Abstract
Alzheimer’s Disease (AD) is the most common neurodegenerative disease and with Americans’ increasing longevity it is becoming an epidemic. There are currently no effective treatments for this disorder. Abnormalities of Tau track more closely with cognitive decline than the most studied therapeutic target in AD, amyloid-beta, but the optimal strategy for targeting Tau has not yet been identified. Based on considerable preclinical data from AD models, we hypothesize that interactions between Tau and the Src-family tyrosine kinase, Fyn, are pathogenic in AD. Genetically reducing either Tau or Fyn is protective in AD mouse models, and a dominant negative fragment of Tau that alters Fyn localization is also protective. Here, we describe a new AlphaScreen assay and a live-cell BRET assay using a novel BRET pair for quantifying the Tau–Fyn interaction. We used these assays to map the binding site on Tau for Fyn to the 5th and 6th PXXP motifs, to show that AD-associated phosphorylation at MARK sites increase the affinity of the Tau–Fyn interaction, and to identify Tau–Fyn interaction inhibitors by HTS. This screen has identified a variety of chemically tractable hits, suggesting that the Tau–Fyn interaction may represent a good drug target for AD.
Keywords: tau, fyn, Alzheimer, sh3, protein-protein interaction
Introduction
The microtubule associated protein Tau is a major pathological marker of Alzheimer’s disease (AD) and plays an important role in facilitating deficits in models of AD. More specifically, Tau reduction has dramatic protective effects in many AD models1–3. Also, Tau reduction is effective in pharmacological and genetic models of epilepsy1,3–5, and Tau reduction remains effective when administered in adulthood6. These studies point to Tau as a key player in mediating AD-related neuronal hyperexcitability, but how to target this aberrant excitability has not been clear. Because Tau has a major role as a scaffolding protein, we looked to Tau interacting partners for therapeutic targets.
Several studies aroused interest in the non-receptor tyrosine kinase Fyn, which interacts with polyproline helices in Tau through its SH3 domain7, and, interestingly, also regulates seizure susceptibility8,9. Tau reduction is protective in a Fyn-dependent model of AD2, but the mechanism for this protection has been unclear until recently. Studies now show that Tau targets Fyn to the dendrites, where Fyn facilitates N-methyl-D-aspartate (NMDA) receptor–mediated dysfunction and aberrant Tau phosphorylation1,10. These studies and others revealed a critical role of dysfunctional, dendritically localized Tau in AD and AD models (reviewed in 11). How might we target this dysfunction? As Fyn kinase activity is increased in AD10, it is logical to consider Fyn kinase inhibitors as a therapeutic possibility. Indeed, this strategy is currently being pursued12. However, Fyn-deficient mice have drastic learning and memory deficits13, indicating that an alternative approach may be needed to capture the beneficial effects of Tau reduction as they relate to Fyn. One alternative approach is suggested by a study demonstrating that overexpression of a dominant negative form of Tau that inhibits endogenous Tau–Fyn interactions was sufficient to prevent behavioral deficits in an AD model1. This study suggests that targeting the Tau–Fyn interaction may be sufficient to capture the beneficial effects of Tau reduction. The Tau–Fyn interaction depends on Fyn’s SH3 domain7,14,15. Because of this, we have conducted a high throughput screen for small molecule inhibitors of the interaction between full-length Tau and the SH3 domain of Fyn.
Materials and Methods
Protein Purification
Bacterial codon-optimized human 4R2N Tau (Life Technologies) was tagged on the N-terminus with a His tag and on the C-terminus with a Strep tag. The human Fyn SH3 domain was tagged on the N-terminus with a GST tag and on the C-terminus with a Strep tag. The constructs were cloned into the pPR-IBA1 vector (IBA Life Sciences) and transformed into BL21(DE3)-RIPL E. Coli (Agilent Technologies). Colonies were picked from freshly streaked LB Agar plates containing 100 μg/mL carbenicillin (Fisher – all chemicals were obtained from Fisher unless otherwise noted) and 25 μg/mL Chloramphenicol into 50 mL LB Broth starter cultures containing 100 μg/mL carbenicillin. Starter cultures were grown at 37°C for 10 hours, then diluted 100x into 2L of LB Broth split between 5 2L flasks containing 100 μg/mL carbenicillin, and growth was allowed to continue at 37°C. Bacteria were then closely monitored, and induced at an OD600 of 0.6 to 1.0 (usually about 2–4 hours of growth) with 1 mM IPTG. PMSF was also added at this stage to a final concentration of 1mM, which we found increased the yield of Tau. Protein expression was allowed to continue for 4 hours at 37°C. Bacteria were then pelleted and immediately purified. Both proteins were bulk purified with 5 mL of Strep-Tactin® Superflow® high capacity resin (IBA Life Sciences) according to the manufacturer’s instructions. The GST–Fyn SH3–strep protein was eluted with 50 mM Tris pH 7.4, 300 mM NaCl, 10 mM TCEP (to reduce disulfide bonds, which we found persisted in purified Tau, even on SDS-page gels with β-mercaptoethanol without this buffer component), 0.2% KATHON™ CG/ICP as an anti-microbial reagent (as sodium azide is not compatible with AlphaScreen), 10 mM D-desthiobiotin, which allows for re-use of Strep columns (IBA Life Sciences) and 2x Halt™ protease inhibitors (Thermo Scientific). This eluate was diluted 1:1 in ethylene glycol as a cryoprotectant for storage at −20°C. The final yield and concentration of GST–Fyn SH3–strep protein was 9.3 mg and 53.5 μM, respectively, as determined by staining compared to BSA standards (Thermo Scientific). We used the Coomassie Fluor™ Orange protein stain (Life Technologies) for concentration determination, which binds to the SDS-coat around proteins, and thus is not susceptible to false concentration determination from differences in the number of aromatic residues present in purified proteins.
The His–Tau4R2N–Strep protein was eluted with 25 mM Bis-Tris pH 6.0, 150 mM NaCl, 200 mM CaCl2, 10 mM TCEP, 0.2% KATHON™ CG/ICP, 10 mM D-desthiobiotin, and 2x Halt™ protease inhibitors. The difference in buffer from the elution buffer for Fyn SH3 was to achieve a pH further from Tau’s isoelectric point and to have the chaotrope CaCl2 present, both as measures against protein aggregation, because high concentration preparations of purified Tau aggregated upon storage at −20°C in the storage buffer described for the Fyn preparation. Since Tau is an intrinsically unstructured protein, the presence of a chaotrope in the storage buffer was not a concern, and the high level of dilution prior to AlphaScreen ensured no effect on AlphaScreen signal. This eluate was diluted 1:1 in ethylene glycol as a cryoprotectant for storage at −20°C. The final yield and concentration of His–Tau 4R2N–Strep protein was 11.5 mg and 5.2 μM, respectively, as determined by staining with Coomassie Fluor™ Orange staining (Life Technologies).
