Abstract
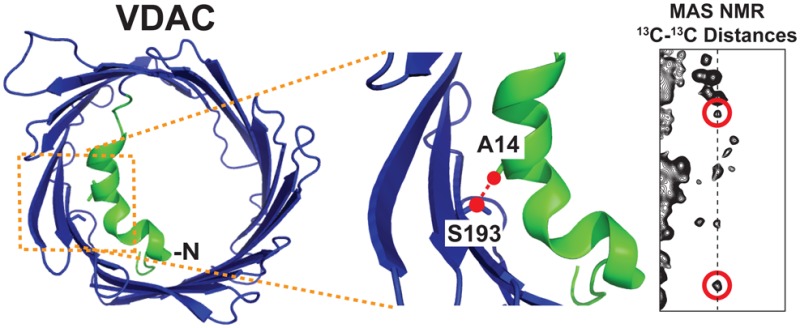
The N-terminus of the voltage-dependent anion channel (VDAC) has been proposed to contain the mechanistically important gating helices that modulate channel opening and closing. In this study, we utilize magic angle spinning nuclear magnetic resonance (MAS NMR) to determine the location and structure of the N-terminus for functional channels in lipid bilayers by measuring long-range 13C–13C distances between residues in the N-terminus and other domains of VDAC reconstituted into DMPC lipid bilayers. Our structural studies show that the distance between A14 Cβ in the N-terminal helix and S193 Cβ is ∼4–6 Å. Furthermore, VDAC phosphorylation by a mitochondrial kinase at residue S193 has been claimed to delay mitochondrial cell death by causing a conformational change that closes the channel, and a VDAC-Ser193Glu mutant has been reported to show properties very similar to those of phosphorylated VDAC in a cellular context. We expressed VDAC-S193E and reconstituted it into DMPC lipid bilayers. Two-dimensional 13C–13C correlation experiments showed chemical shift perturbations for residues located in the N-terminus, indicating possible structural perturbations to that region. However, electrophysiological data recorded on VDAC-S193E showed that channel characteristics were identical to those of wild type samples, indicating that phosphorylation of S193 does not directly affect channel gating. The combination of NMR and electrophysiological results allows us to discuss the validity of proposed gating models.
Membrane proteins can provide essential pathways for transport of a variety of metabolites and ligands through a bilayer leaflet and do so in a switchable fashion. An outstanding example demonstrating this ability is the human voltage-dependent anion channel (VDAC), a 32 kDa integral membrane protein that is known to be the primary avenue for metabolite traffic between the mitochondrion and cytoplasm.1,2 Because of VDAC’s importance, a great deal of effort has and continues to be directed at determining its detailed structure as well as understanding its function. The initial structure reported for VDAC was determined with solution nuclear magnetic resonance (NMR) in lauryldimethylamine oxide (LDAO) micelles and revealed a 19-strand β-barrel with a flexible N-terminal helix.3 Shortly thereafter, additional X-ray and solution NMR structures of VDAC with similar features appeared in the literature.4,5 In the diffraction structure,4 two α-helices are present in the N-terminus and this section of the protein is located adjacent to the wall of the β-barrel. In contrast, in the solution NMR structure, a single helix is resolved,3,5 and while the N-terminus was located inside the β-barrel, the family of calculated structures shows greater conformational heterogeneity for the N-terminal region. Specifically, some of the calculated structures in the bundle of conformers show an N-terminus that protrudes out of the β-barrel into the surrounding medium. Thus, while there is agreement about the general position of the N-terminus, there are significant details that differ among the structures.
The position of the N-terminus is of considerable interest because of its purported critical role in VDAC’s characteristic ability to selectively transport either cations or anions between open or closed states, respectively. Some gating models have proposed that the conformation of VDAC’s N-terminus underlies its ability to switch between transporting either anions or cations. A number of these models consider an independent movement of the N-terminus upon gating.6−10 For example, one model for explaining the gating mechanism proposed that the N-terminal region could be displaced from its position adjacent to the barrel wall out into the middle of the pore to gate between cations and anions.11 However, these models were not supported by more recent cross-linking experiments that demonstrated the N-terminus and barrel did not move independently between open and closed states.12 In the first gating model developed by Colombini and co-workers, the N-terminus is part of a voltage-sensor domain that slides in and out of the channel upon voltage gating.13−15 Although the mechanisms are still under debate, most observations have demonstrated the presence of the N-terminus is required for channel gating,3,16−18 and new models incorporating recent experiments19−21 and computational studies22 are needed to improve our understanding of the structural basis of gating and the role of the N-terminus.
A second important question is how the gating mechanism is regulated in cells as part of signaling pathways that control cellular life and death processes.8,23 VDAC’s N-terminus has been reported to be a critical feature for activating such pathways, but the structural basis of this role is not understood. A potential mechanism for in vivo regulation of VDAC’s N-terminal region is through posttranslational modifications of the surrounding residues. For example, Chen et al. recently reported that the mitochondrial kinase Nek1 phosphorylates VDAC on residue serine 193, which was found to regulate phenotypical responses to DNA damage;24 cells with VDAC phosphorylated at S193 were reported to survive longer after being exposed to lethal amounts of radiation. Similar effects in a cellular context were observed for an S193E VDAC mutant.24 In a follow-up study, the same group reported that S193 phosphorylation or an S193E mutation closed VDAC irreversibly, as suggested by analysis of atomic force microscopy images and cytochrome c leakage assays.25 The S193A mutant was reported to be always open.25
In the crystal structure, S193 is positioned inside the β-barrel with its side chain pointing toward the lumen. In addition, the N-terminus is situated such that A14 Cβ is located adjacent to S193 Cβ approximately 4–6 Å away. In contrast to these publications, a previous model put forth by Colombini and co-workers, based on biochemical and biophysical data, proposed that S192, an analogous residue in VDAC from the organism Neurospora crassa, is positioned at a very different location in a loop outside the path of ion flow and not in a region spanning the membrane.26,27 Because differences between Colombini’s model and the three-dimensional (3D) structures have been the subject of recent discussions,28,29 it is important to establish the position of S193 for VDAC reconstituted in a lipid bilayer environment in which samples have been demonstrated to be fully functional.30
We previously demonstrated reconstitution of VDAC in lipid bilayer two-dimensional (2D) crystals yielded high-resolution NMR spectra and, importantly, also fully functional channels.30 This provided a platform for investigating the structure of VDAC, and the structural basis of channel gating. To determine the location and structure of the N-terminus for functional channels in lipid bilayers, we have used magic angle spinning NMR (MAS NMR) dipolar recoupling methods to measure long-range 13C–13C distances between S193 and residues in the N-terminus for VDAC reconstituted in DMPC bilayers. In particular, 3D heteronuclear experiments permitted unambiguous assignment of several long-range contacts for VDAC in 2D crystals. Our structural studies show that the distance between A14 Cβ in the N-terminal helix and S193 Cβ is ∼4–6 Å. The phosphorylation mimetic mutant S193E, which was supposed to mimic the closed state of VDAC,25 demonstrated minor spectral changes compared to those of the wild-type VDAC and showed virtually no difference in function between the S193E mutant and wild-type protein as investigated by electrophysiological measurements. These high-resolution structural results determined the relative location of the N-terminus for functional channels in lipid bilayers and prompted us to discuss the validity of proposed gating models.
Experimental Procedures
Expression and Purification of Recombinant Human VDAC
The expression and purification of VDAC followed the protocol of Malia and Wagner31 and Hiller et al.32 with some minor modifications. Isoform 1 of human VDAC was expressed in BL21(DE3) cells transformed with the VDAC plasmid containing a six-His C-terminal tag. Overnight cultures were grown in 10 mL of unlabeled LB broth for 12–16 h and then used to inoculate 700 mL of LB broth. Cells were grown at 37 °C until they reached an OD600 of 0.6, pelleted, suspended again in M9 minimal medium, and grown again at 37 °C until they reached an OD600 of 0.8–1.0. Overexpression was then induced with 1 mM isopropyl β-d-1-thiogalactopyranoside for 3–5 h. All isotopically labeled materials were purchased from Cambridge Isotope Laboratories. For [U-13C,15N]VDAC, 3 g of [13C]glucose and 1 g of 15NH4Cl were the sole carbon and nitrogen sources, respectively, in the M9 medium. [U-13C,12C-FLY,15N]VDAC was prepared by adding 12C- and 14N-labeled F, L, and Y (1 mM each) to M9 medium in addition to 3 g of [13C]glucose and 1 g of 15NH4Cl. [U-13C,12C-WHIFY,15N]VDAC was prepared by adding 12C- and 14N-labeled W, H, I, F, and Y (1 mM each) to the M9 medium in addition to 3 g of [13C]glucose and 1 g of 15NH4Cl.
