Abstract
Objectives:
Screening for mutations in large genes is challenging in a molecular diagnostic environment. Sanger-based DNA sequencing methods are largely used; however, massively parallel sequencing (MPS) can accommodate increasing test demands and financial constraints. This study aimed to establish a simple workflow to amplify and screen all coding regions of the BRCA1 and BRCA2 (BRCA1/2) genes by Sanger-based sequencing as well as to assess a MPS approach encompassing multiplex polymerase chain reaction (PCR) and pyrosequencing.
Methods:
This study was conducted between July 2011 and April 2013. A total of 20 patients were included in the study who had been referred to Genetic Health Services New Zealand (Northern Hub) for BRCA1/2 mutation screening. Patients were randomly divided into a MPS evaluation and validation cohort (n = 10 patients each). Primers were designed to amplify all coding exons of BRCA1/2 (28 and 42 primer pairs, respectively). Primers overlying known variants were avoided to circumvent allelic drop-out. The MPS approach necessitated utilisation of a complementary fragment analysis assay to eliminate apparent false-positives at homopolymeric regions. Variants were filtered on the basis of their frequency and sequence depth.
Results:
Sanger-based sequencing of PCR-amplified coding regions was successfully achieved. Sensitivity and specificity of the combined MPS/homopolymer protocol was determined to be 100% and 99.5%, respectively.
Conclusion:
In comparison to traditional Sanger-based sequencing, the MPS workflow led to a reduction in both cost and analysis time for BRCA1/2 screening. MPS analysis achieved high analytical sensitivity and specificity, but required complementary fragment analysis combined with Sanger-based sequencing confirmation in some instances.
Keywords: Massively Parallel Sequencing; BRCA1 Gene; BRCA2 Gene; HBOC Syndrome; Detection, heterozygote
Advances in Knowledge
- The research described in this study allows for capillary-based sequencing as well as massively parallel sequencing (MPS) of the BRCA1 and BRCA2 (BRCA1/2) genes for disease-causing mutations in patients with apparent hereditary breast and ovarian cancer.
- The results of this study have determined parameters for assessing sequence quality in order to reduce the number of false-positive calls using MPS and a pyrosequencing platform.
Application to Patient Care
- The use of MPS enables sequence-detectable mutations to be identified quickly, which is a significant advantage to all patients, regardless of their results.
- Furthermore, the MPS approach used in the current study was found to improve turn-around time for screening BRCA1/2 genes and reduce the cost of diagnostic screening.
Conventional sanger dideoxynucleotide (ddNTP) DNA sequencing is the most commonly used method of routine mutation screening.1–3 This method was developed in the 1970s and has become the gold standard for diagnostic sequencing.1,4 However, the cost of Sanger-based sequencing is relatively high and the procedure is time-consuming, making it impractical for screening large genes such as BRCA1 and BRCA2 (BRCA1/2).
The increasing demand for diagnostic genetic testing in a clinical setting has created the need for an alternative technology that can accommodate high throughput, while reducing costs and turnaround times. The development of massively parallel sequencing (MPS), also known as next-generation sequencing, enables the sequencing of millions of DNA fragments in a single run.5,6 MPS technology has been readily adopted in research settings and has recently moved into the diagnostic environment; however, this has led to issues regarding sequence quality parameters and the need for comprehensive bioinformatic analysis.
In New Zealand, breast cancer is the most frequently registered cancer and the second leading cause of cancer-related deaths among women. Compared to the second half of last century, the incidence of breast cancer has increased in New Zealand.7 Germline mutations in the BRCA1/2 genes account for approximately 10–15% of all breast and ovarian cancers; these are known as hereditary breast and ovarian cancers (HBOC).8,9 The BRCA1/2 genes, which were identified by positional cloning during the 1990s, encode for proteins that are responsible for controlling cellular growth and differentiation.8
The majority of germline mutations in the BRCA1/2 genes are either nonsense or frame-shift mutations, which result in truncated proteins.8,9 Exonic deletions or duplications have also been reported but are rare. In view of the above, together with the large number of reported mutations, screening the coding regions of the BRCA1/2 genes for mutations has largely involved denaturing high-performance liquid chromatography or direct sequencing.10–14 In contrast, targeted mutation analysis using conventional Sanger-based sequencing is a common initial testing strategy for individuals of Ashkenazi Jewish ancestry with HBOC. Three founder mutations have been described in this population: c.68_69delAG and c.5266dupC in the BRCA1 gene and c.5946delT in the BRCA2 gene, with a combined frequency of approximately 2%.15,16 The BRCA1/2 genes are classic examples of the difficulties commonly encountered in a diagnostic setting: they are large autosomal genes with a wide spectrum of mutations, rich in homopolymeric regions and are highly polymorphic, complicating primer designs for polymerase chain reaction (PCR) amplification.17
The objective of this study was to investigate the diagnostic potential of the more cost-effective MPS approach in screening for mutations in the BRCA1/2 genes, compared to conventional Sanger-based sequencing. This strategy involved a pyrosequencing approach in order to develop a rapid, inexpensive and rigorous assay for identifying disease-causing mutations in the BRCA1/2 genes.
Methods
This study was carried out between July and April 2013. A total of 20 patients who had been referred to Genetic Health Services New Zealand (Northern Hub) for BRCA1/2 gene mutation screening were selected for inclusion. Of the patients, 16 carried known disease-causing mutations. These patients carried a range of BRCA1/2 gene variants, including missense, nonsense, duplication, insertion-deletion and deletion variants. The analysis of whole exon deletion events was excluded from the current study as it concerned only the ability to detect intra-exonic and splice site mutations.
A total of 10 patients were randomly assigned to comprise the MPS evaluation cohort. Genomic DNA from the evaluation group was subjected to two sequencing strategies. The first comprised exontargeted amplification and subsequent Sanger-based sequencing of the coding regions of BRCA1/2 while the second comprised the MPS approach described below. A comparison of the latter data against the former allowed MPS analysis parameters to be determined. Identified parameters were then applied to the genomic DNA of the 10 patients in the MPS validation cohort in order to identify sequence variants. Variants were identified using SeqNext, Sequence Pilot software, Version 3.4.2 (JSI Medical Systems GmbH, Kippenheim, Germany). All identified variants, as well as amplicons with insufficient coverage (set at 30x coverage), were subjected to Sanger-based sequencing to determine the sensitivity and specificity of the MPS approach. A fragment analysis approach was also used in conjunction with MPS to reduce the frequency of the false-positive (FP) calls that required Sanger-based sequencing confirmation.
