Abstract
A number of bacterial species rely on compartmentalization to gain specific functionalities that will provide them with a selective advantage. Here, we will highlight several of these modes of bacterial compartmentalization with an eye towards describing the mechanisms of their formation and their evolutionary origins. Spore formation in Bacillus subtilis, outer membrane biogenesis in Gram-negative bacteria and protein diffusion barriers of Caulobacter crescentus will be used to demonstrate the physical, chemical and compositional remodeling events that lead to compartmentalization. In addition, magnetosomes and carboxysomes will serve as models to examine the interplay between cytoskeletal systems and the subcellular positioning of organelles.
Introduction
The cell is crowded place and subcellular organization is vital to regulating its many metabolic processes [1]. In eukaryotes, such organization is prominently achieved through the compartmentalization of biochemical reactions in various intracellular organelles. By limiting diffusion to a confined space, concentrations of enzymes and substrates can be optimized to promote specific enzymatic reactions. In turn, sequestration of activities within compartments protects the cell from toxic byproducts of such reactions. While historically considered to be simple cells with a low degree of subcellular differentiation, compartmentalization in the form of organelles is also a widespread phenomenon amongst bacterial cells [2]. Unlike the eukaryotic endomembrane system, bacterial species are not equipped with a standard set of organelles. Instead, varying combinations of organelles provide unique capabilities to individual bacterial species. One notable class is the protein-bounded bacterial microcompartments, exemplified by the carbon-fixing carboxysomes of cyanobacteria [3]. Lipid-bounded organelles, including the dazzling varieties of photosynthetic membranes found in heterotrophic bacteria and cyanobacteria, constitute another set of bacterial compartments [4–6]. In addition to organelles, other modes of compartmentalization, such as the creation of spores and protein diffusion barriers to subdivide the cytoplasmic space, have also been described in bacterial cells [7,8••].
Despite the impressive list of bacterial compartments and their cytological characterization, the molecular mechanisms that govern their formation, function and segregation are still a major question in bacterial cell biology. Here, we will focus on recent discoveries on the physical, chemical and compositional remodeling of membranes during compartmentalization as well as the mechanisms leading to the spacing and positioning of organelles within the cell. Those interested in acquiring a more in depth knowledge of this fascinating topic, are encouraged to read several recent review articles on the function, diversity and evolution of bacterial organelles [2,9].
Membrane remodeling
A fundamental aspect of organelle formation in any organism is the remodeling of cellular membranes during the compartmentalization process. Remodeling can be physical in nature such as the bending, migration and fusion of lipid bilayers to produce and stabilize organelles. Chemical remodeling of lipids can also produce distinct compartments within the cell. Additionally, compositional remodeling of membrane domains, through protein targeting and/or exclusion, can subdivide a continuous structure into distinct compartments. These types of membrane remodeling have been described in numerous eukaryotic systems. In contrast, almost nothing is known about the methods and molecules used by bacteria to remodel their lipids into a compartment. Here we highlight three cases where mechanisms and evolution of bacterial membrane remodeling events have been recently elucidated.
Physical remodeling: spore formation
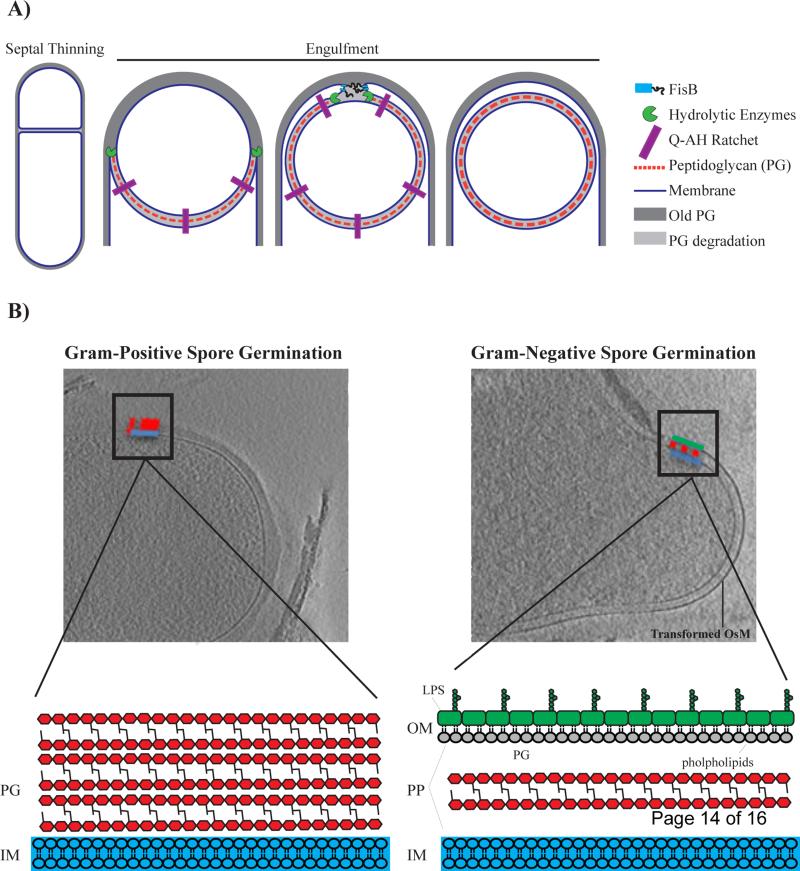
Perhaps the most thoroughly studied example of membrane remodeling in bacteria is the engulfment of forespore during the sporulation process of Bacillus subtilis. When growth is challenged by harsh conditions, B. subtilis cells undergo a unique developmental program to form a highly durable and dormant endospore. During the early stages of sporulation, an asymmetric division event creates a larger “mother” cell that proceeds to engulf the smaller “forespore” cell to form an internal, double-membraned compartment (Figure 1a). A number of mechanisms have been implicated to drive the mother cell membrane around the forespore and have been recently reviewed [10]. These include cell wall synthesis, cell wall degradation and specific protein interaction between SpoIIQ and SpoIIIAH that bridge across the mother and forespore membrane to prevent membrane retraction (Figure 1a)[10–13].