A GST-His fusion protein was also generated, which was expressed in the same manner and purified by both Cobalt resin and Glutathione resin (Thermo Scientific) by bulk purification according to the manufacturer’s instructions. The GST-His protein was then buffer-exchanged using a centrifugal filter unit (Millipore) with a 3 kDa MWCO into 50 mM Tris pH 7.4, 300 mM NaCl, 10 mM TCEP, 0.2% KATHON™ CG/ICP, and 2x Halt™ protease inhibitors. This eluate was diluted 1:1 in ethylene glycol as a cryoprotectant for storage at −20°C. The final yield and concentration of GST-His protein was 2.3 mg and 39.4 μM, respectively, as determined by staining with Coomassie Fluor™ Orange.
Small amounts of Tau and Fyn SH3 containing a C-terminal strep tag, but without N-terminal tags needed for interaction with the AlphaScreen beads, were also generated as competitive protein inhibitors for initial optimization experiments.
For small batches of protein to investigate the effect of Tau modifications on the Tau–Fyn SH3 interaction, proteins were expressed in KRX bacteria (Promega) via autoinduction per the manufacturer’s instructions, purified with strep-tactin spin columns (IBA Life Sciences) per the manufacturer’s instructions, eluted with the same buffer used to elute the GST–Fyn SH3–strep protein, then diluted 1:1 in Milli-Q water (Millipore) containing 0.2% Casein (EMD Chemicals) (to prevent the low amount of purified protein from sticking to the sides of the storage tubes) and 2x Halt™ protease inhibitors for short term storage at 4°C.
AlphaScreen
AlphaScreen was conducted with 10 μg/mL of glutathione donor beads (PerkinElmer #6765302) and Nickel chelate acceptor beads (PerkinElmer #6760619R). The buffer composition was 10 mM Bis-Tris pH 7.0, 1 mM TCEP, 0.02% Casein, and 0.1% Tween-20. All dilutions were made in this buffer unless otherwise specified. The concentration of each protein used was 75 nM for HTS as this was the minimal amount of protein needed to provide an acceptable signal for HTS, and 150 nM for manual optimization and verification plates as this concentration gave a less variable signal when plates were prepared by hand.
Generally, compounds were plated first in experiments where compounds were used (including HTS), followed by simultaneous addition of the proteins. The protein-compound mixture was allowed to incubate for 30 minutes, then the beads were added simultaneously and the final mixture was allowed to incubate for 1–5 hours, over which time the signal remained strong and consistent. All experiments were conducted in 384-well plates (PerkinElmer Catalog #6005350). Miniaturization to 1536-well plates was attempted for HTS, but we observed a complete loss of signal in approximately 1% of the wells that we did not observe in 384-well plates. We speculate that this may have been due to Tau aggregation seeded by increased surface area contact in 1536-well plates. Because of this, HTS was conducted in 384-well plates (PerkinElmer #6005359). HTS results were read on an EnVision plate reader (PerkinElmer). Other results were obtained using a Synergy2 plate reader (BioTek) with excitation at 680/30 nm and emission at 570/100 nm (BioTek).
For small preparations used for investigation of the effects of Tau modifications on Tau–Fyn SH3 interactions where it was important to maintain a consistent concentration in order to ensure that an effect in AlphaScreen signal was due to a difference in Tau–Fyn SH3 interaction affinity and not due to a difference in concentration, 300 nM of each protein was used, as this concentration was at the top of the “hook point” for the AlphaScreen assay (see Fig. 1B), and thus was less susceptible to variations in protein expression between preparations. This “hook point” is a characteristic feature of AlphaScreen assays where saturating protein concentrations can attenuate signal (see Wenham et al.16 for further description). To further control for protein concentration, a protein gel was run as previously described with aliquots of the purified Tau or Fyn, along with lanes of BSA standards (10–300 μg). After running this gel, protein concentration was quantified vs. the BSA standards by staining with Coomassie Fluor™ Orange and analyzed using ImageJ (NIH). The proteins were then diluted to the concentration of the lowest-concentration preparation. These diluted samples were run on a second gel in order to verify equalization of protein concentration, and quantified vs. BSA standards as before. After determination of the average concentration from this second gel, Any preparations with concentration values less than half or more than double the calculated average on the 2nd gel were discarded. Generally, preparations did not vary much more than 20–30% from the mean using this method and it was not usually necessary to discard outliers preparations as described.
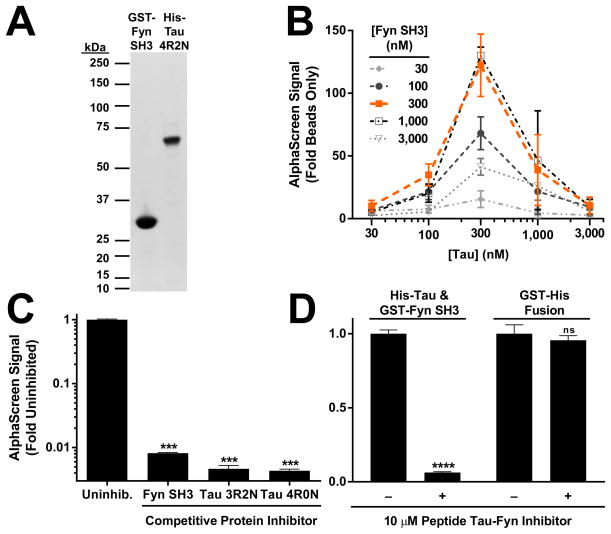
Figure 1.
An AlphaScreen assay provides an HTS-compatible signal for the Tau–Fyn SH3 interaction. (A) Recombinant Tau and Fyn SH3 used in the AlphaScreen assay was high purity. Recombinant protein was purified as described in the Methods. Approximately 1 microgram of each protein was run on a 4–12% gel and stained with SimplyBlue SafeStain (Life Technologies). (B) Optimal concentration for Tau and Fyn SH3 was 300 nM of each protein (n = 2–4 wells per group). Error bars indicate SEM. (C) Interaction between Tau and Fyn SH3, measured by AlphaScreen, can be robustly inhibited using an 8-fold excess of untagged Tau or Fyn SH3. ANOVA, p < 0.0001; *** indicates p < 0.001 by Dunnett’s post hoc vs. uninhibited (n = 6–33 wells per group). Error bars indicate SEM. (D) Addition of 10 μM of a Tat-tagged peptide spanning the 5th and 6th PXXP motifs in Tau robustly inhibits the Tau–Fyn SH3 interaction, but not the control reaction of covalent fusion between a GST and His tag. Inhibitor Effect by two-way ANOVA, p < 0.0001, **** indicates p <0.0001 by Sidak’s post hoc vs. uninhibited (n = 4 wells per group). Error bars indicate SEM.
