Introduction
Hypertrophic cardiomyopathy (HCM) is a genotypically and phenotypically heterogeneous disorder. Clinically defined as cardiac hypertrophy in the absence of extrinsic causes such as hypertension and valvular disease, HCM affects approximately 1 in 500 persons and is one of the most common causes of sudden cardiac death in young athletes [1, 2]. To date, hundreds of mutations in over 15 genes have been implicated in the pathogenesis of HCM. The most common subtype of HCM is myofilament-HCM secondary to mutations in myosin binding protein C (MYBPC3), β-myosin heavy chain (MYH7), cardiac troponin T (TNNT2), α-tropomyosin (TPM1), cardiac troponin I (TNNI3), cardiac actin (ACTC), regulatory myosin light chain (MYL2), and essential myosin light chain (MYL3) [3–9]. Genetic alterations in these eight common myofilament genes have been found in 30 – 61% of HCM patients in different cohorts around the world [10]. In our cohort, the yield of myofilament mutations was approximately 40%, indicating that the pathogenic basis for a significant percentage of clinically diagnosed HCM remains unknown [11–13].
Recently, it has become clear that HCM may also be caused by mutations in Z-disc proteins with the discovery of mutations involving proteins such as telethonin encoded by TCAP, muscle LIM protein encoded by CSRP3, metavinculin and vinculin encoded by VCL, and LIM domain binding 3 encoded by LDB3 (also known as ZASP) [14–18]. Similarly, previous studies have shown glycogen storage diseases mimicking HCM with mutations in PRKAG2-encoding protein kinase gamma 2 and LAMP2-encoding lysosome-associated membrane protein 2 [19]. Compared to myofilament-HCM, both Z-disc- and metabolic-HCM account for a relatively small percentage of HCM leaving a significant proportion of HCM genetically unexplained.
Multiple experimental studies have suggested a pathogenic role for abnormal calcium handling in cardiac hypertrophy and failure. In particular, cellular proteins involved in calcium-induced calcium release, which allows for coordinated myocyte contraction, have been targeted as candidates in the pathogenesis of cardiomyopathy [20–22]. Mutations in promoter and coding region of phospholamban, a regulator of the sarcoplasmic reticulum calcium ATPase (SERCA2a) and a regulator of calcium homeostasis within the cell, have been identified in hypertrophic and dilated cardiomyopathy respectively [23, 24]. These findings suggest that mutations in proteins involved in intracellular calcium homeostasis might be linked to HCM and may provide insight into novel mechanisms of HCM pathogenesis.
Junctophilin type 2, encoded by JPH2, is a cardiac member of the junctophilins family of junctional membrane complex proteins which physically approximates the plasmalemmal L-type calcium channel and the sarcoplasmic reticulum (SR). Junctophilins have multiple membrane occupation and recognition nexus (MORN) domains and a SR transmembrane domain thought to allow for proper localization in the dyadic cleft between the transverse tubules and SR membrane [25, 26]. JPH2 knockout mice demonstrate embryonic lethality, irregular calcium handling, asynchronous and stochastic contractions, and SR segmentation, suggesting a crucial role for junctophilin-2 in facilitating intracellular calcium release and cardiac contractility [25]. In addition, disruption of JPH expression in skeletal muscle results in compromised intracellular calcium homeostasis [27]. JPH2 gene expression is down-regulated in murine models of both dilated and hypertrophic cardiomyopathy [28]. Therefore, we pursued JPH2 as a novel candidate gene for HCM in humans.
Methods
Study population
Between April 1997 and December 2001, 388 consecutive, unrelated patients were referred to the Hypertrophic Cardiomyopathy Clinic at Mayo Clinic, Rochester, Minnesota. Following receipt of written consent for this Mayo Foundation Institutional Review Board-approved protocol, DNA was extracted from peripheral blood lymphocytes using the Purgene DNA extraction kit (Gentra, Inc, Minneapolis, MN). All patients have been previously analyzed for mutations in eight myofilament-associated (MYBPC, MYH7, TNNT2, TPM1, TNNI3, ACTC, MYL2, and MYL3) and five Z disc-associated (ACTN2, VCL, TCAP, CSRP3, and LDB3) genes.
JPH2 Mutational Analysis
All five translated exons, with flanking intronic regions of JPH2, were amplified by PCR using oligonucleotide primers. Exons 1, 2, and 4 were subdivided into two, three, and three fragments of overlapping nucleotides, respectively, for optimal mutational analysis yielding a total of 10 amplicons. The JPH2 gene and the linear topology of junctophilin-2 are schematically represented in Figure 1. Each amplicon was evaluated for mutations using denaturing high performance liquid chromatography (DHPLC, Transgenomic, Omaha, Nebraska), and samples with an abnormal elution profile were directly sequenced (ABI Prism 377; Applied Biosystem, Foster City, California) to characterize the difference between the wild type and variant alleles. Primer sequences, PCR, and DHPLC conditions are available upon request.
Figure 1. Junctophilin-2 linear topology and location of identified mutations.
Approximate location of HCM-associated mutations as well as exons boundaries and putative functional domains of junctophilin-2.
A panel of 1000 ethnically-matched reference alleles, including 400 Caucasian reference alleles derived from 200 Coriell Repository DNA samples and 600 reference alleles from 300 Caucasian subjects with normal screening electro- and echo-cardiograms, were examined for all amino acid variants identified among cases. Absence of these variants in 1000 reference alleles demonstrates with 95% confidence that the true allelic frequency of these variants is less than 0.004. All amino acid variants were denoted using known and accepted nomenclature. To be considered as a putative HCM-susceptibility mutation, minimal molecular/genetic requirements included i) absence in 1000 ethnic-matched reference alleles, ii) non-synonymous amino acid substitutions, and iii) localization to key functional domains.
Construction of expression vectors for complementary mutations
The full-length mouse JPH2 cDNA was amplified by PCR from plasmid pMHMG72, provided by H. Takeshima, using the primers 5′-AAG AAT TCG CCA CCA TGA GCG GGG GCC GCT TTG ACT and 5′-AAT CTA GAT CAA GTC AGG AGG TGA ACA AAT AGG. The resulting PCR product was cloned into the EcoRI and XbaI sites of a modified pCMS-EGFP vector (Invitrogen) called pCMS-RFP. In this modified plasmid, a red fluorescent protein cDNA has been subcloned in place of the green fluorescent protein cDNA at the BamHI and SacII sites which was followed by removal of the SacII site by Klenow fill-in and self-ligation. pCMS-RFP was used as a control plasmid for appropriate experiments.
