Abstract
Behavioral problems (e.g. learning and memory) following developmental exposure to toxicants suggests that dysregulation of the process of synapse formation and function may occur. The ability to assess these changes is thus of value. This protocol describes a method to investigate toxicant-induced changes to synaptic structure formation in primary hippocampal neurons using immunocytochemical labeling of the pre- and post-synaptic markers synaptophysin and PSD-95, confocal imaging, and three-dimensional object analysis. Protocols for the long-term culturing of primary hippocampal neurons and of primary cortical astrocytes, as well as their co-culture are included. While the described methods focus on how astrocytes influence synapse formation and how toxicants may interfere in this process, modifications to the experimental plan can easily be implemented. This would allow for the investigation of the effects of toxicants after treating neurons alone, or both astrocytes and neurons in co-culture. With the common endpoint of synapse structure formation, differences between varying treatment paradigms can expand our understanding of the influence of particular toxicants on these diverse cell types and provide insight into potential mechanisms of effect and the contributions of each to synapse formation.
Keywords: synaptogenesis, astrocytes, sandwich co-culture, 3-dimensional analysis, PSD-95, synaptophysin
INTRODUCTION
Exposure to toxicants at crucial moments during neurodevelopment may result in long-lasting behavioral problems, including alterations of learning and memory, suggesting that dysregulation of synapse formation and synaptic function may occur. Astrocytes, the most abundant cells in the brain, proliferate at a time in development when synapses are forming (Eroglu and Barres, 2010; Bernal, 2007; de Graff-Peters and Hadders-Algra, 2006), and it is increasingly becoming clear that many of the factors that astrocytes release are crucial for the process of synaptogenesis (Nagler et al, 2001; Pfrieger and Barres, 1997; Ullian et al., 2001; Christopherson et al, 2005; Mauch et al., 2001). This newly emerging understanding of the important astrocytic contributions to synaptic development reinforces the idea that astrocytes may also be implicated in various diseases (Barres, 2008; Ransom cite). The protocols described in this UNIT will enable a researcher to investigate how soluble factors released by astrocytes influence synaptic structure formation and how toxicants may modulate their effect.
While this protocol focuses on the treatment of astrocytes with a toxicant and how treatment modulates the astrocytic effect on synapse formation, experimental modifications can be easily made depending on the experimental question, thus also allowing for the testing and the comparison of the effects on synapse formation after toxicant treatment on neurons directly, as well as both neurons and astrocytes simultaneously, using the co-culture system described in this UNIT. From these kinds of manipulations the relative contribution of astrocytes and neurons to synapse formation can be obtained. Furthermore, by investigating other synaptic markers and by using pharmacological agents to activate or inhibit receptors or signaling pathways the mechanisms implicated in physiological, pathological, and toxicant-induced changes in synapse formation can be explored.
This UNIT describes methods for a sandwich co-culture system of primary hippocampal neurons and astrocytes. Neurons are plated on glass coverslips for imaging and for co-culturing, and are grown in the absence of astrocytes for 13 days; during this time in culture they develop axons and dendrites, form extensive networks, and express synaptic proteins (Basic Protocol 1). Primary cortical astrocytes are prepared and grown for two weeks (Basic Protocol 2) after which they are passed for experimental usage, treated with a toxicant, and then co-cultured with neurons for 24 hours (Basic Protocol 3). The co-culture system and neuronal culturing methods described in the present UNIT relies on previously used methods (Banker and Cowan, 1977; Kaech and Banker, 2006; Viviani, 2003; Boraso and Viviani, 2011; Goslin et al., 1998; Roqué, et al, 2011), with a number of necessary modifications in order to address the specific experimental question being asked, specifically the need to maintain a long-term neuronal culture.
To compare differences in the formation of synapses between treatment groups at the end of 24 h co-culture with astrocytes, neurons are fixed and immunocytochemically labeled for the pre- and post-synaptic proteins synaptophysin and PSD-95 (Wiedenmann and Franke, 1985; Okabe, 2002), both of which have been used extensively in assessments of synapse formation (Rao and Craig, 1997; Okabe et al., 1999) (Basic Protocol 4). The method developed for quantification of synaptic structures uses scanning laser confocal microscopy for imaging, deconvolution of the images to re-focus light that inherently scatters as it moves through various optical components and an optimized thresholding method to render the surface of synaptic puncta in three-dimensions. Once rendered, the number of pre- and post-synaptic puncta are automatically determined, allowing the ability to compare how treatment affects the localization of these proteins into pre- and post-synaptic specializations. Finally, those pre- and postsynaptic puncta that are in apposition, or overlapping, are quantified, and used to assess effects on synapse number (Basic Protocol 5). As the UNIT includes protocols for the growth of two primary cultures, each requiring different tasks at varying time points, an overview of the general day-to-day experimental timeline is presented in Table 1, and estimates of timing for hands-on tasks are described in Table 2.
Table 1.
Neuron and Astrocyte Experimental Task List by Day
| Time Relative to Neuronal Prep | Neurons | Astrocytes |
|---|---|---|
| 1.5 – 2 weeks prior | Culture Primary Astrocytes | |
| 2 days prior | Prepare Coverslips | |
| 1 day prior | Wash & Coat Coverslips with Substrate |
|
| Day 0 | Prepare & Plate Primary Neurons | |
| Day 1 | Change neuronal medium | |
| Day 2 | Treat with ARAC | |
| Day 3 | ||
| Day 4 | ||
| Day 5 | Change 1/3 neuronal medium | |
| Day 6 | Pass experimental astrocytes |
|
| Day 7 | Change medium | |
| Day 8 | ||
| Day 9 | Change 1/3 neuronal medium | Change medium |
| Day 10 | Serum Deprive Astrocytes | |
| Day 11 | Treat Astrocytes | |
| Day 12 | Stop Treatment | |
| Co-culture neurons and astrocytes | ||
| Day 13 | Stop co-culture | |
| Fix neurons & store or begin Immunolabeling |
||
| After Day 13 | Complete ICC labeling Mount Image, deconvolve, analyze |
|
Table 2.
Timing by Experimental Task
| Experimental Task |
Hands On Time (in hours) |
Variables Influencing Time |
Overall Time in Culture |
|
|---|---|---|---|---|
|
Coverslip Preparation |
Application of Wax Spacers |
~ 1 – 1.5 hours |
Can be prepared ahead of time |
|
| HCL Sterilization | Overnight | |||
|
Wash & Substrate Coating |
1 hour over 3–3.5 hours (5 washes, 30 minutes each) |
|||
| Neuronal Culture | 13 Days | |||
|
Dissection & Cell Preparation |
3.5 – 4.5 hours |
Number of pups in the litter. |
||
| Medium Changes | ½ hour | |||
|
Astrocyte Cultures |
2 weeks + | |||
|
Primary Cell Dissection & Preparation |
3 – 4 hours | |||
| Medium Changes |
½ to 1 hour every 2 – 3 days |
Number of flasks | ||
|
Astrocyte passage for experiments |
1.5 – 2 hours | |||
| Serum Deprivation |
½ hour to 1 hour |
Number of wells | 24 hours | |
| Treatment | ~ 1 hour |
Treatment preparation |
24 hours * | |
| Co-culture | 24 hours * | |||
|
Neuronal Fixation & Wash |
¾ - 1hour | |||
| Immunolabeling | Day 1 – Primary AB | 1.5 – 2 hours |
Number of coverslips |
Overnight (at least 18 hours) |
|
Day 2 – 2ndary Ab Washes and Incubation |
2 ¾ - 3 hours | |||
| Mounting Slides | 1–1.5 hours |
Number of coverslips |
||
| Imaging | Confocal Imaging |
~ 3 – 4 hours per treatment group |
15 neurons | |
| Thresholding |
1 – 1.5 hours for 15 images |
|||
| 3-D Image Analysis |
1 – 1.5 hours per experiment |
|||
While differences in synaptic structure formation after treatment are of interest, for a broader and more complete understanding of any effect on synapse formation, electrophysiology studies to assess the functionality of synapses formed after co-culture or treatment are strongly recommended. While the electrophysiological methodology is not included in this UNIT, these investigations would both corroborate and expand the finding on structural changes into actual functional effects.
All work prior to the fixation of the hippocampal neurons should be performed in aseptic conditions. All animal work must follow IACUC-approved procedures.
BASIC PROTOCOL 1
NEURONAL PREPARATION AND CULTURE
Primary hippocampal neurons from Sprague-Dawley rats (E21) are dissected, prepared and cultured for 13 days on glass coverslips containing wax spacers for use in co-culturing. This optimized long-term culture assures that most of the neurons survive, develop intricate dendritic and axonal networks, and acquire the ability to form functional synapses. Because the interest here is to assess how astrocyte secreted factors modulate synaptogenesis and how astrocyte exposure to toxicants may alter synapse formation, it is important that the neuronal cultures are relatively astrocyte-free. The use of B-27 supplement in the neuronal medium, and a one-time addition of an anti-mitotic agent, cytosine β-d-arabinofuranoside (ARAC), together work to make these cultures 94% non-astrocytic. Cells are plated at a density of 80,000 cells per coverslip, this neuronal cell density was optimized to maximize the survival of primary neurons during the 13 days in culture in the absence of astrocytes, and to allow for the imaging of individual neurons. With practice, the expected yield of neurons per pup should be approximately 1–1.5 × 106 cells per fetus (See Critical Parameters/Troubleshooting). All work should be performed in aseptic conditions.
Materials
E21 Sprague-Dawley rat fetuses.
Hank’s Balanced Salt Solution with calcium and magnesium containing 10 mM HEPES (HBSS + CM). Keep sterile and store at RT.
Hank’s Balanced Salt Solution (Calcium and magnesium-free) containing 10 mM HEPES (HBSS CM-Free). Keep sterile and store at RT.
Papain from papaya latex.
Magnesium Chloride (1 M), sterile
Deoxyribonuclease-1 from bovine pancreas (DNase) (4 mg/mL) (See Reagents and Solutions)
Neurobasal-A Complete Medium (See Reagents and Solutions)
Cytosine β-d-arabinofuranoside (ARAC) (100 µM working solution) (See Reagents and Solutions)
Sterile surgical tools
Sterile, disposable 5 mL syringe and 22 um filter
40 µm pore nylon mesh filter in a 50 mL conical tube
Rubber bulb
Sterile glass Pasteur pipettes (9”)
Two days prior to neuronal plating, prepare the coverslips (See Support Protocol 1). Perform the following steps in aseptic conditions.
Immediately before the hippocampal dissection, wash the prepared coverslips once with sterile water and replace with HBSS (+CM). Set the plates aside in the sterile hood until cell plating.
- Prepare 4 mL of papain solution (2mgs/mL) for the enzymatic digestion of hippocampal tissue. Add DNase (See Reagents and Solutions) (1:500) and MgCl2 (5 mM). Invert to mix and incubate in a 37°C water bath to solubilize the papain.This solution will be filtered prior to use immediately after the hippocampal dissection. Four mL of papain solution are sufficient to digest one litter, up to 15 fetuses. If additional cells are needed, a second aliquot of the solution should be prepared for effective dissociation.
-
Working with one fetus at a time, decapitate the fetus and place the cerebral hemisphere on sterile filter paper. Dissect the hippocampi from E21 Sprague-Dawley fetuses. Place the hippocampal tissue in a 35 mm dish containing 1.5 mL of HBSS - (CM Free) and stored over ice.