Bioluminescence Resonance Energy Transfer (BRET)
BRET was conducted with the Fyn SH3 domain fused on the C-terminus to Click Beetle Green (CBG) luciferase17 (Promega #E1461) and Tau tagged at each terminus with mKate218 (Evrogen #FP184). CHO cells (Sigma #85051005-1VL) were plated in 24-well opaque white plates (PerkinElmer #6005168), and transfected using Fugene HD (Promega #E2311) using the manufacturer’s instructions. 48 hours later, fluorescence was read by excitation with a 530/25 nm filter and emission at a 645/40 nm filter on a Synergy2 (BioTek) to control for the concentration of Tau. Immediately after fluorescence measurement, D-Luciferin (Promega #E1605) was added to a final concentration of 200 μM to each well. 2–4 hours later, after the signal had stabilized, plates were read with various wavelength filters in the far red region (620/40, 645/40, 665/34, 680/30) and also with various filters in the green region (528/20, 540/45). We found that the relative signal separation between donor only and BRET increased with more red-shifted filters, but that the noise did as well. We also observed a reduction in the amount of green light emitted with fusion of CBG and mKate2, consistent with efficient BRET. For this reason, we conducted initial optimization experiments reading at both red and green filters. However, for the BRET experiments to measure the Tau–Fyn SH3 interaction, measuring the signal at 645/40 nm alone and quantifying it vs. the total amount of light emitted gave the most consistent results that maximized signal separation between donor only and BRET conditions while minimizing noise.
Peptides
Peptides were produced at >95% purity and concentration was verified by mass spectrometry (Peptide 2.0). Peptides used were YGRKKRRQRRRRSRTPSLPTPPTREPKK (a Tat tag [underlined] fused with residues 209–225 of Tau, spanning the 5th and 6th PXXP motifs in Tau), YGRKKRRQRRRRTPPKSPSSAKSR (a Tat tag fused with residues 230–242 from Tau, spanning the 7th PXXP in Tau), and RSRTPSLPTPPTREPKK (the peptide spanning the 5th and 6th PXXP motifs in Tau, without a Tat tag). The peptide without a Tat tag was used for Z′ score19 determination for the pilot screen and as an additional control (in addition to a tool inhibitor identified in the pilot screen) for Z′ determination for HTS.
Statistics
We found that both AlphaScreen and BRET assays yielded normally distributed data. Therefore, we performed parametric one-way ANOVA tests with Dunnett’s post hoc20 for comparison to controls or multiple comparisons, correcting for multiple comparisons in each case. Where appropriate, we performed two-way ANOVA analysis with Sidak’s post hoc21 instead. All statistical tests were performed with Prism 6 (GraphPad).
Results and Discussion
AlphaScreen Assay Development
In order to identify small molecule Tau–Fyn SH3 interaction inhibitors, we developed an AlphaScreen assay amenable for HTS (Suppl. Fig. S1). The assay used purified GST–Fyn SH3 and His-Tau (Fig. 1A), and showed maximal signal with around 300 nM of each protein (Fig. 1B), although 75–150 nM was sufficient to give an HTS-compatible signal. The AlphaScreen signal was robustly inhibited by addition of an 8-fold excess of untagged Fyn SH3 or Tau protein (Fig. 1C). In order to determine if the assay could distinguish between true Tau–Fyn SH3 inhibitors and compounds that simply inhibited the interaction of the protein tags with the AlphaScreen beads, the effect on either the Tau–Fyn SH3 interaction signal or the signal from GST-His alone when adding a peptide tool inhibitor of the Tau–Fyn SH3 interaction was compared. The peptide robustly inhibited the Tau–Fyn SH3 interaction signal, but not the GST-His interaction signal, indicating that identifying inhibitors of the Tau–Fyn SH3 interaction is possible with this system (Fig. 1D).
Potential Fyn SH3 Binding Sites in Tau
Before utilizing this system for HTS, we tested the ability of the assay to report on known effectors of the Tau–Fyn SH3 interaction. One source of information on known effectors of the Tau–Fyn SH3 interaction comes from studies aimed at determining the residues in Tau important for Fyn binding. Deletion mutant studies have implicated both the 5th/6th and the 7th PXXP motifs in mediating Tau–Fyn SH3 binding1,7. However, more targeted peptide and alanine scan studies have implicated the 5th/6th PXXP motifs, but not the 7th, as important for binding15,22.
One approach to identify Fyn SH3 binding sites in Tau is by competitive titration of peptides spanning these regions implicated in binding (Fig. 2A indicates where implicated PXXP motifs lie in Tau). A Tat-tagged peptide spanning the 5th/6th PXXP motifs provided more potent inhibition than a Tat-tagged peptide spanning the 7th (Fig 2B). These data suggest that the 5th/6th PXXP motif in Tau is a stronger binding site for Fyn than the 7th PXXP.
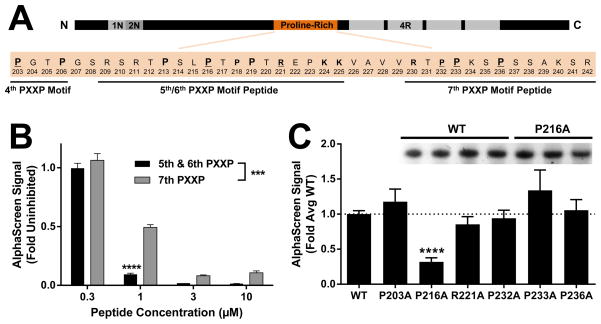
Figure 2.
Fyn SH3 preferentially binds the 5th/6th PXXP motifs in Tau. (A) A portion of the proline-rich domain of Tau containing the 4th–7th PXXPs is shown. Residues potentially involved in polyproline helix binding are in bold and residues that were mutated for alanine scanning in panel C are underlined. Residues present in peptides spanning the 5th/6th or 7th PXXP motifs used in panel B are indicated. Primary prolines in the PXXP motifs are indicated by larger size. (B) Dose-response with Tat-peptides revealed significantly more potent inhibition by a peptide spanning the 5th/6th PXXPs than a peptide spanning the 7th PXXP. *** indicates p < 0.001 peptide effect by two-way ANOVA, and **** indicates p < 0.0001 by Sidak’s post hoc (n = 4 wells per group) vs. alternate peptide. Error bars indicate SEM. (C) An alanine scan revealed significant attenuation of the Tau–Fyn SH3 interaction by mutation of the proline shared by the 5th/6th PXXP motifs (P216A), but not by mutation of several residues in and around the 7th PXXP motif. ANOVA, p < 0.0001; **** indicates p < 0.0001 by Dunnett’s post hoc vs. WT control (n = 3–18 independent protein preparations per group). Error bars indicate SEM. (C, Inset) Representative protein concentration gel showing no difference between the concentration of WT and P216A protein preparations.
An alternative approach to identify Fyn SH3 binding sites in Tau is alanine scanning mutagenesis. Blocking the 5th/6th PXXP motifs by mutating the central proline to alanine (P216A) reduced the Tau–Fyn interaction, but blocking the 3rd/4th PXXP motifs (P203A) or the 7th PXXP motif by mutating either P233 or P236 (or the preceding P232) did not (Fig. 2C). The effect of the P216A in the 5th/6th PXXP motif on the Tau–Fyn interaction was not due to lower protein concentration (Fig. 2C, Inset), suggesting that the inhibition of AlphaScreen signal was due to reduction in the affinity of P216A Tau for the Fyn SH3 domain rather than lower protein levels.