Mutations were introduced into the mouse JPH2 cDNA in pCMS-RFP using a recently developed PCR-based technique that makes use of the unique effects of type II restriction enzymes [29]. For this mutagenesis approach, the primer mJP2 F1 (5′-AAG AAT TCG CCA CCA TGA GCG GGG GCC GCT TTG ACT) was used as the upstream anchor primer, while R585 (5′-AGC AGA GTG AGC GCG AAA CC) and R641 (5′-CCC AGC AGT GTG CCA CGC GTG A) acted as downstream anchor primers. The complementary mutation in r. norvegicus (rat) and m. musculus (mouse) for the human mutation S101R is N101R, thus N101R was introduced for expression in H9C2 and HL-1 cell lines respectively. Two mutagenesis primers were used to produce this mutation, 5′-AAC TCT TCA AGA AGT GGT GCC AAG TAC GAG GGC A and 5′-AAC TCT TCA TCT TGT GCT CTG CCG GAT TCC GTA GCG. For the mutant Y141H, the primers 5′-AAC TCT TCA CAT GGT GTG CGC CAA AGC GTG CC and 5′-AAC TCT TCA ATG GCC ATG GCG CAT GCC GTT GGT were used. Finally, the primers 5′-AAC TCT TCA TTT CTG CGC AGC GAG CAC AGC AAT and 5′-AAC TCT TCA AAA GGA CAG AGA AGT GCG CAG CG were used to generate S165F. All mutations were confirmed by direct sequencing using a primer specific for vector sequence which flanks the JPH2 cDNA. GFP-fusion constructs were generated by subcloning of these fragments into the pEGFP-C1 vector (Clontech).
Cell culture and DNA transfection
H9C2 cells were maintained in Dulbecco’s Modified Eagle’s Media (DMEM, GIBCO) supplemented with 10% Fetal Bovine Serum (FBS, GIBCO) and 1% penicillin/streptomycin (Sigma) in 5% CO2 at 37°C. For experimentation, 2×105 cells were plated on 30 mm2 ΔTC3 dishes (Bioptechs). HL-1 cells maintained in Claycomb media (JRH Biosciences) were supplemented with 10% FBS (JRH Biosciences Lot # 3J0229), 1% penicillin/streptomycin, 0.1 mM norepinephrine and 2 mM L-glutamine. For experimentation, 1×105 cells were plated on 30 mm2 ΔTC3 dishes that were coated with fibronectin (Sigma) and gelatin (Fisher Scientific). Cells were transfected with GeneJammer transfection reagent (Stratagene) per manufacturer’s instructions. During culturing of cells, medium was replaced every 2 days. Cells were washed twice by a balanced salt solution (BSS) (in mM: 140 NaCl, 2.8 KCl, 2 CaCl2, 2 MgCl2, 10 HEPES, pH 7.2) before experiments. Cellular transfection was confirmed through DsRED2 red fluorescence.
Western Blotting
Cells were collected in cell lysis buffer (150 mM NaCl, 1% Trition-X, 0.1% SDS, 50 mM Tris-HCl, pH 8.0) and homogenized by a handheld motorized rotary homogenizer (Kontes). Protein concentrations of whole cell extracts were determined by Dc protein assay (BioRad) and 25 μg per sample was separated by SDS-polyacrylamide gel electrophoresis on 10% gels. After blotting onto PVDF (BioRad) membrane, equivalent loading and efficient transfer were confirmed with Ponceau S staining (Sigma). A rabbit polyclonal antibody raised against mouse JPH2 was applied to the membrane as the primary antibody [27]. The ECL+ kit (Amersham) was used to produce luminescent signal that was detected by exposure to autoradiography film (Denville).
Cell morphology determination and immunocytochemistry
Transfected H9C2 cells were examined using the 40X objective of a Zeiss Axiovert 200 fluorescent microscope equipped with a temperature (37°C) and CO2 (5%) environmental chamber to facilitate live cell imaging. Fluorescent images from 15 randomly selected fields were selected from each dish. The area of all transfected cells (as determined by red fluorescence protein marker) in the field was determined by using AxioVision V4 software (Zeiss). Transfections were performed in duplicate for three separate trials.
Following morphometric analysis, cells were fixed with 100% ethanol at −20° C for 10 minutes then washed three times in phosphate buffered saline (PBS). Cells were blocked in 5% total milk protein in PBS for 1 hour at room temperature. Primary rabbit anti- JPH2 (1:300 dil.) or anti-calreticulin (1:250 dil.) was applied for 2 hours at room temperature. Cells were washed 3 times for 10 minutes in PBS with gentle agitation. Goat anti-rabbit secondary antibody coupled to AlexaFluor488 (1:300 dil., Molecular Probes) was applied for 1 hour at room temperature. Cells were washed 3 times for 10 minutes in PBS with gentle agitation before mounting on glass slides. Mounting media contained 4′,6-diamidino-2-phenylindole, dihydrochloride (DAPI) in some experiments. Images were captured by confocal microscopy as described below.
Confocal microscopy measurement of calcium waves
Transfected HL-1 cells were loaded in the dark with 10 μM Fluo-4-AM calcium indicator (Molecular Probes) for 60 minutes at 37° C. After the loading period, cells were washed three times with PBS. Measurements of intracellular calcium handling were performed at room temperature on a BioRad Radiance-2100 confocal microscope equipped with both argon (488 nm) and helium/neon (543 nm) lasers on a Nikon Eclipse TE2000S inverted microscope using a 60X, 1.4 NA oil immersion objective. Regions of interest (ROI) with equal area were selected and serial x-y images (512×512) were acquired at 3.08 seconds per frame. Transfected cells could be identified by the presence of red fluorescent protein marker. All transfections were performed in duplicate for three experimental trials. Data analysis was conducted using OriginPro 7.0 software (OriginLab).
Statistical analysis
ANOVA analysis was performed to establish differences between experimental groups. A p-value lower than 0.05 was considered statistically significant.
Results
The demographics of our HCM cohort are shown in the first column of Table 1. After mutational analysis for eight myofilament-associated and five Z disc-associated genes, 165 of the 388 patients were found to harbor a putative HCM-associated mutation. The clinical characteristics of the three subgroups (myofilament negative, myofilament positive and Z-disc HCM) are shown in Table 1.