Figure 1.
A) Mechanisms of membrane remodeling during the different stages of engulfment. Engulfment initiates with degradation of septal peptidoglycan, commonly referred to as septal thinning. Peptidoglycan synthesis, peptidoglycan degradation and a specific “ratchet-like” mechanism that is mediated by proteins SpoIIQ and SpoIIIAH (QAH) are all factors that have been shown to be important for driving the mother cell membrane around the forespore. To be released into the mother cell, migrating membranes meet at the cell pole and undergo a fission event that is mediated by FisB. Note that the outer spore membrane (OsM) is derived from the cytoplasmic membrane of the mother cell. B) Spore germination of Gram-positive Bacillus subtilis (left) as compared to Gram-negative Acetonema longum (right). Upon germination, Gram-positive Bacillus subtilis (left) sheds its OsM whereas Gram-negative Acetonema longum (right) retains its OsM. Furthermore, A. longum transforms its OsM into a canonical Gram-negative outer membrane (OM). Cell envelope of Gram-negative vs. Gram-positive bacteria are very different. Gram-positive bacteria have a thick cell wall that is made up of multiple layers of peptidoglycan (PG) that surrounds an inner membrane (IM). On the other hand, Gram-negative bacteria maintain a thin layer of cell wall in-between two membranes. The OM of Gram-negative bacteria is compositionally different from the IM and creates a compartment, the periplasm (PP), that chemically distinct from the cytoplasm. The OM is an asymmetric bilayer with lipopolysaccharide (LPS) distributed in the outer leaflet and phospholipids distributed in the inner leaflet. Copyright (2013) Wiley. Used with permission from [Tocheva et al., Mol Micro, 2013].
The final step of the engulfment process is the joining of the two ends of the migrating membranes to create a completely internalized endospore. Genetic analysis had implicated one protein, SpoIIIE, in membrane fission. However, spoIIIE null mutants also have defects in DNA translocation and septum morphology at earlier stages of sporulation raising the possibility that impaired membrane fission may be an indirect effect [14–16••]. By examining a set of mother cell genes that are expressed at the early stages of engulfment that when mutated, fail to release the forespore into the mother cell cytoplasm, Doan and colleagues recently identified fission protein B (FisB) as a direct catalyst of membrane fission in B. subtilis [16••] (Figure 1a). Fluorescent protein fusions to FisB localize primarily to the edges of the migrating mother cell membrane that serve as the sites of membrane fission. Furthermore, purified FisB promotes membrane fusion in vitro via a specific interaction involving the extracellular domain of FisB and cardiolipin, an anionic phospholipid that is enriched in highly curved membranes including the forespore of B. subtilis [16••–18]. The authors propose that FisB may concentrate cardiolipin and create localized defects in the lipid bilayer that would favor the fission reaction. While these results highlight the importance of FisB in membrane fission, a small percentage of fisB mutants eventually complete engulfment, suggesting that alternative mechanisms may function at a slower rate to complete engulfment [16]. A recent report identified core set of genes shared amongst sporulating bacteria, including three novel genes involved in endospore formation [19]. This genomic signature may be a useful tool to identify additional mechanisms for membrane remodeling.