Data on the primary Tau binding site for Fyn have been somewhat discrepant. This is likely due in part to the different approaches that have been used to address the question, each of which has certain advantages and disadvantages. One possible reason for the discrepancy between prior studies addressing the roles of the 5th/6th vs. 7th PXXP could arise from the fact that polyproline helices are typically flanked by positive charges that participate in binding23. In some cases, deletion of the 7th PXXP motif could also delete key positive charges peripheral to the 5th/6th PXXP motifs that play a role in binding. For example, K224 and K225 were deleted in a prior study that implicated the 7th PXXP, but these residues have been shown to facilitate the 5th/6th PXXP motif interaction1,15. Interaction of polyproline helices in Tau with SH3 domains is likely promiscuous and different PXXP motifs might contribute depending on the conditions. However, it remains interesting that a single point mutation (P216A) of the proline shared by the 5th/6th PXXP motifs in Tau was sufficient for significant attenuation of the Tau–Fyn SH3 signal in AlphaScreen, but attenuation was not observed with P233A or P236A to block the 7th PXXP motif in Tau. This observation is consistent with a previous study that observed a drastic reduction in the amount of Tau pulled down with GST–Fyn SH3 with a P216A, but not with a P233A mutation22. It is important to emphasize, however, that in both that study and in ours, there is some residual Tau–Fyn SH3 interaction signal even with P216A (Fig. 2C). This suggests that at least under some circumstances, Tau can bind Fyn through another PXXP motif, or perhaps through a non-canonical interaction between Tau and the Fyn SH3 domain. Overall, the data favors the interpretation that the 5th/6th PXXP motifs are the strongest site for Tau’s interaction with Fyn, but that other PXXP motifs might contribute to a lesser extent, especially if interaction with the 5th/6th PXXP motifs is blocked.
Effect of Tau Modifications
In order to further validate the AlphaScreen system, AD-related modifications were introduced to ask if these modifications alter the Tau–Fyn SH3 interaction signal. Because the Tau–Fyn SH3 interaction plays a role in the dendrites, we examined Tau pseudophosphorylated at sites that are preferentially phosphorylated in the dendrites24,25 to probe the effects of phosphorylation at these sites on the Tau–Fyn SH3 interaction. These sites include four “KXGS” sites in the microtubule-binding domains that are phosphorylated by microtubule affinity regulating kinase (MARK) and recognized by the 12E8 antibody (S262, S293, S324, S356) and three sites in the proline-rich domain recognized by the AT8 antibody (S199, S202, T205). The construct in which all seven of these sites are pseudophosphorylated is denoted as “Dendritic Tau” or D-Tau (Fig. 3A).
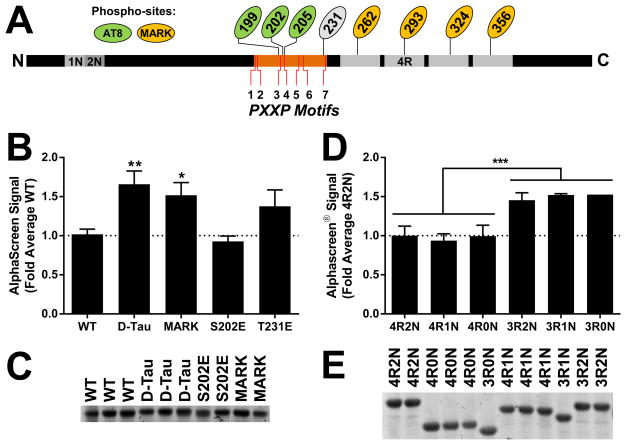
Figure 3.
The AlphaScreen Tau–Fyn SH3 assay signal is increased by Alzheimer’s disease–associated modifications in Tau and by 3R Tau. (A) Schematic diagram of Tau indicating the seven PXXP motifs in the proline-rich domain (orange) and the positions of the phosphorylation sites examined, including the MARK site (yellow) and sites in the AT8 epitope (green). (B) Pseudophosphorylation at MARK sites, alone or in combination with the AT8 epitope, increased the Tau–Fyn interaction. ANOVA, p < 0.0001; * indicates p < 0.05 and ** indicates p < 0.01 by Dunnett’s post hoc vs. WT control (n = 2–12 independent protein preparations per group). Error bars indicate SEM. (C) Representative protein concentration gel showed no difference in concentration between protein preparations used in panel B. (D) 3R isoforms of Tau showed increased Tau–Fyn SH3 AlphaScreen signal. *** indicates p < 0.001 3R vs. 4R effect by two-way ANOVA (n = 1–6 independent protein preparations per group). Error bars indicate SEM. (E) Representative protein concentration gel showing no difference in concentration between protein preparations used in panel D.
Pseudophosphorylation at the MARK sites of Tau, alone or in combination with pseudophosphorylation at the AT8 epitope of Tau, increased the strength of the Tau–Fyn SH3 interaction in the AlphaScreen system (Fig. 3B). Pseudophosphorylation at S202, one of the residues in the AT8 epitope, or at T231 did not significantly change the interaction. Concentrations of the proteins used in the assay were not significantly different, indicating that the changes in AlphaScreen signal were due to increases in the affinity for Fyn of the MARK site pseudophosphorylated constructs rather than differences in protein levels (Fig. 3C).
Tau has six alternatively spliced isoforms expressed in the brain, with either three or four microtubule-binding repeats, denoted 3R or 4R, and either zero, one, or two N-terminal inserts, denoted 0N/1N/2N; thus tau isoforms can be identified as, for example, 4R2N or 3R0N. 3R Tau is known to have a higher affinity for Fyn than 4R Tau14, so we also evaluated all 3R and 4R isoforms of Tau to validate the AlphaScreen system. Corroborating published results, 3R isoforms of Tau showed significantly higher signal in the AlphaScreen system than 4R isoforms (Fig. 3D). The differences were not explained by variation in protein concentration (Fig. 3E). Variation in the number of N-terminal inserts (0N, 1N, or 2N Tau) did not detectably affect the AlphaScreen signal (Fig. 3D).
Together, these results are consistent with the possibility that certain Tau modifications affect the tertiary structure of Tau, which could mediate differences in the affinity of Tau–Fyn SH3 interactions. Indeed, it has been shown that the AT8 Tau modification (one of the Tau modifications prevalent in the dendrites24,25) can affect the tertiary structure of Tau by disrupting intramolecular interactions in Tau26. Furthermore, a β-strand near the 5th–7th PXXP motifs has been implicated in microtubule binding27. It is possible that β-strand–microtubule interactions, or possibly intramolecular β-strand interactions, normally could attenuate Tau–Fyn SH3 interactions through steric hindrance by occluding the 5th–7th PXXP motifs. Disruption of these β-strand interactions (either by phosphorylation or, in 3R Tau, by the absence of a β-strand that is encoded in the exon unique to 4R Tau) may alter the tertiary structure of Tau, increasing exposure of the proline-rich domain and allowing for increased Tau–Fyn SH3 binding (Fig. 4). In any case, the results indicate that Tau–Fyn SH3 interactions may be increased in disease not only through an increase in the total amount of Tau in the dendrites, but also through increased affinity of Tau–Fyn SH3 interactions by phosphorylation of Tau.