Identifying and dissecting out the hippocampi takes practice. Dissection techniques are easily found online or described elsewhere (Giordano and Costa, 2011). At age E21, the rat hippocampus is visible by eye, but if necessary, a stereomicroscope can be used under aseptic conditions to identify the hippocampus. Work rapidly, as the cerebral hemispheres lose their structure relatively quickly, and may become quite gelatinous, making identification and separation of the distinct brain areas difficult. Even with practice, sometime this occurs. If it does, it is best to set the brain aside and not use it, rather than risk contamination of the culture with non-hippocampal cells. Additionally, to minimize contamination of the cultures with non-neuronal cells, it is important to remove the meninges from the hippocampi during the dissection. This can be accomplished by either manually removing the meningeal layer with tweezers, or rolling the tissue on sterile filter paper.
After the hippocampi from all of the fetuses have been collected, sterile filter the enzymatic papain solution using a disposable 5 mL syringe and a 22 um sterile filter.
Remove as much HBSS (CM-Free) as possible from the dish without disturbing hippocampal tissue and discard. Manually cut the tissue into small pieces using a sterile surgical scissor.
Transfer the tissue to a 15 mL tube. Wash the plate with the filtered papain solution numerous times to ensure that all the tissue is collected. Add the remaining papain solution to the tissue.
-
Incubate the minced tissue in papain for 30 minutes in a 37.5 C° water bath.
Check the tissue after ten minutes into the enzymatic incubation. Invert 2X to mix. For the most effective digestion of the tissue, the particles should remain distinctly separate. Depending on the amount of tissue (number of fetuses), the amount of manual dissociation with the scissors, and the strength of the DNase, the tissue may appear coagulated or clumped, due to the release of DNA from the physically damaged cells. If this is observed, add additional DNase (1:500), in sterile conditions. Gently invert to mix and return to the water bath to complete the 30 minute incubation.
Centrifuge the tissue for 5 minutes at 185 × g at 4°C.
- Aspirate off the papain taking care not to disturb the pelleted cells. Add 2 mL of warmed complete medium and DNase (1:500) to the pelleted cells.After this first step, the addition of DNase is optional. If the tissue is dissociating easily and entering solution, the DNase addition at each subsequent trituration step may be skipped.
Cell Dissociation
These young primary neurons are quite fragile and sensitive. The trituration process below proceeds through a series of steps where the tip of the tool used to dissociate the cells starts large and gets successively smaller. This is a gentler form of generating individual cells without causing excessive damage and cell death.
-
11.
Place a rubber bulb on the end of a sterile, 9” long glass Pasteur pipette.
-
12.Triturate the cells by gently drawing the tissue and medium into the pipette. To minimize damage to the cells, release slowly along the side of the 15 mL tube. Repeat for a total of 15 times.As cells enter solution, the tissue particles should decrease in size and the medium will become slightly cloudy.
-
13.
Let the tissue settle by gravity sedimentation for two minutes.
-
14.
Without disturbing the settled tissue, filter the medium, which now contains suspended cells through a 40 µm filter into a 50 mL tube.
-
15.
To the remaining tissue, add 2 mL of complete medium and DNase (if necessary). Trim the end of a 200 µL pipette tip to widen the opening slightly and carefully place it on the end of the glass Pasteur pipette. Repeat steps 12 through 14.
-
16.
To the remaining tissue, add 2 mL of complete medium and DNase (if necessary). Place a 1000 µL pipette tip on the end of a 5 mL sterile disposable pipette and repeat steps 12 through 13. If there is no visible tissue remaining after two minutes of sedimentation, filter the medium through the 40 um filter. If tissue remains, filter the medium without disturbing the tissue, repeat the addition of medium and DNase and continue the trituration until the tissue is no longer visible. Filter the entire medium through the 40 µm filter.
-
17.
Bring the filtered medium volume to 10 mL and transfer to a 15 mL tube.
-
18.
Centrifuge the dissociated cells for 5 minutes at 145 x g at 4°C.
-
19.
Aspirate the medium and add 5 – 10 mL of warmed medium to cells.
If fewer than 5–6 pups were used, bring the final volume to 5 mL instead of 10 mL for a more accurate cell count.
-
20.
Re-suspend the cells gently, but completely.
-
21.
Count the cells using a hemocytometer. Mix a small volume of cells for counting in a 1:1 ratio of cell suspension to trypan blue.
Trypan blue will allow you to quantify the amount of cell death after dissociation. Typically, a 10–15 % cell death is observed. Cell death greater than this usually results in a culture that will not survive. (See Critical Parameters – Neuronal Preparation)
-
22.
Plate neurons at a density of 80,000 per coverslip (0.080 × 106 cells/mL), dispensing cells against the wall of the well to minimize damage to the cells.
-
23.
Use a sterile needle to push the coverslip to the bottom of the well to ensure neuronal adherence to the surface of the coverslip and not the plastic of the well.
-
24.
Incubate overnight at 37°C to allow the neurons to adhere to the coverslips.
-
25.
The next morning, gently wash the cells 1X with warmed HBSS and replace the medium.
-
26.
Add ARAC (2.5 uM final concentration) to each well on the third day in culture to minimize astrocyte survival. (See Reagents and Solutions).
-
27.
Replace 1/3 of the medium with warmed complete medium on the 6th day after initial plating and then every 2–3 days until co-culture with astrocytes.
SUPPORT PROTOCOL 1
NEURONAL COVERSLIP PREPARATION
For confocal imaging, the neurons are plated on 12 mm, round, glass coverslips, in triplicate. To enable the co-culture of astrocytes and neurons, 4 paraffin wax spacers are manually placed on each coverslip, so that the neurons can be inverted over the treated or un-treated astrocyte monolayers without direct contact. Because wax spacers are placed by hand, it is necessary to sterilize the coverslips with additional steps. Coverslips need to be incubated overnight in HCl to sterilize and slightly etch the glass, then washed the following day and coated with poly-l-ornithine substrate. Coverslips with spacers can be prepared ahead of time and stored in a covered box until use, but the coating of the coverslips with substrate should occur the day prior to the neuronal preparation, and requires an overnight incubation. The application of wax spacers of consistent size takes a bit of practice. See Critical Parameters – Wax Spacer Application for important considerations and tips for working with the molten wax. Alternative methods of preparing glass coverslips with wax spacers for co-culturing are also available (Boraso and Viviani, 2011).
Materials
Round, microscope glass coverslips: 12 mm, No. 1.
Paraffin wax, melted
1 mL disposable syringes
Disposable needle (20 gauge, 1 inch)
Hydrochloric Acid (1 M); 1 mL per coverslip
Sterile water
Poly-l-ornithine hydrobromide (PLO, 10 mg/mL stock) (See Reagents and Solutions)
22 µm sterile filter & syringe
Prepare and Coat Glass Coverslips
-
Melt paraffin wax in a 100 mL glass beaker by placing the beaker on a hot plate set on high heat. Cover the beaker with aluminum foil to retain the heat, and when melted, stir constantly.
Take care when working as the molten wax is quite hot. (See Critical Parameters – Wax Spacer Application). The same beaker of wax can be set aside and reheated for reuse for subsequent coverslip preparation, with new wax added when needed.
-
Use a disposable 1 mL syringe and 20-gauge needle to apply 4 small wax dots, each of similar size (~ 1 – 1.5 mm), to each coverslip. Place the dots relatively equidistant apart, along the perimeter of the coverslip.
To minimize wax cooling and potential solidification, pre-warm the syringe and needle with hot wax by pulling up and expelling a small volume of wax a few times. Take up no more than 100–200 µL of wax when applying the wax spacers. (See Critical Parameters for additional tips and warnings for proper application of wax spacers)
Using surgical tweezers check that all the wax dots are sufficiently adhered by lightly touching each one. Add additional dots if necessary.
Place 3 coverslips per treatment group with wax spacers adhered in the wells of a 24-well plate.
Perform the remainder of the steps in a sterile hood
-
5.
Add 1 mL of HCL (1 M) to each well. With a sterile needle or glass pipette tip, gently tap the coverslip down to the well bottom to ensure that it is entirely submerged in HCl. Incubate overnight in sterile hood.
Coverslips with wax spacers attached have a tendency to stay more buoyant in solution. The HCl acts to both sterilize the coverslips and to slightly etch the glass, improving cell adhesion to the glass.
-
6.
The next day, wash the coverslips 5 times with 1 mL sterile water for 30 minutes each wash.
Coat Coverslips with Substrate
-
7.
After the final wash, prepare poly-l-ornithine (15 µg/mL) from stock (See Reagents and Solutions).
-
8.
Add 1 mL of poly-l-ornithine solution to each coverslip. Tap down the coverslips with a sterile needle. Incubate overnight at 37°C.
Wash and coat the coverslips the day prior to the neuronal dissection. Coverslips will be washed just prior to the start of the preparation (See Basic Protocol 1).
BASIC PROTOCOL 2
PRIMARY ASTROCYTE PREPARATION AND CULTURE
Primary cortical astrocytes, prepared from E21 Sprague-Dawley pups, are grown in 75 cm2 flasks for 1.5 to 2 weeks prior to sub-culturing for experimental use.
Materials
HBSS (CM-Free)
Astrocyte Maintenance Medium (DMEM-FBS) (See Reagents and Solutions)
Poly-d-lysine (PDL) Working Solution (40 µg/mL) (See Reagents and Solutions)
Trypsin (0.25%) in dH2O. (See Reagents and Solutions)
Phosphate buffered saline (PBS)
Sterile surgical tools
Sterile filter paper
100 µm pore nylon mesh filter
Flask Preparation
Pre-coat 75 cm2 sterile flasks by adding 10 mL of PDL (40 µg/mL) to each flask for 10 minutes at RT.
Wash 1 X with sterile water.
-
Wash 1X with PBS and aspirate PBS. Set aside until use.
Flasks may be pre-coated prior to the preparation and stored dry in sterile conditions until use.
Astrocyte Preparation
-
4.
Working in aseptic conditions, isolate the cortices, one at a time, from E21 Sprague-Dawley rat fetuses and carefully remove the meninges either with tweezers, or by rolling the tissue on sterile filter paper. Place the tissue in a 100-mm plate containing 5 mL of HBSS (CM – free).
-
5.
Collect the cortices from the entire litter, and cut the tissue into small pieces using sterile surgical scissors.
-
6.
Transfer the tissue into 20 mL of warmed trypsin (0.25%).
-
7.
Incubate at 37°C for 10 minutes.
-
8.
Add 20 mL of warmed Astrocyte Maintenance Medium to stop the enzymatic reaction. Mix well by pipette.
-
9.
Centrifuge at 250 x g at 25°C for 10 minutes.
-
10.
Aspirate the supernatant without disturbing the tissue and re-suspend in 20 mL of DMEM-FBS.
-
11.
Vortex for 1 minute, at maximum speed.
This harsh treatment aids in reducing the number of neurons present in the culture.
-
12.
Re-centrifuge at 250 x g at 25°C for 10 minutes, and re-suspend in 20 mL of DMEM-FBS. Aspirate the supernatant and repeat this step 3 times.
-
13.
After the final resuspension, filter the solution through a 100 µm pore nylon mesh into a 50 mL conical tube to remove aggregated cells.
-
14.
Count the cells using a hemocytometer.
-
15.
In the PDL pre-coated flasks, plate 2.5 × 106 cells per flask in 10 mLs of DMEM-FBS.
-
16.
Incubate at 37°C in a humidified atmosphere of 5% CO2.
-
17.
Wash each flask 2 times with 10 mL of PBS the day after seeding and replace the medium. Replace the medium every 2–3 days.
At each medium change, rock the flasks by hand at least 20 times to remove non-astrocytic cells that have adhered to the astrocyte monolayer (See Critical Parameters – Astrocyte Culture Purity).