Chemical remodeling: Gram-negative outer membrane
Membrane remodeling is not limited to the physical movement and bending of lipid bilayers. Often, extensive modification of the chemical composition of cellular membranes is required for compartment formation and function. In bacteria, the construction of the Gram-negative outer membrane (OM) provides a model for understanding the mechanisms of chemical remodeling in biological membranes. As should be familiar to most students of biology, the Gram stain, developed by Hans Christian Gram in 1884, is a histological method for identifying and classifying bacterial species. Gram-positive bacteria have a single cell membrane that is surrounded by a thick, multi-layered cell wall. Gram-negative bacteria have two membranes separated by a thin cell wall [20] (Figure 1b). The OM displays a number of characteristics that define it as a bacterial compartment. It is an asymmetric lipid bilayer with lipopolysaccharide (LPS), a glycolipid, on its outer leaflet and phospholipids on its inner leaflet. Beta-barrel proteins in the OM form pores that allow for non-specific exchange of small molecules with the extracellular environment while limiting diffusion of many cytoplasmically secreted proteins. Furthermore, the tight packing of LPS on the outer leaflet provides an impermeable barrier to small hydrophobic and hydrophilic molecules that can be harmful to the cell [20]. The OM does not just function as barrier but also creates a chemically distinct periplasmic compartment where important processes, such as certain types of respiration, can occur.
While tremendous progress has been made in defining the chemical nature of the OM and uncovering the molecular mechanisms of its biogenesis [21,22], the evolutionary origins of this structure have remained mysterious. A recent study has searched for the answer to this question by examining the curious mode by which some bacterial cells produce and germinate spores [23••]. Phylogenetically, most endospore forming bacteria fall under the Bacilli and Clostridia classes of the Gram-positive Firmicutes phylum [24]. Interestingly, some members of the Clostridia, such as Acetonema longum, can form spores but are decidedly Gram-negative in their vegetative state, as they possess an inner membrane and an OM that contains LPS. Through high-resolution electron cryo-tomographic (ECT) imaging of A. longum and B. subtilis cells at different stages of sporulation and spore germination, Tocheva and colleagues were able to show that the inner membrane of the mother cell engulfs the forespore and forms the outer spore membrane [23••,25]. During germination in A. longum, however, this membrane acquires the morphological and chemical signatures of an OM, including LPS (Figure 1b). Sequencing of the A. longum genome revealed the presence of homologs of genes known to encode for OM proteins in Gram-negative bacteria as well as those involved in sporulation in Gram-positives. Phylogenetic analysis showed that the OM proteins of A. longum do not cluster with those of other Gram-negatives whereas its sporulation proteins group with those of other Gram-positives. A parsimonious explanation for these results is that A. longum did not acquire the ability to produce an OM due to a recent horizontal gene transfer event, in which case its OM proteins would have been in tighter phylogenetic groups with homologs from Gram-negative organisms. Similarly, sporulation also seems to be an ancient characteristic of this organism. These findings lead to the tantalizing hypothesis that ancestors of Gram-positive and Gram-negative bacteria were able to link spore germination to construction of an OM. Subsequently, the majority of Gram-positives lost the ability to form an OM while the loss of sporulation genes gave rise to present-day Gram-negative organisms.
Compositional remodeling: protein diffusion barriers
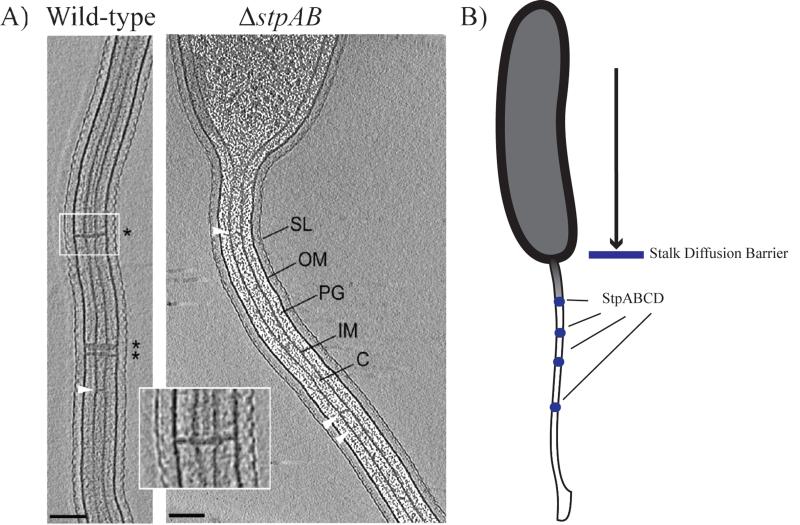
Diffusion gradients are used by cells to control the timing and location of essential cellular processes [26]. Protein[EC1] diffusion barriers had only been described as a feature of eukaryotic cells until their recent discovery by Schlimpert and colleagues in the Gram-negative bacterium, Caulobacter crescentus [8••,27]. At a specific point during its cell cycle, C. crescentus physically remodels its cell envelope to form a polar stalk that functions as a holdfast for attachment to surfaces. During phosphorus limiting growth periods the stalk undergoes a dramatic elongation leading to the hypothesis that it may have a role in phosphorus uptake from the environment. Even though the stalk is a protrusion of the cell envelope, Schlimpert and colleagues found that periplasmic proteins do not move between the stalk and the cell body. Furthermore, they found that the stalk itself was compartmentalized into several segments that restricted protein movement. Early electron [EC2] microscopic examination had found that curious cross band structures were spread through out the C. crescentus stalk. Using a combination of genetic and biochemical approaches, the authors identified a protein complex consisting of four proteins (StpA, StpB, StpC and StpD) that localize to these conspicuous crossband structures. Deletions of the genes encoding the Stp factors result in a loss of cross band structures visualized by ECT and absence of compartmentalization within the stalk (Figure 2a). By preventing the accumulation of newly synthesized proteins into the stalk, these cross band structures essentially lead to a compositional remodeling of the compartment. One consequence of the loss of stalk compartmentalization is that proteins synthesized after stalk formation are no longer restricted to the cell body and are less concentrated in locales where their activity might be needed. This is especially of concern in phosphate starvation conditions in which the stalk elongates to the point that its surface area exceeds that of the cell body. In agreement with this model, the authors demonstrate that mutants missing the stp genes, take longer to accumulate proteins within their cell body and are outcompeted by wild-type cells in co-culturing experiments. Thus, in the case of the C. crescentus stalk, compositional remodeling partially acts to prevent the dilution of important cellular proteins (Figure 2b). Additionally, the authors hypothesize that the compositional remodeling of the stalk may also allow for localized activities that may be used to shuttle phosphate into the cell.