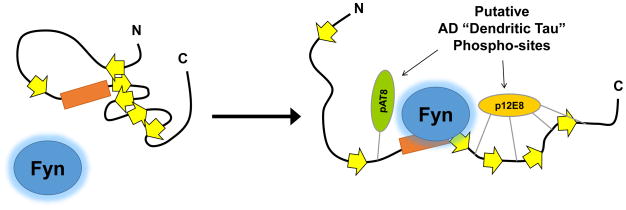
Figure 4.
Model for how disruption of Tau tertiary structure may facilitate Tau–Fyn SH3 interactions. Location of β-strands are approximate based on a published map of Tau secondary structure elements36. Under normal conditions, interactions between β-strands may help maintain Tau in a conformation that reduces access to the proline-rich domain. Phosphorylation may disrupt interactions between β-strands (right), opening the paperclip conformation of Tau and facilitating Fyn binding to the proline-rich domain.
HTS Results
After validating the AlphaScreen assay, HTS was conducted to identify small molecule inhibitors of the Tau–Fyn SH3 interaction. For HTS, we used the D-Tau construct (full-length human Tau pseudophosphorylated at the seven sites found in dendritic Tau, Fig. 3A), since this represents the putative form of Tau found in the dendrites, where the pathogenic Tau–Fyn interaction is believed to occur.
The Z′ score19 was monitored by plate and averaged 0.84 ± 0.05 SD (Suppl. Fig. S2A). We screened 108,138 compounds and set a threshold of three standard deviations above the mean inhibition to advance to dose response (Suppl. Fig. S2B). This cutoff yielded 1,852 compounds showing greater than ~70% inhibition. These 1,852 compounds were then screened in dose-response using four assays: the primary Tau–Fyn SH3 interaction AlphaScreen assay, an AlphaScreen counterscreen with covalently linked GST-His to eliminate compounds that nonspecifically disrupt the AlphaScreen system, an LL47 cell line toxicity assay, and a THP-1 cell line toxicity assay. Compounds were then evaluated based on four criteria: (1) activity in the main Tau–Fyn SH3 dose-response (significant inhibition >50% at any dose), (2) a selectivity index > 2 vs. the GST-His counterscreen (i.e. an IC50 in GST-His at least twice as high as the Tau–Fyn SH3 IC50), and (3,4) a selectivity index > 2 in each cell based toxicity assay (i.e. an EC50 in each cell-based toxicity assay at least twice as high as the Tau–Fyn SH3 IC50). 64 compounds met all four criteria (Suppl. Fig. S2C). Most of the top hits showed much better than 2-fold selectivity in these counterscreens. Indeed, many of the top hits showed no detectable activity at all in these counterscreens. More specifically, of the 64 HTS hits meeting the counterscreen criteria, 23 had undetectable activity in the GST-His assay, 37 had undetectable activity in the THP-1 toxicity assay, and 53 had undetectable activity in the LL47 toxicity assay. 15 had undetectable activity in all 3 counterscreens. 56 compounds out of this set of 64 hits were then selected for further evaluation based on favorable structural properties. These 56 compounds were re-ordered, verified by mass spectrometry, and evaluated in triplicate in dose-response. 39 of the 56 re-ordered compounds were verified to show an IC50 less than 100 μM, with 15 showing an IC50 less than 10 μM.
BRET for Live-Cell Measurement of Tau–Fyn SH3 Interactions
These 39 verified hits from HTS were further evaluated in a cell-based bioluminescence resonance energy transfer (BRET) assay that we developed using a novel BRET pair: click beetle green luciferase (CBG) as the donor and the far-red fluorescent protein mKate2 as the acceptor (Suppl. Fig. S3A,B). The BRET assay complements the primary AlphaScreen assay in several ways, most notably in that it is cell-based and allows evaluation of hits in live cells.
The BRET assay consists of the Fyn SH3 domain tagged with CBG as the donor, and Tau tagged with the far-red fluorescent protein mKate2. When the Fyn SH3 domain interacts with Tau, it brings the donor luciferase in close enough proximity to the acceptor fluorescent proteins to allow for energy transfer, quantified by a shift in the amount of light emitted in the red portion of the spectrum. The combination of the high amount of light emitted by click beetle green28 along with the low noise of the far-red signal from mKate2 allowed for live-cell quantitation of the Tau–Fyn SH3 interaction.
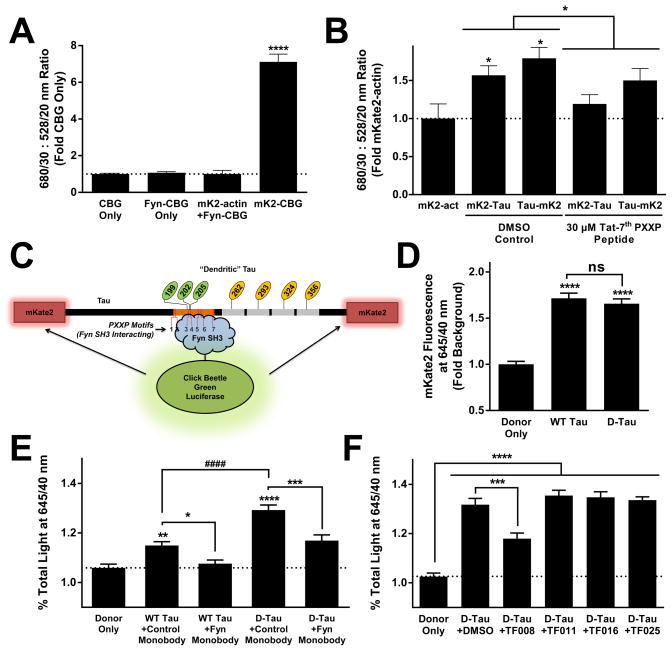
BRET was not detectable with co-expression of mKate2-actin and Fyn-CBG, indicating that there was no non-specific BRET signal between non-interacting proteins (actin and Fyn) (Fig. 5A). However, covalent fusion of the two fluorophores, mKate2-CBG, generated a robust BRET signal in live cells (Fig. 5A). Next, we tested CBG coupled to Fyn SH3 with mKate2 coupled to either the N-terminus or C-terminus of Tau (mKate2-Tau and Tau-mKate2). A significant BRET signal was observed with either form of tagged Tau, and the signal could be inhibited with a Tat-tagged (and thus cell-permeable) peptide inhibitor spanning the 7th PXXP domain (Fig. 5B). To maximize signal, Tau was tagged at both termini with mKate2 for the final embodiment of the assay (Fig. 5C).
Figure 5.
