduces hepatocellular carcinoma cell viability at concentrations where the single drugs have no effect. Introduction of constitutively active AKT reverses this benefit [30].
Although mTOR and HDAC inhibitors, individually, show activity against some breast cancers, and their combination appears to yield added growth inhibiting effects against selected cell line models, preclinical investigation of this therapeutic combination against various subtypes of breast cancer has yet to be reported. In this study, we demonstrate that combining MLN0128 and TSA synergistically inhibits the viability of a panel of phenotypically diverse breast cancer cell lines (ER/PR−/+, HER2−/+), without similarly affecting a non-malignant mammary epithelial cell line. This synergistic interaction in breast cancer cells is associated with decreased AKT S473 and 4eBP1 S65 phosphorylation and impaired polysome assembly.
Materials and methods
Cell culture
SKBR3 cells (American Type Culture Collection (ATCC), Manassas, VA) were cultured in McCoy’s medium with 10 % Fetal Bovine Serum (FBS). MDA-MB-231 cells were cultured in DMEM with 10 % FBS. MCF7/NEO3 (stably transfected with an empty control vector, herein referred to as MCF7) and MCF7/HER2-18 cells, stably overexpressing HER2 [31], were grown in DMEM with 10 % FBS and 1 % insulin. BT-474 cells (ATCC) were grown in RPMI 1640 with 10 % FBS. Non-malignant MCF-10A cells (ATCC) were grown in 50 % DMEM and 50 % Ham’s F12 media with 5 % horse serum, 2 mM l-glutamine, 0.02 mM non-essential amino acids, 10 ng/ml EGF, 0.5 μg/ml hydrocortisone, 0.1 μg/ml cholera toxin, and 10 μg/ml insulin.
Drugs
MLN0128 and GDC0941 (developed by Genentech, South San Francisco, CA) were obtained under a material transfer agreement from Intellikine, Inc. (now Millennium Pharmaceuticals, Cambridge, MA). Trichostatin A (TSA), Suberoylanilide hydroxamic acid (SAHA/Vorinostat), and Adriamycin (doxorubicin) were obtained from Sigma Aldrich (St. Louis, MO).
Cell viability assay
Cells were plated at ~10 % confluency in 96 well plates and treated as indicated the next day. Viability was quantified via CellTiter-Glo (Promega, Madison, WI) per manufacturer’s protocol. Luminescence determined with a Fluoroskan Ascent FL luminometer (Thermo Scientific, Rockford, IL). IC50 values were calculated using GraphPad Prism (La Jolla, CA). Combination indices (CI) were determined using Calcusyn (Biosoft, Great Shelford Cambridge, UK).
Immunoblots
Cells were plated at ~35 % confluency for apoptosis analysis or ~50 % confluency for signaling analysis. Following treatments, cells were washed with Dulbecco’s phosphate-buffered saline (PBS) and harvested in RIPA buffer (50 mM Tris–CL (pH 8.0), 150 mM NaCl, 1 % triton X-100, 0.5 % sodium deoxycholate, 0.1 % SDS) containing complete Mini EDTA-free Protease Inhibitor Cocktail tablets (Roche, Indianapolis, IN) and PhosSTOP Phosphatase Inhibitor Cocktail tablets (Roche). Lysates were cleared by centrifugation.
The Bradford Coomassie Assay (BCA) kit (Pierce, Rockford, IL) was used to determine protein concentrations. Equal amounts of protein were diluted in 2× Laemmli sample buffer. Immunoblots were performed as previously described [32]. The following antibodies were used: PARP, β-tubulin, pS6 S240/244, S6, p4eBp1 S65, 4eBp1, pAKT-S473, and AKT (Cell Signaling, Danvers, MA).
Microscopy and Annexin V analysis
Cells were plated on 8-well chamber slides at 30 % confluency and treated the next day. One day later, Annexin V and propidium iodide (PI) were stained using the Dead Cell Apoptosis Kit with Annexin V Alexa Fluor® 488 & PI from Life Technologies (Grand Island, NY) per manufacturer’s protocol. Cells were fixed with a 1:1 mixture of 8 % PFA in PBS and 2× Annexin binding buffer (Life Technologies) and visualized by bright-field and epifluorescence microscopy.
Flow cytometry and cell cycle analysis
Cells were plated on 10 cm plates at 20 % confluency and treated as indicated. Adherent cells were trypsinized, washed three times with PBS, and fixed with 70 % ethanol at 4 °C overnight. Cells were then washed with PBS three times, resuspended in 100 μl ribonuclease (100 μg/ml) for 5 min, then stained with 400 μl PI (50 μg/ml). Cell cycle analysis was performed using a BD FACSAriaII with BD FACSDiva software (BD Biosciences, San Jose, CA) collecting a minimum of 15,000 events per sample. Flowjo (TreeStar inc, Ashland, OR) was used to analyze data. Debris were eliminated using forward/side scatter criteria, while doublets were excluded using PE-W versus PE-A criteria.
Polysome profiling
SKBR3 cells were plated at ~50 % confluency and drug-treated for 24 h. Fifteen minutes before lysis, cells were treated with 50 μg/ml cycloheximide (Sigma). Cells were washed with PBS, and harvested with lysis buffer (10 mM HEPES, 10 mM KCl, 75 mM NaCl, 10 mM MgCl2, 0.35 % NP40, pH 7.9 with 0.1 M sodium orthovanadate, complete Mini, EDTA-free Protease Inhibitor Cocktail and PhosSTOP Phosphatase Inhibitor Cocktail tablets (Roche), 50 μg/ml cyclohexamide, the SUPERase RNAse inhibitor (Life Technologies)). Lysates were dounced and cleared by centrifugation. Polysome profiles were performed as previously described [33].