BASIC PROTOCOL 3
ASTROCYTE PASSAGE FOR EXPERIMENTAL TREATMENT & CO-CULTURE
This protocol describes the passage of astrocytes from the established cultures that were prepared and grown for 1.5 to 2 weeks (Basic Protocol 2). Astrocytes are plated in 24-well plates, at a density of 250,000/mL or well. Astrocytes are grown until confluency, serum-deprived for 24 h to synchronize astrocytes in the G0/G1 phase of the cell cycle and to eliminate the effects triggered by growth factors present in the serum, and then treated for 24 hours with the pharmacological compound or toxicant of interest. After 24 hours, the treatment is removed and the neurons, grown separately on coverslips (Basic Protocol 1), are inverted over the astrocytes and co-cultured for an additional 24 hours. The average yield of astrocytes obtained from 2-week old established cultures typically ranges from 8 to 10 million cells per flask. Treatment and co-culture times may be modified depending on the experimental question; however, we have not characterized the effects of co-culturing astrocytes and neurons for more than 48 hours.
Materials
Poly-d-lysine (PDL) Working Solution (40 µg/mL) (See Reagents and Solutions)
Phosphate buffered saline (PBS)
Established primary cortical astrocyte culture grown for 1.5 to 2 weeks (See Basic Protocol 2).
Trypsin (0.25%) (See Reagents and Solutions)
Astrocyte Maintenance Medium (DMEM-FBS) (See Reagents and Solutions)
Astrocyte Serum Deprivation Medium (DMEM-BSA) (See Reagents and Solutions)
HBSS + CM
24-well sterile plates
Treatment Solutions
Passage of Experimental Astrocytes
Warm trypsin and astrocyte maintenance medium (DMEM-FBS) in a water bath to 37° prior to use.
In 24-well plates, coat one well for each neuronal coverslip with 1 mL PDL (40 µg/mL) for 10 minutes.
Wash once with sterile water and replace with 1 mL sterile PBS. Set aside until astrocyte plating.
Determine the number of astrocyte flasks to be used for experiment. Two-week astrocyte cultures yield approximately 8–10 million astrocytes per flask; astrocytes are plated at a density of 250,000 per well.
Gently rock each flask 25 times by hand to dislodge any cells on the surface of the astrocyte layer. Aspirate off the medium and wash 2X with 10 mL PBS.
Add 2 mLs of warmed trypsin to each flask and incubate at 37°C for 10 minutes.
-
Remove the flasks from the incubator. Strike the sides of each flask 10–15 times by hand to release the cells from the flask surface.
The clear trypsin will turn cloudy as cells dislodge from the flask surface. At times, large sheets of astrocytes may be visible in solution. Cells will separate after pipetting.
Add 8 mL of warmed DMEM + FBS to each flask to stop the enzymatic reaction.
Suspend the cells by vigorously pipetting and rinse the surface of each flask 5–6 times as you suspend to ensure complete cell collection.
Transfer the contents of each flask to a single 15 mL tube, for one flask, or a 50 mL tube if using more than one flask.
Centrifuge at 250 x g for 10 minutes at RT.
Aspirate off the supernatant and thoroughly re-suspend the pellet in 10 mL of medium. If more than 2 flasks are used re-suspend in a higher volume of medium. Count the cells with a hemocytometer.
Remove the PBS from the pre-coated 24-well plate and plate the cells at a density of 250,000 cells/mL (or well). Incubate overnight at 37°C.
Replace the medium on day 2 and 4 after plating, washing with PBS 1X at the first medium change.
Astrocyte Serum Deprivation and Treatment
-
15.
On the 5th day, or 48 hours prior to co-culture (See Table 1), wash the astrocytes 2X with PBS and replace the medium with serum-free medium (DMEM-BSA). Incubate at 37° for 24 hours.
-
16.
After 24 hours of serum deprivation, treat the cells with the compound of interest, preparing the solutions in warmed DMEM-BSA. Incubate for 24 hours.
-
17.
At treatment end, remove the astrocyte treatment by washing 2X with PBS. Replace with warmed DMEM-BSA.
-
18.
Allow the astrocytes to condition the medium prior to the addition of the neurons by incubating the astrocytes for 3 hours at 37°C.
This conditioning step will allow for the release of astrocyte secreted factors into the medium to aid in neuronal survival when neurons are co-cultured in this shared environment.
Astrocyte-Neuronal Co-culture
Materials
HBSS + CM
Sterile tweezers and needle
-
19.
At the end of 3 hours, gently wash the neurons 1X with warmed HBSS.
-
20.
Using sterile tweezers and a needle, carefully remove the coverslip, by slightly lifting one edge of the coverslip with the needle and grasping the edge with the tweezers. Avoid touching the center of the coverslip, so as not to damage the growing neurons.
-
21.
Invert the coverslip over the monolayer of astrocytes so that the neurons are facing the astrocytes and the wax spacers create a separation between the two cell types.
-
22.
Incubate together for 24 hours at 37°C.
Fixation of neurons
Materials
Warmed HBSS + CM
Paraformaldehyde (4%) in HBSS+CM (See Support Protocol 2)
PBS
Tweezers and Needle
-
After 24 hours of co-culture with astrocytes, re-invert the coverslips, neurons face up into a 24-well plate containing 1 mL of warmed HBSS +CM per well.
Again, grasp the coverslip on the edge as to not disturb the neurons.
Gently wash 1X with warmed HBSS+CM, releasing the solution slowly and on the well wall as to not disrupt the neurons with the force of the solution.
-
Fix the neurons by adding 1 mL of warmed paraformaldehyde (4%) (pH 7.4) (See Support Protocol 2).
NOTE: PFA is toxic and work should be performed in a fume hood. Dispose of PFA as hazardous waste.
Cover and incubate for 20 minutes at 37.5°C.
Remove paraformaldehyde in a fume hood and dispose as hazardous waste.
Wash 2X with PBS for 5 minutes each on a rocker at RT.
-
Remove paraffin wax spacers with tweezers.
Slight force may be necessary to dislodge the spacers. Grasp the wax spacer and be sure to push out towards the coverslip edge to avoid damaging the neurons. Spacers can also be removed just prior to immunolabeling, if followed by 1 PBS wash for 5 minutes.
Wash 1X in PBS for 5 minutes at RT on rocker to remove any debris remaining after wax removal.
-
Store plates containing coverslips wrapped in Parafilm at 4°C for up to a week or begin the 1st day of immunostaining process.
If timing for staining is not appropriate, fixed cells may be wrapped in Parafilm and stored in PBS for up to 1 week at 4°C, though this is not recommended.
SUPPORT PROTOCOL 2
PARAFORMALDEHYDE (PFA) (4%) PREPARATION
Neurons are fixed with 4% paraformaldehyde. This can either be purchased or prepared following the instructions below. Caution: Paraformaldehyde is toxic, so care must be used in its use and preparation. Work must be carried out in a fume hood and PFA disposed of as hazardous waste. Store prepared PFA at 4°C for up to 2 weeks.
Materials
Paraformaldehyde powder
HBSS +CM
Sodium Hydroxide (NaOH) (1 M)
Fume Hood
Heat pad/stirrer
Thermometer
-
In a fume hood, add 4% (w/v) paraformaldehyde powder to HBSS (+CM), stirring constantly.
The solution will appear light pink and opaque.
-
Heat to 70°C and when reached, add 1 drop of NaOH (1 M).
After the addition of NaOH the solution should become clear.
Continue heating until the solution reaches 80°C.
Turn off the heat and continue stirring for 5 minutes.
-
Let PFA cool to room temperature and transfer to a covered glass bottle to avoid the release of toxic fumes.
Remember to label the bottle as toxic.
Keeping covered with aluminum foil, adjust the pH to 7.4.
Store for 2 weeks at 4°C.
Prior to use, warm the solution to 37°C in a water bath and re-adjust the pH to 7.4.
BASIC PROTOCOL 4
NEURONAL IMMUNOSTAINING FOR SYNAPTIC PROTEINS
In this section, neurons fixed in paraformaldehyde are prepared for imaging. For consistency and successful antibody labeling, it is important that the procedure is performed over two consecutive days, so plan accordingly (See Timing Issues and Table 2). Incubation with the primary antibody should occur over the first night, followed by fluorescent labeling with the secondary antibody and mounting on slides the following day. The immunolabeling steps include removal of the wax spacers (if not performed already), blocking to reduce non-specific antibody binding, and permeabilization with a detergent so that the primary and secondary antibodies can enter the cells and find their targets. Pre- and postsynaptic formations will be immunolabeled with antibodies to synaptophysin, a pre-synaptic vesicular protein, and PSD-95, an excitatory receptor postsynaptic scaffolding protein.
Materials
Phosphate buffered saline (PBS)
Bovine serum albumin (BSA)
Iso-octylphenoxy-polyethoxyethanol (Triton-X) (10%) in PBS
Rabbit monoclonal antibody to synaptophysin
Mouse monoclonal antibody to PSD-95
Donkey anti-rabbit Alexa 488 fluorescent secondary antibody
Donkey anti-mouse Alexa 555 fluorescent secondary antibody
Trihydrochloride, trihydrate (Hoechst 33342) (10 mg/mL)
Glass microscope slide: 75 × 25 x 1 mm
Glass microscope cover glass: 22 × 40 mm, No 1 ½
Vectashield mounting medium for fluorescence
Nail polish to seal the coverslips
Day 1 – Primary Antibody Incubation
If coverslips have been stored at 4°C, wash 1X for 5 minutes in PBS to remove settled particles.
To each well of coverslips, add 1 mL of PBS containing 5% BSA and 0.1% Triton-X, to block non-specific binding and permeabilize the cell membrane.
Place plates on an orbital shaker, at a slow speed, for ½ hour at RT.
Wash 1 time with PBS supplemented with Triton-X (0.1%) for 5 minutes at RT on an orbital shaker.
-
Prepare 100 µL per coverslip of the primary antibody dilution buffer by adding the antibody to synaptophysin (1:200) and antibody to PSD-95 (1:250) to PBS supplemented with 1% BSA.
Primary antibodies are quite expensive. To conserve the amount used, the neuronal coverslips will be inverted over a pool of antibody buffer solution. When inverted over the solution, the neurons ride on the pool of the solution and with gentle rocking are always in contact with the antibody solution.
-
Dispense 95 µL of the solution per coverslip into the wells of a clean 24-well plate.
Be sure to mix the solution between aliquots to ensure a consistent concentration of antibodies between wells. Alternatively, if cost is not an issue, 200 µL of primary antibody per coverslip can be prepared and applied directly to the well containing the coverslip. This volume will entirely cover the coverslip, removing the need for manipulation of the coverslip. If this option is chosen, skip step 7.
-
As during the co-culturing step, carefully lift each coverslip with tweezers and a needle. In order to ensure a consistent dilution of the antibody solution, carefully blot the edge of the coverslip on a Kimwipe to remove the excess solution. Invert the coverslip over the antibody solution previously placed in the 24-well plate and place gently, so that the neurons are face down on top of the buffer. Do not tap down, but let them float on the solution.
This requires careful handling. With care, the neurons will not be harmed during this process.
Wrap the plate in Parafilm and place on an orbital shaker set at low speed at 4°C for at least 18 hours.
Day 2 - Secondary Antibody Labeling
-
9.
Prepare Day 2 Wash Buffer (See Reagents and Solutions). Make enough for six washes at 1 mL/coverslip.
-
10.
Re-invert coverslips, neurons face-up, into a clean 24 well plate containing 1 mL of Day 2 Wash Buffer per coverslip.
If 200 µL of antibody dilution buffer was applied directly to the coverslips, do not re-invert, but aspirate off the solution.
-
11.Wash with the Day 2 Wash Buffer 3X for 5 minutes each on a rocker at RT.Perform the remaining steps after turning off the lights as the secondary antibodies contain light sensitive fluorophores. Clearly, it is difficult to perform the following steps completely in the dark, as the glass coverslips are fragile and require manipulation at each step. However, darken the room as much as possible and avoid direct light on the slides to reduce the loss of fluorescent signal.
-
12.
Centrifuge the stock of secondary antibody at the highest speed for 3 minutes to pellet down any large fluorescent aggregates.