VDAC was purified under denaturing conditions [8 M urea, 50 mM Tris-HCl (pH 7.5), 100 mM NaCl, and 20 mM imidazole] over Ni-agarose resin and eluted with the same buffer but containing 250 mM imidazole. Eluted fractions containing VDAC were dialyzed against 4 L of buffer [50 mM Tris-HCl (pH 7.5), 50 mM NaCl, 1 mM EDTA, and 20 mM BME]. Precipitated VDAC was centrifuged and dissolved in 6 M guanidine (GuHCl) buffer and refolded at 4 °C by dropwise dilution of 1 volume of urea buffer containing VDAC into 10 volumes of stirred refolding buffer [25 mM Na·PO4 (pH 7.0), 100 mM NaCl, 1 mM EDTA, 20 mM BME, and 1% (43 mM) LDAO]. After the mixture had been stirred overnight at 4 °C, refolded VDAC was dialyzed against 20 volumes of 25 mM Na·PO4 (pH 7.0), 2–3 mM DTT, and 1 mM EDTA.
For cation exchange chromatography, samples were loaded onto a 50 mL SP sepharose HP column (GE Healthcare) equilibrated with buffer A [25 mM Na·PO4 (pH 7.0), 5 mM DTT, and 0.1% LDAO]. VDAC was eluted over a 25 to 50% gradient with buffer B (buffer A with 1 M NaCl) at approximately 30–35% buffer B. Fractions containing properly folded VDAC were pooled and concentrated using Centricon 10 kDa molecular weight cutoff (MWCO) concentrators.
Mutagenesis
VDAC S193E and S193A mutants were prepared using the QuikChange Site-Directed Mutagenesis Kit from Stratagene (Agilent Technologies). The synthesis of each mutant was verified by DNA sequencing at the Massachusetts Institute of Technology Biopolymers Laboratory using an Applied Biosystems model 3730 capillary DNA sequencer with a Big Dye Terminator Cycle Sequencing Kit.
2D Crystallization
2D crystals were prepared according to the method of Dolder et al.33 with some modifications that have been described previously.30 Purified, refolded VDAC in 0.1% LDAO was dialyzed for 24 h against 2–4 L of 0.6% (w/v) octyl POE (Bachem), 50 mM Tris (pH 8), and 2 mM DTT. Dried DMPC (Avanti Polar Lipids) was added to 1% octyl POE, solubilized, and added to VDAC in equal volumes at a lipid:protein ratio of 1:2 (w/w). The mixture was then dialyzed against buffer containing 150 mM NaCl, 20 mM MgCl2, 10 mM MES (pH 6.5), and 5 mM DTT at room temperature for several days with 2 × 4 L of buffer changes per day in a dialysis cassette (Thermo Scientific) with a 10 kDa MWCO. The solution became cloudy after dialysis for 12–24 h, and crystals appeared fully formed after 36–48 h. Dialysis was allowed to proceed for an additional 48–72 h until the absorbance of VDAC in solution was ≤5%. Samples were then removed and washed against 25 mM Na2HPO4/NaH2PO4 buffer (pH 7) to remove any remaining detergent and to lower salt concentrations for MAS NMR studies. Each sample contained 18 mg of VDAC and 9 mg of lipids. Samples were centrifuged into Bruker 3.2 mm rotors, and the drive tips were sealed with epoxy to prevent dehydration.
VDAC Reconstitution and Conductance Measurements
Planar lipid membranes were formed on a 70–90 μm diameter orifice in the 15 μm thick Teflon partition that separated two compartments as previously described.34 The membrane was formed from two opposed lipid monolayers made from a 5 mg/mL solution of diphytanoylphosphatidylcholine (DPhPC) (Avanti Polar Lipids, Inc.) in pentane. The compartments were filled with 1 M KCl buffered with 5 mM HEPES (pH 7.4). Channel insertion was achieved by adding ∼0.5 μL of VDAC/DMPC 2D crystals diluted at a 1:100 (v/v) ratio in a buffer [10 mM Tris, 50 mM KCl, 1 mM EDTA, 15% (v/v) DMSO, and 2.5% (v/v) Triton X-100 (pH 7.0)] into the 1.2 mL aqueous phase of the cis compartment while being stirred. The potential was defined as positive when it was greater at the side of VDAC addition (cis side). Current recordings were performed as described previously34 using an Axopatch 200B amplifier (Axon Instruments, Inc., Foster City, CA) in voltage clamp mode. Single-channel data were filtered by a low-pass Bessel filter at 10 kHz and directly saved into the computer memory with a sampling frequency of 50 kHz. VDAC ion selectivity was determined from the measurements of reversal potential, the voltage corresponding to zero current at a KCl gradient of 1 M KCl cis versus 0.2 M KCl trans, buffered with 5 mM HEPES (pH 7.4) on single channels. The voltage-dependent properties of a membrane containing many channels were assessed following the protocol devised by Colombini and colleagues35,36 in which gating is inferred from response of VDAC to slowly changing applied periodic voltage waves. A symmetrical 5 mHz triangular voltage wave with an amplitude of ±60 mV from a model 33120A function waveform generator (Hewlett-Packard) was used. Data were acquired with a Digidata 1322A board (Axon Instruments, Inc.) at a sampling frequency of 1 Hz and analyzed using pClamp version 10.2 (Axon Instruments, Inc.). Analysis of VDAC voltage gating was performed as previously described.34 Open probabilities were defined as the ratio (G – Gmin)/(Gmax – Gmin), where Gmax and Gmin are the maximal and minimal conductances, respectively. Two gating parameters, n, defined as the effective gating charge, and V0, the voltage at which half of the channels are open and half are closed, were calculated from the open probability plots as previously described.34,36 Plot fitting was performed with the Boltzmann equation.34
MAS NMR Spectroscopy
Homonuclear 2D 13C–13C correlation spectra were acquired with radio frequency-driven dipolar recoupling (RFDR)37 on a custom-built spectrometer (courtesy of D. Rubin) operating at a 1H Larmor frequency of 750 MHz equipped with a Bruker (Billerica, MA) 3.2 triple-resonance 1H/13C/15N E-free probe. Experiments employed MAS frequencies of 12.5 and 16.667 kHz maintained at ±5 Hz using a Bruker MAS controller. For RFDR spectra, the carrier was placed in the middle of the aliphatic region and 33 kHz (at 12.5 kHz MAS) or 40 kHz (at 16.6 kHz MAS) π-pulses were applied with 100–110 kHz CW 1H decoupling during mixing and 83 kHz TPPM381H decoupling during acquisition and evolution. A recycle delay of 2.5 s was used, and typically, 32–64 scans per t1 value were averaged, yielding a total acquisition time between 16 and 32 h. Samples were cooled with a stream of nitrogen gas at a flow rate of 2500 L/h. The temperature was regulated at 4 °C as monitored by a thermocouple next to the stator, and the internal sample temperature (not including radiofrequency heating) at a given spinning frequency and specified amount of cooling was calibrated using 79Br.39
Experiments for estimating 13C–13C distances were conducted on a Bruker Avance II spectrometer (Bruker Biopsin) equipped with a 3.2 mm E-free MAS probe at 20 kHz MAS and a 900 MHz 1H field strength. Long-range 13C–13C contacts were obtained with a series of 2D proton-assisted recoupling (PAR)40 experiments optimized using a 13C radiofrequency field of ∼50 kHz and a 1H radiofrequency field of ∼55 kHz for mixing times between 8 and 15 ms and employing 83 kHz TPPM 1H decoupling during acquisition and evolution periods.
The 3D NCOCX spectra were recorded using a pulse sequence described previously by Ladizhansky and co-workers.41 Specifically, 1H–15N cross-polarization (CP)42 was optimized for a 1 ms contact time with a radiofrequency field of 50 kHz on 15N and the 1H field ramped linearly around the n = 1 Hartmann–Hahn condition.43 SPECIFIC-CP44 for the 15N–13C′ transfer was optimized with an 8 ms contact time with a radiofrequency field strength of 2.5υr on 15N and 3.5υr (where υr = ωr/2π, spinning frequency) on 13C′ with 100 kHz 1H CW decoupling at a ωr/2π = 12.5 kHz frequency; 83 kHz TPPM 1H decoupling was used during evolution and acquisition periods with a pulse length of 5.8 μs and optimized phases of 0° and 13°. 13C–13C mixing was performed with 100 ms DARR mixing45 (n = 1 condition), RFDR,46 or proton-assisted recoupling (PAR).40 The NCOCX experiment employed a 2 s recycle delay and 16 scans were acquired for a total acquisition time of ∼7 days.
Molecular Dynamics Simulations
The complex structure [Protein Data Bank (PDB) entry 3EMN] was embedded into an explicit lipid bilayer and solvated in 150 mM KCl, and the CHARMM-GUI interface of Im et al. and CHARMM-36 parameters were used for the DOPE lipid bilayer. The TIP3P water model and recently developed ion parameters were used for all MD simulations. The published crystal structure was used to construct the simulation box for WT VDAC. SCWRL version 3.0 was used to construct the S193E mutant system. Each constructed protein–membrane complex was equilibrated at 303.15 K with the NPaT ensemble for 20 ns using periodic boundary conditions in a tetragonal box with dimensions of 84.9 Å × 84.9 Å × 82.5 Å. This was followed with simulation runs of 80 ns. The temperature was maintained with a Lowe-Anderson thermostat as implemented in NAMD. All MD simulations were performed with the program suite NAMD version 2.9.47 Subsequent analysis of the system was performed using the CHARMM program suite (35b1r1).48
Brownian Dynamics Simulations
We used the BROMOC program suite to compute ion conductance with Grand-Canonical Monte Carlo simulations utilizing an algorithm of Im, Seefeld, and Roux.49 The newly developed method of Luo et al.50 and Egwolf et al.51 was used to assess the reaction field component for GCMC/BD simulations. The details of Poisson–Boltzmann computations required for obtaining static and reaction field maps can be found in ref (52). In all BROMOC simulations, the VDAC channel was treated as a rigid structure with a dielectric constant of 2 surrounded by a high-dielectric solvent (εw = 80) and embedded in an implicit membrane (εm = 2) with a thickness of 30 Å. To introduce effects of protein dynamics, 25 evenly spaced frames were selected from an 80 ns MD trajectory. We considered two systems with symmetric (1 M KCl) and asymmetric (1 M/0.1 M) buffers to assess conductance and reversal potentials associated with S193E mutations. The simulation box covered an entire length of protein with GCMC buffers located approximately 15 Å from its boundaries.