Figure 2.
A) Electron cryotomography images of wild-type Caulobacter crescentus cells containing stalk diffusion barriers (* and inset). ΔstpAB cells lack these structures. White arrows indicate unidentified structures that span the interior of the stalk (SL= S layer, OM= outer membrane, PG= peptidoglycan, IM= inner membrane, C= cytoplasm). Scale bar 100 nm. Reprinted from [Schlimpert et al., Cell, 2011] with permission from Elsevier. B) Stalk diffusion barriers: Comprising a protein complex of StpA, StpB, StpC and StpD (blue circles), diffusion barriers limit soluble and membrane protein diffusion into the stalk of C. crescentus.
Organelle positioning
The subcellular positioning of compartments plays a key role in the function of many bacterial organelles. Recent studies have uncovered some of the molecular mechanisms that lead to the organization and positioning of two unique bacterial organelles, the magnetosome compartments of magnetotactic bacteria and the carboxysomes of cyanobacteria [28].
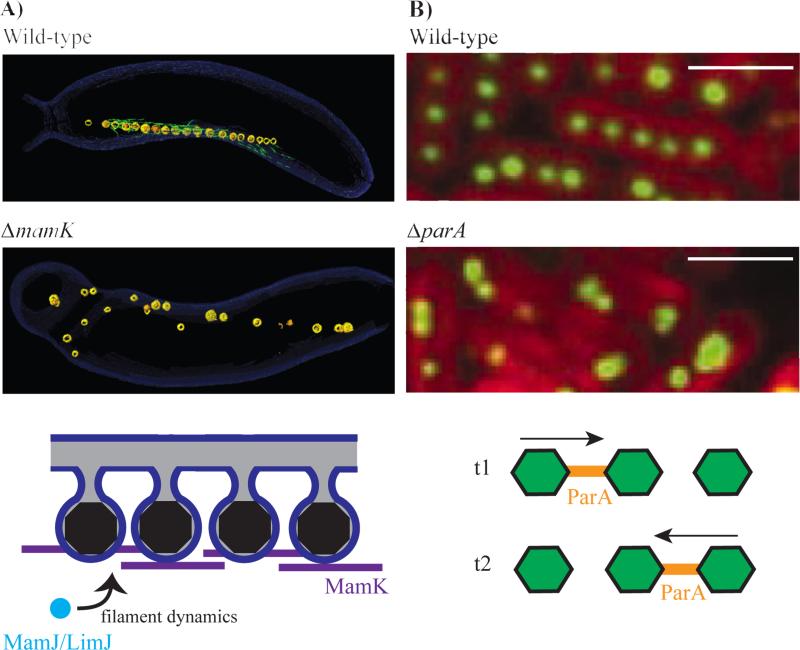
Compartmentalized through an invagination of the inner cell membrane, the magnetosome creates a controlled chemical environment optimal for formation and maturation of magnetite or greigite both of which are iron-based magnetic crystals. Magnetosome formation requires multiple steps utilizing genes that form membrane invaginations, localize proteins, nucleate crystals and align structures into a chain [29–34]. The end product of these processes is a well-aligned magnetosome chain (Figure 3a), which is thought to provide navigational capability towards preferred oxygen concentrations in a stratified water column, a process termed magnetoaerotaxis [35].
Figure 3.