-
13.
Avoiding the bottom of the centrifuged stock tube, prepare 100 µL of the secondary antibody buffer per coverslip by adding each secondary antibody (1:500 each) to PBS.
-
14.
Dispense 95 µL per coverslip of secondary antibody solution per well into a clean 24-well plate.
-
15.
Invert the coverslips as before, blotting the edge on a Kimwipe. Place the coverslip neuron-side down.
Alternatively, as the secondary is less costly than primary antibodies, 200 µL per coverslip of the secondary antibody can be prepared and applied directly to the face of the neurons, avoiding the inversion of the coverslips onto 95 µL of secondary antibody solution, while working in the dark. If this option is chosen, skip steps 14 and 15.
-
16.
Wrap the plate in aluminum foil to protect from light and incubate on an orbital shaker at RT for 1 hour.
-
17.
After 1 hour, carefully remove the coverslips with a needle and tweezers, and re-invert them, (neuron side up) into 1 mL of Day 2 wash buffer, or if directly applied to the coverslips, aspirate off the secondary antibody solution and replace with Day 2 wash buffer.
-
18.
Wash 3X in Day 2 wash buffer, for 5 minutes each on a rocker at RT. Protect from light.
Nuclei Counterstain
-
19.
Prepare 20 mL of Hoechst 333421 solution (1 µg/mL in ddH20) to counterstain the nuclei.
-
20.
Add 200 µL of Hoechst solution to each well.
-
21.
Cover with foil and rock for 10 minutes at RT.
-
22.
Wash 2X with PBS for 5 minutes each, on the rocker.
Coverslip Mounting
Work in the dark. Work with only one treatment group at a time, keeping the others covered and protected from light.
-
23.
Carefully remove the coverslips and blot excess PBS on a Kimwipe as before.
-
24.
Place the coverslips of one treatment group, neuron side up, on the working area of the slide.
Stagger the three coverslips of one treatment group so that all will be completely covered by the slide cover glass.
-
25.
Place a small drop of Vectashield on each neuron containing coverslip.
-
26.
Carefully place the rectangular, top cover glass over the three neuronal coverslips.
The Vectashield will spread. To reduce air bubble formation, place one edge of the cover glass down and then lay it over the remaining coverslips, as opposed to placing it right on top. Align the top cover glass at placement, so that subsequent adjustment is not necessary, as the neurons may be damaged with movement.
-
27.
Use nail polish to seal the perimeter of the cover glass.
Apply the polish carefully. Brushing that is too harsh may cause the top cover glass to move, and may potentially harm the neurons. To avoid this, apply four drops of polish at the corners of the cover glass, protect from light and let dry while mounting the next slide. This polish will set and stabilize the cover glass, minimizing movement of the cover glass during completion of the sealing process. Once set, completely seal the perimeter of the cover glass.
-
28.
Store the slides in the dark at 4°C. Image as soon as possible after mounting to minimize loss of the fluorescent signal.
BASIC PROTOCOL 5
SYNAPTIC STRUCTURE IMAGING & ANALYSIS
To determine the effects of astrocyte treatments on synaptic structure formation, confocal microscopy is used to image immunolabeled neurons, allowing for the collection of pre- and post-synaptic signals throughout the depth of the neuronal extension, or in three-dimensions. The images are acquired using a 60X oil immersion objective and the confocal should be equipped with excitation lasers that allow the visualization of the nuclei (405 nm for blue emission) and each synaptic probe (488 nm for green emission and 561 or 568 nm for red emission). Once collected, the images are deconvolved to refocus light that inherently scatters during imaging, and intensity threshold is determined with ImageJ and used in the three-dimensional rendering of the synaptic structures; the relative number of synapses is quantified using Huygens Professional’s Object Analysis software. While the protocol described here is specific to these software programs, there are other imaging programs that will accomplish the same tasks. The general concepts can be applied and adapted for use depending on software available in your laboratory.
After neurons have been immunolabeled and the slides mounted, image all the slides of an experiment as close in time as possible to ensure that loss of fluorescent signal over time does not influence analysis and a fair comparison between groups can be made. Ideally, an experiment should be completed in one sitting. Plan accordingly, as imaging is time-consuming; to collect images from 15 neurons of one treatment group takes approximately 3–4 hours. If imaging an entire experiment in one sitting is not practical, it is recommended to image one coverslip of each treatment group, alternating between them until imaging of the experiment is complete. Pawley’s Handbook of Biological Confocal Microscopy (Pawley, 2006) is a valuable resource for information about numerous aspects of fluorescent confocal microscopy and image analysis.
Materials
Scanning Confocal Microscope
60X oil immersion objective
ImageJ Software (http://rsbweb.nih.gov/ij/index.html)
Huygens Professional (Scientific Volume Imaging, Hilversum, The Netherlands, http://www.svi.nl).
Object Analyzer from Huygens Professional, Scientific Volume Imaging
Confocal Imaging
-
Set the imaging parameters of a confocal microscope to obtain images with a resolution of 1024 × 1024 pixels with a 2X zoom. These settings will result in a final resolution of 0.103 µm/pixel when a 60X oil objective is used.
These parameters will allow you to capture an image that contains one neuron per field, and at a resolution that is appropriate for thresholding and analysis.
To minimize bleed-through of overlapping spectra, set the imaging acquisition software to acquire the images from each channel sequentially, using a 488 nm laser to excite the labeled synaptophysin protein (green) and a 561 or 568 nm laser to excite the labeled PSD-95 signal (red).
Set the z-series step size to 0.30 µm.
With the lasers set to the lowest possible intensity and working rapidly to minimize excessive exposure of the slides to light, identify the slide or treatment group with the brightest intensity. (See Critical Parameters – Neuronal Imaging).
-
To set the microscope parameters for the experiment, use the identified slide, and visualize the neurons in grayscale mode. Following the instructions particular to the microscope being used, determine the appropriate gain and threshold of each channel using one random neuron from each coverslip from the slide with the greatest intensity. Hold these settings constant while imaging subsequent treatment groups of the same experiment so comparisons between groups can be made.
The setting of the gain and threshold is an important part of the imaging process. If incorrectly set, important information about differences in signal will be lost. (See Critical Parameters – Neuronal Imaging; Pawley, 2006; Glynn and McAllister, 2006).
-
To reduce selection bias, identify a neuron to be imaged by first visualizing only the nuclei, or blue channel, using epifluorescence. Locate a healthy cell for imaging that is at least two cell bodies away from another and will result in an image of one cell per field.
For consistency, attempt to select neurons within an experiment that are in similar neighborhoods on the coverslip, that is, not excessively clustered, nor seemingly far from other neurons, as these are not typical environments. You will gain familiarity as you image multiple coverslips.
-
Once a neuron is identified, set the z-series distance by selecting the start and end location for the z-plane. Start the series just below the point where the extensions are visible on the coverslip. Move up through the extension to one step above where the last signal in the extension disappears and set the “end of series” location.
These parameters will capture the necessary extension information through the z-plane. Nuclei signal will be visible throughout the series, although the extension will no longer be visible at the end setting. Using the 0.3 µm step size and these z-series parameters, a 12–18 optical sections will be collected for each channel of one neuronal image.
Sequentially acquire the z-series image from both the 488 green channel and the 568 red channel and save.
-
Without using the green and red channels, turn on the 405 laser (blue emission) and acquire one plane, without z-series depth, of the labeled nuclei, and save.
Images of the nuclei will not be used in analysis, but only when presenting images. Acquiring one plane, as opposed to entire z-series, saves a significant amount of time in this time consuming portion of the experiment.
-
Obtain 5 images per coverslip, for a total of 15 images per treatment group.
In addition to ensuring that cells are of similar neighborhoods, attempt to image neurons from 5 different areas of the coverslip, the center, and one in each of the four quadrants.
These are large image files. Be sure to have sufficient storage and back-up capacity to retain the images.
Deconvolution
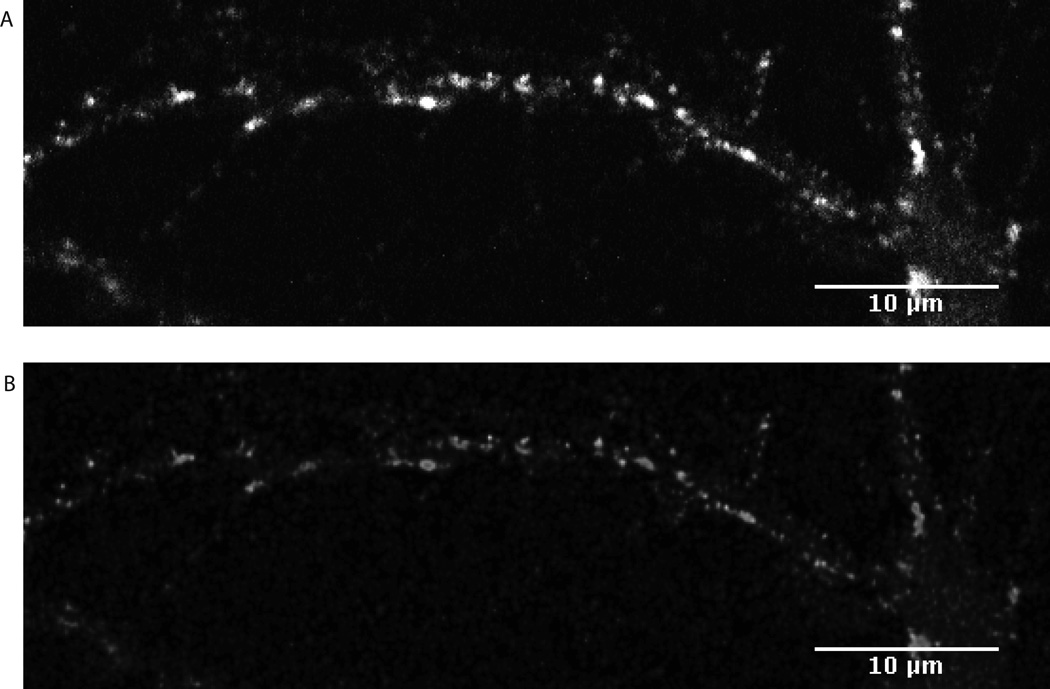
During imaging, as light waves move through various media, it inherently scatters, resulting in a slight blurring of the actual signal. The process of deconvolution improves the resolution of the signal by reducing the noise generated by scattering and refocuses the light through an iterative algorithm that considers the acquired signal, parameters of particular microscope being used and the refractive indices of materials that light is moving through (Cannell, et al., 2006). Deconvolved images yield a sharper image and so a more accurate assessment of signal, which improves the quantitative results (Figure 2). While Huygens Professional software is used here to deconvolve the confocal images, alternative deconvolution programs could be used. Most programs will require information about the imaging parameters, including the type of microscopy performed, the size of the sampling interval, the numerical aperture of the objective, and the refractive indices of both the mounting and immersion medium (Cannell, 2006). For more information about confocal microscopy, immunocytochemistry, deconvolution or image processing, Pawley’s Handbook of Confocal Microscopy (J.B. Pawley, 2006) is an excellent resource.
Figure 2. Effect of Deconvolution.
Hippocampal neurons (13 DIC) were directly treated with high-density lipoprotein (10 µg/mL cholesterol) for 24 hours, immunocytochemically labeled for the synaptic protein synaptophysin, and imaged using confocal microscopy. Prior to deconvolution (A) the signal perimeter appears cloudy and somewhat blurred. After deconvolution (B) the signal is refined and refocused without a loss of relative intensity differences. Shown is the greyscale representation of a portion of the maximum intensity z-projected image taken from the green channel and enlarged to 150%.
Using deconvolution software, enter the requested parameters of the microscope.
Choose a “theoretical” algorithm and set the number of iterations to 40.