Results
MLN0128 and TSA inhibit the proliferation of breast cancer cells more than non-malignant mammary epithelial cells
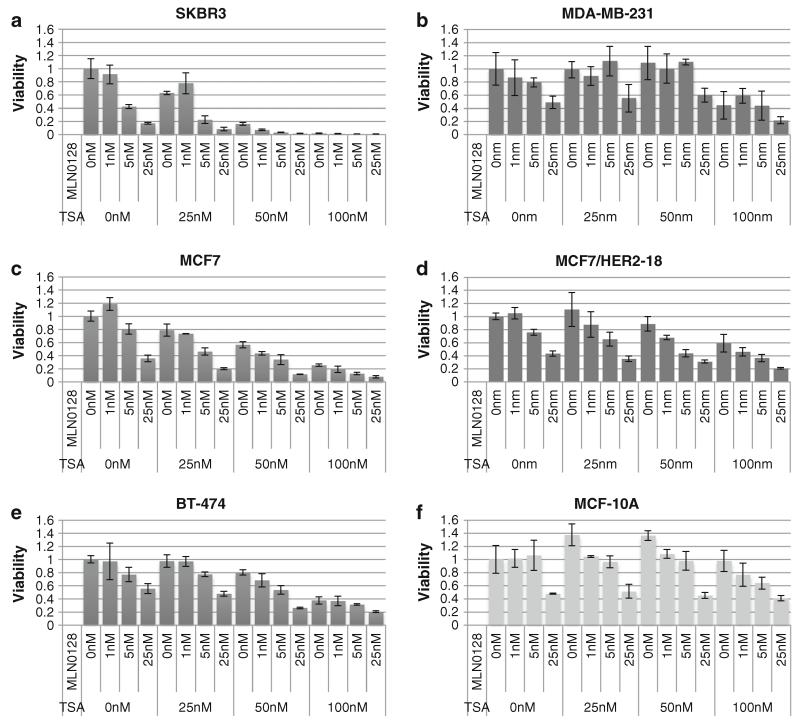
To access effects combined mTORC1/2/pan-HDAC inhibition on breast cancer cell viability, CellTiter-Glo assays were performed on a panel that included ER/PR negative/positive and HER2 negative/positive cell lines. SKBR3, MDA-MB-231, MCF7, and MCF7/HER2-18 cells were the most sensitive to the single agent MLN0128 treatment, with IC50 values ranging from 4.36 to 18.6 nM (Fig. 1a-d; Table 1). In contrast, BT-474 breast cancer cells and non-malignant MCF-10A mammary epithelial cells had MLN0128 IC50 values >24.5 nM (Fig. 1e, f; Table 1). There was no correlation between the ER, PR, HER2, PIK3CA, or PTEN status and MLN0128 sensitivity (Table 1). SKBR3, MCF7, and MCF7/HER2-18 cells were more sensitive to MLN0128 than to RAD001 or the pan-PI3K inhibitor, GDC0941 (Supplementary Fig. S1).
Fig. 1.
MLN0128 and TSA inhibit the proliferation of breast cancer cells more than non-transformed mammary epithelial cells. Viability assays were carried out after 72 h of treatment with the indicated doses of MLN0128 and TSA on the following cell lines: a SKBR3, b MDA-MB-231, c MCF7, d MCF7/HER2-18, e BT-474, and f MCF-10A. Error bars represent the standard deviation calculated from three triplicate wells. Each cell line was analyzed at least twice, and a representative experiment is shown
Table 1.
IC50 values (3 day treatment) for MLN0128 and TSA as single agents in breast cancer cells and non-transformed mammary epithelial cells
| Cell Line | ER | PR | HER2 | PIK3CA | PTEN protein expression/mutation | MLN0128 IC50 (nM) | TSA IC50 (nM) |
|---|---|---|---|---|---|---|---|
| SKBR3 | − | − | + | WT | +/WT | 4.36 | 30.9 |
| MDA-MB-231 | − | − | − | WT | ++++/WT | 11.0 | 93.3 |
| MCF7 | + | + | − | E545K | +++/WT | 11.8 | 60.3 |
| MCF7/HER2-18 | + | + | + | E545K | +++/WT | 18.6 | >100 |
| BT-474 | − | + | + | WT | ++++/WT | >25.0 | 93.9 |
| MCF-10A | − | − | − | WT | +++/WT | 24.5 | >100 |
IC50 values were calculated for MLN0128 and TSA from each cell line following a 72 h cell viability assay. Values were calculated from at least six independent wells from two independent experiments. Overexpression status for ER, PR, and HER2 is indicated along with PIK3CA mutation status and PTEN loss
SKBR3 and MCF7 cells were the most sensitive to TSA with IC50 values of 30.9 and 60.3 nM, respectively (Fig. 1a, c; Table 1). MDA-MB-231 and BT-474, with IC50 values of 93.3 and 93.9 nM, respectively, were slightly less sensitive to TSA (Fig. 1b, e; Table 1). MCF7 HER2-18 and MCF-10A cells were the least sensitive with TSA IC50 values >100 nM (Fig. 1d, f; Table 1). As with MLN0128, no pattern emerged between expression/mutation of ER, PR, HER2, PIK3CA, or PTEN and TSA IC50’s (Table 1).
In all cell lines tested, the MLN0128/TSA combination inhibited viability more than single agents (Fig. 1). MCF-10A mammary epithelial cells were less affected by the combined MLN0128/TSA treatment than all breast cancer cell lines tested; dual treatment of non-malignant MCF-10A cells with 25 nM of MLN0128 and 100 nM of TSA reduced viability by about 60 %, whereas viability was reduced by >80 % in all breast cancer cell lines tested (Fig. 1).
MLN0128 and TSA synergistically inhibit breast cancer cell proliferation
CI’s were calculated to determine whether the combination of MLN0128 and TSA is synergistic, additive, or antagonistic. A CI ~1 represents additivity, while <1 represents synergy and >1 represents antagonism. Synergism resulted from almost all of the doses tested in SKBR3, MCF7, MCF7/HER-18, and BT474 cells. In MDA-MB-231 cells, antagonism resulted with 5 nM MLN0128 and 50 nM TSA. However, with 25 nM MLN0128 and 100 nM TSA strong synergy resulted (Table 2). Non-transformed MCF-10A was not sensitive enough to either drug for CalcuSyn to determine a CI, since the algorithm requires growth inhibiting doses of each drug alone that cause a similar effect as the dual treatment. Furthermore, the clinically relevant pan-HDAC inhibitor SAHA/Vorinostat (IC50 = 2,512 nM) proved less potent than TSA as a single agent, but in combination with MLN0128, synergistically reduced MCF7 viability to a slightly greater degree than TSA (Supplementary Fig. S2).
Table 2.