Organization of bacterial organelles by cytoskeletal filaments. A) The bacterial actin protein MamK is responsible for alignment of magnetosomes into a chain in Magentotactic Bacteria. Reconstructed electron cryotomography images of wild-type Magnetospirillum magneticum AMB-1 with an aligned magnetosome chain (top panel) and ΔmamK cells with disorganized magnetosomes (middle panel). The magnetosome (yellow) chain contains iron-based crystals (orange) and is flanked by filamentous structures (green) that disappear in the ΔmamK mutant. MamK dynamic filament behavior is influenced by the presence of MamJ and LimJ (bottome panel). The precise mechanisms that govern this process remain elusive. From [Komeili et al. Sceince, 2006]. Reprinted with permission from AAAS. B) Alignment and segregation of carboxysomes in cyanobacteria is dependent on ParA, another bacterial cytoskeletal protein. Carboxysomes are evenly distributed throughout the cell in wild-type Synechococcus elongatus PCC 7942 (top panel), but not in a mutant lacking parA (middle panel). Rubisco protein (RbcL) fused to YFP indicates the localization of carboxysomes (green) and thylakoid membrane fluorescence is shown in red. ParA oscillates from pole to pole and is found distributed in between carboxysomes (bottom panel). T1 and T2 represent time points that that are roughly 30 minutes apart. From [Savage et al., Science, 2010]. Reprinted with permission from AAAS. Scale bar 2 μm.
Subcellular organization of magnetosomes into a chain is dependent on MamK, a bacterial actin homolog that is present in all magnetotactic bacteria. High resolution imaging of Magnetospirillum magneticum AMB-1 by ECT revealed that organized chains of magnetosomes are flanked by filaments that disappear in a mamK deletion mutant (Figure 3a)[34]. Much like other actin-like proteins, MamK forms filaments that are dynamic in vivo [36•] and in vitro [37,38] in a manner that is dependent on the ATPase activity of the protein. The dynamic behavior of MamK in vivo also relies on the redundant action of acidic proteins MamJ and LimJ (Figure 3a)[EC3][36•]. In their absence, MamK filaments are static as seen in a fluorescence recovery after photobleaching assay. In addition, the loss of mamJ and limJ results in multiple gaps within the magnetosome chain and a clustering of putative MamK filaments within these spaces. One interpretation of these results is that MamK dynamics may be acting to pull magnetosomes together. Alternatively, the loss of MamJ and LimJ could change the localization of MamK in the cell leading to altered dynamics and an inability to form a coherent chain. [EC4]
Despite the clear advances in understanding the behavior and properties of MamK as an actin-like protein, the specific mechanism by which it acts on the magnetosome chain remains unknown. The phenotypes of the mamK deletion strains are consistent with its action in either establishing or maintaining the magnetosome chain. Furthermore, in a related species, Magnetospirillum gryphiswaldense MSR-1, deletion of mamK results in ectopic chain localization and failure of partitioning the chain between daughter cells [39,40]. It has even been suggested that MamK acts to link physical forces on the magnetosome chain into changes in cell motility [41]. Regardless of the specific mechanism of its action, MamK is clearly important for proper navigation in magnetic fields [42]. A recent study using an agar plate magnetoaerotaxis assay found that mamK mutants had a significantly wider path around a magnetic field as compared to the wild type strain. In the environment, such a difference could translate into a longer travel period, and subsequently higher energy expenditure, for reaching a desired locale.
Similar to magnetosomes, the carboxysome organelles of cyanobacteria are also arranged linearly within a cell with a consistent spacing pattern. This arrangement relies on the action of the cytoskeletal protein, ParA, without which carboxysomes are unequally distributed amongst daughter cells (Figure 3b)[43••]. ParA filaments are also highly dynamic within the cell and undergo pole-to-pole oscillations that may be important in carboxysome positioning (Figure 3b). Given its crucial role in carbon fixation, the failure to acquire carboxysomes results in a significantly longer doubling time. ParA and its homologs belong to the Walker type family of ATPases and are often associated with segregation of plasmid or chromosomal DNA in diverse bacterial species. However, recent work has shown that a ParA homolog, PpfA, can also be used to segregate chemotaxis protein clusters during cell division in Rhodobacter sphaeroides [44]. In this case, nonspecific interactions of PpfA with DNA as well as specific interactions with its chemotaxis protein cargo provide the means for separating the protein complexes [45]. These findings leave open the possibility that in cyanobacteria, carboxysomes interact with chromosomes and that ParA is primarily directing the localization and segregation of DNA. However, deletion of parA does not affect the spacing of chromosomal DNA in cyanobacteria while disrupting the organization of carboxysomes [46]. Thus, while DNAParA interactions may be important, specific interactions of this cytoskeletal protein with the carboxysome are likely needed for the proper positioning of this bacterial organelle.
Conclusion
The discovery of cytoskeletal elements, organelles and means of sophisticated spatio-temporal regulation in bacteria have transformed the simplistic views of this complex and metabolically diverse domain of life. The most fundamental roadblock in understanding compartmentalization comprehensively in bacteria is the scarcity of reliable model systems that can help to illuminate specific facets of the organelle formation process. As such, isolated examples of compartmentalization in genetically tractable organisms have carried the bulk of the weight in defining the mechanistic basis of membrane remodeling, protein localization and organelle segregation in bacteria. The diversity of these organelles and the specifications in their morphology and function makes it unlikely that a universal mechanism for compartmentalization exists amongst all bacteria. For instance, membrane remodeling in sporulation relies in part on the use of peptidoglycan as a guide while no such cues will be available during magnetosome formation. Likewise, while magnetosomes and carboxysomes rely on cytoskeletal elements for their subcellular positioning, the proteins involved are not related hinting at distinct molecular activities in these two systems. Clearly, an expansion of available model organisms and a more thorough cataloging of bacterial organelles are needed to truly revolutionize this field. If successful, such endeavors will open new avenues of research in microbiology and uncover novel mechanisms of cell organization and compartmentalization.