Results
Assignment of S193 and Surrounding Residues
To probe the effect of phosphorylation of S193, it was first necessary to identify this residue in the WT construct and confirm its location relative to the N-terminus. To identify and unambiguously assign S193, we conducted a 3D NCOCX correlation experiment with [U-13C,15N]WT VDAC. The chemical shift of S193 Ca and Cb and the sequential triplet G192-S193-I194 permitted unambiguous identification of this peak. Figure 1 shows two slices through the 3D NCOCX experiment with assignments for residues 192–194. Cross-peaks are well-resolved, yielding <0.5 ppm line widths for both 15N and 13C. The chemical shifts of S193 Ca and Cb are consistent with a β-barrel secondary structure, and the intensity of the S193 cross-peak is well above the noise in cross-polarization experiments; thus, this residue is relatively rigid at ambient temperature. Many additional assignments were obtained from this experiment and other 3D experiments on multiple samples, and these assignments and experiments will be discussed in detail in a forthcoming publication.
Figure 1.
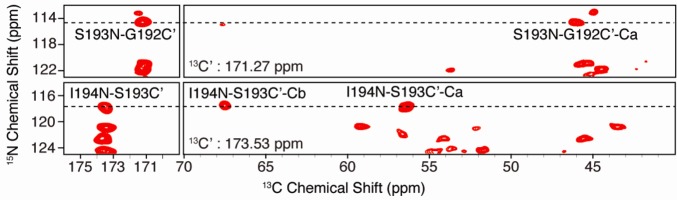
Typical 2D planes from a 3D NCOCX experiment with [U-13C,15N]VDAC in 2D DMPC crystals. Selected assignments are shown for residues G192, S193, and I194, allowing confirmation of the S193 Ca–Cb peak in 2D correlation spectra. Experimental details are described in Experimental Procedures. The total experimental time was ∼7 days.
Assignments taken from the 3D spectra permitted unambiguous identification of S193 in 2D 13C–13C correlation spectra. An example 2D 13C–13C RFDR correlation spectrum obtained with [U-13C,12C-FLY,15N]VDAC is shown in Figure 2 with S193 assigned. Assignments of the N-terminal portion of VDAC in 2D crystals were previously achieved via 2D and 3D experiments.30 The chemical shift for S193 Cb is unique in the direct dimension; no other peaks exist at 67.8 ppm.
Figure 2.
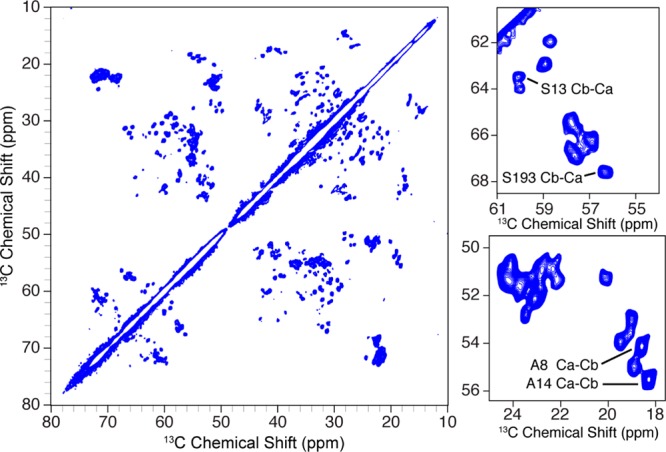
13C–13C RFDR MAS correlation spectrum of [U-13C,12C-FLY,15N]VDAC in DMPC lipid bilayer 2D crystals. Assignments for A8, S13, A14, and S193 are labeled in expansions of the spectrum (right). Experimental parameters were as follows: τmix = 1.3 ms, ωr/2π = 12.5 kHz, ω0H/2π = 750 MHz, 32 scans per t1 slice, and a 2.5 s recycle delay for a total experiment time of ∼16 h.
13C–13C Long-Range Distance Measurements
To confirm the proximity of residue 193 to the surrounding residues requires long-range contacts. Arguably, one of the better methods for doing this is with proton-assisted recoupling (PAR),40 which permits long-range, weak contacts in the presence of many stronger couplings. Assignment of S193 and its unique chemical shift allowed us to look for contacts between S193 Cb and sites adjacent in space. Any cross-peaks appearing along 67.8 ppm could then be unambiguously attributed to contacts with S193. Figure 3 shows a long mixing 13C–13C PAR spectrum in red overlaid with a short mixing RFDR experiment in blue. The RFDR experiment shows almost exclusively one-bond contacts and demonstrates that the S193 Cb resonance is unique. The only other possible resonances that occur at the A14 Cb chemical shift arise from isoleucine Cg2 resonances. We therefore reverse labeled isoleucine as well as WHFY residues with 12C- and 14N-labeled amino acids to unambiguously assign S193 to A14 contacts in the PAR experiments.
Figure 3.
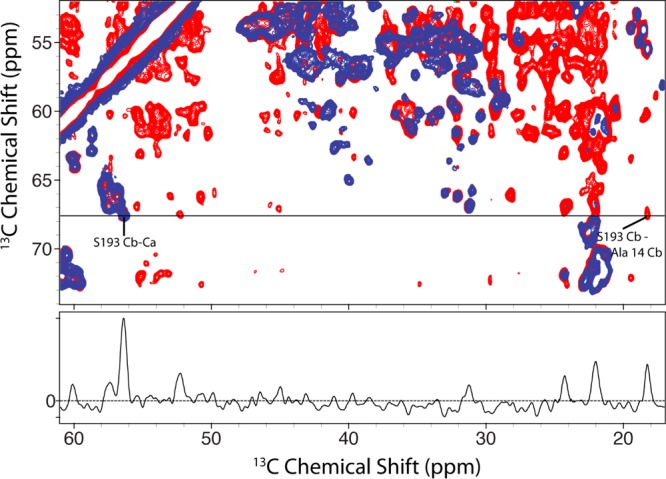
Fourteen millisecond 13C–13C PAR (red) of [U-13C,12C-WHIFY,15N]VDAC overlaid on a 1.8 ms RFDR (blue) spectrum of [U-13C,15N]VDAC showing the unambiguous contact between S193 Cb and A14 Cb. Both spectra were acquired at ω0H/2π = 900 MHz and ωr/2π = 20 kHz.
Figure 4 shows an expansion of the previous PAR spectrum with several additional assignments. PAR transfer efficiencies depend not only on the distance between recoupled spins but also on the local 1H geometry surrounding those spins. Thus, while it cannot be used in this way to extract precise internuclear distances, determining the maximal cross-peak intensity as a function of PAR mixing time can put an upper limit on a specified contact. The maximal peak intensity was observed for 10 ms PAR mixing and is consistent with a 4–6 Å contact.40
Figure 4.
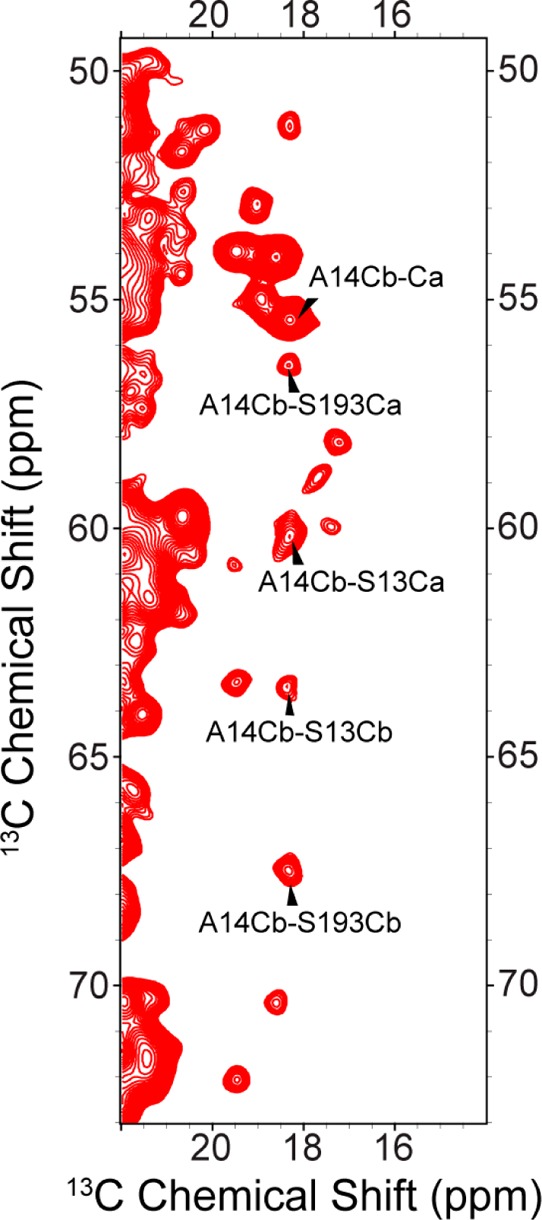
Expansion of the PAR spectrum of Figure 3. Long mixing 13C–13C experiment on [U-13C,12C-WHIFY,15N]VDAC obtained with 14 ms CC PAR. Selected assignments are shown for A14–S13 and A14–S193 contacts.