A BRET assay allows measuring the Tau–Fyn interaction in living cells. (A) To account for the possibility of non-specific BRET, mKate2-actin (which should not interact with Fyn SH3) was tested as a negative control. BRET signal for Fyn SH3–CBG only or Fyn SH3–CBG plus mKate2-actin did not differ from CBG donor only, but there was a robust BRET signal with the covalent fusion of mKate2 and CBG. ANOVA, p < 0.0001; **** indicates p < 0.0001 by Dunnett’s post hoc vs. CBG Only (n = 8–16 independent transfections per group). Error bars indicate SEM. (B) Various mKate2-tagged constructs were cotransfected with Fyn SH3–CBG. Tau tagged with mKate2 on either the N-terminus or C-terminus produce BRET signal, while mKate2-actin did not. ANOVA, p < 0.05; * indicates p < 0.05 by Dunnett’s post hoc vs. mKate2-actin control. The BRET signal between Tau and Fyn SH3 can be inhibited by addition of 30 μM of a Tat-peptide spanning the 7th PXXP in Tau. * indicates significant peptide effect by two-way ANOVA (n = 4–12 independent transfections per group). Error bars indicate SEM. (C) The final embodiment of the BRET assay was conducted with Tau tagged on both termini with mKate2, and Fyn SH3 tagged with CBG luciferase. (D–F) Wild-type (WT) or dendritically pseudophosphorylated (D-Tau) forms of Tau were cotransfected with Fyn SH3–CBG donor and compared to transfection of Fyn SH3–CBG donor only. BRET signal was measured by measuring the percentage of total light emitted through a 645/40 nm filter compared to the total amount of unfiltered light emitted. The effects of various inhibitors were tested. (D) mKate2 fluorescence of mKate2–WT Tau and mKate2–D-Tau did not differ; both were significantly above background (Donor Only). ANOVA, p < 0.0001; **** indicates p < 0.0001 vs. Donor Only by Dunnett’s post hoc vs. Donor Only (n = 20–40 independent transfections per group). Error bars indicate SEM. (E) WT Tau interacted with Fyn SH3 in living cells, as measured by BRET. The interaction between D-Tau and Fyn SH3 was stronger than with WT Tau. These signals were inhibited by the G9 monobody that specifically binds Fyn SH3; FN3 indicates the control monobody that does not bind Fyn SH3. ANOVA, p < 0.0001; ** indicates p < 0.01 and **** indicates p < 0.0001 by Dunnett’s post hoc vs. Donor Only, **** indicates p < 0.0001 by Dunnett’s post hoc for comparison of uninhibited WT and D-Tau signal, * indicates p < 0.05 and *** indicates p < 0.001 by Dunnett’s post hoc for comparison between the Fyn-binding monobody (G9) and the non–Fyn-binding monobody control (FN3) for either WT Tau or D-Tau (n = 41–80 independent transfections per group). Error bars indicate SEM. (F) BRET with D-Tau to screen HTS hits for inhibitory activity in a stringent live-cell assay. TF008, but not three other hits, inhibited the Tau–Fyn interaction in living cell using this assay. ANOVA, p < 0.0001; **** indicates p < 0.0001 by Dunnett’s post hoc for all groups vs. Donor Only. *** indicates p < 0.001 by Dunnett’s post hoc comparison to DMSO control for TF008 (n = 12–20 independent transfections per group). Error bars indicate SEM.
Before evaluating compounds for activity using this assay, we first asked if the BRET assay could detect the increased affinity induced by mimicking dendritic phosphorylation sites in Tau that we had observed in AlphaScreen (Fig. 3B). To address this question, we compared Tau pseudophosphorylated at the MARK sites and AT8 epitope (D-Tau) to unmodified Tau as we did with the AlphaScreen assay. To ensure that differences in protein expression between WT and D-Tau did not confound the results, we measured the amount of mKate2 fluorescence prior to addition of the luciferase substrate and verified that the fluorescent signals for unmodified Tau and D-Tau were not different (Fig 5D). We then proceeded to measure BRET. WT Tau showed a small but significant signal in BRET (Fig. 5E), consistent with the published observation that unmodified Tau has a low affinity for Fyn14. The BRET signal was indeed potentiated when Tau was pseudophosphorylated at MARK and 12E8 sites (Fig 5E), consistent with the AlphaScreen findings.
To further validate this assay, we also co-transfected with a “monobody” (clone G9) that had been selected by phage display to bind selectively to the Fyn SH3 domain29. This G9 monobody has a KD of 166 nM for Fyn SH3 and competitively inhibits proline-rich–Fyn SH3 interactions29. In the BRET assay, the G9 monobody completely inhibited the WT Tau–Fyn SH3 interaction (Fig. 5E). Interestingly, it only partially inhibited the D-Tau–Fyn SH3 interaction (Fig. 5E). There are at least two possible explanations for why this tool inhibitor only achieved partial inhibition with D-Tau: (1) The G9 monobody and D-Tau may have similar KD for Fyn SH3 interaction, consistent with the idea that D-Tau modifications increase the affinity of Tau for Fyn SH3 (Figs. 3B and 5E). Thus, if the proteins are expressed at approximately equal levels in cells, one may expect that the signal may be attenuated by about half, as observed. (2) D-Tau may unmask an alternate mode of Tau interaction with the Fyn SH3 domain. We do not have any data to refute this possibility, which is intriguing because it raises the possibility that inhibitors could be developed that target pathogenic Tau–Fyn SH3 interactions while leaving endogenous interactions intact.
Evaluation of Hits with BRET
Of the 39 compounds evaluated, seven showed significant activity in the BRET assay (representative example, compound TF008, shown in Fig. 5F). Though this hit rate may seem low, the G9 monobody inhibitor, which has an excellent KD for Fyn SH3 (166 nM)29, inhibited the D-Tau–Fyn SH3 interaction to a similar degree. Therefore, the fact that several compounds with distinct chemotypes show activity in this stringent assay that requires cell permeability, resistance to cellular degradation, non-toxicity, and high potency is very promising. Furthermore, performance in this stringent assay helped to prioritize the pool of candidate compounds for further counterscreening and evaluation in animal models of AD. Finally, it is possible that other hits may be optimized for good cell-based performance with medicinal chemistry effort.
Properties of Lead Compounds
In summary, we developed and validated two assays to measure the Tau–Fyn SH3 interaction and used these assays in a screening campaign to identify top hits that can inhibit this interaction. Many of the top hits are low molecular weight and have a low polar surface area, making them desirable candidates for further development (Fig. 6A). Furthermore, seven of the top hits have already shown activity in the stringent cell-based BRET assay, even before medicinal chemistry optimization (Fig. 6A). 13 of the top hits have particularly desirable chemical properties and low toxicity, including five that have shown activity in the BRET assay. Of these 13, four have polar surface areas below 90 Å2, which is an indicator of good potential for CNS penetration30. Therefore, we have identified several promising hits, plus others that may be improved by medicinal chemistry optimization.
Figure 6.