Combination indexes of MLN0128 and TSA in breast cancer cells
| Drug | Drug | Cell line | Cell line | Cell line | Cell line | Cell line |
|---|---|---|---|---|---|---|
| MLN0128 (nM) |
TSA (nM) |
SKBR3 (CI) |
MCF7 (CI) |
MCF7/ HER2-18 (CI) |
BT-474 (CI) |
MDA- MB-231 (CI) |
| 5 | 50 | 0.701 | 0.711 | 0.742 | 0.778 | 4.328 |
| 25 | 50 | 0.566 | 0.758 | 0.903 | 0.73 | 0.895 |
| 5 | 100 | 0.762 | 0.637 | 0.873 | 0.898 | 0.598 |
| 25 | 100 | 0.672 | 0.771 | 0.811 | 0.879 | 0.264 |
| ||||||
Combination indices (CIs) were calculated for breast cancer cell lines using the Calcusyn software. Antagonism is indicated by a CI>1.10, additivity is indicated by a CI ranging from 0.90 to 1.10, and synergism is indicated by a CI ranging from 0.1 to 0.90
MLN0128 and TSA treatments enhance apoptosis in SKBR3 and MCF7 cells but not in MDA-MB-231 and MCF-10A cells
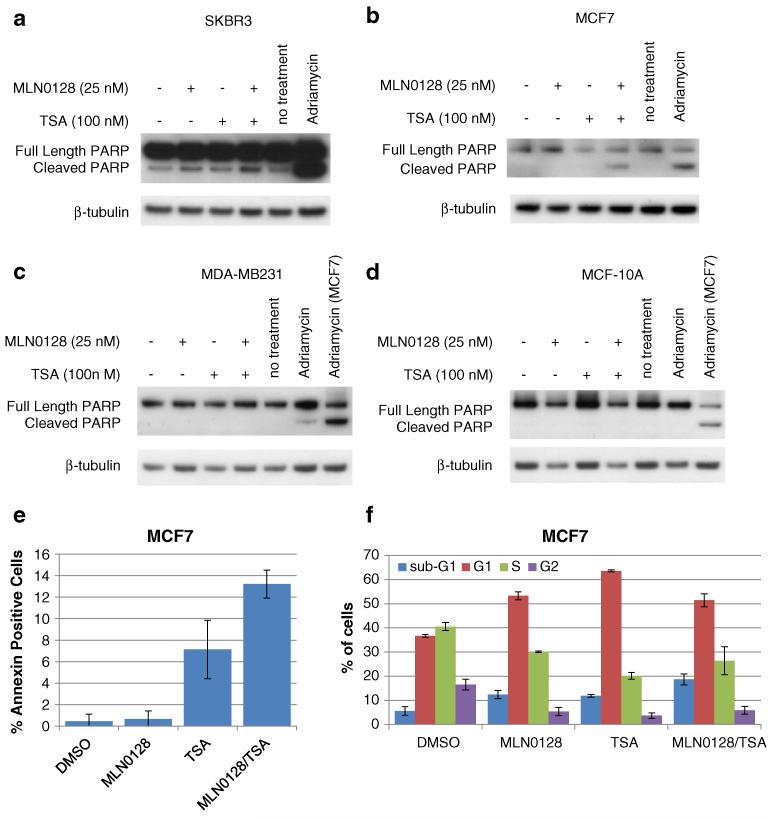
To determine if the growth inhibitory effects of MLN0128/TSA result from apoptosis, PARP cleavage, Annexin V, and sub-G1 analysis were performed. In SKBR3 cells, single treatments increased cleaved PARP. Cleaved PARP levels were further increased by the dual MLN0128/TSA treatment (Fig. 2a). Dual treatment also induced PARP cleavage in MCF7 cells, although single treatments had no obvious effect (Fig. 2b). In contrast, neither single nor dual MLN0128/TSA treatments induced PARP cleavage in MDA-MB-231 cells (Fig. 2c). MDA-MB-231 cells also showed less PARP cleavage when treated with the Adriamycin (0.5 μg/ml) positive apoptosis control than MCF7 cells (two right-most lanes Fig. 2c), indicating that MDA-MB-231 cells are generally more resistant to drug-induced apoptosis. Neither MLN0128 nor TSA increased PARP cleavage in MCF-10A cells despite induction of PARP cleavage by Adriamycin (Fig. 2d). Consistent with PARP cleavage, 24 h combination MLN0128/TSA treatment increased Annexin V staining of MCF7 cells (Fig. 2e; Supplementary Fig. S3). Single TSA treatment increased Annexin V in MCF7 cells, albeit to a lesser extent than the dual MLN0128/TSA treatment, even though PARP cleavage was not detected in MCF7 cells given the single dose TSA treatment, suggesting more sensitive apoptosis detection by Annexin V. Furthermore, cell cycle analysis revealed that, after 48 h of treatment, single MLN0128 or TSA treatments increased the percentage of cells in sub-G1 (indicative of apoptotic cells) to 12.4 and 11.9 %, respectively (compared to 5.6 % in the control) (Fig. 2f). Dual MLN0128/TSA treatment further increased the percentage of cells in sub-G1 to 18.6 %. The MLN0128/TSA combination also increased the percentage of cells in G1 and decreased the percentage of cells in S and G2 compared to the control, indicating that G1 arrest contributes to the loss of viable cell growth following this therapeutic combination. In summary, effects of the dual MLN0128/TSA treatment on apoptosis induction in SKBR3, MCF7, MDA-MB-213, and MCF-10A cells correlated with the degree to which this treatment combination reduced cell viability (Fig. 1), with SKBR3 showing the greatest sensitivity and the non-malignant MCF-10As showing the least sensitivity to the MLN0128/TSA combination.
Fig. 2.