Highlights.
Bacterial cells contain lipid- and protein-bounded organelles.
FisB mediates membrane fission during spore formation in Bacillus subtilis.
Outer membrane of Gram-negatives may have originated in a sporulating bacterium.
A protein complex forms a diffusion barrier in the stalks of Caulobacter crescentus.
Magnetosomes and carboxysomes are positioned within the cell by cytoskeletal proteins.
Acknowledgements
Elias Cornejo is supported by award number R25GM090110 from the National Institute of General Medical Sciences and the National Science Foundation under Grant No. DGE 110640. Nicole Abreu is funded through the American Society for Microbiology's Robert D. Watkins Fellowship, UC Berkeley's Chancellors Fellowship and Mentored Research Awards. AK is supported by grants from the Office of Naval Research (N000141310421) and the National Institutes of Health (R01GM084122).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Spitzer J, Poolman B. How crowded is the prokaryotic cytoplasm? FEBS Lett. 2013;587:2094–2098. doi: 10.1016/j.febslet.2013.05.051. [DOI] [PubMed] [Google Scholar]
- 2.Murat D, Byrne M, Komeili A. Cell biology of prokaryotic organelles. Cold Spring Harb. Perspect. Biol. 2010;2:a000422. doi: 10.1101/cshperspect.a000422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kerfeld C a, Heinhorst S, Cannon GC. Bacterial microcompartments. Annu. Rev. Microbiol. 2010;64:391–408. doi: 10.1146/annurev.micro.112408.134211. [DOI] [PubMed] [Google Scholar]
- 4.Hsin J, Chandler DE, Gumbart J, Harrison CB, Sener M, Strumpfer J, Schulten K. Self-assembly of photosynthetic membranes. Chemphyschem. 2010;11:1154–9. doi: 10.1002/cphc.200900911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Oostergetel GT, van Amerongen H, Boekema EJ. The chlorosome: a prototype for efficient light harvesting in photosynthesis. Photosynth. Res. 2010;104:245–55. doi: 10.1007/s11120-010-9533-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vothknecht UC, Westhoff P. Biogenesis and origin of thylakoid membranes. Biochim. Biophys. Acta. 2001;1541:91–101. doi: 10.1016/s0167-4889(01)00153-7. [DOI] [PubMed] [Google Scholar]
- 7.Errington J. Regulation of endospore formation in Bacillus subtilis. Nat. Rev. Microbiol. 2003;1:117–26. doi: 10.1038/nrmicro750. [DOI] [PubMed] [Google Scholar]
- 8••.Schlimpert S, Klein E a, Briegel A, Hughes V, Kahnt J, Bolte K, Maier UG, Brun YV, Jensen GJ, Gitai Z, et al. General protein diffusion barriers create compartments within bacterial cells. Cell. 2012;151:1270–82. doi: 10.1016/j.cell.2012.10.046. [Large protein complexes were discovered that limit protein diffusion, providing a fitness advantage in Caulobacter crescentus cells.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Diekmann Y, Pereira-Leal JB. Evolution of intracellular compartmentalization. Biochem. J. 2013;449:319–31. doi: 10.1042/BJ20120957. [DOI] [PubMed] [Google Scholar]
- 10.Higgins D, Dworkin J. Recent progress in Bacillus subtilis sporulation. FEMS Microbiol. Rev. 2012;36:131–48. doi: 10.1111/j.1574-6976.2011.00310.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Abanes-De Mello A, Sun Y, Aung S, Pogliano K. A cytoskeleton-like role for the bacterial cell wall during engulfment of the Bacillus subtilis forespore. Genes Dev. 2002;16:3253–64. doi: 10.1101/gad.1039902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Broder DH, Pogliano K. Forespore engulfment mediated by a ratchet-like mechanism. Cell. 2006;126:917–28. doi: 10.1016/j.cell.2006.06.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Meyer P, Gutierrez J, Pogliano K, Dworkin J. Cell wall synthesis is necessary for membrane dynamics during sporulation of Bacillus subtilis. Mol. Microbiol. 2010;76:956–70. doi: 10.1111/j.1365-2958.2010.07155.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sharp MD, Pogliano K. An in vivo membrane fusion assay implicates SpoIIIE in the final stages of engulfment during Bacillus subtilis sporulation. Proc. Natl. Acad. Sci. U. S. A. 1999;96:14553–14558. doi: 10.1073/pnas.96.25.14553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sharp MD, Pogliano K. The Membrane Domain of SpoIIIE Is Required for Membrane Fusion during Bacillus subtilis Sporulation. J. Bacteriol. 2003;185:2005–2008. doi: 10.1128/JB.185.6.2005-2008.