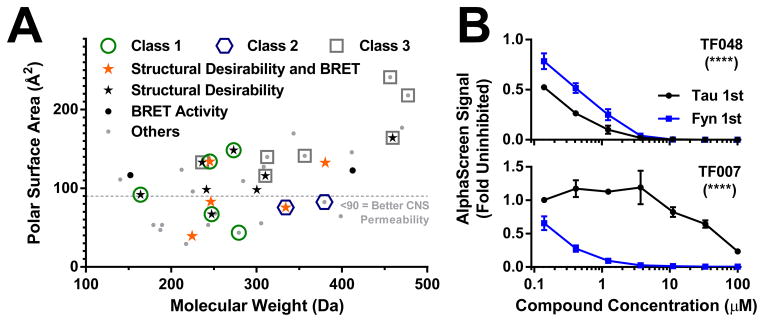
Lead compounds show desirable properties. (A) Out of the top 37 hits, 10 have particularly desirable properties as determined by the medicinal chemistry team. 7 compounds showed activity in the stringent live-cell BRET assay. Also, many of the leads have low molecular weight and polar surface area, particularly desirable properties for CNS drug development. (B) Because the screen could potentially pick up compounds binding to either Tau or Fyn, top hits were tested for binding to Tau or Fyn with a pre-incubation assay to determine the most probable binding partner. Some compounds had significantly more potent IC50 values when pre-incubated with Tau, indicating potential binding to Tau (TF048 shown) while other compounds had significantly more potent IC50 values when pre-incubated with Fyn, indicating potential binding to Fyn (TF007 shown). **** indicates p < 0.0001 pre-incubation partner effect by two-way ANOVA (n = 2–3 wells per group). Error bars indicate SEM.
Interestingly, a pre-incubation assay reveals that some of the hit compounds appear to bind to the Fyn SH3 domain, while others may bind to Tau (Fig. 6B). This is intriguing, because Tau is in complex with other SH3 domain–containing proteins in mice, including the synaptic scaffolding protein PSD-95 and the late-onset Alzheimer’s risk gene BIN11,31. Therefore, a small molecule targeting the Tau side of the Tau–Fyn SH3 interaction has the potential to inhibit these other interactions as well, which may improve the therapeutic potential if Tau aberrantly interacts with multiple SH3 domain–containing proteins in disease.
We plan to move these hits forward into primary neuron and animal models of AD, as well as into more in-depth specificity and liability screening. Furthermore, three chemical classes have been identified in the top hits. Classes 1 and 2 have members with desirable properties and/or BRET activity with low polar surface area, making these classes particularly desirable for medicinal chemistry follow-up. Class 3 may provide promise with medicinal chemistry effort as well, and many singletons that do not have obvious chemical similarity with other compounds may also provide starting points for medicinal chemistry modification.
Implications for Targeting Protein-Protein Interactions with Small Molecules
Inhibition of protein-protein interactions has been discussed as a rich source of potential drug targets32. Here we have presented evidence that proline-rich–SH3 interactions can be targeted with small molecules. Furthermore, we identified several structural classes, suggesting that we may have identified inhibitors of multiple contact points between Tau’s polyproline helix and Fyn’s SH3 domain. Indeed, proline-rich–SH3 interactions have been shown to consist of three contacts with SH3 domains (two from prolines and one from flanking positive charges contacting an acidic region of SH3 domains)23, so this is certainly a possibility. The fact that proline-rich–SH3 interactions have multiple contact points may seem to make targeting proline-rich–SH3 interactions more difficult, but our data in combination with other studies targeting this interaction class with small molecules shows that it is indeed feasible to target either polyproline helices33 or SH3 domains34 with small molecules. Finally, the presence of multiple potential target sites supports the possibility that high specificity between related proteins can be achieved. This is consistent with published studies showing that it is possible to develop peptoid35 or monobody29 ligands that are highly selective for the Fyn SH3 domain over related Src-family kinase SH3 domains. This has implications not only for the Tau–Fyn SH3 interaction, but also for targeting other proline-rich–SH3 interactions that may be involved in disease pathways.
In conclusion, we provide primary evidence that the Tau–Fyn interaction can be targeted with small molecules, which presents a new opportunity for developing Tau-directed therapies for neurodegenerative disease.
Supplementary Material
Acknowledgments
We thank Andy West for various plasmids, Alicia Gross and Scott Wilson for the use of equipment, and members of the Roberson lab for critical reading of the manuscript. We also thank Maaike Everts for facilitating collaboration between the University of Alabama at Birmingham and Southern Research Institute through the Alabama Drug Discovery Alliance.
Funding
This work was supported by NIH grants R01NS075487, T32NS061788, UL1TR00165, P30NS057098, P30NS047466, P30AI027767, the Alabama Drug Discovery Alliance, and the Howard Hughes Medical Institute through the Med into Grad Initiative, HHMI-56006768.
Footnotes
Declaration of Conflicting Interests
E.D.R. is an owner of intellectual property related to Tau.
References
- 1.Ittner LM, Ke YD, Delerue F, et al. Dendritic function of tau mediates amyloid-beta toxicity in Alzheimer’s disease mouse models. Cell. 2010;142:387–397. doi: 10.1016/j.cell.2010.06.036. [DOI] [PubMed] [Google Scholar]
- 2.Roberson ED, Halabisky B, Yoo JW, et al. Amyloid-β/Fyn–induced synaptic, network, and cognitive impairments depend on tau levels in multiple mouse models of Alzheimer’s disease. J Neurosci. 2011;31:700–711. doi: 10.1523/JNEUROSCI.4152-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Roberson ED, Scearce-Levie K, Palop JJ, et al. Reducing endogenous tau ameliorates amyloid β–induced deficits in an Alzheimer’s disease mouse model. Science. 2007;316:750–754. doi: 10.1126/science.1141736. [DOI] [PubMed] [Google Scholar]
- 4.Holth JK, Bomben VC, Reed JG, et al. Tau loss attenuates neuronal network hyperexcitability in mouse and Drosophila genetic models of epilepsy. J Neurosci. 2013;33:1651–1659. doi: 10.1523/JNEUROSCI.3191-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li Z, Hall AM, Kelinske M, et al. Seizure resistance without parkinsonism in aged mice after tau reduction. Neurobiol Aging. 2014 doi: 10.1016/j.neurobiolaging.2014.1005.1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.DeVos SL, Goncharoff DK, Chen G, et al. Antisense reduction of tau in adult mice protects against seizures. J Neurosci. 2013;33:12887–12897. doi: 10.1523/JNEUROSCI.2107-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee G, Newman ST, Gard DL, et al. Tau interacts with src-family non-receptor tyrosine kinases. J Cell Sci. 1998;111(Pt 21):3167–3177. doi: 10.1242/jcs.111.21.3167. [DOI] [PubMed] [Google Scholar]