Table 1.
Clinical Characteristics of HCM cohort
| Clinical Characteristic | HCM Cohort | Myofilament Negative HCM | Myofilament Positive HCM | Z-Disc Positive HCM | JPH2 |
|---|---|---|---|---|---|
| No. of individuals | 388 | 224 | 129 | 13 | 3 |
| Sex, male/female | 215/173 | 125/99 | 71/58 | 7/6 | 2/1 |
| Age at Dx | 41.2 ± 19 | 45.0 ± 19 | 36.4 ± 17 | 45.4 ± 20 | 26.8 ± 3 |
| Cardiac Symptoms | 55% | 55% | 54% | 83% | 100% |
| Max LVWT (mm) | 21.5 ± 7 | 20.7 ± 6 | 22.8 ± 6 | 20.8 ± 8 | 26 ± 10.6 |
| Resting LVOTO (mm Hg) | 46.6 ± 42 | 47.7 ± 42 | 43.8 ± 43 | 53.5 ± 48 | 20.3 ± 18.4 |
| Pos. FH for HCM | 147 (31%) | 54 (24%) | 61 (47%) | 2 (15%) | 1 (33%) |
| Pos. FH for SCD | 54 (14%) | 41 (18%) | 32 (25%) | 0 (0%) | 0 (0%) |
| Surgical myectomy | 160 (41%) | 87 (39%) | 56 (43%) | 8 (62%) | 1 (33%) |
| Pacemaker | 68 (18%) | 38 (17%) | 27 (21%) | 2 (15%) | 1 (33%) |
| ICD | 60 (15%) | 22 (10%) | 31 (24%) | 0 (0%) | 2 (66%) |
Values are mean ± SD or n (%). Dx indicates diagnosis; FH, family history; HCM, hypertrophic cardiomyopathy; ICD, implantable cardioverter-defibrillator; LVOTO, left ventricular outflow tract obstruction; LVWT, left ventricular wall thickness. The 19 patients harboring compound mutations with either multiple myofilament mutations or a myofilament and Z-disc mutation have been excluded from this comparison.
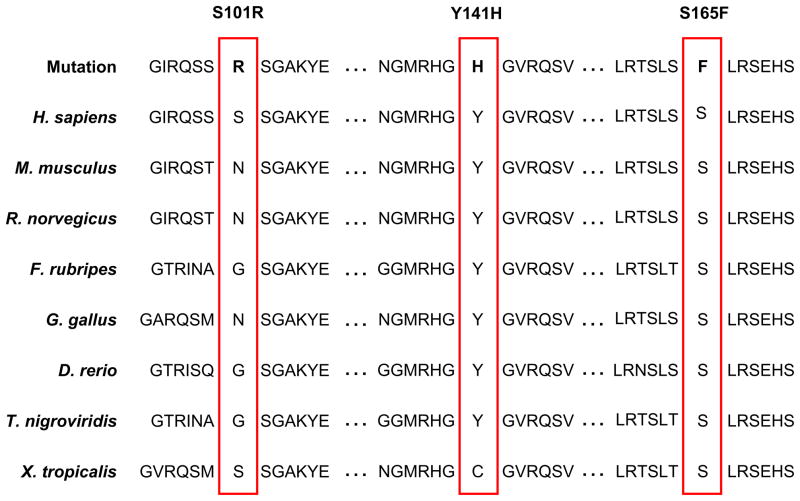
Following criteria outlined in the Methods section, three novel putative HCM mutations were discovered: S101R, Y141H, and S165F, in 3/388 unrelated, white patients with HCM (Figure 1). Y141H and S165F mutant residues are conserved across multiple species while the S101R mutation localized to the conserved MORN motif as shown in Figure 2. The clinical characteristics of these three patients are listed in Table 1. The absence of these variants in 1000 ethnic-matched Caucasian reference alleles demonstrates with 95% confidence that the true allelic frequency of these variants is less than 0.004. In addition, no other mutations were detected in these three patients following comprehensive open reading frame/splice site mutational analysis of 13 known HCM-susceptibility genes.
Figure 2. JPH2 primary sequence mutations are conserved across species or may effect protein phosphorylation.
Sequence conservation analysis for the observed JPH2 mutations demonstrates that Y141H and S165F are conserved between multiple divergent species, while S101R localizes to the conserved MORN motif of JPH2.
As summarized in Table 2, the JPH2 genotype positive subjects were diagnosed with HCM at 27, 24, and 30 years with septal wall thicknesses of 22, 38, and 18 mm for each mutation respectively. Case 1 has a positive family history of familial HCM involving three first-degree relatives and one second-degree relative. There is no apparent family history of HCM among either first- or second-degree relatives for cases 2 or 3 suggesting the possibility of a sporadic de novo mutation or incomplete penetrance. To date, relatives of all 3 families have declined participation precluding a molecular determination of co-segregation or sporadicity.
Table 2.
Clinical Phenotype of JPH2 Genotype Positive Patients with HCM
| Case | Mutation | Age (y) Sex | Age at Dx (y) | Race | Symptoms at Presentation | Subsequent symptoms | AF | Max LVWT (mm) | Resting LVOTO (mmHg) | FH of HCM | FH of SCD | Treatment |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | S101R | 40/M | 27 | C | Dyspnea | Dyspnea | No | 22 | 0 | Yes | No | ICD |
| 2 | Y141H | 33/M | 24 | C | Dyspnea, palpitations, angina, non-Q-wave MI | Dyspnea, palpitations | No | 38 | 25 | No | No | Pacemaker ICD |
| 3 | S165F | 58/F | 30 | C | Dyspnea, SBE | Dyspnea | No | 18 | 36 | No | No | Myectomy |
HCM, hypertrophic cardiomyopathy; Dx, diagnosis; C, Caucasian; SBE, subacute bacterial endocarditis; AF, atrial fibrillation; LVWT, left ventricular wall thickness; LVOTO, left ventricular outflow tract obstruction; FH, family history; SCD, sudden cardiac death; ICD, implantable cardioverter-defibrillator.