It is interesting to compare these results with the known structure for what is purportedly the open state of VDAC. The diffraction structure of VDAC shows the N-terminal region close in space to the inner barrel wall. Figure 5 shows a top-down view of the VDAC pore and the proximity of residue S193 to the N-terminus in the crystal structure of murine VDAC (PDB entry 3EMN).53 The crystal structure of murine VDAC shows S193 to be close in space to the N-terminus. The S193 side chain points toward the N-terminus and is likely making a hydrogen bond with N-terminal residues. The reported distance in the crystal structure from S193 Cb to A14 Cb is 4.0 Å.
Figure 5.

Top-down view of the crystal structure of murine VDAC (PDB entry 3EMN).53 Residues A14 and S193 are colored green and labeled at the left. The right panel shows an expansion of the N-terminal region and nearby β-strands. The distance between A14 Cb and S193 Cb is highlighted with a dashed line and measures 4.0 Å in the crystal structure. The images were created in Pymol.54
Characterization of S193E and S193A VDAC Mutants
During refolding and cation exchange chromatography, S193E and S193A behaved exactly like WT samples. Samples eluted at approximately 30–35% buffer B, and ion exchange chromatograms between WT and mutant samples appeared to be very similar. Samples of each mutant were reconstituted into DMPC lipid bilayers at a protein:lipid ratio of 2:1 (w/w) (∼1:25 protein:lipid molar ratio). We recorded 2D 13C–13C correlation spectra of the mutant samples to verify that the protein refolded and was properly reconstituted into lipid bilayers. Figure 6 shows the aliphatic region of 13C–13C RFDR correlation spectra of S193E and S193A. Both spectra exhibit well-resolved resonances and good spectral dispersion, indicating that both mutants are properly folded in the bilayer. The spectral resolution and dispersion appeared to be nearly identical to those of wild-type samples (<0.5 ppm 13C line widths at ω0H/2π = 750 MHz). As expected, the S193 Ca–Cb cross-peak is missing for both mutants. Identification of the new residue, E or A, was difficult in 2D spectra as those peaks did not appear to have chemical shifts consistent with loop or helical regions and were presumably in regions of the spectra that were too overlapped to resolve with 2D experiments alone.
Figure 6.
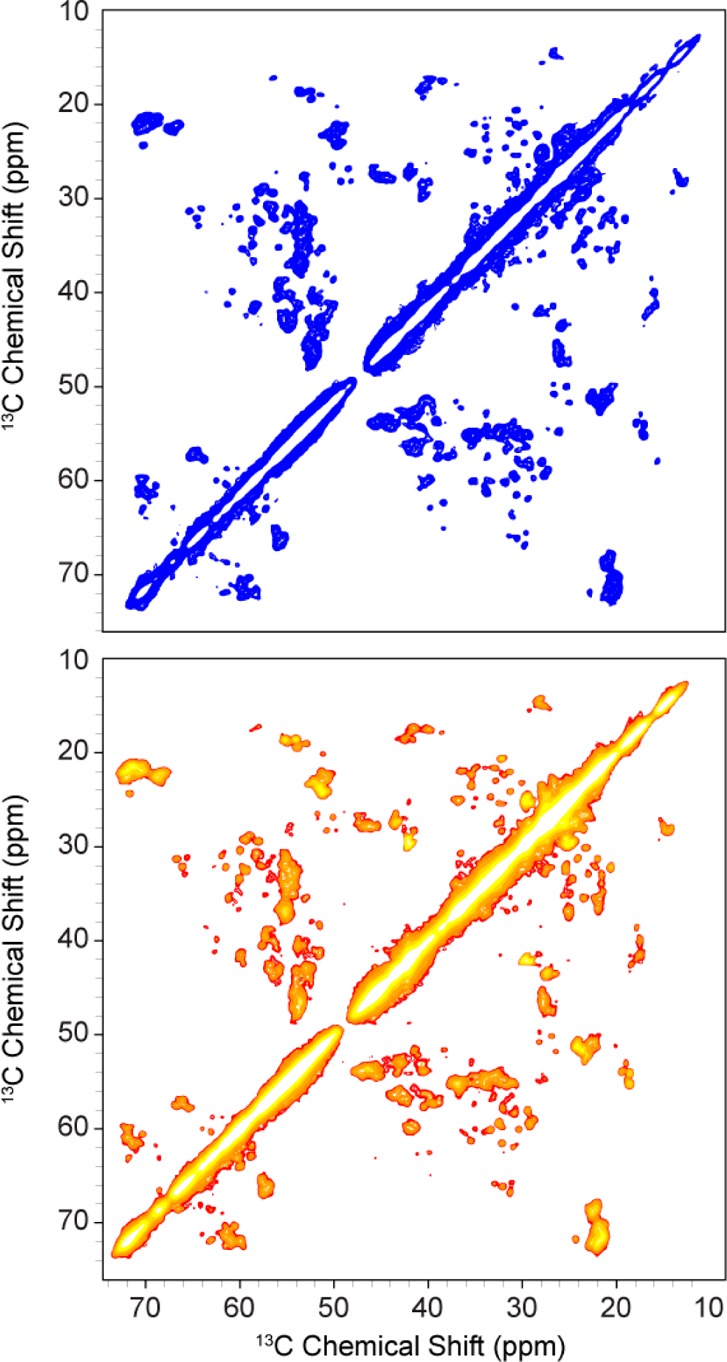
13C–13C RFDR correlation spectra with τmix = 1.3 ms acquired on [U-13C,15N]S193E (top, blue) and [U-13C,15N]S193A (bottom, red-orange). Both spectra were recorded at ωr/2π = 12.5 kHz MAS, ω0H/2π = 750 MHz, and T = 4 °C. Each experiment averaged 32 scans in the t1 dimension and took ∼16 h.
Comparison of NMR Spectra of Mutant and Wild-Type Samples
Figures 7 and 8 show overlay comparisons of WT VDAC with S193A and with S193E. Chemical shifts and peak intensities for both the WT and S193A appeared relatively unperturbed, indicating that the secondary structures of WT VDAC and S193A are similar. All cross-peaks arising from the N-terminal region of VDAC are present in both mutant spectra, indicating that the N-terminus is still relatively rigid and remains in an α-helical conformation. On closer inspection, comparison of both spectra reveals some perturbation of chemical shifts for both mutants compared to WT samples. Those chemical shift changes appear for a sequential set of residues in the N-terminus. Specifically, 13C chemical shifts for residues 7–14 are shifted by ∼0.5 ppm on average. This indicates that while the secondary structure of S193A as a whole appears to be very similar to that of WT samples, there does appear to be a localized structural change for those residues.
Figure 7.
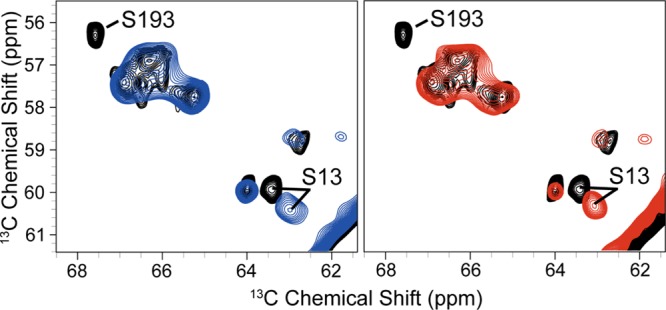
Overlay of 13C–13C RFDR correlation spectra of WT VDAC (black), S193E (blue), and S193A (red). All spectra were acquired at ω0H/2π = 750 MHz and used the same experimental parameters.
Figure 8.
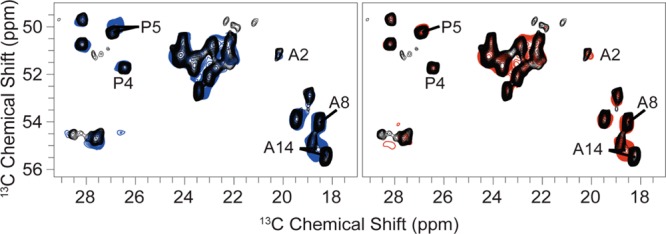
Overlay of 13C–13C RFDR correlation spectra of WT VDAC (black), S193E (blue), and S193A (red). All spectra were acquired at ω0H/2π = 750 MHz and used identical experimental parameters.