-
Load the images to be deconvolved and set the batch processor to begin.
Because it is iterative, it takes some time to complete the process on a complete set of images from one experiment. In Huygens Professional software, the entire batch of images can be loaded for analysis and run overnight to minimize wait time.
Thresholding
Synapses have three-dimensional structure. As opposed to other methods that use immunocytochemistry and confocal imaging to determine the overlap of synaptic puncta, three-dimensional object analysis uses all of the confocal data to render the surface of an object. To do this, Huygens Professional relies on an intensity threshold to draw borders around those voxels that are at this threshold or higher to generate an object, making its determination an important part of the analytical process (See Critical Parameters – Thresholding). To generate that threshold, the mean intensity of the puncta of each channel is determined by sampling multiple puncta from the images of those neurons used to set the confocal imaging parameters. This method uses the NIH open-source software ImageJ (http://rsbweb.nih.gov/ij/index.html). When a representative sampling of puncta is selected for analysis this method results in three-dimensional renderings that are consistently reflective of the deconvolved images. The method described below includes specific commands from ImageJ, version 1.47v for Windows. These commands may change with later versions of the program; however, the principles should not, and should be easily adaptable if changes have occurred. Be sure to download the plugin that allows the importation of images saved in microscope specific file formats unique to the confocal used for imaging.
Open ImageJ
-
From the treatment group used to determine the confocal imaging settings, import one deconvolved stack of images as grayscale (not RGB), separating the channels when prompted, so a distinct threshold for each channel can be obtained.
Each channel will open in a separate window and will include the stack of images acquired for that channel. Using the brightest group of coverslips to identify the intensity threshold will enable comparisons to those groups that are less bright. See Critical Parameters for a discussion about outcomes if the threshold is incorrectly determined).
On each set of channel images, scroll to the middle optical section.
Enlarge the images to 200% for more accurate puncta selection.
Double click on the oval selection tool to open the settings dialog box. Select the brush option and set the brush size to 3 pixels.
-
Open the Region of Interest (ROI) Manager window.
(Analyze > Tools > ROI Manager)
-
Set the measurement parameters to acquire the image name, selection size and mean intensity.
(Analyze > Set Measurements > Select “Area”, “Mean gray value”, and “Display label.”)
-
Select the round selection tool to begin using it. Starting approximately 5 µm from the cell body, manually select 10 puncta each on three extensions for a total of 30 puncta per imaged field, moving over the entire length of the extension. Hold down the “ctrl” key to select multiple puncta.
Select puncta that are well clustered and distinct. Be sure to select a representative sample of intensities, not simply those that are the most intense (See Critical Parameters – Thresholding).
After 30 puncta have been selected, add them to the Region of Interest (ROI) manager. (In the ROI window, select “Add”). Save the selection (More > Save) to retain a record of analysis, renaming it to reflect the image name and channel.
-
Select the renamed file in the ROI Manager. Split the ROI containing the 30 selected puncta into 30 separate ROIs (More > Split). Delete the original ROI (first named file on top of the list).
Splitting the ROI will allow us to obtain the mean intensity of each individual puncta, and so retaining any variability of puncta intensities between neurons.
-
Choose “Measure.”
A results window containing the mean intensities of each selected puncta will be generated.
Save the results file, naming it to retain the image and channel information of the particular neuron.
Repeat the process on the second channel. Continue until all the neurons of the treatment group used to set the confocal parameters have been completed and the results saved.
Combine the files obtained for each neuron and determine the threshold of each channel by averaging the puncta intensity values. Use this value during three-dimensional analysis to set the threshold.
Three-Dimensional Analysis
Finally, to assess potential changes to synaptic structure formation in hippocampal neurons after treatment, this portion of the protocol describes the process of three-dimensional object analysis using Huygens Professional Object Analyzer software. Using the deconvolved images and the calculate threshold, this software will integrate all of the confocal information to render the surface of synaptic puncta in three dimensions. Counts of the number of pre- and postsynaptic puncta, as well as counts of those puncta that are overlapping will automatically be obtained. While this section generally applies to Huygens Professional the concepts can be easily applied to alternative programs capable of analyzing three-dimensional structure.
Open a deconvolved confocal file in Huygens Professional and select the Object Analyzer.
Set the threshold for each channel using the values obtained above.
Confirm by eye that the rendered image generated after thresholding is representative of the raw image (See Critical Parameters – Thresholding, Figure 3).
Record the number of starting puncta in each channel. This number will represent the number of pre- and postsynaptic puncta.
Select the filter option again and choose the command to remove the non-overlapping objects. Record the number of objects remaining in each channel.
Of the overlapping puncta that remain, use the filter that presents those objects of the first channel that “pair” with puncta from the second channel. Record this value, as it reflects the number of synaptophysin and PSD-95 structures that are in apposition to one another, or the number of structural synapses.
Holding the threshold value constant for all treatment groups, repeat the process until all neurons of the experiment have been analyzed.
Determine the average number of starting puncta for each channel in each treatment group, as well as the average number of overlapping pre and post-synaptic pairs.
Graph and compare the ratios of each treatment group relative to control. Perform statistical analysis.
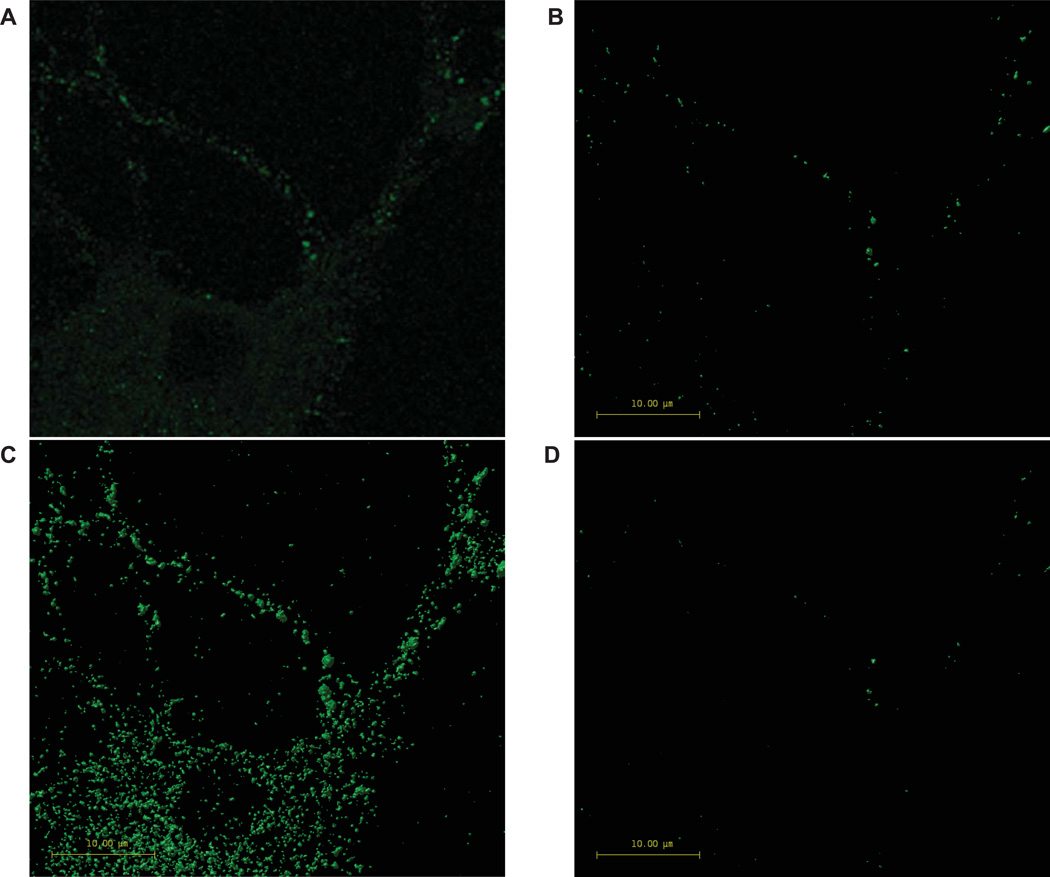
Figure 3. Comparison of deconvolved image vs. surface rendering after thresholding.
The method used to generate a threshold for each channel results in a three-dimensional surface rendering of puncta (B) that is representative of punctate structures evident in the deconvolved image (A). Shown is one slice of a deconvolved confocal z-series channel (A) of hippocampal neurons grown in culture for 13 days and treated for 24 hours with high density lipoproteins (10 µg/mL cholesterol), and immunocytochemically labeled for synaptophysin. A total of 450 puncta per channel were manually selected from 15 images and the threshold value was determined by averaging the mean intensities. Selecting a representative sample of puncta results in a threshold and a rendering (B) that accurately reflects the deconvolved image (A). Setting the threshold too high (D), or too low (C), results in an inaccurate three-dimensional rendering.
REAGENTS AND SOLUTIONS
Poly-L-ornithine hydrobromide stock solution
Dissolve poly-L-ornithine hydrobromide in sterile water at a final concentration of 10 mg/mL. To avoid excessive freeze-thaw cycles, and loss of substrate, store 100–200 µL aliquots at −20°C until use. Prepare substrate solution (15 µg/mL) fresh at time of use and sterile filter.
Neuronal Preparation and Culture
Deoxyribonuclease-1 (DNase)
Dissolve deoxyribosnuclease-1 from bovine pancreas (4 mg/mL) in HBSS – CM Free and sterile filter. Prepare a 2 mL aliquot for repeated use and store at −20°C. Excessive thawing between uses may inactivate the enzyme. This would be noticeable if neuronal tissue does not effectively dissociate during trituration. Make a fresh aliquot if effectiveness of DNase is decreased.
Papain Solution for Neuronal Enzymatic Digestion
Make fresh just prior to the start of the neuronal dissection. Dissolve 2 mg/mL of papain from papaya latex in 4 mL of HBSS – CM Free. Add DNase (1:500) and 5 mM magnesium chloride. Gently invert to mix and place in water bath (37°C) to solubilize the papain. Sterile- filter immediately prior to use.
Complete Neuronal Medium
Neurobasal-A Medium (Gibco)
30 mM D – (+) glucose solution
1% B-27 Supplement
3 mM GlutaMax
0.5% fungizone
100 µg/mL gentamicin
Keep sterile and store up to 3 weeks at 4°C. To minimize oxidation of the medium and changes to the pH, warm volumes of medium needed for each task prior to use.
ARAC
ARAC is an anti-mitotic agent. Follow appropriate handling and disposal of this toxic material.
Prepare a 1 mM stock of cytosine β-d-arabinofuranoside (ARAC) in HBSS + HEPES. Prepare a working stock of 100 uM ARAC in HBSS + HEPES and sterile filter. Store both stocks at −20°C.
Trypsin Stock
Prepare trypsin (0.25%) in dH2O. Sterile filter and store at −20°C in 20 mL aliquots. Successive repeated freeze-thaw cycles may inactivate trypsin.
Astrocyte Maintenance Medium
Dulbecco’s Modified Eagle’s Medium (DMEM) with low glucose
10% fetal bovine serum
100 U/mL penicillin
100 µg/mL streptomycin
Keep sterile and store at 4°C for up to 1 month.
Astrocyte Serum Deprivation Medium
DMEM with low glucose
0.1% bovine serum albumin
100 U/mL penicillin
100 µg/mL streptomycin
Keep sterile and store at 4°C for up to 1 month.
Paraformaldehyde (4%) (See Support Protocol 2)
COMMENTARY
Background Information
The ability to assess disruption of synapse formation and function after exposure to toxicants may be relevant, as quantifying changes in structural synapse formation after treatment with a toxicant may characterize the site of toxicity and identify the underlying mechanisms. Numerous methods have been used to co-culture neurons and astrocytes and to assess changes in synaptic structures (Goslin et al., 1998; Viviani, 2003; Kaech and Baker, 2006; Boraso and Viviani, 2011; Roqué et al, 2011). The protocols described in this UNIT incorporate many of these previously used techniques with modifications and optimization to address the question at hand.