To further investigate the molecular/cellular effects of these HCM-susceptibility JPH2 mutations, complementary mutations to all three JPH2 missense mutations were engineered into mouse JPH2 cDNA rather than human to facilitate functional studies in cultured cells of rodent muscle origin. The mouse and human JPH2 share greater than 98% identity over the MORN motif region 1 and 85% identity over the length of the protein. These constructs were transfected into both H9c2 and HL-1 homologous muscle cell lines. HL-1 and H9c2 cells provide a rodent muscle cell background for expression studies that either express, or do not endogenous JPH2 respectively. Additionally, HL-1 cells display spontaneous calcium oscillations and contractions in vitro. Western blot analysis confirmed that H9c2 cells did not express native JPH2 while transfection of wild type and or mutant forms of JPH2 vectors produced robust expression of JPH2 (Figure 3A). HL-1 cardiomyocytes express abundant endogenous JPH2 and expression of the mutant GFP-fusion constructs could be observed by Western blot (data not shown). Furthermore, the in vitro assays revealed altered arrangement of the JPH2 protein, abnormal calcium signaling, and marked cardiomyocyte hypertrophy (Figures 3 and 4).
Figure 3. JPH2 mutants are mis-localized and induce cardiomyocyte hypertrophy.
(A) Western blot analysis for JPH2 expression in H9c2 cells. H9c2 cells do not express endogenous JPH2 (vector lane); however, expression of all mutants was found when cells were harvested 24 hours or 72 hours post transfection. Markers indicate molecular weight in kiloDaltons (kDa).
(B) Confocal images of H9c2 cells expressing full-length wild type and mutant JPH2 stained with anti-JPH2 antibody. Intracellular vacuolization and disrupted localization of the JPH2 gene product are present in cells expressing mutant JPH2. Scale bar indicates 10 μm.
(C) Confocal images of H9c2 cells expressing full-length wild type and mutant JPH2 stained with anti-calreticulin antibody to indicate the presence of an intact SR network (green). Red indicates expression of red fluorescent protein from a separate promoter on the plasmid that servers to mark transfected cells as well as to stain the cytsolic compartment. Blue indicates nuclei as stained by DAPI. Scale bar indicates 10 μm.
(D, E) Live-cell confocal images of HL-1 cells transfected with JPH2-GFP fusion proteins at 24 (C) and 48 (D) hours. These fusion proteins do not contain the C-terminal transmembrane domain (TM) which normally allows the JPH2 gene product to association with the SR. Deletion of the TM allows for JPH2-GFP fusion proteins (JPH2-ΔTM-GFP) to associate with the plasma membrane via the MORN motifs. Expression of JPH2-ΔTM-GFP and mutant constructs did not have any obvious detrimental effects on cell morphology. Scale bar indicates 20 μm.
(F) Hypertrophy of H9c2 cells transfected with mutant JPH2 was observed when cell size was measured at 5 days post-transfection. Results are representative from one trial of three conducted. In this trial control H9c2 cells, (n=148, mean=63.3 μm2, SE=5.3 μm2), JPH2-WT (n=182, mean=66.4 μm2, SE=4.0 μm2), JPH2-N101R (n=118, mean=78.1 μm2, SE=7.2 μm2), JPH2-Y141H, (n=108, mean=96.6 μm2, SE=8.5 μm2), and JPH2-S165F (n=132, mean=97.7 μm2, SE=16.0 μm2) transfected cells were examined. Arrows indicate vacuolization of intracellular structure. Data listed as mean ± SE. * p<0.05
Figure 4. JPH2 mutants disrupt calcium signaling in HL-1 cells.
(A) HL-1 cells were loaded with Fluo-4-AM (top panel) at 5 days post-transfection. Transfected cells were selected by the presence of a red fluorescent protein marker (bottom panel overlay image) from a separate promoter of the expression vector backbone. Scale bar indicates 10 μm. Boxed areas are representative regions of interest (ROI) used for analysis.
(B) ROI were selected in transfected cells and signal was collected by xy scan for 5–10 minutes. Representative traces for each mutant construct are provided. Scale bars indicate 1.0 for the ratio of fluorescence change (ΔF/F0) for the y-axis and 200 seconds for the x-axis.
(C) The ratio of fluorescence change (ΔF/F0), as an indication of global calcium flux, is significantly attenuated in HL-1 cells expressing JPH2 mutants. The ΔF/F0 was measured in HL-1 (n=29, mean=0.933 ΔF/F0, SE=0.142), JPH2-WT (n=11, mean=0.884 ΔF/F0, SE=0.0831), JPH2-N101R (n=5, mean=0.474 ΔF/F0, SE=0.124), JPH2-Y141H, (n=7, mean=0.200 ΔF/F0, SE=0.0309), and JPH2-S165F (n=7, mean=0.269 ΔF/F0, SE=0.0971) transfected cells were examined. Data listed as mean ± SE. * p<0.05.
Since these membrane association studies required the deletion of the JPH2 transmembrane domain, the localization of full-length JPH2 and mutants were examined. Localization of exogenously expressed wild type and mutant JPH2 in H9c2 cells was determined by immunohistochemistry, since these cells did not express endogenous JPH2. Only cells with similar intensity of JPH2 staining were compared to minimize the effect of expression level on protein localization. All JPH2 mutants exhibited an altered localization pattern with respect to the SR when compared to wild type JPH2 in H9c2 cells (Figure 3B). H9c2 cells expressing mutant JPH2 frequently displayed alteration of intracellular localization and apparent vacuolization of intracellular structures, reminiscent of the JPH2 knockout mouse (Figure 3B) [25]. However, the SR remained intact for the majority of H9c2 cells expressing mutant JPH2 (Figure 3C). In addition, H9c2 cells transfected with mutant JPH2 do not display large inclusion bodies under light microscopy, suggesting that the hypertrophy was not secondary to an alteration of metabolic status (data not shown).
Since the mutants identified appear in the MORN motif-mediated cell membrane interaction, we explored whether the missense mutations disrupted the localization to the cell membrane. For this purpose, GFP-fusion proteins were generated by linking GFP to the carboxy terminus of residues 1–430 of JPH2 and the JPH2 mutants. This construct resulted in removal of the transmembrane domain of JPH2 that targets JPH2 to the SR membranes, thereby enabling a precise assessment of the effect of the missense mutations that reside in the MORN motifs on localization of JPH2 to the cell membrane. Following transfection of each mutant JPH2 in to HL-1 cells, there was no detectible alteration in the MORN-motif mediated interaction with the cell membrane (Figure 3D and 3E).