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16••.Doan T, Coleman J, Marquis K a, Meeske AJ, Burton BM, Karatekin E, Rudner DZ. FisB mediates membrane fission during sporulation in Bacillus subtilis. Genes Dev. 2013;27:322–34. doi: 10.1101/gad.209049.112. [Identifies the first protein that catalyzes membrane fission, FisB, in Bacillus subtilis. Show by fluorescence microscopy that FisB-GFP and FisB-YFP localize to the site of membrane fission. Use biochemical methods to show that FisB fuses membranes together and that this action is dependent on cardiolipin.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kawai F, Shoda M, Harashima R, Sadaie Y, Hara H, Matsumoto K. Cardiolipin Domains in Bacillus subtilis Marburg Membranes. J. Bacteriol. 2004;186:1475–1483. doi: 10.1128/JB.186.5.1475-1483.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Matsumoto K, Kusaka J, Nishibori A, Hara H. Lipid domains in bacterial membranes. Mol. Microbiol. 2006;61:1110–7. doi: 10.1111/j.1365-2958.2006.05317.x. [DOI] [PubMed] [Google Scholar]
- 19.Abecasis AB, Serrano M, Alves R, Quintais L, Pereira-Leal JB, Henriques AO. A genomic signature and the identification of new sporulation genes. J. Bacteriol. 2013;195:2101–15. doi: 10.1128/JB.02110-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ruiz N, Kahne D, Silhavy TJ. Transport of lipopolysaccharide across the cell envelope: the long road of discovery. Nat. Rev. Microbiol. 2009;7:677–83. doi: 10.1038/nrmicro2184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rigel NW, Silhavy TJ. Making a beta-barrel: assembly of outer membrane proteins in Gram-negative bacteria. Curr. Opin. Microbiol. 2012;15:189–93. doi: 10.1016/j.mib.2011.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Okuda S, Freinkman E, Kahne D. Cytoplasmic ATP hydrolysis powers transport of lipopolysaccharide across the periplasm in E. coli. Science. 2012;338:1214–7. doi: 10.1126/science.1228984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23••.Tocheva EI, Matson EG, Morris DM, Moussavi F, Leadbetter JR, Jensen GJ. Peptidoglycan remodeling and conversion of an inner membrane into an outer membrane during sporulation. Cell. 2011;146:799–812. doi: 10.1016/j.cell.2011.07.029. [Electron cryotomography on a rare, Gram-negative endospore forming bacterium, Acetonema longum, shows that the cytoplasmic membrane is transformed into an LPS-containing outer membrane during endospore formation.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McKenney PT, Driks A, Eichenberger P. The Bacillus subtilis endospore: assembly and functions of the multilayered coat. Nat. Rev. Microbiol. 2013;11:33–44. doi: 10.1038/nrmicro2921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tocheva EI, López-Garrido J, Hughes HV, Fredlund J, Kuru E, Vannieuwenhze MS, Brun YV, Pogliano K, Jensen GJ. Peptidoglycan transformations during Bacillus subtilis sporulation. Mol. Microbiol. 2013;88:673–86. doi: 10.1111/mmi.12201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Endres RG. Intracellular chemical gradients: morphing principle in bacteria. [Internet]. BMC Biophys. 2012;5:18. doi: 10.1186/2046-1682-5-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Caudron F, Barral Y. Septins and the lateral compartmentalization of eukaryotic membranes. Dev. Cell. 2009;16:493–506. doi: 10.1016/j.devcel.2009.04.003. [DOI] [PubMed] [Google Scholar]
- 28.Komeili A. Molecular mechanisms of compartmentalization and biomineralization in magnetotactic bacteria. FEMS Microbiol. Rev. 2012;36:232–55. doi: 10.1111/j.1574-6976.2011.00315.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Murat D, Quinlan A, Vali H, Komeili A. Comprehensive genetic dissection of the magnetosome gene island reveals the step-wise assembly of a prokaryotic organelle. Proc. Natl. Acad. Sci. U. S. A. 2010;107:5593–8. doi: 10.1073/pnas.0914439107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lohsse A, Ullrich S, Katzmann E, Borg S, Wanner G, Richter M, Voigt B, Schweder T, Schüler D. Functional analysis of the magnetosome island in Magnetospirillum gryphiswaldense: the mamAB operon is sufficient for magnetite biomineralization. PLoS One. 2011;6:e25561. doi: 10.1371/journal.pone.0025561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Quinlan A, Murat D, Vali H, Komeili A. The HtrA/DegP family protease MamE is a bifunctional protein with roles in magnetosome protein localization and magnetite biomineralization. Mol. Microbiol. 2011;80:1075–87. doi: 10.1111/j.1365-2958.2011.07631.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Uebe R, Junge K, Henn V, Poxleitner G, Katzmann E, Plitzko JM, Zarivach R, Kasama T, Wanner G, Pósfai M, et al. The cation diffusion facilitator proteins MamB and MamM of Magnetospirillum gryphiswaldense have distinct and complex functions, and are involved in magnetite biomineralization and magnetosome membrane assembly. Mol. Microbiol. 2011;82:818–35. doi: 10.1111/j.1365-2958.2011.07863.x. [DOI] [PubMed] [Google Scholar]