- 8.Kojima N, Ishibashi H, Obata K, et al. Higher seizure susceptibility and enhanced tyrosine phosphorylation of N-methyl-D-aspartate receptor subunit 2B in fyn transgenic mice. Learn Mem. 1998;5:429–445. [PMC free article] [PubMed] [Google Scholar]
- 9.Cain DP, Grant SG, Saucier D, et al. Fyn tyrosine kinase is required for normal amygdala kindling. Epilepsy Res. 1995;22:107–114. doi: 10.1016/0920-1211(95)00029-1. [DOI] [PubMed] [Google Scholar]
- 10.Larson M, Sherman MA, Amar F, et al. The complex PrP(c)-Fyn couples human oligomeric Aβ with pathological tau changes in Alzheimer’s disease. J Neurosci. 2012;32:16857–16871a. doi: 10.1523/JNEUROSCI.1858-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cochran JN, Hall AM, Roberson ED. The dendritic hypothesis for Alzheimer’s disease pathophysiology. Brain Res Bull. 2013 doi: 10.1016/j.brainresbull.2013.1012.1004. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nygaard HB, van Dyck CH, Strittmatter SM. Fyn kinase inhibition as a novel therapy for Alzheimer’s disease. Alzheimers Res Ther. 2014;6:8. doi: 10.1186/alzrt238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grant SG, O’Dell TJ, Karl KA, et al. Impaired long-term potentiation, spatial learning, and hippocampal development in fyn mutant mice. Science. 1992;258:1903–1910. doi: 10.1126/science.1361685. [DOI] [PubMed] [Google Scholar]
- 14.Bhaskar K, Yen SH, Lee G. Disease-related modifications in tau affect the interaction between Fyn and Tau. J Biol Chem. 2005;280:35119–35125. doi: 10.1074/jbc.M505895200. [DOI] [PubMed] [Google Scholar]
- 15.Reynolds CH, Garwood CJ, Wray S, et al. Phosphorylation regulates tau interactions with Src homology 3 domains of phosphatidylinositol 3-kinase, phospholipase Cgamma1, Grb2, and Src family kinases. J Biol Chem. 2008;283:18177–18186. doi: 10.1074/jbc.M709715200. [DOI] [PubMed] [Google Scholar]
- 16.Wenham D, Illy C, Pierre JAS, et al. Development of high-throughput screening assays for kinase drug targets using alphascreen™ technology. Handbook of Assay Development in Drug Discovery. 2006:53–70. [Google Scholar]
- 17.Wood KV, Lam YA, Seliger HH, et al. Complementary DNA coding click beetle luciferases can elicit bioluminescence of different colors. Science. 1989;244:700–702. doi: 10.1126/science.2655091. [DOI] [PubMed] [Google Scholar]
- 18.Shcherbo D, Murphy CS, Ermakova GV, et al. Far-red fluorescent tags for protein imaging in living tissues. Biochem J. 2009;418:567–574. doi: 10.1042/BJ20081949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang JH, Chung TD, Oldenburg KR. A Simple Statistical Parameter for Use in Evaluation and Validation of High Throughput Screening Assays. J Biomol Screen. 1999;4:67–73. doi: 10.1177/108705719900400206. [DOI] [PubMed] [Google Scholar]
- 20.Dunnett CW. A multiple comparison procedure for comparing several treatments with a control. Journal of the American Statistical Association. 1955;50:1096–1121. [Google Scholar]
- 21.Abdi H. The Bonferonni and Šidák corrections for multiple comparisons. Encyclopedia of measurement and statistics. 2007;3:103–107. [Google Scholar]
- 22.Usardi A, Pooler AM, Seereeram A, et al. Tyrosine phosphorylation of tau regulates its interactions with Fyn SH2 domains, but not SH3 domains, altering the cellular localization of tau. FEBS J. 2011;278:2927–2937. doi: 10.1111/j.1742-4658.2011.08218.x. [DOI] [PubMed] [Google Scholar]
- 23.Kay BK, Williamson MP, Sudol M. The importance of being proline: the interaction of proline-rich motifs in signaling proteins with their cognate domains. FASEB J. 2000;14:231–241. [PubMed] [Google Scholar]
- 24.Jin M, Shepardson N, Yang T, et al. Soluble amyloid β-protein dimers isolated from Alzheimer cortex directly induce Tau hyperphosphorylation and neuritic degeneration. Proc Natl Acad Sci USA. 2011;108:5819–5824. doi: 10.1073/pnas.1017033108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zempel H, Thies E, Mandelkow E, et al. Aβ oligomers cause localized Ca(2+) elevation, missorting of endogenous Tau into dendrites, Tau phosphorylation, and destruction of microtubules and spines. J Neurosci. 2010;30:11938–11950. doi: 10.1523/JNEUROSCI.2357-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jeganathan S, Hascher A, Chinnathambi S, et al. Proline-directed pseudo-phosphorylation at AT8 and PHF1 epitopes induces a compaction of the paperclip folding of Tau and generates a pathological (MC-1) conformation. J Biol Chem. 2008;283:32066–32076. doi: 10.1074/jbc.M805300200. [DOI] [PubMed] [Google Scholar]
- 27.Mukrasch MD, von Bergen M, Biernat J, et al. The “jaws” of the tau-microtubule interaction. J Biol Chem. 2007;282:12230–12239. doi: 10.1074/jbc.M607159200. [DOI] [PubMed] [Google Scholar]
- 28.Miloud T, Henrich C, Hammerling GJ. Quantitative comparison of click beetle and firefly luciferases for in vivo bioluminescence imaging. J Biomed Opt. 2007;12:054018. doi: 10.1117/1.2800386. [DOI] [PubMed] [Google Scholar]
- 29.Huang R, Fang P, Kay BK. Isolation of monobodies that bind specifically to the SH3 domain of the Fyn tyrosine protein kinase. N Biotechnol. 2012;29:526–533. doi: 10.1016/j.nbt.2011.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pajouhesh H, Lenz GR. Medicinal chemical properties of successful central nervous system drugs. NeuroRx. 2005;2:541–553. doi: 10.1602/neurorx.2.4.541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chapuis J, Hansmannel F, Gistelinck M, et al. Increased expression of BIN1 mediates Alzheimer genetic risk by modulating tau pathology. Mol Psychiatry. 2013;18:1225–1234. doi: 10.1038/mp.2013.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mullard A. Protein-protein interaction inhibitors get into the groove. Nat Rev Drug Discov. 2012;11:173–175. doi: 10.1038/nrd3680. [DOI] [PubMed] [Google Scholar]
- 33.Oneyama C, Agatsuma T, Kanda Y, et al. Synthetic inhibitors of proline-rich ligand-mediated protein-protein interaction: potent analogs of UCS15A. Chem Biol. 2003;10:443–451. doi: 10.1016/s1074-5521(03)00101-7. [DOI] [PubMed] [Google Scholar]
- 34.Inglis SR, Stojkoski C, Branson KM, et al. Identification and specificity studies of small-molecule ligands for SH3 protein domains. J Med Chem. 2004;47:5405–5417. doi: 10.1021/jm049533z. [DOI] [PubMed] [Google Scholar]
- 35.Li H, Lawrence DS. Acquisition of Fyn-selective SH3 domain ligands via a combinatorial library strategy. Chem Biol. 2005;12:905–912. doi: 10.1016/j.chembiol.2005.06.007. [DOI] [PubMed] [Google Scholar]
- 36.Mukrasch MD, Bibow S, Korukottu J, et al. Structural polymorphism of 441-residue tau at single residue resolution. PLoS Biol. 2009;7:e34. doi: 10.1371/journal.pbio.1000034. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.