Immunostaining also suggested qualitatively that JPH2 mutant-expressing H9c2 cells were larger than cells expressing either wild type JPH2 or no JPH2 at all. Cell imaging was used to quantitative myocyte size. Indeed, H9c2 cells expressing JPH2 mutants Y141H and S165F displayed significant hypertrophy (2 – 3 fold) when compared to cells expressing wild type JPH2 or cells expressing only marker fluorescent protein from control plasmid (Figure 3F). The N101R JPH2 mutant did not induce significant hypertrophy, perhaps due to genetic variation in the mouse and human protein sequence at this residue or suggesting a different pathogenic mechanism due to the loss of a phosphorylation site. As a whole, these results indicate an association between these JPH2 mutations and induction of myocyte hypertrophy.
Since JPH2 ablation has been linked to defects in calcium handling in embryonic cardiomyocytes and adult skeletal muscle, we investigated the effects of these human JPH2 mutations on intracellular calcium signaling when expressed in HL-1 cardiomyocytes [25, 27]. Here, HL-1 cells were ideally suited for this approach since they express endogenous JPH2 and display spontaneous calcium wave activity. To determine if calcium signaling was disrupted in HL-1 cardiomyocytes expressing JPH2 mutants, cells were transfected and 5 days later were loaded with Fluo-4-AM calcium indicator dye. Transfected cells were identified by the presence of red fluorescent protein marker during confocal microscopy (Figure 4A). The amplitude of spontaneous calcium waves, which represent relative global cytosolic calcium concentrations, were markedly attenuated in JPH2 mutant expressing cells compared to cells transfected with either wild type JPH2 or control vector plasmid (Figure 4B and C). This represents a decreased spontaneous calcium release from the SR, which suggests that excitation-contraction process is disrupted in cells expressing the JPH2 mutant constructs.
Discussion
Since the identification of the first mutation in MYH7, HCM has been viewed conceptually as a disease of the sarcomere, with the typical gross and molecular characteristics of the disease considered to result from inadequate or dysregulated force generation [30]. Recent discoveries have implicated several genes encoding components of the Z-disc as well as genes previously associated with dilated cardiomyopathy in the pathogenesis of HCM [19, 31, 32]. This paper reports the discovery of S101R, Y141H, and S165F mutations in JPH2 as a novel genetic basis for HCM. Junctophilin-2 is involved in the calcium signaling apparatus of cardiomyocytes, and the discovery of these mutations supports a key role for calcium perturbations in a pathway that culminates in the clinically expressed disease phenotype of HCM.
Examination of the known structure-function domains in junctophilin-2 shows that all three missense mutations involve residues localizing to key domains. S101R localizes to part of the conserved MORN motif region I, specifically, MORN domain IV. Substitution of a basic amino acid, arginine, for the hydrophilic amino acid serine caused a charge alteration in a MORN repeat hypothesized to interact with the plasma membrane. The Y141H mutation in MORN domain VI also caused a substitution of a basic amino acid for a hydrophilic amino acid in the plasma membrane binding domain. The S165F mutation abolished a putative PKC-phosphorylation site, substituting a hydrophobic amino acid for the native hydrophilic amino acid, thereby losing a hydroxyl group required for phosphorylation. There has been significant interest in understanding the molecular and cellular adaptations leading to hypertrophy caused by HCM, hypertension, and valvular disease. Several studies have shown that although the inward calcium current (ICa) through the L-type calcium channel is not altered in hypertrophic or failing rat and murine cardiomyocytes, there is a decrease in the intracellular calcium transient, [Ca]I [33, 34]. In post-myocardial infarction cardiomyocytes, this normal ICa and decreased intracellular calcium transient, [Ca]i, was shown to be independent of SR calcium concentration [35]. Gomez et al showed that receptor concentrations, receptor properties, and SR calcium concentrations remained the same between control and hypertrophied myocytes and consecutively demonstrated that the decrease in [Ca]i was most consistent with partial uncoupling of the L-type calcium channel and ryanodine receptor through increased inter-receptor distance [33]. This process of ‘orphaned ryanodine receptors’ and disruption of EC coupling was described more recently by Song et al. as a model for Ca2+ dyssynchrony in heart failure [36]. Such a change in the micro-architecture of the dyad would lead to ineffective signaling and result in hypertrophy. As junctophilin-2 was first discovered as a protein bridging the dyad junction, allowing proper positioning of the L-type calcium channel and ryanodine receptor to allow normal calcium signaling [25], disruption of membrane coupling may contribute to the pathogenesis observed with mutation in JPH2. This hypothesis is supported by Xu et al. who recently demonstrated that the expression of JPH2 was reduced by nearly half in rats with pressure overload-induced hypertrophy. This observation was made in the setting of early, compensatory hypertrophy in which Ca2+ transient, cardiac myocyte contractility, and myocyte ultrastructure were normal [37].
HCM often displays the histological pattern of disorganization and fibrosis among cardiomyocytes, thought to be caused by activation of stress-induced trophic and nuclear factors, with vasoactive and hormonal factors implicated in the process [21, 38]. Efficient calcium handling is essential in myocyte organization as the correct t-tubular system orientation is dependent on calcium signaling during development for example [39–41]. Therefore, disrupted calcium signaling may predispose to myocyte disarray. A recent study provides a link at the level of the myofilament between calcium signaling and force generation for HCM patients with myosin heavy chain mutations [42]. Here, individual muscle fibers showed substantial variability in their responsiveness to calcium and the authors hypothesize that this could potentially lead to imbalanced force generation and myocyte disarray. Further, alterations in calcium signaling through ryanodine receptor mutations have been shown to cause sudden cardiac death and catecholaminergic polymorphic ventricular tachycardia. Therefore, calcium signaling irregularities might be a component of the frequent sudden cardiac death seen in HCM [2, 43–47].
Consistent with these aforementioned hypotheses, the HCM-associated JPH2 mutations discovered in this study localized to key, conserved MORN motifs or potential sites of phosphorylation sites. Our data may suggest a pivotal role of MORN motifs in plasma membrane and SR protein complexes scaffolding with the formation of large JPH2 multimeric polypeptide aggregates. This is supported by our functional expression of mutant JPH2 which resulted in mis-localization of JPH2 in both heterologous cells and homologous cells of muscle origin as well as significant perturbations in myocyte intracellular calcium signaling and marked hypertrophy. Furthermore, disruption of the C-terminal transmembrane domain allows for localization of JPH2 protein to the plasma membrane, potentially due to the interaction of the MORN motifs. Since HL-1 cardiocytes express endogenous JPH2, our experiments suggest a dominant-negative mechanism of action whereby the JPH2 HCM-associated mutants hinder the normal function of the normal protein derived from the wild type allele, consistent with the patient’s heterozygous status at JPH2.