MLN0128 and TSA treatments enhance apoptosis in SKBR3 and MCF7 cells but not in MDA-MB-231 and MCF-10A cells. a SKBR3, b MCF7, c MDA-MB-231, and d MCF-10A cells were treated with MLN0128 (25 nM), TSA (100 nM), or Adriamycin (0.5 μg/mL) for 48 h. Whole cell lysates were collected and immunoblots were performed to detect cleaved PARP and a β-tubulin loading control. e MCF7 cells were treated with MLN0128 (25 nM) and/or TSA (100 nM) for 24 h. Annexin V was stained for and visualized by microscopy. The percentage of Annexin V-positive cells relative to total cells is expressed for four biological replicates. Error bars represent standard deviation. f MCF7 cells were treated with MLN0128 (25 nM) and/or TSA (100 nM) for 48 h. Cell cycle analysis was performed as described in materials and methods. Error bars represent the standard deviation from three biological replicates
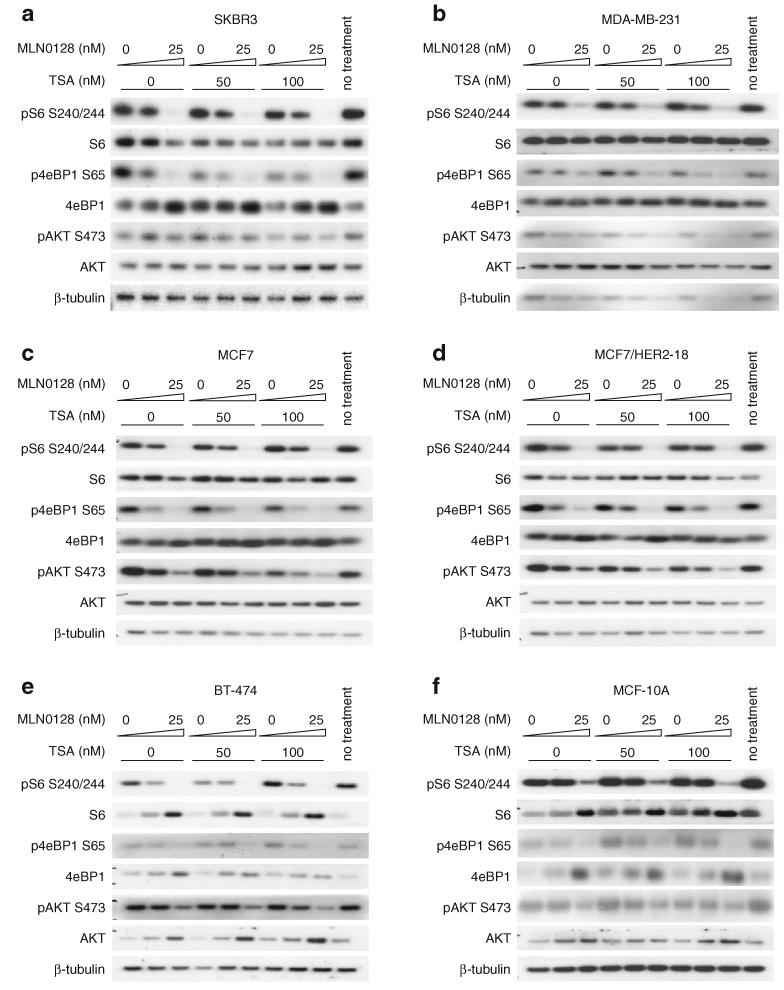
MLN0128 reduces AKT, S6, and 4eBP1 phosphorylation, while the combination with TSA further reduces AKT phosphorylation
To elucidate signaling pathway inhibition by the MLN0128/TSA combination, PI3K/AKT/mTOR pathway events were examined 8 h after drug treatment. As expected, MLN0128, alone or in combination with TSA, reduced phosphorylation of AKT S473, S6 S240/244, and 4eBP1 S65 (Fig. 3). TSA treatment alone did not affect S6 S204/244 or 4eBP1 S65 phosphorylation at doses/time points tested, except for reduced 4eBP1 S65 phosphorylation by TSA treatment in SKBR3 (Fig. 3a). TSA did not enhance the effect of MLN0128 on S6 S204/244 phosphorylation in any of the cell lines tested (Fig. 3). However, the MLN0128/TSA combination reduced 4eBP1 S65 phosphorylation in SKBR3, MDA-MB-231, and MCF7/HER2-18 cells relative to single treatments (Fig. 3a, b, d). Single 100 nM TSA treatment reduced AKT S473 phosphorylation in all cell lines tested except BT-474. Adding TSA to MLN0128 further reduced AKT S473 phosphorylation in all cell lines tested, even in BT-474 cells where single agent TSA had no effect (Fig. 3). Interestingly, AKT S473 phosphorylation was inhibited less by the highest doses of MLN0128 (25 nM) and TSA (100 nM) in MCF-10A than in any of the breast cancer cell lines (second rightmost lanes of Fig. 3f vs. a-e). In contrast to reduced AKT S473 phosphorylation in SKBR3 cells, MLN0128 increased AKT T308 phosphorylation. TSA alone did not affect AKT T308 phosphorylation, but adding it to MLN0128 abrogated MLN0128-induced increase in AKT T308 phosphorylation (Supplementary Fig. S4).
Fig. 3.
MLN0128 reduces AKT, S6, and 4eBP1 phosphorylation, while the combination with TSA further reduces AKT phosphorylation. Cells were treated with MLN0128 and TSA for 8 h. Whole cell lysates were collected and immunoblots were performed using the indicated antibodies. The following cell lines were analyzed: a SKBR3, b MDA-MB-231, c MCF7, d MCF7/HER2-18, e BT-474, and f MCF-10A
The reduction of AKT phosphorylation by MLN0128 and TSA is accompanied by reduced 4eBP1 phosphorylation and impaired polysome formation
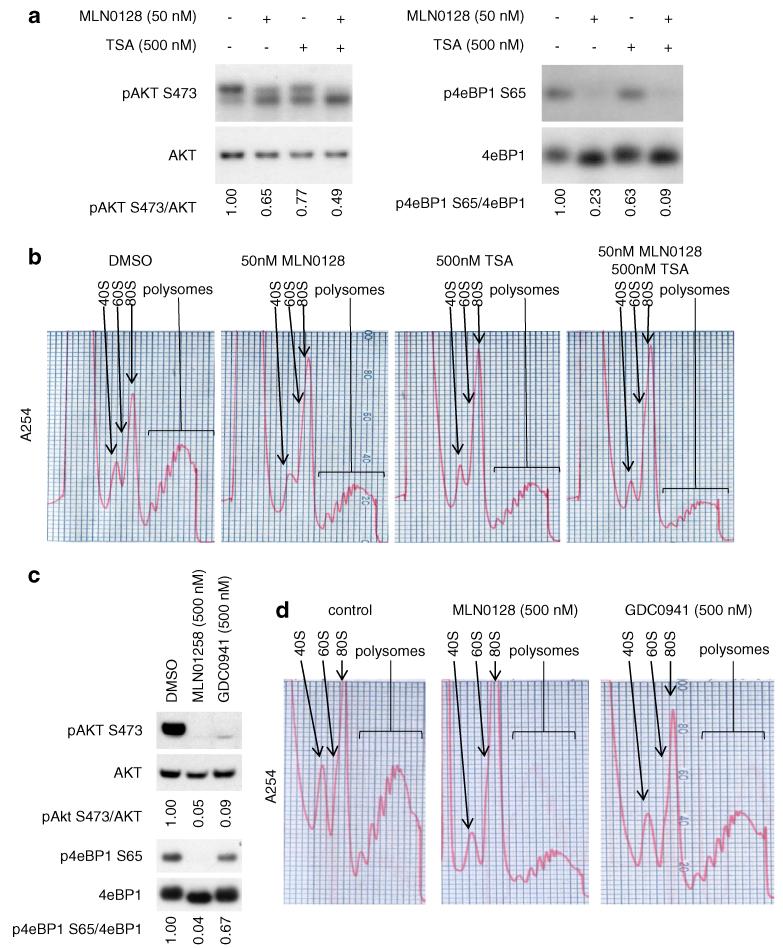
Since dual MLN0128/TSA treatment reduced AKT phosphorylation to a greater extent than single agents at the 8 h time point (Fig. 3a), we asked if extended treatment sustains AKT phosphorylation inhibition. In SKBR3 cells, 24 h of single 50 nM MLN0128 or 500 nM TSA treatment reduced AKT S473 phosphorylation to 65 and 77 % of control values, respectively. Dual MLN0128/TSA treatment further reduced AKT S473 phosphorylation to 49 % of the control (Fig. 4a). A similar pattern was observed for 4eBP1 phosphorylation at S65, with MLN0128 having an even greater effect on this phosphorylation event than on AKT S473 phosphorylation. Since PI3K/AKT/mTOR pathway signaling regulates polysome formation [5, 6], we asked if MLN0128 and/or TSA treatment affect polysome formation. We found that 24 h treatment with 50 nM MLN0128 or 500 nM TSA modestly reduced polysome formation, while combined treatment further reduced polysome formation (Fig. 4b).