Synapse formation as an endpoint requires long-term neuronal cultures, in which interneuronal networks are well developed. The use of high concentrations of poly-d-lysine (PDL) as a growth substrate is quite common in protocols of neuronal cultures. However, while neurons adhere and grow extensions quickly with PDL as a substrate, we have had a hard time maintaining the cultures past 7 days. At that time point, while neuronal networks with extensive interconnecting extensions are visible, the postsynaptic labeling of PSD-95 is diffused and not clustered in post-synaptic puncta, suggesting a time in development not appropriate for investigating synaptic structure by the methodology used here. We have instead had success using a lower concentration of poly-l-ornithine as a growth substrate (Viviani, 2003).
In most of the astrocyte-neuron co-culture models neurons are co-cultured with astrocytes immediately after isolation as astrocytes promote both neuronal survival and development. Here, however, because the interest is in investigating the effect of astrocyte exposure to toxicant on synapse formation, highly enriched neuronal cultures are maintained in a medium, which sustains neuronal survival and neurite extension, for 13 days before their co-culture with astrocytes. The neuronal enrichment is achieved by the one-time addition of an anti-mitotic (ARAC) to inhibit astrocytic growth in the neuronal cultures and also by the supplementation of the medium with the neuronal specific supplement mixture, B-27 (Gibco/Invitrogen), which also restrict astrocyte proliferation throughout the first 13 days in culture. These culture conditions result in 94% pure neuronal cultures after 13 days in culture. An additional method by which we prevent the proliferation of astrocytes in neuronal cultures is by eliminating the use of FBS from the initial steps of neuron isolation (which many protocols suggest). Alternatively, because astrocytes proliferate post-natally in rodents (Dobbings and Sands, 1979; Bernal, 2007; de Graff-Peters and Hadders-Algra, 2006; Eroglu and Barres, 2010) many protocols use E18 fetuses for culturing primary hippocampal neurons. At this time in development, the ratio of astrocytes to neurons is quite low and so neuronal purity is enhanced.
The astrocyte culture is also an important aspect of this system. It should be noted, that astrocytes are heterogeneous and have distinct properties depending on the brain region or environment where they reside (Oberheim et al., 2012; Matyash and Kettenmann, 2010; Zhang and Barres, 2010). Because we are interested in the astrocytic effect on hippocampal neurons, it would be ideal to culture astrocytes obtained from the hippocampus. We have previously shown that hippocampal and cortical neurons behave very similarly (Guizzetti et al, 2008; Zhang et al., 2014).
Hippocampal and cortical astrocytes are more commonly prepared from PD 1–3 animals. The use of E21 animals, as presented in the protocols here, is a practical, mid-point solution, since one litter can be used for both the neuronal and astrocyte cultures, and if performed carefully, the hippocampi and the cortices can be isolated from the same pup. This then allows for two distinct cultures, and a reduction in the number of experimental animals used. Furthermore, the astrocytes yield is not a limiting factor on embryonic day 21. It should be noted that there are questions about whether the long-term growth of astrocytes using traditional FBS-containing medium protocols result in cells that are reflective of the actual starting in vivo milieu (Foo et al, 2011). The experimental plan ultimately chosen should consider these issues, the particular experimental question, and your laboratory resources. Regardless of the method ultimately chosen, transparency is always important when reporting methodology.
Additionally, because these protocols investigate how astrocytes modulate synapse formation and how toxicant exposure interferes, it is important to maintain the purity of the astrocyte cultures. To accomplish this in this system, primary astrocytes are initially plated at a low density, which helps to minimize neuronal survival; flasks are rocked at each medium change to dislodges cells, such as microglia or oligodendrocytes that have adhered to the surface of the astrocyte monolayer; and cultures are grown for 1.5 – 2 weeks before experimental use, which helps to retain a consistent level of astrocyte maturity. Cultures should not be maintained post-3 weeks, as fibroblasts tend to develop in the culture. Alternatively, for highly pure astrocyte cultures, immunopanning methods have been developed to isolate astrocytes prior to culturing (Foo et al. 2011; Foo, 2013; Barres).
In this protocol, antibodies are used to label the synaptophysin and PSD-95 proteins, both of which are unique to their pre and post-synaptic locations and have been used extensively in immunocytochemistry to label each specific synaptic puncta (Rao and Craig, 1997; Okabe et al, 1999). Once labeled, the use of laser scanning confocal microscopy enables the identification of fluorescent signal throughout the neuronal extensions and the localization of synaptic proteins into synaptic structures, with a higher concentration of protein yielding a more intense and punctate fluorescent signal. There are numerous scientific questions where optimizing the imaging parameters for each image is appropriate, but because the endpoint here is a quantitative comparison of different treatment groups, it becomes important to keep the imaging settings constant throughout an experiment (Glynn and McAllister, 2006).
It is clear that the process of synaptogenesis is highly regulated. Distinct synaptic components are trafficked to the pre- and postsynaptic sites, and assembled into pre- and postsynaptic specializations. The precise matching of those structures, or the apposition of the pre- and postsynaptic puncta, is required to constitute a synapse (Lardi-Studler and Fritschy, 2007; Craig et al., 2006). In determining appropriate methods to analyze synaptic structure using immunocytochemistry, it is important to consider the experimental question being asked. The co-localization of signals from two channels used to measure the extent of overlap of fluorescent signals provides information about the overlap of individually labeled proteins, but not about the clustering or aggregation of those signals into structures. More commonly used methods to assess synapse formation project the three-dimensional confocal data down into one or multiple, two-dimensional planes prior to analysis to determine if the signals of two channels overlap. However, it is possible that signals that are not near each other in vertical space may appear to be overlapping once projected into one plane. The methodology developed and optimized in this UNIT uses all of the confocal data to render the surface of the puncta in three-dimensions, and allows for determination of where the structures reside in a three-dimensional space and whether the pre- and postsynaptic puncta are apposed.
Huygens Professional software relies on an intensity threshold to define objects and uses that threshold to automatically render their surface. A border is drawn, connecting pixels that are determined be at the intensity of the threshold or higher with adjacent pixels of the same or higher intensity, generating objects which represents a cluster of synaptic proteins. Because the threshold has an effect on the dimension of the objects generated, the determination of a threshold that provides and accurate rendered representation of the actual neuron is an important part of the analytical process (Wouterlood et al, 2008). Setting a threshold too low will aberrantly capture adjacent pixels, resulting in objects that may be larger than what is actually represented; setting the threshold too high may exclude signal that may actually be a component of the puncta present. The thresholding method optimized here relies on the manual selection of representative puncta in each experiment. The sample size is high enough that it results in the consistent three-dimensional reconstruction of objects that are reflective of what is seen in the confocal image. Additionally, the accuracy of the threshold can be confirmed by comparing both the rendered image after thresholding with its deconvolved source (Figure 3). (See Critical Parameters).
While the methods and protocols described here allow for the quantification of synaptic structures, additional experiments that assess functionality should be performed to corroborate structural change results, or provide insights into changes in functionality using electrophysiological approaches.
Critical Parameters
Within these protocols, there are steps and techniques that are important for success of the cultures and analysis. This section provides information about important aspects of this methodology and includes tips, as well as rationale for completing some of them. Additional suggestions are included in brief in Table 3 – Troubleshooting.
Table 3.
– Troubleshooting
| Problem | Possible Cause | Solution | |
|---|---|---|---|
| Neuronal Culture | |||
| Yield too low | Technique too harsh |
|
|
| Ineffective DNase |
|
||
| Incomplete Dissociation |
|
||
| Issue with reagents |
|
||
| Neurons die early in Culture | Technique too harsh |
|
|
| Wax Spacer Application | |||
| Spacers not adhere | Wax not hot enough | ||
| Spacers lie flat | Wax too hot | ||
| Astrocyte Culture | |||
| Excessive non-astrocytic cells | |||
|
|||
| Thresholding | |||
|
Rendering results in Larger puncta generating fewer objects than observed in the deconvolved image |
Threshold is too low. |
|
|
|
Rendering results in Smaller and greater number of puncta than observed |
Threshold is too high. |
|
|
Neuronal Preparation and Culture
Primary neurons are quite sensitive, so it is important to be gentle with these young neurons both during the preparation and their time in culture to increase the yield of the cells and maintain a healthy culture. With practice, the approximate yield of the hippocampal dissection should be approximately 1 – 1.5 × 106 neurons per pup. If after completing the neuronal preparation the yield is higher, be sure that you are dissecting out only the hippocampus and not tissue from the surrounding structures. At E21, the hippocampus is visible by eye, but you may want practice locating and removing the hippocampus using a stereo microscope.
More likely, however, when first starting, the number of cells obtained may be too low. The most common reasons for this include an increase in cell death during the preparation steps and while in culture, as well as incomplete dissociation of the tissue. During the preparation, harsh trituration or direct washes may cause neuronal death, decreasing the yield and impacting the health of the neurons. Be sure to slowly and gently triturate the tissue, using the side of the tube to release the medium. To ensure that cell death is minimized after initial plating, add solutions slowly down the side of the well wall during wash steps and medium changes, and never directly on the coverslip, as the force will damage and dislodge the growing neurons. If the yield of the cells is still low after these techniques have been mastered, there may be a problem with the solutions or reagents used. Confirm that the solutions used during the preparation process contain the appropriate concentrations of reagents, and that these reagents have been kept sterile. Check the temperature of the water bath used for the incubation step, as excess heat may contribute to increased cell death at this early stage. The use of Trypan blue when counting will allow you to quantify the amount of cell death after dissociation. This can help you determine whether your technique is too harsh, or if there was an issue with a reagent used in your preparation. Cell death greater than 15% usually results in a culture that will not survive.
Additionally, incomplete dissociation can cause a reduction in cell number as tissue may be lost when the cell-containing medium is filtered. Potential reasons for this include incomplete trituration, improperly prepared papain solution, or issues with the effectiveness and use of DNase. During the trituration, allow the tissue to completely settle before filtering the medium. Use a timer if necessary. Confirm that the papain solution is properly prepared and that the papain was solubilized before sterile filtering. A loss of papain at this early step will make dissociation of the tissue more difficult. Finally, the ease of dissociation may be affected by the amount of tissue collected, the extent of the manual mincing of the tissue using the scissors, and the effectiveness of the DNase. The addition of DNase to each step of the trituration process after the first is optional, and with practice, it is easy to tell by eye whether more is needed to completely digest the tissue. If the tissue easily breaks down and the medium becomes cloudy, it may not be necessary, but if the tissue retains its size, DNase should be added. The stock of enzymatic DNase may also lose its effectiveness after repeated freeze-thaw cycles. If this is suspected, prior to the subsequent neuronal preparation, make a smaller volume of fresh stock. When there are issues with either the papain or DNase, undisrupted tissue will still be visible after three trituration steps. If this occurs, repeat the trituration to increase the yield.
Finally, while it is necessary to work gently when triturating the neurons, it is also necessary to work quickly, particularly during the dissection steps. With increased time, the tissue of the brain may begin to break down, becoming gel-like, making it difficult to identify distinct structures like the hippocampus. If this occurs, for accuracy, do not use this tissue, and move on to the next pup to avoid contaminating the hippocampal culture with neurons from other brain regions.