Taken together, the observed mutations in JPH2 may result in vacuolization of the sarcoplasmic reticulum and perturbations in calcium handling. A defect in calcium-induced calcium-release and ineffective calcium signaling may result in a loss of contractile strength, compensatory cellular hypertrophy, and myocyte disarray – hallmarks of hypertrophic cardiomyopathy. Despite the compelling molecular and in vitro functional analyses of the newly discovered JPH2 mutations in patients with HCM, the causal link between genetic perturbations in JPH2 and the pathogenesis of HCM in humans would be bolstered further by co-segregation studies. However, such co-segregation studies require a pedigree of sufficient size and a willingness of family members from mutation-positive index cases to participate. Unfortunately, neither element exists for the three putative HCM-associated JPH2 mutations described herein. Alternatively, recapitulation of the HCM phenotype in a suitable animal model, such as a knock-in mouse heterozygous for one of these mutations, would further strengthen the causal link.
Conclusion
We report the initial discovery of mutations in JPH2-encoded junctophilin-2 resulting in protein mis-localization, impaired calcium-handling, and cardiomyocyte hypertrophy as a novel pathogenic basis for HCM in humans. Unexplained cardiac hypertrophy in humans, such as HCM, is the first human disease associated with mutations in JPH2. It remains to be seen whether the pathway for HCM involves calcium dysregulation either primarily as shown here for JPH2-HCM or secondarily in either myofilament- or Z-disc-HCM. Whether susceptibility for other cardiomyopathies, such as dilated cardiomyopathy, is conferred by mutations in JPH2 warrants further investigation.
Acknowledgments
Funding: MJA’s research program is supported by the Mayo Foundation, the Dr. Scholl Foundation, the CJ Foundation for SIDS, the Doris Duke Charitable Foundation (clinical scientist development award), the American Heart Association (established investigator award), and the National Institutes of Health (NIH-HD42569). JM is supported by the NIH (AG15556, HL69000, CA95739). NW is supported by an American Heart Association Postdoctoral Fellowship.
We thank H. Takeshima for providing the plasmid DNA and PeiHui Lin for her help with the immunocytochemistry experiments.
ABBREVIATIONS
- CICR
Calcium-induced calcium release
- DHPLC
Denaturing high performance liquid chromatography
- HCM
Hypertrophic cardiomyopathy
- JPH2
Junctophilin 2
- MORN
Membrane occupation and recognition nexus
- SR
Sarcoplasmic reticulum
Footnotes
Conflict of Interest: There are no conflicts of interest to disclose.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Maron BJ, Gardin JM, Flack JM, Gidding SS, Kurosaki TT, Bild DE. Prevalence of hypertrophic cardiomyopathy in a general population of young adults. Echocardiographic analysis of 4111 subjects in the CARDIA Study. Coronary Artery Risk Development in (Young) Adults. Circulation. 1995;92:785–9. doi: 10.1161/01.cir.92.4.785. [DOI] [PubMed] [Google Scholar]
- 2.Maron BJ. Hypertrophic cardiomyopathy: A systematic review. JAMA. 2002;287:1308–20. doi: 10.1001/jama.287.10.1308. [DOI] [PubMed] [Google Scholar]
- 3.Thierfelder L, Watkins H, MacRae C, Lamas R, McKenna W, Vosberg HP, et al. Alpha-tropomyosin and cardiac troponin T mutations cause familial hypertrophic cardiomyopathy: a disease of the sarcomere. Cell. 1994;77:701–12. doi: 10.1016/0092-8674(94)90054-x. [DOI] [PubMed] [Google Scholar]
- 4.Geisterfer-Lowrance AA, Kass S, Tanigawa G, Vosberg H, McKenna W, Seidman CE, et al. A molecular basis for familial hypertrophic cardiomyopathy: A beta cardiac myosin heavy chain gene missense mutation. Cell. 1990;62:999–1006. doi: 10.1016/0092-8674(90)90274-i. [DOI] [PubMed] [Google Scholar]
- 5.Watkins H, Conner D, Thierfelder L, Jarcho JA, MacRae C, McKenna WJ, et al. Mutations in the cardiac myosin binding protein-C gene on chromosome 11 cause familial hypertrophic cardiomyopathy. Nat Genet. 1995;11:434–7. doi: 10.1038/ng1295-434. [DOI] [PubMed] [Google Scholar]
- 6.Kimura A, Harada H, Park JE, Nishi H, Satoh M, Takahashi M, et al. Mutations in the cardiac troponin I gene associated with hypertrophic cardiomyopathy. Nat Genet. 1997;16:379–82. doi: 10.1038/ng0897-379. [DOI] [PubMed] [Google Scholar]
- 7.Mogensen J, Klausen IC, Pedersen AK, Egeblad H, Bross P, Kruse TA, et al. Alpha-cardiac actin is a novel disease gene in familial hypertrophic cardiomyopathy. J Clin Invest. 1999;103:R39–R43. doi: 10.1172/JCI6460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Olson TM, Doan TP, Kishimoto NY, Whitby FG, Ackerman MJ, Fananapazir L. Inherited and de novo mutations in the cardiac actin gene cause hypertrophic cardiomyopathy. J Mol Cell Cardiol. 2000;32:1687–94. doi: 10.1006/jmcc.2000.1204. [DOI] [PubMed] [Google Scholar]
- 9.Poetter K, Jiang H, Hassanzadeh S, Master SR, Chang A, Dalakas MC, et al. Mutations in either the essential or regulatory light chains of myosin are associated with a rare myopathy in human heart and skeletal muscle. Nat Genet. 1996;13:63–9. doi: 10.1038/ng0596-63. [DOI] [PubMed] [Google Scholar]