Fig. 4.
The reduction of AKT phosphorylation by MLN0128 and TSA is accompanied by reduced 4eBP1 phosphorylation and impaired polysome formation: a SKBR3 cells were treated with 50 nM of MLN0128 and/or 500 nM of TSA for 24 h in single agent and in combination treatment. Whole cell lysates were collected and western blots were performed with the indicated antibodies. Ratios of phosphorylated to total protein were quantified using ImageJ. b Polysome profiling was performed from the same set of samples that were treated in (a). c SKBR3 cells were treated with 500 nM of MLN0128 or GDC0941 for 24 h. Whole cell lysates were collected and western blots and quantification were performed as in (a). d Polysome profiles were performed from samples treated in part (c)
We also determined if additional PI3K/AKT/mTOR pathway inhibitors that reduce AKT S473 phosphorylation can also impair polysome formation. The pan-PI3K inhibitor GDC0941, which has a higher IC50 than MLN0128 against SKBR3, MCF7, and MCF7/HER2-18 cell viability (Supplementary Fig. S1), reduced AKT S473 phosphorylation, albeit to a lesser extent than the same concentration of MLN0128 in SKBR3 (Fig. 4c). GDC0941 also reduced 4eBP1 S65 phosphorylation to a lesser extent than its effect on AKT phosphorylation and consistently reduced polysome formation to a lesser extent than the same dose of MLN0128 (Fig. 4d). These data suggest that modulation of AKT phosphorylation at S473 and 4eBP1 phosphorylation at S65 could be key mechanisms by which MLN0128, TSA, the combination of MLN0128 and TSA, or GDC0941 impair polysome assembly.
Discussion
We demonstrate synergy between the mTORC1/2 inhibitor, MLN0128, and a well-studied hydroxamic acid pan-HDAC inhibitor, TSA, in reducing viability of a phenotypically diverse panel of breast cancer cells (Tables 1, 2; Fig. 1). To our knowledge, this is the first study investigating any combination of mTOR and HDAC inhibitors in breast cancer cells, and the first to evaluate MLN0128 with an HDAC inhibitor in any cancer model. Against HER2+ SKBR3 and ER+ MCF7 cells, synergistic growth inhibition caused by the MLN0128/TSA combination was due to enhanced apoptosis as well as cell cycle arrest (Fig. 2). In general, the degree to which dual MLN0128/TSA treatment reduced viability correlated with the degree to which they underwent PARP cleavage, with SKBR3 being the most sensitive to the dual treatment and MDA-MB-231 being the least sensitive. However, for MDA-MB-231 and MCF-10A, the MLN0128/TSA combination did not induce PARP cleavage at the doses tested (Fig. 2); yet for MDA-MB-231 cells, those dual drug doses synergistically reduced cell viability (Table 2), likely by arresting cell growth and proliferation. In contrast, non-malignant MCF-10A mammary epithelial cells not only failed to undergo apoptosis, but also proved most resistant to both drugs individually and in combination (Table 1; Fig. 1). This latter observation suggests that this combination therapy may produce enhanced anticancer effects with minimal host organ toxicity. Our finding that use of a TORC1/C2 inhibitor in combination with a pan-HDAC inhibitor reduces AKT phosphorylation at S473 (Figs. 3, 4) supports the rationale that this combination would be more effective than one using a TORC1 inhibiting rapalog like everolimus (RAD001), which paradoxically increases AKT S473 phosphorylation [9, 10]. This important mechanistic difference could especially benefit breast cancer patients with tumors with high pAKT levels, since elevated tumor AKT S473 phosphorylation is correlated with poor outcome [11].
We provide new evidence that pan-HDAC inhibitors can potentially augment the anticancer activity of PI3K/AKT/mTOR pathway inhibition by further reducing AKT activation (Figs. 3, 4a). Previous studies suggest that the mechanism by which TSA inhibits AKT S473 phosphorylation involves preventing HDAC1 and HDAC6 from interacting with the phosphatase PP1, allowing PP1 to dephosphorylate on AKT S473 [34]. In that study, TSA was most potent in down-regulating AKT phosphorylation compared to the pan-HDAC inhibitors HDAC42 and SAHA, or the class I HDAC inhibitor MS-275 (entinostat) [34]. High HDAC inhibitor doses are also known to reduce total AKT protein levels through heat shock protein 90-mediated AKT destabilization [35]. The importance of modulating AKT phosphorylation by mTOR/HDAC inhibitors is further illustrated by a study showing that constitutively active AKT1 reduces the efficacy of PP242 (an mTORC1/C2 inhibitor) in combination with either SAHA or LBH589 against hepatocellular carcinoma cells [30]. We extend these earlier observations by showing that the combination of potent mTORC1/C1 and pan-HDAC inhibitors synergistically inhibit growth of five phenotypically diverse breast cancer cell lines while also reducing AKT S473 phosphorylation, emphasizing the potential role of AKT S473 phosphorylation as a companion biomarker of tumor responsiveness to this novel breast cancer combination treatment strategy.
We also introduce the novel observation that reducing AKT S473 and 4eBP1 S65 phosphorylation, whether by mTORC1/C2 (MLN0128), pan-HDAC (TSA), or pan-PI3K (GDC0941) inhibitors, is accompanied by impaired polysome formation (Figs. 4, 5). Previous studies illustrate the value of understanding polysome function as a cancer drug mechanism of action. For example, mTOR signaling regulates prostate cancer invasiveness by increasing polysome association of mRNAs involved in invasion [8]. Furthermore, haploinsufficiency of the ribosomal protein RPL24 improves survival of mice bearing AKT-driven tumors by decreasing cap-dependent polysome-driven protein synthesis [36]. The decrease in breast cancer cell polysome formation caused by TSA represents the first such observation and raises the question of the mechanistic contribution of impaired polysome formation as part of the overall anticancer consequences of pan-HDAC inhibitor therapy.
Fig. 5.