- 33.Komeili A, Li Z, Newman DK, Jensen GJ. Magnetosomes are cell membrane invaginations organized by the actin-like protein MamK. Science. 2006;311:242–5. doi: 10.1126/science.1123231. [DOI] [PubMed] [Google Scholar]
- 34.Murat D, Falahati V, Bertinetti L, Csencsits R, Körnig A, Downing K, Faivre D, Komeili A. The magnetosome membrane protein, MmsF, is a major regulator of magnetite biomineralization in Magnetospirillum magneticum AMB-1. Mol. Microbiol. 2012;85:684–99. doi: 10.1111/j.1365-2958.2012.08132.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Frankel RB, Bazylinski D a, Johnson MS, Taylor BL. Magneto-aerotaxis in marine coccoid bacteria. Biophys. J. 1997;73:994–1000. doi: 10.1016/S0006-3495(97)78132-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36•.Draper O, Byrne ME, Li Z, Keyhani S, Barrozo JC, Jensen G, Komeili A. MamK, a bacterial actin, forms dynamic filaments in vivo that are regulated by the acidic proteins MamJ and LimJ. Mol. Microbiol. 2011;82:342–54. doi: 10.1111/j.1365-2958.2011.07815.x. [In this research the dynamic behavior of the bacterial actin homolog MamK is investigated and genes within the Magnetosome Island are required for filament turnover.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ozyamak E, Kollman J, Agard D a, Komeili A. The bacterial actin MamK: in vitro assembly behavior and filament architecture. J. Biol. Chem. 2013;288:4265–77. doi: 10.1074/jbc.M112.417030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sonkaria S, Fuentes G, Verma C, Narang R, Khare V, Fischer A, Faivre D. Insight into the assembly properties and functional organisation of the magnetotactic bacterial actin-like homolog, MamK. PLoS One. 2012;7:e34189. doi: 10.1371/journal.pone.0034189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Katzmann E, Müller FD, Lang C, Messerer M, Winklhofer M, Plitzko JM, Schüler D. Magnetosome chains are recruited to cellular division sites and split by asymmetric septation. Mol. Microbiol. 2011;82:1316–29. doi: 10.1111/j.1365-2958.2011.07874.x. [DOI] [PubMed] [Google Scholar]
- 40.Katzmann E, Scheffel A, Gruska M, Plitzko JM, Schüler D. Loss of the actin-like protein MamK has pleiotropic effects on magnetosome formation and chain assembly in Magnetospirillum gryphiswaldense. Mol. Microbiol. 2010;77:208–24. doi: 10.1111/j.1365-2958.2010.07202.x. [DOI] [PubMed] [Google Scholar]
- 41.Philippe N, Wu L-F. An MCP-like protein interacts with the MamK cytoskeleton and is involved in magnetotaxis in Magnetospirillum magneticum AMB-1. J. Mol. Biol. 2010;400:309–22. doi: 10.1016/j.jmb.2010.05.011. [DOI] [PubMed] [Google Scholar]
- 42.Sakaguchi S, Taoka A, Fukumori Y. Analysis of Magnetotactic Behavior by Swimming Assay. Biosci. Biotechnol. Biochem. 2013;77:940–947. doi: 10.1271/bbb.120880. [DOI] [PubMed] [Google Scholar]
- 43••.Savage DF, Afonso B, Chen AH, Silver P a. Spatially ordered dynamics of the bacterial carbon fixation machinery. Science. 2010;327:1258–61. doi: 10.1126/science.1186090. [Savage and colleagues demonstrate that a cytoskeletal protein ParA is important for aligning carboxysomes in Synechococcus elongatus.] [DOI] [PubMed] [Google Scholar]
- 44.Roberts MAJ, Wadhams GH, Had KA, Tickner S, Armitage JP. ParA-like protein uses nonspeci fi c chromosomal DNA binding to partition protein complexes. Proc. Natl. Acad. Sci. U. S. A. 2012;109:6698–6703. doi: 10.1073/pnas.1114000109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Thompson SR, Wadhams GH, Armitage JP. The positioning of cytoplasmic protein clusters in bacteria. Proc. Natl. Acad. Sci. U. S. A. 2006;103:8209–14. doi: 10.1073/pnas.0600919103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jain IH, Vijayan V, Shea EKO. Spatial ordering of chromosomes enhances the fidelity of chromosome partitioning in cyanobacteria. Proc. Natl. Acad. Sci. U. S. A. 2012;109:13638–43. doi: 10.1073/pnas.1211144109. [DOI] [PMC free article] [PubMed] [Google Scholar]