Chemical shifts for both the WT and S193E samples also overlap, suggesting as with S193A that the structures of WT VDAC and S193E are very similar. No changes in chemical shifts above ∼0.5 ppm were observed between S193E and WT VDAC. Detailed comparison of both spectra reveals some perturbation of chemical shifts for S193E. Paralleling S193A, those chemical shift changes appear for the same sequential set of residues in the N-terminus. Specifically, chemical shifts of residues 7–16 are again shifted by ∼0.5 ppm on average. This suggests a structural change in those same residues for S193E.
MD Simulations of WT VDAC and S193E
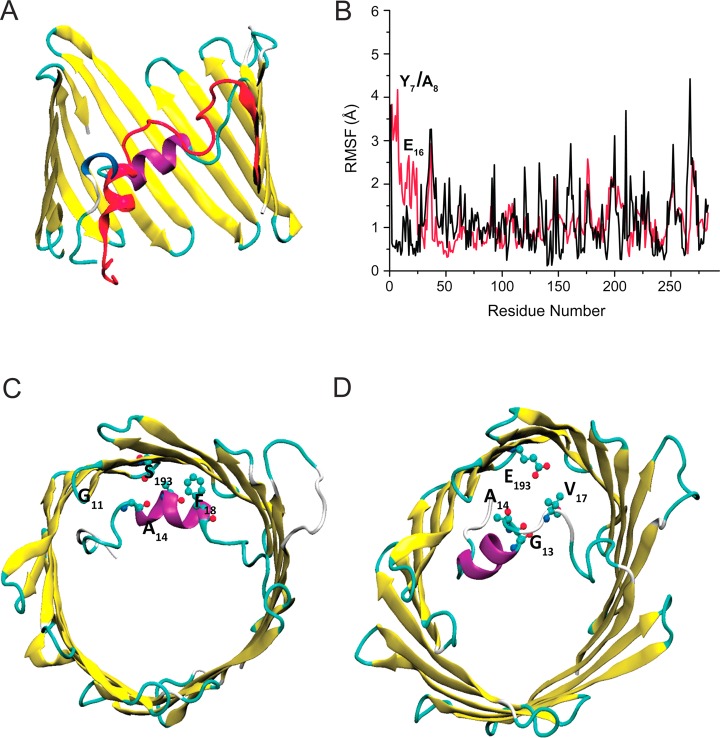
To assess the extent to which chemical shift changes observed for S193E are due to structural changes in the N-terminus, we performed equilibrium MD simulations on WT and S193E systems. MD simulations of a protein embedded in the lipid bilayer display stable protein structures. The overall root-mean-square (rms) values of the backbone atoms relative to original crystallographic coordinates are 1.9 and 2.6 Å for WT and S193E, respectively. The per-residue decompositions of rms values for both systems are shown in Figure 9 with a superposition of two average structures extracted from MD simulations.
Figure 9.
(A) Superposition of the N-terminus for average structures extracted from MD simulations for WT VDAC (purple) and S193E (red). (B) Per-residue rms fluctuations of heavy atoms in WT (black) and S193E (red) channels with respect to the crystal structure. (C and D) Representative states showing the key contacts from MD simulations between S/E193 and the N-terminal helix in WT (left) and S193E (right). A heavy atom–heavy atom cutoff distance of 4.5 Å was used.
Data collected in Figure 9 (panels A and B) show that the barrel region remains unperturbed by the mutation. The cross-sectional area of S193E is ∼94% of that for the WT channel, and positional fluctuations are on the average scale previously observed in MD simulations of other proteins. However, residues 4–16 positioned in the N-terminus are considerably affected by introduction of a negative charge at residue 193. According to the MD simulations, the partial unwinding and refolding of the helical region of the N-terminus (residues 8–16) occur several times in the course of the simulation of S193E, while this is absent in the WT. This finding is consistent with MAS NMR data suggesting the S193E mutation does not perturb the β-strands of the channel but does cause a potential structural change in the N-terminal region. The A14–S193 contact seen in both the crystal structure and MAS NMR data shown above is a long-lived contact in WT VDAC and is present in >50% of all frames over the last 80 ns of simulation. The amphipathic side chain of S193 also forms short-lived contacts with the carbonyl oxygen of G11 (<7%) and aromatic ring of F18 (<5%) (Figure 9C). For the S193E mutant, most of the same contact surface between the N-terminus and channel’s wall is retained in the simulation. However, most of the contacts between E193 and the N-terminus become much more short-lived. It is important to stress that E193 is shielded from the main permeation pathway by N-terminal residues for VDAC-S193E, which has consequences for the functional studies of S193E, yet the S193–A14 stable contact present in WT is formed in only ∼18% of all frames in the S193E mutant. Unwinding of the N-terminal helical region led to numerous contacts (bifurcating) between E193 and the backbone of G13 and V17 (Figure 3D). We can conclude that surface charge on the β-barrel wall of VDAC is an important determinant of the packing and stability of the N-terminus.
Comparison of Channel Properties of WT VDAC and the S193E Mutant Reconstituted into Planar Membranes
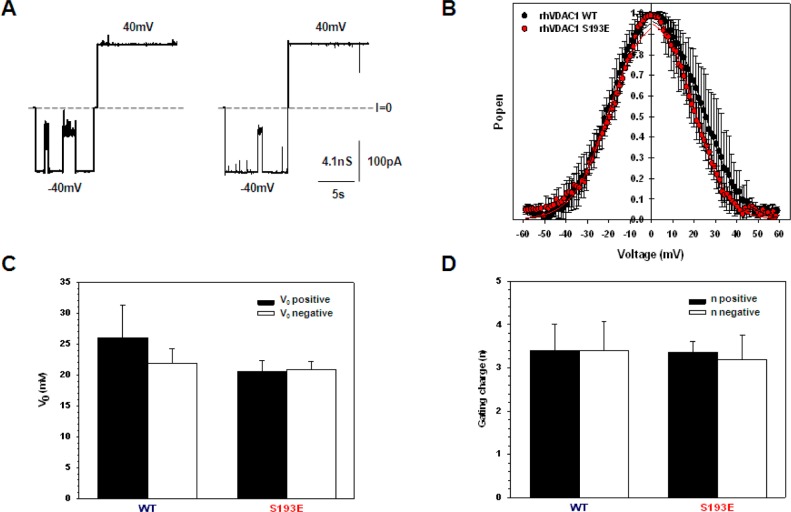
If residue S193 is positioned inside the β-barrel (Figure 5), its phosphorylation mimetic S193E introduces an extra negative charge inside the channel lumen. The additional charge could significantly change VDAC channel properties such as single-channel conductance, voltage gating, and ion selectivity. To test for such functional changes, we performed a comparative study of WT VDAC and S193E reconstituted into planar lipid membranes. Figure 10 shows the results of an electrophysiological study of S193E. S193E forms functional channels with a typical single-channel conductance of 4.1 ± 0.1 nS in 1 M KCl (Figure 10A). Both WT VDAC and the S193E mutant also show similar ion selectivity measured in a 1 M/0.2 M (cis/trans) KCl gradient. The voltages corresponding to zero current at this KCl concentration gradient, the so-called reversal potentials, were 8.8 ± 1.0 and 9.8 ± 1.9 mV for WT and S193E, respectively. The calculated Cl–/K+ permeability ratios for WT VDAC and S193E were 1.6 ± 0.1 and 1.7 ± 0.2, respectively.
Figure 10.
(A) Representative single-channel current traces of WT VDAC (left trace) and S193E (right trace) reconstituted into planar lipid membranes showing the same single-channel conductance and characteristic VDAC gating at −40 mV. Dashed lines indicate zero current level. The membrane bathing solution contained 1 M KCl buffered with 5 mM HEPES (pH 7.4). Records were digitally filtered at 100 Hz using a Bessel algorithm. (B) Probability to be open at different voltages on multichannel membranes containing WT VDAC (black circles) or S193E (red circles). Data are means of three to five independent experiments ± the standard deviation. (C and D) Voltage gating parameters obtained from the open probability plots in panel B. The voltage at which half of the channels are open and half are closed, V0 (C), and effective gating charge, n (D), of WT VDAC and S193E were calculated at positive (black bars) and negative (white bars) potentials. The error bars correspond to the standard deviation.
Voltage gating is the most characteristic property of VDAC15 and could be seen in Figure 10A as transitions from one high-conducting “open” state to the variety of low-conducting “closed” states at −40 mV of applied voltage. To quantitatively characterize voltage gating, we performed experiments using multichannel membranes as was previously described.34−36 Analysis of the probability of VDAC to be open at varying potentials showed similar voltage gating for WT VDAC and S193E with characteristic bell-shaped plots (Figure 10B). Thus, phosphorylation mimetic S193E does not show permanent VDAC closure. Two gating parameters were calculated from the open probability plots: V0, the voltage required to close half of the channels (Figure 10C), and n, the effective gating charge (Figure 10D). Although at positive potentials V0 appeared to be slightly higher for WT VDAC than for S193E (Figure 10C), this difference is not statistically significant (p = 0.08). These results show that although possible structural changes may be observed for S193E, the introduction of the phosphorylation mimetic at residue S193 does not measurably affect VDAC channel properties.