DNA was extracted from peripheral ethylene-diaminetetraacetic acid (EDTA) blood samples using the Gentra® Puregene® Blood Kit (3 mL) (Qiagen, Venlo, Limburg, Netherlands), according to the manufacturer’s instructions. These DNA samples were assessed by an accredited overseas laboratory to validate the local gene screening strategy used in the study.
In order to offer local BRCA1/2 gene screening, primers were designed for the simple amplification of all coding regions of the two genes. The design process used, which allows for amplification using a standard reaction condition and cycling protocol, has been previously described.18 Critically, the primer designs involved extensive analysis of the Single Nucleotide Polymorphism Database (dbSNP)19 and an iterative design process in order to ensure that primers did not overlie known single nucleotide polymorphisms (SNPs) using SNPCheck (National Genetics Reference Laboratory, Manchester, UK).20 In the case of Sanger-based sequencing, 70 pairs of primers were designed to flank all of the coding and adjacent splice junction regions of the BRCA1/2 genes for PCR amplification [Appendix 1].21,22 In total, 140 fragments were generated with amplicon lengths of 218–745 base pairs (bp). The amplicon lengths were short enough to allow complete bi-directional sequencing of the coding regions of the BRCA1/2 genes. In order to allow the unambiguous sequencing of amplicons containing repetitive sequences, eight nested primers were also designed [Appendix 2].
The coding regions of the BRCA1/2 genes were then amplified in a reaction volume of 25 μL containing 50 ng of genomic DNA, 0.4 mM of ddNTP (GE Healthcare Ltd., Little Chalfont, UK), 0.8 μM each of forward and reverse primers, 2 mM of magnesium chloride (MgCl2) and one unit of PCR reaction buffer without MgCl2 together with one unit of FastStart Taq DNA Polymerase (Roche Applied Science, Penzberg, Upper Bavaria, Germany). The thermal cycling conditions consisted of a denaturation step of 95 °C for five minutes, 35 cycles of 94 °C for 45 seconds, 60 °C for 30 seconds and 72 °C for 30 seconds, followed by a final extension of 72 °C for 10 minutes. Purification of each 5 μL of PCR product was performed using USB® ExoSAP-IT® (Affymetrix Inc., Santa Clara, California, USA). The purified PCR product was then diluted to 2.5 ng per 100 bp and sequenced bi-directionally using M13 forward and reverse primers and BigDye® Terminator Version 3.1 Cycle Sequencing Kit (Applied Biosystems, Life Technologies, Thermo Fisher Scientific Corp., Carlsbad, California, USA). Sequenced products (5 μL) were purified using the BigDye® XTerminator™ Purification Kit (Applied Biosystems) and then subjected to capillary electrophoresis using a 3130xl Genetic Analyser (Applied Biosystems) with a 50 cm capillary array. Sequence traces were analysed using KB™ Basecaller Sequencing Analysis Software, Version 1.4, and Variant Reporter™ Software, Version 1.0 (Applied Biosystems), with a minimum trace score of 35, which corresponds to an average FP base call frequency of 0.031%.
MPS library amplification involved the amplification of the coding regions of the BRCA1/2 genes using the BRCA MASTR™ Dx assay (Multiplicom, Niel, Belgium) in a reaction volume of 15 μL containing 50 ng of genomic DNA. This molecular diagnostic assay allows for multiplex PCR amplification of 93 amplicons in five separate PCRs for each patient. The thermal cycling conditions consisted of a denaturation step of 95 °C for 10 seconds, 20 cycles consisting of 95 °C for 45 seconds, 60 °C for 45 seconds and 68 °C for two minutes, followed by a final extension of 72 °C for 10 minutes.
Secondary PCR used the 454 MID Dx kit (Multiplicom) to tag the amplicons with patient-specific molecular barcodes and 454 sequencing adaptors (A and B) for downstream sequencing. The multiplex identifier (MID) sequences consist of six unique nucleotides and are used to index each BRCA library, allowing multiple patient libraries to be multiplexed in a single MPS run. The primary PCR products were diluted in sterile water (1:1000) and 1 μL of this product was used as a template for the secondary PCR with indexing adapters in a total reaction volume of 25 μL. The thermal cycling conditions consisted of a denaturation step at 98 °C for 10 seconds, 20 cycles consisting of 95 °C for 45 seconds, 64 °C for 45 seconds and 68 °C for two minutes, followed by a final extension of 72 °C for 10 minutes.
Subsequent pooling of tagged amplicons for each patient was followed using a predefined mixing protocol (Multiplicom). Each patient’s amplicon-pooled library was purified using Agencourt AMPure XP (Beckman Coulter Inc., Beverly, Massachusetts, USA) to remove small residual DNA fragments such as primer dimers. The concentration of each patient’s amplicon-pooled library was determined using QuantiT ™ PicoGreen® dsDNA Reagent and Assay Kit (Invitrogen, Life Technologies) and normalised to 10 nM in Tris-EDTA buffer (pH 8). In each experiment, five patients’ amplicon-pooled libraries were pooled in an equimolar ratio to generate a sequencing library, which contained a total of 465 amplicons for sequencing on the MPS platform.
Emulsion PCR using the GS Junior Titanium emPCR Lib-A Kit (Roche Diagnostics Corp., Indianapolis, Indiana, USA) was performed to clonally amplify single DNA molecules in their own micro-reactors. This was achieved by hybridising the DNA library onto primer-coated beads together with emulsification through vigorous shaking of an oil-water mixture and amplification reagents to achieve emulsion micro-reactors. After subsequent PCR amplification, the micro-reactors were broken and DNA-positive beads were deposited onto a picotiter plate for subsequent pyrosequencing.
Data processing was carried out using a predefined amplicon pipeline program, Genome Sequencer Run Processor (Roche Applied Science). Sequence data were aligned against the reference sequences NC_000017.10 (NM_007294.3; BRCA1) and NC_000013.10 (NM_000059.3; BRCA2) from the Human Genome Assembly (HG19; Genome Bioinformatics Group, University of California Santa Cruz, USA). Nomenclature from the Human Genome Variation Society (HGVS), Version 2.0,23 was used to describe all variants with nucleotide numbering starting from the first nucleotide of the translated sequence. Amplicons with flanking regions of 3–20 bp upstream and downstream were analysed using SeqNext. Settings were customised to achieve a Phred quality score equivalent to 33, as described in others studies.24–26 SeqNext sorts the sequence data for each patient according to the attached MID and variant calling is achieved based on a defined variant frequency, which was set at 20%. As pyrosequencing errors commonly arise in homopolymeric regions, a secondary threshold for these regions of 35% was set to filter out apparent FPs. Currently, a minimum read depth of 20x is widely adopted in research settings as not all targeted regions are evenly sequenced by MPS.25,27,28 This threshold, termed the minimum base coverage threshold, is necessary in order to avoid possible variant miscalling.29 The minimum base coverage threshold was set at 30x (combined forward and reverse reads) [Table 1]. An initial variant frequency (VF) of 20% allowed reliable variant calling. Subsequent VF ranges of 40–60% and 90–100% were used for calling heterozygotes and homozygotes, respectively.