- 10.Van Driest SL, Ommen SR, Tajik AJ, Gersh BJ, Ackerman MJ. Sarcomeric genotyping in hypertrophic cardiomyopathy. Mayo Clin Proc. 2005;80:463–9. doi: 10.1016/S0025-6196(11)63196-0. [DOI] [PubMed] [Google Scholar]
- 11.Van Driest SL, Jaeger MA, Ommen SR, Will ML, Gersh BJ, Tajik AJ, et al. Comprehensive analysis of the beta-myosin heavy chain gene in 389 unrelated patients with hypertrophic cardiomyopathy. J Am Coll Cardiol. 2004;44:602–10. doi: 10.1016/j.jacc.2004.04.039. [DOI] [PubMed] [Google Scholar]
- 12.Van Driest SL, Ellsworth EG, Ommen SR, Tajik AJ, Gersh BJ, Ackerman MJ. Prevalence and spectrum of thin filament mutations in an outpatient referral population with hypertrophic cardiomyopathy. Circulation. 2003;108:445–51. doi: 10.1161/01.CIR.0000080896.52003.DF. [DOI] [PubMed] [Google Scholar]
- 13.Van Driest SL, Vasile VC, Ommen SR, Will ML, Gersh BJ, Nishimura RA, et al. Myosin binding protein C mutations and compound herterozygosity in hypertrophic cardiomyopathy. J Am Coll Cardiol. 2004;44:1903–10. doi: 10.1016/j.jacc.2004.07.045. [DOI] [PubMed] [Google Scholar]
- 14.Theis JL, Martijn Bos J, Bartleson VB, Will ML, Binder J, Vatta M, et al. Echocardiographic-determined septal morphology in Z-disc hypertrophic cardiomyopathy. Biochem Biophys Res Commun. 2006;351:896–902. doi: 10.1016/j.bbrc.2006.10.119. [DOI] [PubMed] [Google Scholar]
- 15.Hayashi T, Arimura T, Itoh-Satoh M, Ueda K, Hohda S, Inagaki N, et al. Tcap gene mutations in hypertrophic cardiomyopathy and dilated cardiomyopathy. J Am Coll Cardiol. 2004;44:2192–201. doi: 10.1016/j.jacc.2004.08.058. [DOI] [PubMed] [Google Scholar]
- 16.Bos JM, Poley RN, Ny M, Tester DJ, Xu X, Vatta M, et al. Genotype-phenotype relationships involving hypertrophic cardiomyopathy-associated mutations in titin, muscle LIM protein, and telethonin. Mol Genet Metab. 2006;88:78–85. doi: 10.1016/j.ymgme.2005.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vasile VC, Will ML, Ommen SR, Edwards WD, Olson TM, Ackerman MJ. Identification of a metavinculin missense mutation, R975W, associated with both hypertrophic and dilated cardiomyopathy. Mol Genet Metab. 2006;87:169–74. doi: 10.1016/j.ymgme.2005.08.006. [DOI] [PubMed] [Google Scholar]
- 18.Vasile VC, Ommen SR, Edwards WD, Ackerman MJ. A missense mutation in a ubiquitously expressed protein, vinculin, confers susceptibility to hypertrophic cardiomyopathy. Biochem Biophys Res Commu. 2006;345:998–1003. doi: 10.1016/j.bbrc.2006.04.151. [DOI] [PubMed] [Google Scholar]
- 19.Arad M, Maron BJ, Gorham JM, Johnson WH, Jr, Saul JP, Perez-Atayde AR, et al. Glycogen storage diseases presenting as hypertrophic cardiomyopathy. N Engl J Med. 2005;352:362–72. doi: 10.1056/NEJMoa033349. [DOI] [PubMed] [Google Scholar]
- 20.Rottbauer W, Baker K, Wo ZG, Mohideen MA, Cantiello HF, Fishman MC. Growth and function of the embryonic heart depend upon the cardiac-specific L-type calcium channel alpha1 subunit. Dev Cell. 2001;1:265–75. doi: 10.1016/s1534-5807(01)00023-5. [DOI] [PubMed] [Google Scholar]
- 21.Molkentin JD, Lu JR, Antos CL, Markham B, Richardson J, Robbins J, et al. A calcineurin-dependent transcriptional pathway for cardiac hypertrophy. Cell. 1998;93:215–28. doi: 10.1016/s0092-8674(00)81573-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lowes BD, Gilbert EM, Abraham WT, Minobe WA, Larrabee P, Ferguson D, et al. Myocardial gene expression in dilated cardiomyopathy treated with beta-blocking agents. N Engl J Med. 2002;346:1357–65. doi: 10.1056/NEJMoa012630. [DOI] [PubMed] [Google Scholar]
- 23.Schmitt JP, Kamisago M, Asahi M, Li GH, Ahmad F, Mende U, et al. Dilated cardiomyopathy and heart failure caused by a mutation in phospholamban. Science. 2003;299:1410–3. doi: 10.1126/science.1081578. [DOI] [PubMed] [Google Scholar]
- 24.Minamisawa S, Sato Y, Tatsuguchi Y, Fujino T, Imamura S-i, Uetsuka Y, et al. Mutation of the phospholamban promoter associated with hypertrophic cardiomyopathy. Biochemical and Biophysical Research Communications. 2003;304:1–4. doi: 10.1016/s0006-291x(03)00526-6. [DOI] [PubMed] [Google Scholar]
- 25.Takeshima H, Komazaki S, Nishi M, Iino M, Kangawa K. Junctophilins: a novel family of junctional membrane complex proteins. Mol Cell. 2000;6:11–22. doi: 10.1016/s1097-2765(00)00003-4. [DOI] [PubMed] [Google Scholar]
- 26.Nishi M, Mizushima A, Nakagawara K, Takeshima H. Characterization of human junctophilin subtype genes. Biochem Biophys Res Commun. 2000;273:920–7. doi: 10.1006/bbrc.2000.3011. [DOI] [PubMed] [Google Scholar]
- 27.Hirata Y, Brotto M, Weisleder N, Chu Y, Lin P, Zhao X, et al. Uncoupling store-operated Ca2+ entry and altered Ca2+ release from sarcoplasmic reticulum through silencing of junctophilin genes. Biophys J. 2006;90:4418–27. doi: 10.1529/biophysj.105.076570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Minamisawa S, Oshikawa J, Takeshima H, Hoshijima M, Wang Y, Chien KR, et al. Junctophilin type 2 is associated with caveolin-3 and is down-regulated in the hypertrophic and dilated cardiomyopathies. Biochem Biophys Res Commun. 2004;325:852–6. doi: 10.1016/j.bbrc.2004.10.107. [DOI] [PubMed] [Google Scholar]