Discussion
Previous MAS NMR experiments with VDAC in liposomes indirectly identified contacts between the N-terminus and barrel wall.55 The previous approach identified possible contacts by observing the disappearance of relatively weak cross-peaks for a truncated construct in which the first 20 residues were removed. While the observation was consistent with the previously published diffraction structure of murine VDAC, the spectral degeneracy for VDAC would have made interpretation of these data very challenging without a priori knowledge of the structure. Here long-range contacts were observed with PAR,40 a homonuclear recoupling sequence based on the third spin-assisted recoupling (TSAR) mechanism,56 which was designed for the observation of long-range contacts at high MAS frequencies57 and magnetic field strengths. Several distance estimates were obtained with PAR between residues in the N-terminus and S193, establishing the location of the N-terminus in lipid bilayer preparations. In contrast to the previous study, the signal-to-noise ratio in the PAR measurements presented here is very high, and through selective isotopic labeling, we have unambiguously identified contacts between S193 and residues in the N-terminus.
On the basis of MAS NMR data, we have determined that WT VDAC and VDAC mutants S193E and S193A adopt similar structures when they are reconstituted in lipid bilayers. 13C–13C correlation spectra of both S193A and S193E show well-resolved and -dispersed cross-peaks, indicating both mutants were folded and properly inserted into lipid bilayers. Comparison of 13C–13C correlation spectra of S193A, S193E, and WT VDAC shows relatively subtle differences in chemical shifts among the three constructs, permitting us to conclude that all three constructs have a very similar overall secondary structure. Assuming that WT VDAC is present in the open state in our NMR samples, S193A and S193E also exist in the open state under the conditions present in our NMR measurements, i.e., with no applied voltage across the bilayer.
Electrophysiological measurements presented here demonstrate that S193E forms typical VDAC channels (Figure 10A) and exhibits gating behavior that is nearly identical to that of WT VDAC (Figure 10B–D). This is consistent with MAS NMR results showing very similar secondary structures between WT VDAC and S193E for the presumably open state. While AFM data from Chen et al.25 claim to show that S193E is always closed and the channel does not gate, AFM measurements do not directly probe channel gating. The well-established and more widely used experimental approach for measuring channel gating is to study a current response to the applied voltage of single or multiple channels reconstituted into the lipid membrane.
Functional studies reported here show that S193E has the same ion selectivity as WT VDAC. One potential explanation for not observing an effect on channel properties by introducing an additional negative charge for S193E would be that residue S193 is not exposed to the VDAC pore lumen. This idea is consistent with a structural model proposed by Colombini and co-workers developed from a number of electrophysiological experiments in combination with point-directed mutagenesis.58,59 In Colombini’s model, the analogous residue from fungal and human VDAC is positioned in a loop outside the transmembrane region and outside the pore.26,60 Therefore, channel ion selectivity and gating would remain unaffected by modification of this residue. This model differs from the structure of the presumed open state of murine VDAC and human VDAC, where S193 is positioned near the N-terminus and the side chain. Furthermore, Colombini’s model differs from our MAS NMR results presented here that show that the chemical shift for S193 is consistent with a β-strand topology and the distance between S193 and N-terminal residue A14 is 4–6 Å. A second interpretation of both the functional and MAS NMR data is that the N-terminal domain screens or obstructs the charged residue of S193E from contributing to the channel’s interior surface charge. It would then follow that during channel gating, the N-terminus does not change its position significantly relative to the barrel wall. This hypothesis is supported by VDAC functional studies of cross-linked samples,12 where cross-linking residue L10C from the α-helical region of the N-terminus to residue A170C from the β-barrel wall of the pore in an engineered double Cys mutant of murine VDAC did not prevent voltage gating of the channel reconstituted into planar lipid membranes. Therefore, it was suggested that the N-terminus is located inside the channel pore attached to β-strand 11 when VDAC is open and does not move independently of A170 during voltage gating. Considering the proximity of the two pairs of residues in the N-terminal helix, L10 and A14, and the two residues in β-strands, A170 and S193, we propose that E193 could be screened by the N-terminal region and therefore not affect channel selectivity.
Modeling of Ion Conductance and Selectivity of WT VDAC and S193E
To explore the extent to which E193 is screened by the N-terminus, we employed Brownian dynamics (BD) simulations for WT VDAC and S193E. The current–voltage (I–V) relation in the symmetric 1.0 M KCl solution from GCMC/BD simulations is shown in Figure 11. The I–V relation appears to be symmetric, and the estimated conductance is 4.3 ± 0.3 and 3.8 ± 0.4 nS for WT and S193E, respectively. All simulations were performed with the inclusion of a hydrodynamic correction as proposed in our previous work. The standard error in BD computations is ∼10%, and therefore, for all practical purposes, both channels (WT and S193E) exhibit very similar open-pore conductance.
Figure 11.
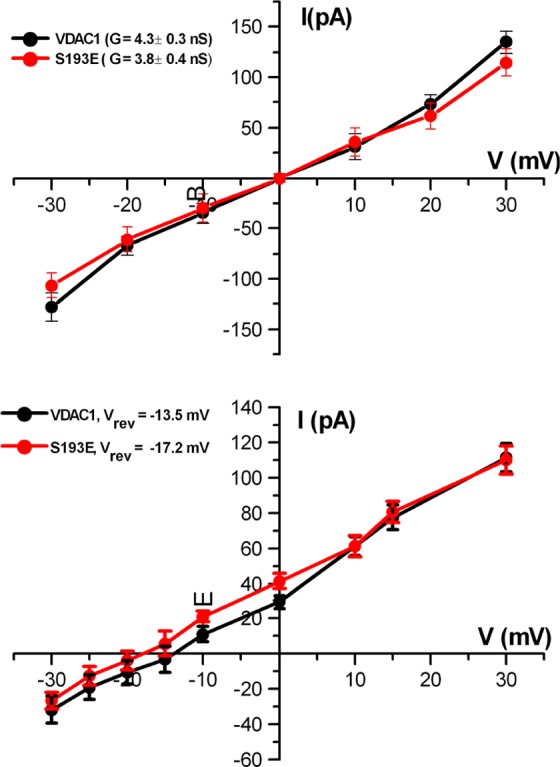
Results of BD simulations for WT VDAC (black) and S193E (red). The I–V plot in the top graph was obtained in symmetric 1 M KCl and in the bottom graph in a 1 M/0.1 M KCl gradient. Calculated channel conductance, G, and reversal potential, Vrev, are shown in the top and bottom panels, respectively. The error bars represent the standard deviation estimated from 25 independent BD simulation runs with different starting conformations of the channel extracted from MD trajectories for WT VDAC and S193E.
Next, we used BD simulations to assess reversal potentials and thus to determine channel selectivity. All simulations were performed with a cis chamber concentration [C]i of 1 M KCl and a trans chamber concentration [C]o of 0.1 M KCl. The resulting reversal potential displayed well-defined anion selectivity of WT VDAC with a reversal potential of −13.5 mV. S193E displayed even higher anion selectivity with a shift of reversal potential to −17.2 mV. The standard error in the reversal potential extracted from BD simulations that use small voltages is generally high, and it is difficult to define small reversal potentials more accurately than ±3–4 mV. However, the reversal potentials from BD simulations marked both pores as anion-selective and computed open-pore conductance in excellent agreement with experimental measurements. Thus, while the conformation of the N-terminus may be affected by the S193E mutation, this perturbation does not result in a change in the selective properties due to E193 shielding by the N-terminus.
Physiological Significance of Mutations and Phosphorylation of VDAC
Our NMR and electrophysiology results suggest that phosphorylation of VDAC alone by Nek1 at S193 is not sufficient to cause channel closure, in contrast to those of Chen et al. reported by AFM. Electrophysiological experiments presented here show that S193E voltage-gating behavior is nearly identical to that of WT VDAC. Isolated S193E channels exhibit characteristic properties of WT VDAC and are found to be in a closed or low-conducting state only at voltages exceeding ±30 mV. MAS NMR results demonstrated that the 3D topologies of S193E and S193A are very similar to those of WT VDAC and that any perturbations in channel structure occur around the N-terminal region. Our results, however, cannot completely rule out the possibility that Nek1 still phosphorylates VDAC and that this event helps to regulate cellular apoptosis. Conditions present in the cell and outside of the scope of this study could explain the discrepancies; for example, interactions between VDAC and other proteins in the presence of Nek1 may contribute to controlling cell survivability through phosphorylation of other proteins. Another possible explanation is that VDAC closure is not required to regulate cell death61 and that phosphorylation of VDAC coincides with other events that delay cell death.
Conclusions
In this study we have shown that unambiguous contacts between the N-terminal region of VDAC with residue S193 in the β-barrel are within 4–6 Å, providing further support that the structure of human VDAC in lipid bilayers is similar to the diffraction structure of murine VDAC. These measurements also demonstrate that valuable structural information can be obtained with MAS NMR on large integral membrane proteins in a reasonable amount of instrument time.
A phosphorylation mimetic, S193E, and a control sample, S193A, both form properly refolded functional channels. Chemical shift differences in MAS NMR spectra of S193E and S193A are relatively small (∼0.5 ppm and less) and are observed only for residues in the N-terminal region. This indicates that the overall secondary structures of WT VDAC, S193E, and S193A are very similar. Furthermore, cross-peak intensities for all three samples are also very similar, indicating that there are no large differences in micro- to millisecond dynamics among these samples.