Table 1:
Custom settings applied to SeqNext, Sequence Pilot Software, Version 3.4.2 (JSI Medical Systems GmbH, Kippenheim, Germany) to identify sequence variants in the genomic DNA of 10 patients
| Setting | Parameter |
|---|---|
| Analyse/ignore region: minimum absolute coverage | Off |
| Low coverage warning | 30 |
| Mutations: minimum absolute coverage | Off |
| Mutations: minimum % coverage | 20% per direction |
| Mutations sorting: distinct/other % coverage | 20% per direction |
| Mutations sorting: distinct/homopolymer coverage | 35% per direction |
BRCA1/2 genes are homopolymer (HP)-rich and sequencing homopolymeric regions is a known limitation of pyrosequencing technology which can hamper its effective use.24,30,31 For this reason, a commercially available fragment analysis assay was used (BRCA HP kit, Version 2.0, Multiplicom) to screen for variants in targeted homopolymeric regions in the BRCA1/2 genes. A fragment analysis approach was used in conjunction with MPS to reduce the frequency of the FP calls that required Sanger-based sequencing confirmation. This assay targets 29 homopolymeric regions (stretches of 6 bp or more) within the coding regions of the BRCA1/2 genes. Two multiplex PCRs were set up in reaction volumes of 15 μL containing 30 ng of genomic DNA. The thermal cycling conditions consisted of a denaturation step at 98 °C for 10 minutes, 24 cycles consisting of 95 °C for 45 seconds, 60 °C for 45 seconds and 68 °C for two minutes, followed by a final extension of 72 °C for 10 minutes. The PCR products were electrophoresed in the 3130xl Genetic Analyzer and the data were analysed using MAQ-S software, Version 1.4.0 (Multiplicom).
Three strategies were established to reduce the number of FP calls. First, MPS using a pyrosequencing approach required a complementary strategy to screen homopolymeric regions. Regions not covered by the fragment analysis assay were therefore subjected to Sanger-based sequencing. Second, literature searches and bioinformatic splice site analyses using the Berkeley Drosophila Genome Project Program (BDGP)32,33 were used in the case of intronic single base insertions or deletions occurring in homopoly-meric tracts of ≥6 bp or outside of ±3 bp of the exon-intron boundaries. These putative variants would be filtered out if they were determined highly unlikely to be of pathogenic significance, as reporting them would be of no clinical significance. Third, it was decided not to report variants in the following untranslated regions (UTRs) of the BRCA2 gene: c.-26G>A (5′ UTR) and c.*105A>C (3′ UTR). While these variants are listed in the Breast Cancer Information Core (BIC) database,34 they have no known clinical significance and thus fall outside the regions of interest (ROI) of the current study. These exclusion strategies were applied to the MPS analysis of the validation cohort.
All patients gave informed consent to be included in this study.
Results
Sanger-based sequencing was applied to the evaluation cohort (n = 10) who carried a range of BRCA1/2 gene variants, including missense, nonsense, insertion-deletion and deletion variants. Table 2 summarises the variants that were detected. External laboratory reports on the same DNA samples confirmed the mutations reported.
Table 2:
Variants detected in the BRCA1 and BRCA2 genes of 20 patients using Sanger-based sequencing
| Nucleotide | Codon | Type | Patient | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | |||
| BRCA1 | ||||||||||||
| c.140G>A | p.Cys47Tyr | M | Het* | |||||||||
| c.1067A>G | p.Gln356Arg | M | Het | Het | Het | |||||||
| c.1175_1214del40 | p.Leu392Glnfs*5 | F | Het* | |||||||||
| c.2077G>A | p.Asp693Asn | M | Het | |||||||||
| c.2082C>T | p.Ser694Ser | S | Het | Het | Het | Hom | Hom | |||||
| c.2311T>C | p.Leu771Leu | S | Het | Het | Het | Hom | Hom | |||||
| c.2315T>C | p.Val772Ala | M | Het | |||||||||
| c.2612C>T | p.Pro871Leu | M | Het | Het | Het | Hom | Hom | |||||
| c.3113A>G | p.Glu1038Gly | M | Het | Het | Het | Hom | Hom | |||||
| c.3548A>G | p.Lys1183Arg | M | Het | Het | Het | Hom | Hom | |||||
| c.3756_3759delGTCT | p.Ser1253Argfs*10 | F | Het* | |||||||||
| c.4065_4068delTCAA | p.Asn1355Lysfs*10 | F | Het* | |||||||||
| c.4308T>C | p.Ser1436Ser | S | Het | Het | Het | Hom | Hom | |||||
| c.4837A>G | p.Ser1613Gly | M | Het | Het | Het | Hom | Hom | |||||
| BRCA2 | ||||||||||||
| c.68-7T>A | - | I | Het | |||||||||
| c.865A>C | p.Asn289His | M | Het | |||||||||
| c.1114A>C | p.Asn372His | M | Het | Het | Het | Hom | Het | Het | Het | |||
| c.1365A>G | p.Ser455Ser | S | Het | |||||||||
| c.1395A>C | p.Val465Val | S | Het | |||||||||
| c.2229T>C | p.His743His | S | Het | |||||||||
| c.2971A>G | p.Asn991Asp | M | Het | |||||||||
| c.3396A>G | p.Lys1132Lys | S | Het | Het | Het | Het | ||||||
| c.3807T>C | p.Val1269Val | S | Het | Het | Het | |||||||
| c.7242A>G | p.Ser2414Ser | S | Het | Het | Het | Het | ||||||
| c.7655_7658delTTAA | p.Ile2552Thrfs*95 | F | Het* | |||||||||
| c.7762_7764delATAinsTT | p.Ile2588Phefs*60 | F | Het* | |||||||||
| c.7806-14T>C | - | I | Het | Het | Hom | Het | Het | Het | Het | Hom | ||
| c.8297delC | p.Thr2766Asnfs*11 | F | Het* | |||||||||
| c.8575delC | p.Gln2859Lysfs*4 | F | Het* | |||||||||
M = missense; Het = heterozygote; Het* = heterozygote disease-causing mutation; FS = frame-shift; S = synonymous; Hom = homozygous; I = intronic.