- 29.Ko JK, Ma J. A rapid and efficient PCR-based mutagenesis method applicable to cell physiology study. Am J Physiol Cell Physiol. 2005;288:C1273–8. doi: 10.1152/ajpcell.00517.2004. [DOI] [PubMed] [Google Scholar]
- 30.Seidman JG, Seidman C. The genetic basis for cardiomyopathy: from mutation identification to mechanistic paradigms. Cell. 2001;104:557–67. doi: 10.1016/s0092-8674(01)00242-2. [DOI] [PubMed] [Google Scholar]
- 31.Hayashi T, Arimura T, Ueda K, Shibata H, Hohda S, Takahashi M, et al. Identification and functional analysis of a caveolin-3 mutation associated with familial hypertrophic cardiomyopathy. Biochem Biophys Res Commu. 2004;313:178–84. doi: 10.1016/j.bbrc.2003.11.101. [DOI] [PubMed] [Google Scholar]
- 32.Geier C, Perrot A, Ozcelik C, Binner P, Counsell D, Hoffmann K, et al. Mutations in the human muscle LIM protein gene in families with hypertrophic cardiomyopathy. Circulation. 2003;107:1390–5. doi: 10.1161/01.cir.0000056522.82563.5f. [DOI] [PubMed] [Google Scholar]
- 33.Gomez AM, Valdivia HH, Cheng H, Lederer MR, Santana LF, Cannell MB, et al. Defective excitation-contraction coupling in experimental cardiac hypertrophy and heart failure. Science. 1997;276:800–6. doi: 10.1126/science.276.5313.800. [DOI] [PubMed] [Google Scholar]
- 34.Zheng M, Dilly K, Dos Santos Cruz J, Li M, Gu Y, Ursitti JA, et al. Sarcoplasmic reticulum calcium defect in Ras-induced hypertrophic cardiomyopathy heart. Am J Physiol Heart Circ Physiol. 2004;286:H424–33. doi: 10.1152/ajpheart.00110.2003. [DOI] [PubMed] [Google Scholar]
- 35.Gomez AM, Guatimosim S, Dilly KW, Vassort G, Lederer WJ. Heart failure after myocardial infarction: altered excitation-contraction coupling. Circulation. 2001;104:688–93. doi: 10.1161/hc3201.092285. [DOI] [PubMed] [Google Scholar]
- 36.Song LS, Sobie EA, McCulle S, Lederer WJ, Balke CW, Cheng H. Orphaned ryanodine receptors in the failing heart. Proc Natl Acad Sci U S A. 2006;103:4305–10. doi: 10.1073/pnas.0509324103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xu M, Zhou P, Xu S-M, Liu Y, Feng X, Bai S-H, et al. Intermolecular Failure of L-type Ca2 Channel and Ryanodine Receptor Signaling in Hypertrophy. PLoS Biology. 2007;5:e21. doi: 10.1371/journal.pbio.0050021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yano M, Ikeda Y, Matsuzaki M. Altered intracellular Ca2+ handling in heart failure. J Clin Invest. 2005;115:556–64. doi: 10.1172/JCI24159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Flucher BE, Andrews SB, Daniels MP. Molecular organization of transverse tubule/sarcoplasmic reticulum junctions during development of excitation-contraction coupling in skeletal muscle. Mol Biol Cell. 1994;5:110–18. doi: 10.1091/mbc.5.10.1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Protasi F, Sun XH, Franzini-Armstrong C. Formation and maturation of the calcium release apparatus in developing and adult avian myocardium. Dev Biol. 1996;173:265–78. doi: 10.1006/dbio.1996.0022. [DOI] [PubMed] [Google Scholar]
- 41.Takekura H, Flucher BE, Franzini-Armstrong C. Sequential docking, molecular differentiation, and positioning of T-Tubule/SR junctions in developing mouse skeletal muscle. Dev Biol. 2001;239:204–14. doi: 10.1006/dbio.2001.0437. [DOI] [PubMed] [Google Scholar]
- 42.Kirschner SE, Becker E, Antognozzi M, Kubis HP, Francino A, Navarro-Lopez F, et al. Hypertrophic cardiomyopathy-related beta-myosin mutations cause highly variable calcium sensitivity with functional imbalances among individual muscle cells. Am J Physiol Heart Circ Physiol. 2005;288:H1242–51. doi: 10.1152/ajpheart.00686.2004. [DOI] [PubMed] [Google Scholar]
- 43.Laitinen PJ, Brown KM, Piippo K, Swan H, Devaney JM, Brahmbhatt B, et al. Mutations of the cardiac ryanodine receptor (RyR2) gene in familial polymorphic ventricular tachycardia. Circulation. 2001;103:485–90. doi: 10.1161/01.cir.103.4.485. [DOI] [PubMed] [Google Scholar]
- 44.Priori SG, Napolitano C, Tiso N, Memmi M, Vignati G, Bloise R, et al. Mutations in the cardiac ryanodine receptor gene (hRyR2) underlie catecholaminergic polymorphic ventricular tachycardia. Circulation. 2001;103:196–200. doi: 10.1161/01.cir.103.2.196. [DOI] [PubMed] [Google Scholar]
- 45.Wehrens XH, Lehnart SE, Huang F, Vest JA, Reiken SR, Mohler PJ, et al. FKBP12. 6 deficiency and defective calcium release channel (ryanodine receptor) function linked to exercise-induced sudden cardiac death. Cell. 2003;113:829–40. doi: 10.1016/s0092-8674(03)00434-3. [DOI] [PubMed] [Google Scholar]
- 46.Tester DJ, Spoon DB, Valdivia HH, Makielski JC, Ackerman MJ. Targeted mutational analysis of the RyR2-encoded cardiac ryanodine receptor in sudden unexplained death: a molecular autopsy of 49 medical examiner/coroner’s cases. Mayo Clin Proc. 2004;79:138–4. doi: 10.4065/79.11.1380. [DOI] [PubMed] [Google Scholar]
- 47.George CH, Higgs GV, Lai FA. Ryanodine receptor mutations associated with stress-induced ventricular tachycardia mediate increased calcium release in stimulated cardiomyocytes. Circ Res. 2003;93:531–40. doi: 10.1161/01.RES.0000091335.07574.86. [DOI] [PubMed] [Google Scholar]