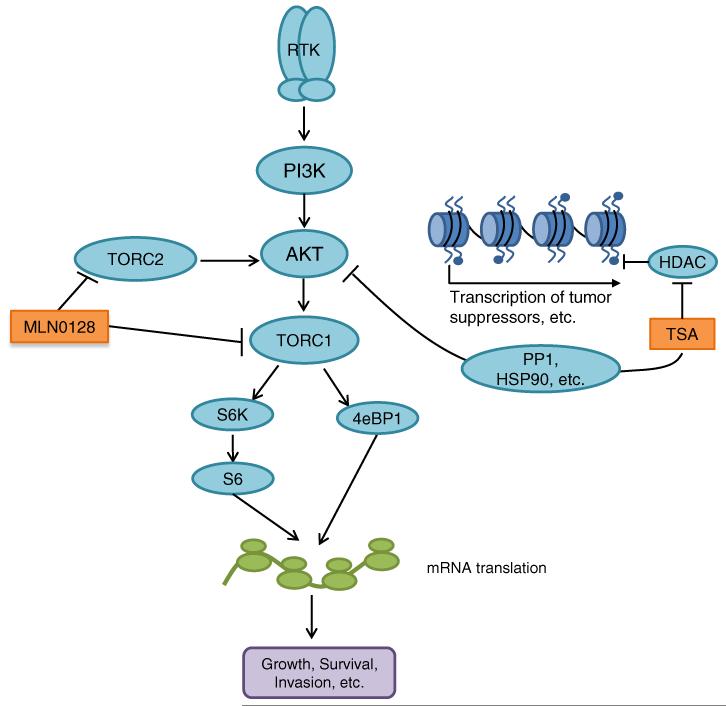
Schematic of signaling pathways targeted by MLN0128 and TSA. AKT is activated in many breast cancers through dysregulation of receptor tyrosine kinases (RTKs) and/or PI3K. MLN0128 inhibits AKT activation by inhibiting TORC2, which activates AKT. MLN0128 also inhibits signaling downstream of AKT by inhibiting TORC1. Furthermore, TSA inhibits AKT through mechanisms involving increased dephosphorylation of AKT via activation of the PP1 phosphatase or increased AKT destabilization through HSP90 inhibition. Down-regulation of AKT by MLN0128 and TSA results in decreased polyribosome function and decreased translation of mRNAs involved in carcinogenesis. TSA also induces the acetylation of histones, thus increasing the transcription of various tumor suppressors
In sum, our findings support a model whereby synergistic growth inhibition caused by combining mTORC1/C2 and pan-HDAC-inhibitors reflects a mechanistic convergence onto the PI3K/AKT/mTOR pathway, inhibiting AKT S473 phosphorylation to a greater extent than that caused by the individual drugs, decreasing 4eBP1 S65 phosphorylation and polysome formation (Fig. 5). This novel therapeutic strategy warrants further preclinical and clinical evaluation.
Supplementary Material
Acknowledgments
This research was supported by NIH/NCI Grants R21-CA155679, R01-CA071468, and U24-CA14358; by NIH/NIA T32-AG000266; and by Hazel P. Munroe memorial funding to the Buck Institute. The drugs INK0128 (MLN0128) and GDC0941 were kindly provided by Intellikine Pharmaceuticals, Inc.; and we thank Christian Rommel for his encouragement and early advice. We are appreciative of technical assistance from Ingrid Hanson from the Benz lab and Taki Te Koi (Te Manu Whakataki) from the Buck Institute Morphology and Imaging Core. We also thank members of the Benz lab for insightful discussions, and Buck Institute scientist Patrick Li for assistance with the polysome profiling.
Footnotes
Conflicts of interest The authors declare no potential conflicts of interest.
Electronic supplementary material The online version of this article (doi:10.1007/s10549-014-2877-y) contains supplementary material, which is available to authorized users.
Contributor Information
Kathleen A. Wilson-Edell, Buck Institute for Research on Aging, 8001 Redwood Boulevard, Novato, CA 94945, USA
Mariya A. Yevtushenko, Buck Institute for Research on Aging, 8001 Redwood Boulevard, Novato, CA 94945, USA; Master of Science in Biology Program, Dominican University, San Rafael, CA, USA
Daniel E. Rothschild, Buck Institute for Research on Aging, 8001 Redwood Boulevard, Novato, CA 94945, USA
Aric N. Rogers, Buck Institute for Research on Aging, 8001 Redwood Boulevard, Novato, CA 94945, USA
Christopher C. Benz, Buck Institute for Research on Aging, 8001 Redwood Boulevard, Novato, CA 94945, USA, cbenz@buckinstitute.org
References
- 1.Cidado J, Park BH. Targeting the PI3K/Akt/mTOR pathway for breast cancer therapy. J Mammary Gland Biol Neoplasia. 2012 doi: 10.1007/s10911-012-9264-2. doi:10.1007/s10911-012-9264-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yuan TL, Cantley LC. PI3K pathway alterations in cancer: variations on a theme. Oncogene. 2008;27(41):5497–5510. doi: 10.1038/onc.2008.245. doi:10.1038/onc.2008.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stemke-Hale K, Gonzalez-Angulo AM, Lluch A, Neve RM, Kuo WL, Davies M, Carey M, Hu Z, Guan Y, Sahin A, Symmans WF, Pusztai L, Nolden LK, Horlings H, Berns K, Hung MC, van de Vijver MJ, Valero V, Gray JW, Bernards R, Mills GB, Hennessy BT. An integrative genomic and proteomic analysis of PIK3CA, PTEN, and AKT mutations in breast cancer. Cancer Res. 2008;68(15):6084–6091. doi: 10.1158/0008-5472.CAN-07-6854. doi:10.1158/0008-5472.CAN-07-6854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chacon RD, Costanzo MV. Triple-negative breast cancer. Breast Cancer Res. 2010;12(Suppl 2):S3. doi: 10.1186/bcr2574. doi:10.1186/bcr2574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mayer C, Grummt I. Ribosome biogenesis and cell growth: mTOR coordinates transcription by all three classes of nuclear RNA polymerases. Oncogene. 2006;25(48):6384–6391. doi: 10.1038/sj.onc.1209883. doi:10.1038/sj.onc.1209883. [DOI] [PubMed] [Google Scholar]
- 6.Thoreen CC, Chantranupong L, Keys HR, Wang T, Gray NS, Sabatini DM. A unifying model for mTORC1-mediated regulation of mRNA translation. Nature. 2012;485(7396):109–113. doi: 10.1038/nature11083. doi:10.1038/nature11083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hsu PP, Kang SA, Rameseder J, Zhang Y, Ottina KA, Lim D, Peterson TR, Choi Y, Gray NS, Yaffe MB, Marto JA, Sabatini DM. The mTOR-regulated phosphoproteome reveals a mechanism of mTORC1-mediated inhibition of growth factor signaling. Science. 2011;332(6035):1317–1322. doi: 10.1126/science.1199498. doi:10.1126/science.1199498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hsieh AC, Liu Y, Edlind MP, Ingolia NT, Janes MR, Sher A, Shi EY, Stumpf CR, Christensen C, Bonham MJ, Wang S, Ren P, Martin M, Jessen K, Feldman ME, Weissman JS, Shokat KM, Rommel C, Ruggero D. The translational landscape of mTOR signalling steers cancer initiation and metastasis. Nature. 