Application of Wax Spacers to Coverslips
The application of wax spacers, while not difficult, does take practice. Familiarity with the material and its properties at various temperatures will come with time. For consistency of results both within and between experiments, it is important when applying the wax spacers to aim for a consistent size (~1–1.5 mm), and for secure adherence of the wax spacers so that they will remain throughout the entire culture. However, it should be remembered that they must be removed prior to mounting the coverslips for imaging. If too much wax remains on the coverslip after their removal, it may interfere with level mounting of the top cover glass on the slide and cause problems when imaging. Clean removal of these spacers is most easily accomplished by aiming for a round shape, which minimizes the amount of surface area that the wax dot and coverslip share. This takes practice as there is a fine balance between wax dots adhered with minimal surface area and assuring that they remain adhered throughout the culturing process. This is normally accomplished by finding the right temperature of the wax. Wax that is too hot, will pool, increasing the surface area and attachment to the cover glass, making it difficult to remove cleanly. Wax that is too cool will be nicely round and adhere initially but may easily be dislodged during washes and medium changes. This is important, as the loss of more than one spacer prior to the co-culture may differentially influence the results, as the coverslip will not lie level in the culture, causing some neurons to be closer to or even in contact with the astroctyes. Be sure test the adherence of the spacers before use by lightly touching each with tweezers. Discard those coverslips with spacers of significantly inconsistent size or those that do not maintain a roundish shape.
To attain the round shape, the following techniques may be of help when first learning. Be aware that the molten wax is quite hot, so work with care and wear safety glasses. Keep the needle warm while working so the wax does not cool; work with small volumes (~100 µL or less when learning), and work relatively quickly. To warm the needle, pull approximately 100 µL of molten wax into the needle and release three or four times. Dispense a small drop at the tip of needle and tap the needle onto the coverslip. The wax should move easily from the syringe when pressure is applied. If it does not, the wax has started to cool and solidify. Do not apply pressure. Forcing the wax may cause the needle to be expelled from the syringe. If the wax does solidify, dispose of the syringe and needle and start with a new one. A working volume of 100 – 150 µLs of wax will be sufficient to prepare about 1 – 2 coverslips, depending on the temperature of the wax. As the wax cools, you will notice that the applied spacers may not be adhering well. Rewarm the needle and continue until all necessary coverslips are complete. Coverslips can be prepared ahead of time, stored in a covered box, and plated in 24-well plates when needed.
Astrocyte Culture Purity
If performing the co-culture experiment, the interest is to assess how astrocytes influence synapse formation and how that effect is modulated by toxicants. It is therefore important that measures are taken to ensure the purity of the astrocyte cultures. During the primary culture preparation, the harshness of the trypsin and the vortexing step aid in reducing the numbers of surviving neurons. Initial plating at a low density will minimize neuronal contamination in these astrocytes cultures, as the large flask with minimal numbers of astrocytes are less supportive of any neurons plated at this early time point. However, some neurons may still be visible early on in culture, either surviving the steps of the astrocyte primary cell preparation, or possibly developing from progenitor cells. In an environment with low initial astrocyte numbers, and the use of medium optimized for astrocytes, neuronal survival is minimized. As astrocytes proliferate to confluence, the ratio of neurons to astrocytes is minimal. Additionally, because neurons are post-mitotic, most of those that do remain after two weeks of culturing do not survive after the trypsinization step when passed for experimental use. Consistent medium changes and rocking the flask with each change will dislodge microglia and other cell types that have adhered to the surface of the astrocyte monolayer, and so will help in maintaining a relatively pure astrocyte working culture. If contaminating cells continue to grow, increase either the force of rocking or the number of times each flask is rocked.
Neuronal Imaging
One of the most crucial aspects of this methodology is proper imaging. There are many experimental questions where adjusting the imaging parameters to obtain the best possible image with each new field is appropriate. However, here we compare relative changes to synapse formation across treatment groups. Any differences can only be observed if the laser intensity and gain settings of the microscope remain constant for each treatment group of the experiment (Glynn and McAllister, 2006), making the setting of these parameters crucial. Inappropriate settings will result in the loss of data, either leaving out signal on the low end of the intensities, or saturating signal at the high end of intensities (Pawley, 2006). Follow the instructions specific to the microscope being used to attain the appropriate settings. Importantly, these settings then should remain constant throughout the experiment.
Using the slide with the brightest intensity to set these parameters ensures that every subsequent treatment group will have signal within the appropriate detectable range and so will allow for comparison of signal, or synaptic structures between groups. If a less-intense slide is used to determine the settings, the result would be oversaturation of signal when a treatment group with a more intense signal is imaged, resulting in the loss of data and difficulty in quantifying differences between groups.
It is also important to reduce bias in selecting neurons for imaging. Be sure to choose healthy cells, visualizing only the nuclei for selection. For consistency, attempt to select neurons within an experiment that are in similar neighborhoods on the coverslip, that is, not excessively clustered, nor too far from other neurons, as these are not typical environments. You will gain familiarity as you image multiple coverslips. If a coverslip presents with environments that are outside the norm, for example, excessive apoptotic cells, or neurons that are highly clustered, it is possible that that the culture was not healthy.
Generating the Intensity Threshold
As in imaging acquisition, the threshold is generated from the same slide that is used to set the confocal parameters, and once determined, it too is held constant throughout an experiment. Because the value of the threshold has important implications for the rendering of three-dimensional objects, its determination is important. The methodology developed here to obtain this threshold value has been optimized from the synthesis of concepts previously described (Glynn and McAllister, 2006). The threshold is calculated by acquiring the mean intensities of 30 puncta from each channel in 15 different images, for a total of 450 measurements per experiment. Despite the fact that the puncta are manually selected, and may introduce the possibility of user bias (Wouterlood, 2008), the large sample size, and the ability to confirm the accuracy of the threshold by comparing the three-dimensional image rendered after thresholding with the deconvolved image, minimizes the potential for subjective selection.
In addition to the intensity component, this methodology also uses a size component when selecting puncta, which ensures that only structures of the appropriate size are selected for inclusion. In the thresholding protocol, a three pixel selection tool size in ImageJ is used to select puncta. The images have a resolution of 0.103 µm/pixel, and so using this tool size would result in the selection of puncta that are approximately 306 nm in diameter. Scanning electron microscopy serial reconstructions have estimated that the size of the postsynaptic density ranges from 200 – 500 nm (Okabe, 2007; Harris, et al., 1992; Spacek and Harris, 1998), placing the manually selected puncta in the appropriate size range. Including this size parameter is important to obtaining accurate thresholding values, as the use of an incorrectly sized selection tool may negatively influence the intensity values obtained (Glynn and McAllister, 2006). If the selection size is too large, dark portions of the image background will be included, resulting in an average intensity that is lower than the actual puncta signal. If the selection size is too small for the actual puncta, the mean intensity will be abnormally high. The selection tool size used here is sufficient to encapsulate the number of pixels that would fit the average of the smallest sized puncta of the two channels and results in generating a threshold that consistently produces a rendering that is indicative of the deconvolved image.
Again, some key tips are important to remember when determining the threshold. It is important to select only puncta that are reflective of the range of intensities across the length of each extension, not just those that present the most intense signal. Do not select diffusely labeled signal or indistinct structures, which can often be seen in the cell body and in the early extension outcroppings. While diffuse signal is indicative of labeled protein, lack of punctate staining suggests a lack of protein clustering, and so raises doubt as to whether this signal is actually indicative of a pre- or postsynaptic structure. It is possible that this diffuse signal may be protein that is either in the process of being produced, concentrated in the cell body awaiting transport, or protein being trafficked and so not yet part of a structure. With experience the ability to discriminate actual puncta from indistinct signal will become easier.
Additionally, it is helpful to verify the accuracy of the thresholding method and puncta selection criteria by comparing the deconvolved image with the automatic surface rendering obtained using three-dimensional analysis software. If the threshold includes a representative sample of the range of intensities, the surface rendering should reflect what is actually present in the deconvolved image.
Anticipated Results
As previously mentioned, hippocampal neurons are plated at a density of 80,000 neurons per coverslip or mL, with one litter of pups (~10–15 pups) yielding between 10 and 15 million neurons. Astrocytes are plated at a density of 250,000 cells per mL in 24-well plates, and approximately 8–10 million astrocytes can be obtained from one 75 cm2 flask, after two weeks in culture.
For confocal imaging, the parameters used in acquiring signals from each channel will vary only slightly between experiments, if the protocols are followed consistently. These differences should not be entirely unexpected as each experiment is unique and any differences most likely derive from small variations in immunolabeling parameters, such as amount of time in primary antibody incubation, or the length of time that has passed after mounting and before imaging. Imaging is optimized for each individual experiment and if set properly, and held constant throughout that experiment, any slight differences will not affect the ultimate results.
On the other hand, large variations may be indicative of significant problems, which are usually clear early on in the imaging process. Noticeable differences in signal from one experiment to the next, little signal in one channel relative to the other, or highly apoptotic cultures may point to issues with immunolabeling, such as, improper neuronal fixation or permeabilization, improperly prepared antibody buffers, ineffectiveness of the primary or secondary antibodies, or even problems with the health of the cultures themselves. If they do occur, imaging and analysis will not be successful.
Finally, analysis of synaptic structures in hippocampal neurons using the three-dimensional object analysis methods, produce consistent results over numerous experiments. The number of synaptic structures in untreated control neurons, never exposed to astrocytes, results in a mean of 23.1 synapses per neuron (SEM 2.21; n = 3 independent experiments). In control neurons that have shared the environment with astrocytes for 24 hours, the number of neurons increases to about 38.7 synapses per neuron (SEM 2.7; n = 5 independent experiments), a 1.7 fold increase (p = 0.0069). Synaptic structures have been observed to both increase and decrease in those neurons co-cultured with treated astrocytes, depending on the treatment. We have observed increases as large as 4.5 fold relative to those neurons co-cultured with untreated astrocytes and attenuation of increases to control levels after incubation with various inhibitors, suggesting that this method of analysis is responsive to changes in synaptic structure formation. As would be expected, variability in the number of individual puncta and the number of synapses have been observed within treatment groups of one experiment, however, analysis of 15 images per treatment group is capable of capturing statistically significant differences between treatment groups, if they are present.
Time Considerations
The total amount of time in culture before experimental endpoints can be assayed is 14 days. Neurons are grown for a total of 13 days with astrocytes passed from working cultures a week prior to the co-culturing of astrocytes and neurons. Within this time frame, various tasks are performed to maintain the two distinct cultures, and these are summarized in Table 1. Treatments and co-culture time-points can be modified to address particular experimental questions, but here we use a 24 hour astrocyte treatment. This requires that the co-culturing and neuronal fixation steps occur at the same time over the subsequent two days, so plan the starting treatment time accordingly.
With experience, to complete the primary neuron and astrocyte preparations, from set up to the final plating of cells, takes approximately 3–4 hours for one litter of pups (~10–15 pups). This time is only slightly shortened when the planned experiments require the use of fewer pups, as dissection time is the one variable that affects overall timing. There is little down time within this primary cell preparation and it must be completed in one sitting. The preparation of the astrocyte primary cultures will take a similar amount of time, and also must be completed in one sitting, although there are numerous 10 minute centrifugation steps.
Immunocytochemical antibody labeling of neurons occurs over two days, which must be consecutive. Primary antibody incubation steps on the first day will take between 1–1.5 hours, which includes a half an hour blocking and permeabilization step, along with numerous, shorter wash steps. Incubation of the neurons with the primary antibodies requires at least 18 hours, and so an overnight incubation is recommended. Tasks on the second day will take approximately 3–5 hours from the first wash step through completion, which ends with the mounting of slides. These time estimates may vary depending on the number of coverslips in each experiment, and are based on four treatment groups, or 12 coverslips. The time will vary during those steps that require the manipulation and inversion of the coverslips and during mounting of the slides when more coverslips are included.
Confocal imaging is the most time laden portion of this method. The imaging of 15 neurons from the three coverslips of one treatment group will take 3–4 hours. All treatment groups should be imaged close in time to reduce inconsistent loss of signal between imaging sessions. Once imaged, however, with experience, thresholding and three-dimensional object analysis takes approximately 2 – 3 hours per experiment, with each task taking about 1 – 1.5 hours.