In total, 128 variants were identified, of which 80 variants were true-positives (TPs) and 48 variants were apparent FPs. The latter were identified by comparing the variant calls against the Sanger-based sequencing data. Of these 48 apparent FPs, 43 lay in homopolymeric regions. Of the 43 apparent FP variants in homopolymeric regions, 17 were in exons (13 were covered by the BRCA HP kit and four lay in homoploymeric tracts of five bases only) and 26 were in introns. Of the remaining five apparent FPs, three corresponded to the same intronic deletion of one base in a homopolymeric tract that was three bases upstream of a splice acceptor site and two were sequenced at insufficient read depth.
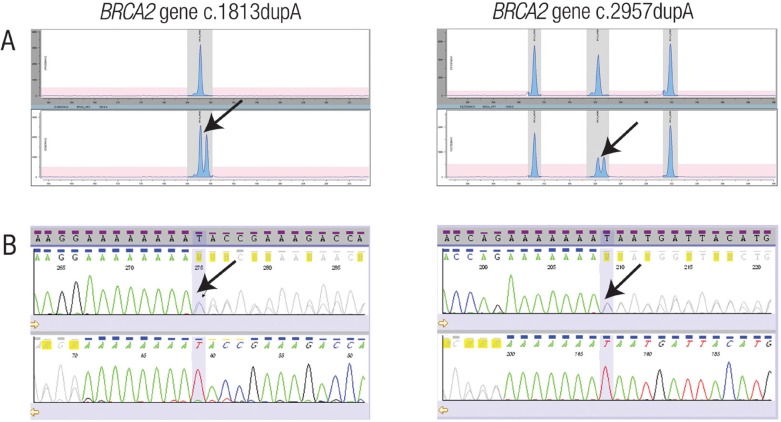
The MPS analysis parameters and the exclusion strategies derived from the evaluation cohort were applied to the MPS analysis of the validation cohort (n = 10). A total of 115 variants were identified using the MPS analysis parameters; however, 66 calls remained after applying the exclusion strategies. Sanger sequencing confirmed 62 of these 66 calls to be TPs. The remaining four calls were localised to homopolymeric regions not covered by the fragment analysis assay. Importantly, two pathogenic mutations in the BRCA2 gene (c.1813dupA and c.2957dupA) were not detected by MPS analysis. These mutations occurred in the homopolymeric regions comprising eight and seven adenosines, respectively. However, these two duplications were clearly identified using the fragment analysis assay and MAQ-S software [Figures 1A & B]. The sensitivity and specificity of the combined MPS/HP protocol was determined to be 100% and 99.5%, respectively.
Figure 1 Panel A & B:
Fragment analysis of homopolymeric regions. A: Electropherograms of selected fragments amplified using a commercially available fragment analysis assay. Heterozygotes are identified by arrows. B: Sequence electropherograms of the heterozygous regions of the BRCA1 and BRCA2 genes shown in panel A. The arrows indicate that length heterozygosity is due to a differing number of adenosines.
Combining the results of the two cohorts, there were 17 true mutations, 11 FPs that were not filtered out using the exclusion criteria, four with a VF out of the acceptable range and two with insufficient base coverage of less than 30 reads. As two of these confirmations occurred in the same amplicon of one patient, a total of 33 amplicons required sequencing confirmation. A summary of the variants detected in all 20 patients is presented in Appendix 3, together with conclusions regarding their pathogenic status based on the BIC database and the Human Genome Mutation Database (BIOBASE HGMD® Professional, BIOBASE Biological Databases, Beverly, Massachusetts, USA).23,34,35 Overall, the sequence data showed variations of approximately 10,000–26,700 total read counts per patient. In terms of read counts per amplicon, the variation was 30–1,900. The Sanger-based sequencing approach achieved a Phred score of 35 with a base call accuracy of 99.97%.
The MPS/HP mutation screening strategy for BRCA1/2 genes resulted in significant savings in terms of both the cost of consumables and time by three-fold and two-fold, respectively [Appendix 4].
Discussion
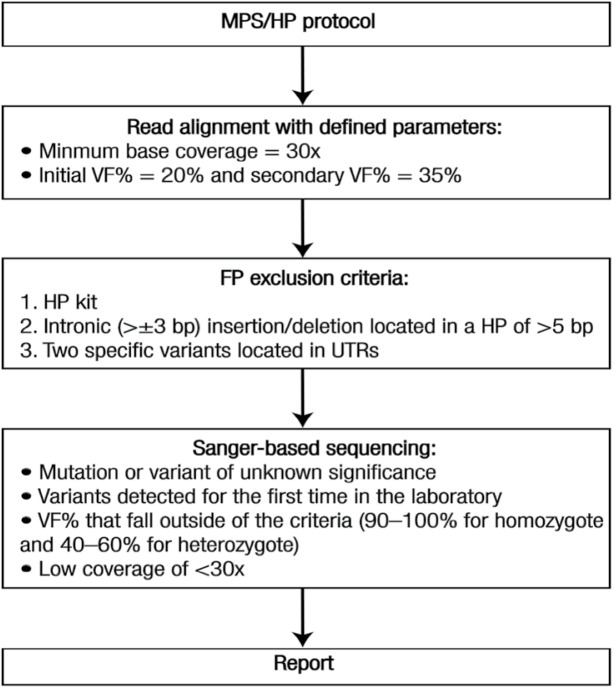
In this study, a simple workflow was established using a benchtop MPS platform to perform comprehensive mutation screening of the BRCA1/2 genes [Figure 2]. The MPS analysis achieved high analytical sensitivity and specificity, but required complementary fragment analysis combined with Sanger-based sequencing confirmation in some instances. This complementary approach eliminated some of the concerns about the inherent limitations of pyrosequencing technology. The MPS/HP mutation screening strategy for BRCA1/2 genes resulted in significant savings in terms of both the cost of consumables and time. These advantages are important criteria for diagnostic laboratories.36 An added advantage of this study was the identification of all variants within the ROI, which comprised the coding regions and flanking 20 bp of the BRCA1/2 genes.
Figure 2:
Flowchart of massively parallel sequencing/homopolymer workflow for BRCA1 and BRCA2 referrals.
MPS = massively parallel sequencing; HP = homopolymer; VF = variant frequency; FP = false-positive; UTR = untranslated region.