2012;485(7396):55–61. doi: 10.1038/nature10912. doi:10.1038/nature10912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Baselga J, Campone M, Piccart M, Burris HA, Rugo HS, Sahmoud T, Noguchi S, Gnant M, Pritchard KI, Lebrun F, Beck JT, Ito Y, Yardley D, Deleu I, Perez A, Bachelot T, Vittori L, Xu Z, Mukhopadhyay P, Lebwohl D, Hortobagyi GN. Everolimus in postmenopausal hormone-receptor-positive advanced breast cancer. N Engl J Med. 2012;366(6):520–529. doi: 10.1056/NEJMoa1109653. doi:10.1056/NEJMoa1109653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bachelot T, Bourgier C, Cropet C, Ray-Coquard I, Ferrero JM, Freyer G, Abadie-Lacourtoisie S, Eymard JC, Debled M, Spaeth D, Legouffe E, Allouache D, El Kouri C, Pujade-Lauraine E. Randomized phase II trial of everolimus in combination with tamoxifen in patients with hormone receptor-positive, human epidermal growth factor receptor 2-negative metastatic breast cancer with prior exposure to aromatase inhibitors: a GI-NECO study. J Clin Oncol. 2012;30(22):2718–2724. doi: 10.1200/JCO.2011.39.0708. doi:10.1200/JCO.2011.39.0708. [DOI] [PubMed] [Google Scholar]
- 11.Perez-Tenorio G, Stal O. Activation of AKT//PKB in breast cancer predicts a worse outcome among endocrine treated patients. Br J Cancer. 2002;86(4):540–545. doi: 10.1038/sj.bjc.6600126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ozes ON, Akca H, Mayo LD, Gustin JA, Maehama T, Dixon JE, Donner DB. A phosphatidylinositol 3-kinase/Akt/mTOR pathway mediates and PTEN antagonizes tumor necrosis factor inhibition of insulin signaling through insulin receptor substrate-1. Proc Natl Acad Sci USA. 2001;98(8):4640–4645. doi: 10.1073/pnas.051042298. doi:10.1073/pnas.051042298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tabernero J, Rojo F, Calvo E, Burris H, Judson I, Hazell K, Martinelli E, Ramon y, Cajal S, Jones S, Vidal L, Shand N, Macarulla T, Ramos FJ, Dimitrijevic S, Zoellner U, Tang P, Stumm M, Lane HA, Lebwohl D, Baselga J. Dose- and schedule-dependent inhibition of the mammalian target of rapamycin pathway with everolimus: a phase I tumor pharmacodynamic study in patients with advanced solid tumors. J Clin Oncol. 2008;26(10):1603–1610. doi: 10.1200/JCO.2007.14.5482. doi:10.1200/JCO.2007.14.5482. [DOI] [PubMed] [Google Scholar]
- 14.Janes MR, Vu C, Mallya S, Shieh MP, Limon JJ, Li LS, Jessen KA, Martin MB, Ren P, Lilly MB, Sender LS, Liu Y, Rommel C, Fruman DA. Efficacy of the investigational mTOR kinase inhibitor MLN0128/INK128 in models of B-cell acute lymphoblastic leukemia. Leukemia. 2013;27(3):586–594. doi: 10.1038/leu.2012.276. doi:10.1038/leu.2012.276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pourdehnad M, Truitt ML, Siddiqi IN, Ducker GS, Shokat KM, Ruggero D. Myc and mTOR converge on a common node in protein synthesis control that confers synthetic lethality in Myc-driven cancers. Proc Natl Acad Sci. 2013 doi: 10.1073/pnas.1310230110. doi:10.1073/pnas.1310230110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gokmen-Polar Y, Liu Y, Toroni RA, Sanders KL, Mehta R, Badve S, Rommel C, Sledge GW., Jr Investigational drug MLN0128, a novel TORC1/2 inhibitor, demonstrates potent oral antitumor activity in human breast cancer xenograft models. Breast Cancer Res Treat. 2012;136(3):673–682. doi: 10.1007/s10549-012-2298-8. doi:10.1007/s10549-012-2298-8. [DOI] [PubMed] [Google Scholar]
- 17.Wagner JM, Hackanson B, Lubbert M, Jung M. Histone deacetylase (HDAC) inhibitors in recent clinical trials for cancer therapy. Clin Epigenetics. 2010;1(3-4):117–136. doi: 10.1007/s13148-010-0012-4. doi:10.1007/s13148-010-0012-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Richon VM, Sandhoff TW, Rifkind RA, Marks PA. Histone deacetylase inhibitor selectively induces p21WAF1 expression and gene-associated histone acetylation. Proc Natl Acad Sci USA. 2000;97(18):10014–10019. doi: 10.1073/pnas.180316197. doi:10.1073/pnas.180316197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Witt O, Deubzer HE, Milde T, Oehme I. HDAC family: what are the cancer relevant targets? Cancer Lett. 2009;277(1):8–21. doi: 10.1016/j.canlet.2008.08.016. doi:10.1016/j.canlet.2008.08.016. [DOI] [PubMed] [Google Scholar]
- 20.Tate CR, Rhodes LV, Segar HC, Driver JL, Pounder FN, Burow ME, Collins-Burow BM. Targeting triple-negative breast cancer cells with the histone deacetylase inhibitor panobinostat. Breast Cancer Res. 2012;14(3):R79. doi: 10.1186/bcr3192. doi:10.1186/bcr3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vigushin DM, Ali S, Pace PE, Mirsaidi N, Ito K, Adcock I, Coombes RC. Trichostatin A is a histone deacetylase inhibitor with potent antitumor activity against breast cancer in vivo. Clin Cancer Res. 2001;7(4):971–976. [PubMed] [Google Scholar]