Figure 1. Immunocytochemical Labeling.
Hippocampal neurons (13 DIC) were co-cultured for 24 hours with astrocytes that were pre- treated with carbachol (1 mM) or ethanol (75 mM). Neurons were fixed, immunocytochemically labeled for the pre-synaptic protein, synaptophysin, and the post-synaptic protein, PSD-95. Shown are maximum intensity projected images after confocal imaging. (Physin: synaptophysin, green; PSD-95, red; merge, yellow.)
ACKNOWLEDGEMENT
The authors acknowledge Glen MacDonald, University of Washington Center on Human Development and Disability Microscopy Core; and Gennaro Giordano and Khoi Dao. Research by the authors is supported by grants from NIH (F31 AA-019868, R01 AA-08154, R21 ES-022611, R01 ES-022949, P30ES-07033, P50 ES-09601, P42 ES-04696, R01 AA-17180).
Contributor Information
Pamela J. Roque, Email: proque@uw.edu, Department of Environmental and Occupational Health Sciences, University of Washington, Box 354695, 4225 Roosevelt Way, NE Suite 100, Seattle, WA 98105, 206.543.8644.
Marina Guizzetti, Email: mguizzetti@psych.uic.edu, Department of Psychiatry, University of Illinois at Chicago, Jesse Brown VA Medical Center, Chicago, IL, 312.569.8684.
Lucio G. Costa, Email: lgcosta@uw.edu, Department of Environmental and Occupational Health Sciences, University of Washington, Box 354695, 4225 Roosevelt Way, NE Suite 100, Seattle, WA 98105, 206.543.2831.
LITERATURE CITED
- Banker GA, Cowan M. Rat hippocampal neurons in dispersed cell culture. Brain Research. 1977;126:397–425. doi: 10.1016/0006-8993(77)90594-7. [DOI] [PubMed] [Google Scholar]
- Barres BA. The mystery and magic of glia: a perspective on their roles in health and disease. Neuron. 2008;60:430–440. doi: 10.1016/j.neuron.2008.10.013. [DOI] [PubMed] [Google Scholar]
- Bernal J. Thyroid hormone receptors in brain development and function. Nature Reviews Endrocrinology. 2007;3:249–259. doi: 10.1038/ncpendmet0424. [DOI] [PubMed] [Google Scholar]
- Boraso M, Viviani B. Glia-neuron sandwich cocultures: An in vitro approach to evaluate cell-to-cell communication in neuroinflammation and neurotoxicity. In: Costa LG, Giordano G, Guizzetti M, editors. In Vitro Neurotoxicology: Methods and Protocols. New York: Springer; 2011. pp. 135–152. [DOI] [PubMed] [Google Scholar]
- Cannell MB, McMorland A, Soeller C. Image enhancement by deconvolution. In: Pawley JB, editor. Handbook of Biological Confocal Microscopy. 3rd ed. New York: Springer Science + Business Media; 2006. pp. 488–497. [Google Scholar]
- Christopherson KS, Ullian EM, Stokes CCA, et al. Thrombospondins are astrocyte-secreted proteins that promote CNS synaptogenesis. Cell. 2005;120:421–433. doi: 10.1016/j.cell.2004.12.020. [DOI] [PubMed] [Google Scholar]
- Craig AM, Graf ER, Linhoff MW. How to build a central synapse: clues from cell culture. Trends in Neurosciences. 2006;29:8–20. doi: 10.1016/j.tins.2005.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Graff-Peters VB, Hadders-Algra M. Ontogeny of the human central nervous system: what is happening when? Early Human Development. 2006;82:257–266. doi: 10.1016/j.earlhumdev.2005.10.013. [DOI] [PubMed] [Google Scholar]
- Dobbing J, Sands J. Comparative aspects of the brain growth spurt. Early Human Development. 1979;3:79–83. doi: 10.1016/0378-3782(79)90022-7. [DOI] [PubMed] [Google Scholar]
- Eroglu C, Barres BA. Regulation of synaptic connectivity by glia. Nature. 2010;468:223–231. doi: 10.1038/nature09612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foo LC, Allen NJ, Bushong EA, Ventura PB, Chung WS, Zhou L, Cahoy JD, Daneman R, Zong H, Ellisman MH, Barres BA. Development of a method for purification and culture of rodent astrocytes. Neuron. 2011;71:799–811. doi: 10.1016/j.neuron.2011.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foo LC. Purification of rat and mouse astrocytes by immunopanning. Cold Spring Harbor Protocols. 2013 doi: 10.1101/pdb.prot074211. [DOI] [PubMed] [Google Scholar]
- Giordano G, Costa LG. Primary Neurons in culture and neuronal cell lines for in vitro neurotoxicological studies. In: Costa LG, Giordano G, Guizzetti M, editors. In Vitro Neurotoxicology: Methods and Protocols. New York: Springer; 2011. pp. 13–27. [DOI] [PubMed] [Google Scholar]
- Glynn MW, McAllister J. Immunocytochemistry and quantification of protein colocalization in cultured neurons. Nature Protocols. 2006;1:1287–1296. doi: 10.1038/nprot.2006.220. [DOI] [PubMed] [Google Scholar]
- Goslin K, Asmussen H, Banker G. Rat hippocampal neurons in low-density culture. In: Banker G, Goslin K, editors. Culturing Nerve Cells. 2nd ed. Massachusetts: The MIT Press; 1998. pp. 339–370. [Google Scholar]
- Guizzetti M, Moore NH, Giordano G, Costa LG. Modulation of neuritogenesis by astrocyte muscarinic receptors. The Journal of Biological Chemistry. 2008;283:31884–31897. doi: 10.1074/jbc.M801316200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris KM, Jensen FE, Tsao B. Three-dimensional structure of dendritic spines and synapses in rat hippocampus (CA1) at postnatal day 15 and adult ages: implications for the maturation of synaptic physiology and long-term potentiation. Journal of Neuroscience. 1992;12:2685–2705. doi: 10.1523/JNEUROSCI.12-07-02685.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaech A, Banker G. Culturing hippocampal neurons. Nature Protocols. 2006;5:2406–2415. doi: 10.1038/nprot.2006.356. [DOI] [PubMed] [Google Scholar]
- Lardi-Studler B, Fritschy JM. Matching of pre- and postsynaptic specializations during synaptogenesis. Neuroscientist. 2007;13:115–126. doi: 10.1177/1073858406296803. [DOI] [PubMed] [Google Scholar]
- Matyash V, Kettenmann H. Heterogeneity in astrocyte morphology and physiology. Brain Research Reviews. 2010;63:2–10. doi: 10.1016/j.brainresrev.2009.12.001. [DOI] [PubMed] [Google Scholar]
- Mauch DH, Nagler K, Schumacher S, et al. CNS synaptogenesis promoted by glia-derived cholesterol. Science. 2001;294:1354–1357. doi: 10.1126/science.294.5545.1354. [DOI] [PubMed] [Google Scholar]
- Nagler KD, Mauch DH, Pfrieger FW. Glia-derived signals induce synapse formation in neurons of the rat central nervous system. Journal of Physiology-London. 2001;533:665–679. doi: 10.1111/j.1469-7793.2001.00665.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberheim, Goldman, Nedergaard Heterogeneity of astrocytic form and function. Methods in Molecular Biology. 2012;814:23–45. doi: 10.1007/978-1-61779-452-0_3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okabe S, Kim H, Miwa A, et al. Continual remodeling of postsynaptic density and its regulation by synaptic activity. Nature Neuroscience. 1999;2:804–811. doi: 10.1038/12175. [DOI] [PubMed] [Google Scholar]
- Okabe S. Birth, growth and elimination of a single synapse. Anatomical Science International. 2002;77:203–210. doi: 10.1046/j.0022-7722.2002.00030.x. [DOI] [PubMed] [Google Scholar]
- Okabe S. Molecular anatomy of the postsynaptic density. Molecular and Cellular Neuroscience. 2007;34:503–518. doi: 10.1016/j.mcn.2007.01.006. [DOI] [PubMed] [Google Scholar]
- Pawley JB, editor. Handbook of Biological Confocal Microscopy. 3rd Edition. New York: Springer Science +Business Media; 2006. [Google Scholar]
- Pfrieger FW, Barres BA. Synaptic efficacy enhanced by glial cells in vitro. Science. 1997;277:1684–1687. doi: 10.1126/science.277.5332.1684. [DOI] [PubMed] [Google Scholar]
- Ransom B, Behar T, Nedergaard M. New roles for astrocytes (stars at last) Trends in Neurosciences. 2003;26:520–522. doi: 10.1016/j.tins.2003.08.006. [DOI] [PubMed] [Google Scholar]
- Rao A, Craig AM. Activity regulates the synaptic localization of the NMDA receptor in hippocampal neurons. Neuron. 1997;19:801–812. doi: 10.1016/s0896-6273(00)80962-9. [DOI] [PubMed] [Google Scholar]
- Roqué PJ, Guizzetti M, Giordano G, Costa LG. Quantification of synaptic structure formation in co-cultures of astrocytes and hippocampal neurons. In: Costa LG, Giordano G, Guizzetti M, editors. In Vitro Neurotoxicology: Methods and Protocols. New York: Springer; 2011. pp. 361–390. [Google Scholar]
- Spacek J, Harris KM. Three-dimensional organization of cell adhesion junctions at synapses and dendritic spines in area CA1 of the rat hippocampus. Journal of Computational Neurology. 1998;393:58–68. doi: 10.1002/(sici)1096-9861(19980330)393:1<58::aid-cne6>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- Sheppard CJR, Gan X, Gu, et al. Signal-to-noise ratio in confocal microscopes. In: Pawley JB, editor. Handbook of Biological Confocal Microscopy. 3rd Edition. New York: Springer Science + Business Media; 2006. pp. 442–451. [Google Scholar]
- Ullian EM, Sapperstein KS, Christopherson KS, Barres BA. Control of synapse number by glia. Science. 2001;291:657–661. doi: 10.1126/science.291.5504.657. [DOI] [PubMed] [Google Scholar]
- Waites CL, Craig AM, Garner CC. Mechanisms of vertebrate synaptogenesis. Annual Review of Neuroscience. 2005;28:251–274. doi: 10.1146/annurev.neuro.27.070203.144336. [DOI] [PubMed] [Google Scholar]
- Weidenmann B, Franke WW. Identification and localization of synaptophysin, and integral membrane glycoprotein of M, 38,000 characteristic of presynaptic vesicles. Cell. 1985;41:1017–1028. doi: 10.1016/s0092-8674(85)80082-9. [DOI] [PubMed] [Google Scholar]
- Wouterlood FG, Boekel AJ, et al. Counting contacts between neurons in 3D in confocal laser scanning images. Journal of Neuroscience Methods. 2008;171:296–308. doi: 10.1016/j.jneumeth.2008.03.014. [DOI] [PubMed] [Google Scholar]
- Viviani B. Coculturing neurons and glial cells. Current Protocols in Toxicology. 2003;(Supplement 15) doi: 10.1002/0471140856.tx1210s15. 12.10.1–12.10.17. [DOI] [PubMed] [Google Scholar]
- Zhang X, Bhattacharyya S, Kusumo H, Goodlett CR, Tobacman JK, Guizzetti M. Arylsulfatase B modulates neurite outgrowth via astrocyte chondroitin-4-sulfate: dysregulation by ethanol. Glia. 2014;62:259–271. doi: 10.1002/glia.22604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Barres BA. Astrocyte heterogeneity: an underappreciated topic in neurobiology. Current Opinions in Neurobiology. 2010;20:588–594. doi: 10.1016/j.conb.2010.06.005. [DOI] [PubMed] [Google Scholar]