The quality of sequencing data is the major concern when implementing MPS in a diagnostic setting. The Phred score system has been widely adopted as a quality measure for automated sequencing approaches.24 Phred scores of 20 for bi-directional sequence data and 30 for uni-directional data have been recommended as rigorous quality control parameters.37 The Sanger-based sequencing approach utilised had a Phred score of 35 and a 99.97% base call accuracy; it is crucial for a diagnostic laboratory to maintain this score quality when implementing MPS. In order to achieve a similar MPS Phred score, a depth of coverage of 30x and a VF of 20% were implemented. These measures were chosen based on the results of the evaluation cohort and the findings of De Leeneer et al.25
Also referred to as the allelic fraction, VF is the proportion of individual reads containing the variant call.25 A VF of 100% for homozygotes and 50% for heterozygotes is ideal. However, cut-off frequencies for heterozygous and homozygous calls have been reported from 23–74% and 78–100%, respectively.25 According to Jones et al., variant calls of <85% for homozygous variants and <40% for heterozygous variants should be discarded.38 From the MPS data in the current study, a homozygous variant with a VF of 81% was observed; Sanger-based sequencing determined that this variant was a true heterozygote. Based on Jones et al.’s findings, a false-negative result would have been generated if this call was discarded.38 As a result, cut-off ranges were defined as 90–100% and 40–60% for homozygotes and heterozygotes, respectively. Genotypes of variants that fell outside of the stated thresholds were confirmed by Sanger-based sequencing.
In the current study, a total of 33 amplicons required sequencing confirmation. However, this sequence load reflects samples that were biased towards positive mutations. In the case of a mutation-negative patient, a Sanger-based sequence load of approximately one amplicon would be expected, which would include the confirmation of any non-pathogenic or missense variants with unknown clinical significance detected for the first time.
The ability to multiplex samples is one of the main advantages of MPS, allowing for cost-effective sequencing with a high sample throughput. The current study was largely confined to analysing five patients per single experiment. An experiment where seven samples were multiplexed was performed to improve cost savings; however, both sensitivity and specificity decreased with less than 10,000 reads generated for each sample. This is lower than the manufacturer’s recommendation of 11,000 reads per sample. In addition, the average base coverage was 106 ± 58 bp, which indicated that multiplexing seven samples decreased the confidence of variant calls due to lower coverage across the ROI.
MPS technology is rapidly evolving and further developments and improvements will continue to reduce costs. While the current study has presented an introduction to MPS in the context of a small diagnostic laboratory, the possibility of whole exome sequencing and even whole genome sequencing is attractive, albeit requiring a significant increase in data storage and processing. However, these developments suggest that screening a defined number of genes in a targeted approach, as shown in this study, will be superseded by filtering sequence data to identify variants in a defined gene list that is relevant to the clinical referral.
The validation of the approach described here was limited in scope due to the number of patients analysed; hence, the current study could be considered under-powered.39 In order to mitigate this feature, further work has been undertaken assessing an additional 70 patients referred for BRCA1/2 gene sequencing by MPS. These data (not shown here) comprise the identification of approximately 500 variants, all of which are either known exonic and intronic SNPs (averaging seven per patient), known mutations (eight patients) or novel mutations (two patients). All mutations were confirmed as correct by Sanger-based sequencing. Importantly, the exon-specific primer designs have provided a critical resource for the confirmation of variants detected by MPS and for predictive testing. The fact that the primers do not overlie known SNPs has reduced the likelihood of allelic drop-out.
Conclusion