- 22.Heiser LM, Sadanandam A, Kuo WL, Benz SC, Goldstein TC, Ng S, Gibb WJ, Wang NJ, Ziyad S, Tong F, Bayani N, Hu Z, Billig JI, Dueregger A, Lewis S, Jakkula L, Korkola JE, Durinck S, Pepin F, Guan Y, Purdom E, Neuvial P, Bengtsson H, Wood KW, Smith PG, Vassilev LT, Hennessy BT, Greshock J, Bachman KE, Hardwicke MA, Park JW, Marton LJ, Wolf DM, Collisson EA, Neve RM, Mills GB, Speed TP, Feiler HS, Wooster RF, Haussler D, Stuart JM, Gray JW, Spellman PT. Subtype and pathway specific responses to anticancer compounds in breast cancer. Proc Natl Acad Sci USA. 2012;109(8):2724–2729. doi: 10.1073/pnas.1018854108. doi:10.1073/pnas.1018854108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Scott GK, Marden C, Xu F, Kirk L, Benz CC. Transcriptional repression of ErbB2 by histone deacetylase inhibitors detected by a genomically integrated ErbB2 promoter-reporting cell screen. Mol Cancer Ther. 2002;1(6):385–392. [PubMed] [Google Scholar]
- 24.Scott GK, Marx C, Berger CE, Saunders LR, Verdin E, Schafer S, Jung M, Benz CC. Destabilization of ERBB2 transcripts by targeting 3′ untranslated region messenger RNA associated HuR and histone deacetylase-6. Mol Cancer Res. 2008;6(7):1250–1258. doi: 10.1158/1541-7786.MCR-07-2110. doi:10.1158/1541-7786.MCR-07-2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Verheul HM, Salumbides B, Van Erp K, Hammers H, Qian DZ, Sanni T, Atadja P, Pili R. Combination strategy targeting the hypoxia inducible factor-1 alpha with mammalian target of rapamycin and histone deacetylase inhibitors. Clin Cancer Res. 2008;14(11):3589–3597. doi: 10.1158/1078-0432.CCR-07-4306. doi:10.1158/1078-0432.CCR-07-4306. [DOI] [PubMed] [Google Scholar]
- 26.Wedel S, Hudak L, Seibel JM, Juengel E, Tsaur I, Wiesner C, Haferkamp A, Blaheta RA. Inhibitory effects of the HDAC inhibitor valproic acid on prostate cancer growth are enhanced by simultaneous application of the mTOR inhibitor RAD001. Life Sci. 2011;88(9-10):418–424. doi: 10.1016/j.lfs.2010.12.017. doi:10.1016/j.lfs.2010.12.017. [DOI] [PubMed] [Google Scholar]
- 27.Dong LH, Cheng S, Zheng Z, Wang L, Shen Y, Shen ZX, Chen SJ, Zhao WL. Histone deacetylase inhibitor potentiated the ability of MTOR inhibitor to induce autophagic cell death in Burkitt leukemia/lymphoma. J Hematol Oncol. 2013;6(1):53. doi: 10.1186/1756-8722-6-53. doi:10.1186/1756-8722-6-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nishioka C, Ikezoe T, Yang J, Koeffler HP, Yokoyama A. Blockade of mTOR signaling potentiates the ability of histone deacetylase inhibitor to induce growth arrest and differentiation of acute myelogenous leukemia cells. Leukemia. 2008;22(12):2159–2168. doi: 10.1038/leu.2008.243. doi:10.1038/leu.2008.243. [DOI] [PubMed] [Google Scholar]
- 29.Erlich RB, Kherrouche Z, Rickwood D, Endo-Munoz L, Cameron S, Dahler A, Hazar-Rethinam M, de Long LM, Wooley K, Guminski A, Saunders NA. Preclinical evaluation of dual PI3K-mTOR inhibitors and histone deacetylase inhibitors in head and neck squamous cell carcinoma. Br J Cancer. 2012;106(1):107–115. doi: 10.1038/bjc.2011.495. doi:10.1038/bjc.2011.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shao H, Gao C, Tang H, Zhang H, Roberts LR, Hylander BL, Repasky EA, Ma WW, Qiu J, Adjei AA, Dy GK, Yu C. Dual targeting of mTORC1/C2 complexes enhances histone deacetylase inhibitor-mediated anti-tumor efficacy in primary HCC cancer in vitro and in vivo. J Hepatol. 2012;56(1):176–183. doi: 10.1016/j.jhep.2011.07.013. doi:10.1016/j.jhep.2011.07.013. [DOI] [PubMed] [Google Scholar]
- 31.Benz CC, Scott GK, Sarup JC, Johnson RM, Tripathy D, Coronado E, Shepard HM, Osborne CK. Estrogen-dependent, tamoxifen-resistant tumorigenic growth of MCF-7 cells transfected with HER2/neu. Breast Cancer Res Treat. 1992;24(2):85–95. doi: 10.1007/BF01961241. [DOI] [PubMed] [Google Scholar]
- 32.Wilson KA, Colavito SA, Schulz V, Wakefield PH, Sessa W, Tuck D, Stern DF. NFBD1/MDC1 regulates Cav1 and Cav2 independently of DNA damage and p53. Mol Cancer Res. 2011;9(6):766–781. doi: 10.1158/1541-7786.MCR-10-0317. doi:10.1158/1541-7786.MCR-10-0317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rogers AN, Chen D, McColl G, Czerwieniec G, Felkey K, Gibson BW, Hubbard A, Melov S, Lithgow GJ, Kapahi P. Life span extension via eIF4G inhibition is mediated by post-transcriptional remodeling of stress response gene expression in C. elegans. Cell Metab. 2011;14(1):55–66. doi: 10.1016/j.cmet.2011.05.010. doi:10.1016/j.cmet.2011.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen C-S, Weng S-C, Tseng P-H, Lin H-P, Chen C-S. Histone acetylation-independent effect of histone deacetylase inhibitors on Akt through the reshuffling of protein phosphatase 1 complexes. J Biol Chem. 2005;280(46):38879–38887. doi: 10.1074/jbc.M505733200. doi:10.1074/jbc.M505733200. [DOI] [PubMed] [Google Scholar]
- 35.Bali P, Pranpat M, Bradner J, Balasis M, Fiskus W, Guo F, Rocha K, Kumaraswamy S, Boyapalle S, Atadja P, Seto E, Bhalla K. Inhibition of histone deacetylase 6 acetylates and disrupts the chaperone function of heat shock protein 90: a novel basis for antileukemia activity of histone deacetylase inhibitors. J Biol Chem. 2005;280(29):26729–26734. doi: 10.1074/jbc.C500186200. doi:10.1074/jbc.C500186200. [DOI] [PubMed] [Google Scholar]
- 36.Hsieh AC, Costa M, Zollo O, Davis C, Feldman ME, Testa JR, Meyuhas O, Shokat KM, Ruggero D. Genetic dissection of the oncogenic mTOR pathway reveals druggable addiction to translational control via 4EBP-eIF4E. Cell. 2010;17(3):249–261. doi: 10.1016/j.ccr.2010.01.021. doi:10.1016/j.ccr.2010.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.