Electrophysiological experiments show that the VDAC-S193E mutant gates properly and its voltage-gating behavior is nearly identical to that of the WT channel. Thus, S193E is not always closed, contradicting previous claims based on AFM measurements. Furthermore, functional data provide a context for interpreting chemical shift perturbations for S193E. The absence of significant changes in the functional parameters of the WT compared to the mutant VDAC indicates that perturbations in the secondary structures of S193E do not affect VDAC voltage gating. This provides further evidence that suggests the N-terminus alone is not responsible for voltage gating, although it may be involved with other regions of the protein. Because our results on isolated VDAC do not probe VDAC within a cell, it is possible that VDAC phosphorylation coincides with or triggers additional events or molecular interactions that delay cell death.
Acknowledgments
M.T.E. acknowledges Dr. Patrick van der Wel for insightful discussions. O.T., T.K.R., and S.Y.N. acknowledge Dr. Sergey Bezrukov for fruitful discussions. L.A. acknowledges the laboratory of James Chou for space to prepare the VDAC mutants.
Glossary
Abbreviations
- MAS NMR
magic angle spinning nuclear magnetic resonance
- VDAC
isoform 1 of the voltage-dependent anion channel
- WT
wild type
- RFDR
radio frequency-driven recoupling
- DMPC
1,2-dimyristoyl-sn-glycero-3-phosphocholine
- CW
continuous wave
- LDAO
lauryldimethylamine oxide
- DTT
dithiothreitol
- TSAR
third spin-assisted recoupling
- PAR
proton-assisted recoupling.
- PDB
protein data bank
Author Contributions
L.A. and O.T. contributed equally to this work.
This research was supported by National Institutes of Health (NIH) Grants EB001960, EB002026 (R.G.G.), and GM075879 (G.W.). S.Y.N. was supported by the National Sciences and Engineering Research Council of Canada (NSERC) (Discovery Grant RGPIN-315019) and the Alberta Innovates Technology Futures (AITF) Strategic Chair in BioMolecular Simulations (Centre for Molecular Simulation). The Intramural Research Program of the Eunice Kennedy Shriver National Institute of Child Health and Human Development (NIH) supported O.T. and T.K.R.
The authors declare no competing financial interest.
Funding Statement
National Institutes of Health, United States
References
- Rostovtseva T.; Colombini M. (1997) VDAC channels mediate and gate the flow of ATP: Implications for the regulation of mitochondrial function. Biophys. J. 72, 1954–1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodge T.; Colombini M. (1997) Regulation of metabolite flux through voltage-gating of VDAC channels. J. Membr. Biol. 157, 271–279. [DOI] [PubMed] [Google Scholar]
- Hiller S.; Garces R. G.; Malia T. J.; Orekhov V. Y.; Colombini M.; Wagner G. (2008) Solution structure of the integral human membrane protein VDAC-1 in detergent micelles. Science 321, 1206–1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ujwal R.; Cascio D.; Colletier J.-P.; Faham S.; Zhang J.; Toro L.; Ping P.; Abramson J. (2008) The crystal structure of mouse VDAC1 at 2.3 Å resolution reveals mechanistic insights into metabolite gating. Proc. Natl. Acad. Sci. U.S.A. 105, 17742–17747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayrhuber M.; Meins T.; Habeck M.; Becker S.; Giller K.; Villinger S.; Vonrhein C.; Griesinger C.; Zweckstetter M.; Zeth K. (2008) Structure of the human voltage-dependent anion channel. Proc. Natl. Acad. Sci. U.S.A. 105, 15370–15375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Pinto V.; Reina S.; Guarino F.; Messina A. (2008) Structure of the voltage dependent anion channel: State of the art. J. Bioenerg. Biomembr. 40, 139–147. [DOI] [PubMed] [Google Scholar]
- De Pinto V.; Tomasello F.; Messina A.; Guarino F.; Benz R.; La Mendola D.; Magrì A.; Milardi D.; Pappalardo G. (2007) Determination of the Conformation of the Human VDAC1 N-Terminal Peptide, a Protein Moiety Essential for the Functional Properties of the Pore. ChemBioChem 8, 744–756. [DOI] [PubMed] [Google Scholar]
- Shoshan-Barmatz V.; De Pinto V.; Zweckstetter M.; Raviv Z.; Keinan N.; Arbel N. (2010) VDAC, a multi-functional mitochondrial protein regulating cell life and death. Mol. Aspects Med. 31, 227–285. [DOI] [PubMed] [Google Scholar]
- Reymann S.; Florke H.; Heiden M.; Jakob C.; Stadtmuller U.; Steinacker P.; Lalk V. E.; Pardowitz I.; Thinnes F. P. (1995) Further evidence for multitopological localization of mammalian porin (VDAC) in the plasmalemma forming part of a chloride channel complex affected in cystic fibrosis and encephalomyopathy. Biochem. Mol. Med. 54, 75–87. [DOI] [PubMed] [Google Scholar]
- Guo X. W.; Smith P. R.; Cognon B.; D’Arcangelis D.; Dolginova E.; Mannella C. A. (1995) Molecular design of the voltage-dependent, anion-selective channel in the mitochondrial outer membrane. J. Struct. Biol. 114, 41–59. [DOI] [PubMed] [Google Scholar]
- Törnroth-Horsefield S.; Neutze R. (2008) Opening and closing the metabolite gate. Proc. Natl. Acad. Sci. U.S.A. 105, 19565–19566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teijido O.; Ujwal R.; Hillerdal C. O.; Kullman L.; Rostovtseva T. K.; Abramson J. (2012) Affixing N-terminal α-Helix to the Wall of the Voltage-dependent Anion Channel Does Not Prevent Its Voltage Gating. J. Biol. Chem. 287, 11437–11445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J. M.; Midson C.; Blachly-Dyson E.; Forte M.; Colombini M. (1998) The topology of VDAC as probed by biotin modification. J. Biol. Chem. 273, 24406–24413. [DOI] [PubMed] [Google Scholar]
- Blachly-Dyson E.; Peng S. Z.; Colombini M.; Forte M. (1990) Selectivity changes in site-directed mutants of the VDAC ion channel: Structural implications. Science 247, 1233–1236. [DOI] [PubMed] [Google Scholar]
- Colombini M., Blachly-Dyson E., and Forte M. (1996) VDAC, a channel in the outer mitochondrial membrane. In Ion Channels (Narashi T., Ed.) pp 169–202, Plenum Press, New York. [DOI] [PubMed] [Google Scholar]
- Choudhary O. P.; Ujwal R.; Kowallis W.; Coalson R.; Abramson J.; Grabe M. (2010) The Electrostatics of VDAC: Implications for Selectivity and Gating. J. Mol. Biol. 396, 580–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mannella C. A. (1998) Conformational changes in the mitochondrial channel protein, VDAC, and their functional implications. J. Struct. Biol. 121, 207–218. [DOI] [PubMed] [Google Scholar]
- Popp B.; Benz R.; Neupert W.; Lill R. (1996) The role of the N and C termini of recombinant Neurospora mitochondrial porin in channel formation and voltage-dependent gating. J. Biol. Chem. 271, 13593–13599. [DOI] [PubMed] [Google Scholar]
- Villinger S.; Giller K.; Bayrhuber M.; Lange A.; Griesinger C.; Becker S.; Zweckstetter M. (2014) Nucleotide Interactions of the Human Voltage-dependent Anion Channel. J. Biol. Chem. 289, 13397–13406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villinger S.; Briones R.; Giller K.; Zachariae U.; Lange A.; de Groot B. L.; Griesinger C.; Becker S.; Zweckstetter M. (2010) Functional dynamics in the voltage-dependent anion channel. Proc. Natl. Acad. Sci. U.S.A. 107, 22546–22551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zachariae U.; Schneider R.; Briones R.; Gattin Z.; Demers J.-P.; Giller K.; Maier E.; Zweckstetter M.; Griesinger C.; Becker S.; Benz R.; De Groot B. L.; Lange A. (2012) β-Barrel mobility underlies closure of the voltage-dependent anion channel. Structure (Oxford, U.K.) 20, 1540–1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhary O. P.; Paz A.; Adelman J. L.; Colletier J.-P.; Abramson J.; Grabe M. (2014) Structure-guided simulations illuminate the mechanism of ATP transport through VDAC1. Nat. Struct. Mol. Biol. 21, 626–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abu-Hamad S.; Arbel N.; Calo D.; Arzoine L.; Israelson A.; Keinan N.; Ben-Romano R.; Friedman O.; Shoshan-Barmatz V. (2009) The VDAC1 N-terminus is essential both for apoptosis and the protective effect of anti-apoptotic proteins. J. Cell Sci. 122, 1906–1916. [DOI] [PubMed] [Google Scholar]