In comparison to traditional Sanger-based sequencing, the MPS workflow led to a reduction in cost, analysis time and turn-around time for the screening of BRCA1/2 genes. The analysis used achieved high analytical sensitivity and specificity, but required complementary fragment analysis combined with Sanger-based sequencing confirmation in some instances. MPS technology is rapidly evolving and further developments and improvements will continue to reduce costs. The current study presented an introduction to MPS in the context of a small diagnostic laboratory.
Appendices
Primer sequences, nested primers for sequencing selected exons, BRCA1/2 gene variants detected and a brief cost-efficiency analysis of the methods used are available in four supplementary tables included in the online version of this article.
Appendix 1: Primer sequences for amplifying the coding regions of the BRCA1 and BRCA2 genes
| Exon | Forward Primer Sequences | Reverse Primer Sequences |
|---|---|---|
| BRCA1 | ||
| 2 | TGTAAAACGACGGCCAGTGGACGTTGTCATTAGTTCTTTGG | CAGGAAACAGCTATGACCAGAGGCAGAGTGGATGGAGA |
| 3 | TGTAAAACGACGGCCAGTAAATATTGAACGAACTTGAGGC | CAGGAAACAGCTATGACCGGTGTTTCCTGGGTTATGAAG |
| 4 | TGTAAAACGACGGCCAGTGGCTCTTAAGGGCAGTTGTG | CAGGAAACAGCTATGACCAAACTTTCAGGAAAATAACTTTGG |
| 5 | TGTAAAACGACGGCCAGTTTGATTATAGAGGTTTTCTACTGTTGC | CAGGAAACAGCTATGACCAAAAGGTCTTATCACCACGTCATAG |
| 6 | TGTAAAACGACGGCCAGTTGTCTTAACACAACAAAGAGCATAC | CAGGAAACAGCTATGACCGAGGACTGCTTCTAGCCTGG |
| 7 | TGTAAAACGACGGCCAGTTTCTCTTCAGGAGGAAAAGCAC | CAGGAAACAGCTATGACCTTGGCAAAACTATAAGATAAGGAATC |
| 8 | TGTAAAACGACGGCCAGTCTAGCATTGTACCTGCCACAG | CAGGAAACAGCTATGACCTGCACATACATCCCTGAACC |
| 9 | TGTAAAACGACGGCCAGTCCCAGCAACCATTTCATTTC | CAGGAAACAGCTATGACCAACGAAAGGGCAACAATCAG |
| 10-1 | TGTAAAACGACGGCCAGTGGTTGATTTCCACCTCCAAG | CAGGAAACAGCTATGACCCAGACTCCCCATCATGTGAG |
| 10-2 | TGTAAAACGACGGCCAGTTGCCATGCTCAGAGAATCC | CAGGAAACAGCTATGACCAGCTTTCGTTTTGAAAGCAG |
| 10-3 | TGTAAAACGACGGCCAGTCAAGAAAGCAGATTTGGCAG | CAGGAAACAGCTATGACCTTTCTTCTCTTGGAAGGCTAGG |
| 10-4 | TGTAAAACGACGGCCAGTGAGCCAAGAAGAGTAACAAGCC | CAGGAAACAGCTATGACCTTGTTTCTTTAAGGACCCAGAG |
| 10-5 | TGTAAAACGACGGCCAGTGCAGAATACATTCAAGGTTTCAAAG | CAGGAAACAGCTATGACCTCATTAATACTGGAGCCCACTTC |
| 10-6 | TGTAAAACGACGGCCAGTACAGTGAGCACAATTAGCCG | CAGGAAACAGCTATGACCAAGCAGGGAAGCTCTTCATC |
| 10-7 | TGTAAAACGACGGCCAGTGGAGTCCTAGCCCTTTCACC | CAGGAAACAGCTATGACCTGCTCCCCAAAAGCATAAAC |
| 11 | TGTAAAACGACGGCCAGTAATCCAGTCCTGCCAATGAG | CAGGAAACAGCTATGACCAATGCAAAGGACACCACACA |
| 12 | TGTAAAACGACGGCCAGTTTAAAAGGTGTTCAGCTAGAACTTG | CAGGAAACAGCTATGACCGGACAAGAACCAAGGCTCC |
| 13 | TGTAAAACGACGGCCAGTTTGTGTATCATAGATTGATGCTTTTG | CAGGAAACAGCTATGACCGCAATAAAAGTGTATAAATGCCTG |
| 14 | TGTAAAACGACGGCCAGTTCTGATCTCTCTGACATGAGCTG | CAGGAAACAGCTATGACCACACCAAGACTCCCTCATCC |
| 15 | TGTAAAACGACGGCCAGTAATTCTTAACAGAGACCAGAACTTTG | CAGGAAACAGCTATGACCTGACAATACCTACATAAAACTCTTTCC |
| 16 | TGTAAAACGACGGCCAGTCACTTTAAATAGTTCCAGGACACG | CAGGAAACAGCTATGACCCGCCTCATGTGGTTTTATGC |
| 17 | TGTAAAACGACGGCCAGTTCCAGATTGATCTTGGGAGTG | CAGGAAACAGCTATGACCTGGTAACTCAGACTCAGCATCAG |
| 18 | TGTAAAACGACGGCCAGTAGGGAAGGACCTCTCCTCTG | CAGGAAACAGCTATGACCGGTGCATTGATGGAAGGAAG |
| 19 | TGTAAAACGACGGCCAGTTGTCTGCTCCACTTCCATTG | CAGGAAACAGCTATGACCTGGAATACAGAGTGGTGGGG |
| 20 | TGTAAAACGACGGCCAGTGCAGCAGAAATCATCAGGTG | CAGGAAACAGCTATGACCTTCAGCAATCTGAGGAACCC |
| 21 | TGTAAAACGACGGCCAGTACCATTGTCCTTTGGAGCAG | CAGGAAACAGCTATGACCATGGAGTCTTTTGGCACAGG |
| 22 | TGTAAAACGACGGCCAGTTGAAGTGACAGTTCCAGTAGTCC | CAGGAAACAGCTATGACCAAACCAAACCCATGCAAAAG |
| 23 | TGTAAAACGACGGCCAGTAGGACCCTGGAGTCGATTG | CAGGAAACAGCTATGACCAAATAATGAATCAGCATCTTGCTC |
| BRCA2 | ||
| 2 | TGTAAAACGACGGCCAGTAATGCATCCCTGTGTAAGTGC | CAGGAAACAGCTATGACCAGCAACACTGTGACGTACTGG |
| 3 | TGTAAAACGACGGCCAGTTGCCTTAACAAAAGTAATCCATAGTC | CAGGAAACAGCTATGACCGATTTGCCCAGCATGACAC |
| 4 | TGTAAAACGACGGCCAGTAAACACTTCCAAAGAATGCAAA | CAGGAAACAGCTATGACCTCTACCAGGCTCTTAGCCAAA |
| 5/6 | TGTAAAACGACGGCCAGTCCAGCAGCTGAAATTTGTGAG | CAGGAAACAGCTATGACCTCTCAGGGCAAAGGTATAACG |
| 7 | TGTAAAACGACGGCCAGTAGCGTTATACCTTTGCCCTG | CAGGAAACAGCTATGACCTTGACACCACTGGACTACCAC |
| 8 | TGTAAAACGACGGCCAGTTTCACTGTGTTGATTGACCTTTC | CAGGAAACAGCTATGACCGACAGCAGAGTTTCACAGGAAG |
| 9 | TGTAAAACGACGGCCAGTTCCATTTCCATTTTCCTTTCC | CAGGAAACAGCTATGACCTCACGACCATTTGAGACCAG |
| 10-1 | TGTAAAACGACGGCCAGTTGTTTCTATGAGAAAGGTTGTGAGA | CAGGAAACAGCTATGACCACAGGCCAAAGACGGTACAA |
| 10-2 | TGTAAAACGACGGCCAGTGGAACCAAATGATACTGATCC | CAGGAAACAGCTATGACCTTTCCAGTCCACTTTCAGAGG |
| 10-3 | TGTAAAACGACGGCCAGTGTGGCTTCTTCATTTCAGGG | CAGGAAACAGCTATGACCCAGAAGGAATCGTCATCTATAAAAC |
| 11-A | TGTAAAACGACGGCCAGTCACTGTGCCCAAACACTACC | CAGGAAACAGCTATGACCGCACATACATCTTGATTCTTTTCC |
| 11-B | TGTAAAACGACGGCCAGTCAAGGATGTTCTGTCAAACCTAGTC | CAGGAAACAGCTATGACCTTGGACCTAAGAGTCCTGCC |
| 11-C | TGTAAAACGACGGCCAGTAAGTAGCTAATGAAAGGAATAA | CAGGAAACAGCTATGACCGTAAATGTGCAGATACAGTATTA |
| 11-D | TGTAAAACGACGGCCAGTACAAATGGGCAGGACTCTTAGGTC | CAGGAAACAGCTATGACCGATCAGCATCTCTGCATTCCTCAGAAG |
| 11-E | TGTAAAACGACGGCCAGTCAGATGTTATTTTCCAAGCAGG | CAGGAAACAGCTATGACCCCCCTAAACCCCACTTCATT |
| 11-F | TGTAAAACGACGGCCAGTGAGGAATGCAGAGATGCTGA | CAGGAAACAGCTATGACCTTGAATCACTGCCATCAAATTC |
| 11-G | TGTAAAACGACGGCCAGTTCTTCAAGTAAATGTCATGATTCTG | CAGGAAACAGCTATGACCTCCATTTTGTTCTTTCTTATGTCAG |
| 11-H | TGTAAAACGACGGCCAGTGCCAGTTTATGAAGGAGGG | CAGGAAACAGCTATGACCGTCACTAGTTGATTTCCAGTACC |
| 11-I | TGTAAAACGACGGCCAGTTCAGACTGCAAGTGGGAAAA | CAGGAAACAGCTATGACCAAGTTGCAGGACTTTTTGCTG |
| 11-J | TGTAAAACGACGGCCAGTAAAACCTTGTTTCTATTGAGACTGTG | CAGGAAACAGCTATGACCCAGTTTGTGGGTATGCATTTG |
| 11-K | TGTAAAACGACGGCCAGTAATGATTCAGGATATCTCTCAAA | CAGGAAACAGCTATGACCACTTTCTCCAATCCAGACATAT |
| 11-L | TGTAAAACGACGGCCAGTGGCCACCTGCATTTAGGATA | CAGGAAACAGCTATGACCCTTGCGTTTTGTAATGAAGCA |
| 11-M | TGTAAAACGACGGCCAGTTGCCAAACGAAAATTATGGC | CAGGAAACAGCTATGACCAGATGAATTTACCACATTATATG |
| 11-N | TGTAAAACGACGGCCAGTGCTTCATAAGTCAGTCTCATCTGC | CAGGAAACAGCTATGACCGCTCTGGGTTTCTCTTATC |
| 11-O | TGTAAAACGACGGCCAGTAATACTGCTATACGTACTCCAGA | CAGGAAACAGCTATGACCCCAAAACATGAATGTTCTCA |
| 11-P | TGTAAAACGACGGCCAGTTTCACCTACGTCTAGACAAAATGTATC | CAGGAAACAGCTATGACCACACTTTAAAAATAGTGATTGGCAAC |
| 12 | TGTAAAACGACGGCCAGTCTGACTTTACTCTTTCAAACATTAGG | CAGGAAACAGCTATGACCAAATGCTCTTTTAGGTCCTCAGT |
| 13 | TGTAAAACGACGGCCAGTTGCTGATTTCTGTTGTATGCTTG | CAGGAAACAGCTATGACCTTGGCTTCCAAACTTTTGTTG |
| 14 | TGTAAAACGACGGCCAGTATGAGGGTCTGCAACAAAGG | CAGGAAACAGCTATGACCGGGAAAACCATCAGGACATT |
| 15 | TGTAAAACGACGGCCAGTGCCTCAGCCTGCTGAATA | CAGGAAACAGCTATGACCCCATTCCTGCACTAATGTGTTC |
| 16 | TGTAAAACGACGGCCAGTTTTGGTAAATTCAGTTTTGGTTTG | CAGGAAACAGCTATGACCGAGAAGAAAGAGGGATGAGGG |
| 17 | TGTAAAACGACGGCCAGTACCATGCTCAGCAATGAAG | CAGGAAACAGCTATGACCCACTGACAACTGGCTTGTGC |
| 18 | TGTAAAACGACGGCCAGTGTTTAAACAGTGGAATTCTAGAGTCA | CAGGAAACAGCTATGACCCAGTACATCTAAGAAATTGAGCATCC |
| 19 | TGTAAAACGACGGCCAGTGGCAGTTCTAGAAGAATGAAAACTC | CAGGAAACAGCTATGACCGGCAAGAGACCGAAACTCC |
| 20 | TGTAAAACGACGGCCAGTGCCTGGCCTGATACAATTAAC | CAGGAAACAGCTATGACCTGTCCCTTGTTGCTATTCTTTG |
| 21 | TGTAAAACGACGGCCAGTTGCTTGGTTCTTTAGTTTTAGTTGC | CAGGAAACAGCTATGACCCCTTGAATAATCATCAAGCCTCA |
| 22 | TGTAAAACGACGGCCAGTGCTCTAATTTTGTTGTATTTGTCCTG | CAGGAAACAGCTATGACCAATCATTTTGTTAGTAAGGTCATTTTT |
| 23/24 | TGTAAAACGACGGCCAGTGCAAAATCCACTACTAATGCCC | CAGGAAACAGCTATGACCTGCCAACTGGTAGCTCCAAC |
| 25 | TGTAAAACGACGGCCAGTAAGCAACAGGTCATTTTGGAA | CAGGAAACAGCTATGACCCCCCATCTCCTGAGGTTCAT |
| 26 | TGTAAAACGACGGCCAGTTCCCTATCAGCTAGATTCCCC | CAGGAAACAGCTATGACCCCCAGAGTTTCATATCTTGCTTC |
| 27-1 | TGTAAAACGACGGCCAGTTAGGGGAGGGAGACTGTGTG | CAGGAAACAGCTATGACCACCAGACAAAAGAGCTTGGG |
| 27-2 | TGTAAAACGACGGCCAGTACAGAAGGCATTTCAGCCAC | CAGGAAACAGCTATGACCAAACGCTGAGGTAAATTTGAAAC |
Appendix 2: Nested primers for sequencing selected exons of the BRCA1 and BRCA2 genes
| Exon | Primer | Sequence |
|---|---|---|
| BRCA1 | ||
| 6 | Internal reverse | GAAGAAGAAAACAAATGG |
| 8 | Internal forward | ACCCTTTTAATTAAGAAA |
| BRCA2 | ||
| 3 | Internal forward | GATTTAGGACCAATAAG |
| 9 | Internal reverse | CAACAACAACAAAAAAACC |
| 10-3 | Internal reverse | GTACTATTTACAAAAAAAAAAA |
| 13 | Internal reverse | GGGAAGTGTTAACTTCT |
| 15-1 | Internal forward | GCTAAGTATTTATTCTTTG |
| 15-1 | Internal reverse | CCATCAGTATTGTAGAC |
Appendix 3: BRCA1 and BRCA2 gene variants* detected in 20 DNA samples
| Mutation | Protein | Mutation type | Variant class† | Clinical importance‡ | Detection method | Count§ |
|---|---|---|---|---|---|---|
| BRCA1 | ||||||
| c.140G>A | p.Cys47Tyr | Missense | DM | - | MPS | 1 |
| c.212+1G>T | Intronic | IVS | DM | Yes | MPS | 1 |
| c.213-11T>G | Intronic | IVS | DM | Yes | MPS | 1 |
| c.1067A>G | p.Gln356Arg | Missense | DP | Unknown | MPS | 3 |
| c.1175_1214del40 | p.Leu392GlnfsX5 | Frame-shift | DM | Yes | MPS | 1 |
| c.2077G>A | p.Asp693Asn | Missense | DP | No | MPS | 2 |
| c.2082C>T | p.Ser694Ser | Synonymous | - | Unknown | MPS | 9 |
| c.2311T>C | p.Leu771Leu | Synonymous | - | Unknown | MPS | 9 |
| c.2315T>C | p.Val772Ala | Missense | DM | Unknown | MPS | 1 |
| c.2612C>T | p.Pro871Leu | Missense | DFP | No | MPS | 8 |
| c.2612delCinsTT | p.Pro871LeufsX32 | Frame-shift | DM | - | MPS | 1 |
| c.3113A>G | p.Glu1038Gly | Missense | DP | No | MPS | 9 |
| c.3119G>A | p.Ser1040Asn | Missense | DM? | Unknown | MPS | 1 |
| c.3548A>G | p.Lys1183Arg | Missense | DP | No | MPS | 9 |
| c.3706_3707delAA | p.Asn1236TyrfsX7 | Frame-shift | DM | Yes | MPS | 1 |
| c.3756_3759delGTCT | p.Ser1253ArgfsX10 | Frame-shift | DM | Yes | MPS | 1 |
| c.4065_4068delTCAA | p.Asn1355LysfsX10 | Frame-shift | DM | Yes | MPS | 1 |