- Chen Y.; Craigen W. J.; Riley D. J. (2009) Nek1 regulates cell death and mitochondrial membrane permeability through phosphorylation of VDAC1. Cell Cycle 8, 257–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y.; Gaczynska M.; Osmulski P.; Polci R.; Riley D. J. (2010) Phosphorylation by Nek1 regulates opening and closing of voltage dependent anion channel 1. Biochem. Biophys. Res. Commun. 394, 798–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J.; Midson C.; Blachly-Dyson E.; Forte M.; Colombini M. (1998) The topology of the VDAC channel as probed by a biotin/streptavidin method. Biophys. J. 74, 24406–24413. [Google Scholar]
- Song J.; Midson C.; Blachly-Dyson E.; Forte M.; Colombini M. (1998) The sensor regions of VDAC are translocated from within the membrane to the surface during the gating processes. Biophys. J. 74, 2926–2944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombini M. (2009) The published 3D structure of the VDAC channel: Native or not?. Trends Biochem. Sci. 34, 382–389. [DOI] [PubMed] [Google Scholar]
- Hiller S.; Abramson J.; Mannella C.; Wagner G.; Zeth K. (2010) The 3D structures of VDAC represent a native conformation. Trends Biochem. Sci. 35, 514–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eddy M. T.; Ong T.-C.; Clark L.; Teijido O.; Van Der Wel P. C. A.; Garces R.; Wagner G.; Rostovtseva T. K.; Griffin R. G. (2012) Lipid Dynamics and Protein-Lipid Interactions in 2D Crystals Formed with the β-barrel Integral Membrane Protein VDAC1. J. Am. Chem. Soc. 134, 6375–6387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malia T. J.; Wagner G. (2007) NMR structural investigation of the mitochondrial outer membrane protein VDAC and its interaction with antiapoptotic Bcl-xL. Biochemistry 46, 514–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiller S.; Garces R. G.; Malia T. J.; Orekhov V. Y.; Colombini M.; Wagner G. (2008) Solution structure of the integral human membrane protein VDAC-1 in detergent micelles. Science 321, 1206–1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolder M.; Zeth K.; Tittmann P.; Gross H.; Welte W.; Wallimann T. (1999) Crystallization of the human, mitochondrial voltage-dependent anion-selective channel in the presence of phospholipids. J. Struct. Biol. 127, 64–71. [DOI] [PubMed] [Google Scholar]
- Rostovtseva T. K.; Kazemi N.; Weinrich M.; Bezrukov S. M. (2006) Voltage Gating of VDAC Is Regulated by Nonlamellar Lipids of Mitochondrial Membranes. J. Biol. Chem. 281, 37496–37506. [DOI] [PubMed] [Google Scholar]
- Zizi M.; Byrd C.; Boxus R.; Colombini M. (1998) The voltage-gating process of the voltage-dependent anion channel is sensitive to ion flow. Biophys. J. 75, 704–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombini M. (1989) Voltage Gating in the Mitochondrial Channel VDAC. J. Membr. Biol. 111, 103–111. [DOI] [PubMed] [Google Scholar]
- Bennett A.; Rienstra C.; Griffiths J.; Zhen W.; Lansbury P.; Griffin R. (1998) Homonuclear radio frequency-driven recoupling in rotating solids. J. Chem. Phys. 108, 9463–9479. [Google Scholar]
- Bennett A. E.; Rienstra C. M.; Auger M.; Lakshmi K. V.; Griffin R. G. (1995) Heteronuclear decoupling in rotating solids. J. Chem. Phys. 1–8. [Google Scholar]
- Thurber K. R.; Tycko R. (2008) Biomolecular solid state NMR with magic-angle spinning at 25 K. J. Magn. Reson. 195, 179–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Paëpe G.; Lewandowski J. R.; Loquet A.; Böckmann A.; Griffin R. G. (2008) Proton assisted recoupling and protein structure determination. J. Chem. Phys. 129, 245101–245121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi L.; Ahmed M. A. M.; Zhang W.; Whited G.; Brown L. S.; Ladizhansky V. (2009) Three-dimensional solid-state NMR study of a seven-helical integral membrane proton pump: Structural insights. J. Mol. Biol. 1–9. [DOI] [PubMed] [Google Scholar]
- Pines A.; Gibby M.; Waugh J. (1973) Proton-enhanced NMR of dilute spins in solids. J. Chem. Phys. 59, 569–590. [Google Scholar]
- Hartmann S.; Hahn E. (1962) Nuclear Double Resonance in Rotating Solidss. Phys. Rev. 128, 2042. [Google Scholar]
- Baldus M.; Petkova A. T.; Herzfeld J.; Griffin R. G. (1998) Cross Polarization in the Tilted Frame: Assignment and Spectral Simplification in Heteronuclear Spin Systems. Mol. Phys. 95, 1197–1207. [Google Scholar]
- Takegoshi K.; Nakamura S.; Terao T. (2001) 13C-1H dipolar-assisted rotational resonance in magic-angle spinning NMR. Chem. Phys. Lett. 344, 631–637. [Google Scholar]
- Bennett A. E.; Rienstra C. M.; Griffiths J. M.; Zhen W.; Lansbury P. T. Jr.; Griffin R. G. (1998) Homonuclear radio frequency-driven recoupling in rotating solids. J. Chem. Phys. 108, 9463. [Google Scholar]
- Phillips J. C.; Braun R.; Wang W.; Gumbart J.; Tajkhorshid E.; Villa E.; Chipot C.; Skeel R. D.; Kalé L.; Schulten K. (2005) Scalable molecular dynamics with NAMD. J. Comput. Chem. 26, 1781–1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks B. R.; Brooks C. L. III; Mackerell A. D. Jr.; Nilsson L.; Petrella R. J.; Roux B.; Won Y.; Archontis G.; Bartels C.; Boresch S.; Caflisch A.; Caves L.; Cui Q.; Dinner A. R.; Feig M.; Fischer S.; Gao J.; Hodoscek M.; Im W.; Kuczera K.; Lazaridis T.; Ma J.; Ovchinnikov V.; Paci E.; Pastor R. W.; Post C. B.; Pu J. Z.; Schaefer M.; Tidor B.; Venable R. M.; Woodcock H. L.; Wu X.; Yang W.; York D. M.; Karplus M. (2009) CHARMM: The biomolecular simulation program. J. Comput. Chem. 30, 1545–1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Im W.; Seefeld S.; Roux B. (2000) A Grand Canonical Monte Carlo-Brownian dynamics algorithm for simulating ion channels. Biophys. J. 79, 788–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Y.; Egwolf B.; Walters D. E.; Roux B. (2010) Ion Selectivity of α-Hemolysin with a β-Cyclodextrin Adapter. I. Single Ion Potential of Mean Force and Diffusion Coefficient. J. Phys. Chem. B 114, 952–958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egwolf B.; Luo Y.; Walters D. E.; Roux B. (2010) Ion Selectivity of α-Hemolysin with β-Cyclodextrin Adapter. II. Multi-Ion Effects Studied with Grand Canonical Monte Carlo/Brownian Dynamics Simulations. J. Phys. Chem. B 114, 2901–2909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Biase P. M.; Solano C. J. F.; Markosyan S.; Czapla L.; Noskov S. Y. (2012) BROMOC-D: Brownian Dynamics/Monte-Carlo Program Suite to Study Ion and DNA Permeation in Nanopores. J. Chem. Theory Comput. 8, 2540–2551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ujwal R.; Cascio D.; Colletier J. P.; Faham S.; Zhang J.; Toro L.; Ping P. P.; Abramson J. (2008) The crystal structure of mouse VDAC1 at 2.3 Å resolution reveals mechanistic insights into metabolite gating. Proc. Natl. Acad. Sci. U.S.A. 105, 17742–17747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The PyMOL Molecular Graphics System, version 1.5.0.4 (2012) Schrödinger, LLC, Portland, OR. [Google Scholar]
- Schneider R.; Etzkorn M.; Giller K.; Daebel V.; Eisfeld J.; Zweckstetter M.; Griesinger C.; Becker S.; Lange A. (2010) The native conformation of the human VDAC1 N terminus. Angew. Chem., Int. Ed. 49, 1882–1885. [DOI] [PubMed] [Google Scholar]
- Lewandowski J. R.; De Paëpe G.; Griffin R. G. (2007) Proton assisted insensitive nuclei cross polarization. J. Am. Chem. Soc. 129, 728–729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewandowski J. R.; De Paëpe G.; Eddy M. T.; Struppe J.; Maas W.; Griffin R. G. (2009) Proton assisted recoupling at high spinning frequencies. J. Phys. Chem. B 113, 9062–9069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blachly-Dyson E.; Peng S.; Colombini M.; Forte M. (1990) Selectivity changes in site-directed mutants of the VDAC ion channel: Structural implications. Science 247, 1233–1236. [DOI] [PubMed] [Google Scholar]
- Thomas L.; Blachly-Dyson E.; Colombini M.; Forte M. (1993) Mapping of residues forming the voltage sensor of the voltage-dependent anion-selective channel. Proc. Natl. Acad. Sci. U.S.A. 90, 5446–5449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombini M. (2004) VDAC: The channel at the interface between mitochondria and the cytosol. Mol. Cell. Biochem. 256–257, 107–115. [DOI] [PubMed] [Google Scholar]
- Baines C. P.; Kaiser R. A.; Sheiko T.; Craigen W. J.; Molkentin J. D. (2007) Voltage-dependent anion channels are dispensable for mitochondrial-dependent cell death. Nat. Cell Biol. 9, 550–555. [DOI] [PMC free article] [PubMed] [Google Scholar]