| c.4308T>C | p.Ser1436Ser | Synonymous | - | Unknown | MPS | 9 |
| c.4837A>G | p.Ser1613Gly | Missense | DM? | No | MPS | 9 |
| BRCA2 | ||||||
| c.68-7T>A | Intronic | IVS | DM | Unknown | MPS | 1 |
| c.865A>C | p.Asn289His | Missense | DP | No | MPS | 1 |
| c.1114A>C | p.Asn372His | Missense | DFP | C>A | MPS | 11 |
| c.1365A>G | p.Ser455Ser | Synonymous | - | No | MPS | 1 |
| c.1395A>C | p.Val465Val | Synonymous | - | Unknown | MPS | 1 |
| c.1813dupA | p.Ile605AsnfsX11 | Frame-shift | DM | Yes | HP kit | 1 |
| c.1938C>T | p.Ser646Ser | Synonymous | - | No | MPS | 1 |
| c.2229T>C | p.His743His | Synonymous | - | No | MPS | 1 |
| c.2957dupA | p.Asn986LysfsX2 | Frame-shift | DM | Yes | HP kit | 1 |
| c.2971A>G | p.Asn991Asp | Missense | DM? | No | MPS | 1 |
| c.3396A>G | p.Lys1132Lys | Synonymous | - | No | MPS | 10 |
| c.3807T>C | p.Val1269Val | Synonymous | - | No | MPS | 5 |
| c.4478_4481delAAAG | p.Glu1493ValfsX10 | Frame-shift | DM | Yes | MPS | 1 |
| c.7242A>G | p.Ser2414Ser | Synonymous | - | No | MPS | 6 |
| c.7655_7658delTTAA | p.Ile2552ThrfsX95 | Frame-shift | DM | Yes | MPS | 1 |
| c.7762_7764delATAinsTT | p.Ile2588PhefsX60 | Frame-shift | DM | - | MPS | 1 |
| c.7806-14T>C | Intronic | IVS | - | Unknown | MPS | 17 |
| c.7977-1G>C | Intronic | IVS | DM | Yes | MPS | 1 |
| c.8149G>T | p.Ala2717Ser | Missense | DM? | No | MPS | 1 |
| c.8297delC | p.Thr2766AsnfsX11 | Frame-shift | DM | Yes | MPS | 1 |
| c.8575delC | p.Gln2859LysfsX4 | Frame-shift | DM | Yes | MPS | 1 |
| c.9257-16T>C | Intronic | IVS | - | Unknown | MPS | 1 |
| c.9976A>T | p.Lys3326X | Nonsense | DP | No | MPS | 1 |
DM = disease-causing mutation; - = not reported; MPS = massively parallel sequencing; IVS = intervening sequence; DP = disease-associated polymorphism; DFP: disease-associated polymorphism with additional supporting functional evidence; DM? = potential disease-causing mutation; C>A = reported as base pair change from cytosine to adenine; HP = homopolymer.
Variants are named according to Human Genome Variation Society nomenclature, Version 2.0.23
According to the Human Genome Mutation Database (BIOBASE HGMD® Professional, BIOBASE Biological Databases, Beverly, Massachusetts, USA) in February 2013.35
According to the 2014 Breast Cancer Information Core database.34
Number of times the variant was observed in the DNA samples.
Appendix 4: Cost-efficiency of massively parallel sequencing/homopolymer mutation screening strategy for BRCA1 and BRCA2 genes
| Sanger sequencing | Multiplexing five samples sequenced by MPS | Savings | |
|---|---|---|---|
| Cost per sample in NZD | 2,700 | 820 | 1,880 |
| Time per sample in minutes | 510 | 284 | 226 |
NZD = New Zealand Dollars; MPS = massively parallel sequencing.
Footnotes
CONFLICT OF INTEREST
The authors declare no conflicts of interest.
References
- 1.França LT, Carrilho E, Kist TB. A review of DNA sequencing techniques. Q Rev Biophys. 2002;35:169–200. doi: 10.1017/S0033583502003797. [DOI] [PubMed] [Google Scholar]
- 2.Metzker ML. Sequencing technologies: The next generation. Nat Rev Genet. 2010;11:31–46. doi: 10.1038/nrg2626. [DOI] [PubMed] [Google Scholar]
- 3.Zhou X, Ren L, Meng Q, Li Y, Yu Y, Yu J. The next-generation sequencing technology and application. Protein Cell. 2010;1:520–36. doi: 10.1007/s13238-010-0065-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sanger F, Nicklen S, Coulson AR. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977;74:5463–7. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wheeler DA, Srinivasan M, Egholm M, Shen Y, Chen L, McGuire A, et al. The complete genome of an individual by massively parallel DNA sequencing. Nature. 2008;452:872–6. doi: 10.1038/nature06884. [DOI] [PubMed] [Google Scholar]
- 6.Voelkerding KV, Dames SA, Durtschi JD. Next-generation sequencing: From basic research to diagnostics. Clin Chem. 2009;55:641–58. doi: 10.1373/clinchem.2008.112789. [DOI] [PubMed] [Google Scholar]
- 7.Cunningham R, Shaw C, Blakely T, Atkinson J, Sarfati D. Ethnic and socioeconomic trends in breast cancer incidence in New Zealand. BMC Cancer. 2010;10:674. doi: 10.1186/1471-2407-10-674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Taylor MR. Genetic testing for inherited breast and ovarian cancer syndromes: Important concepts for the primary care physician. Postgrad Med J. 2001;77:11–15. doi: 10.1136/pmj.77.903.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Michils G, Hollants S, Dehaspe L, Van Houdt J, Bidet Y, Uhrhammer N, et al. Molecular analysis of the breast cancer genes BRCA1 and BRCA2 using amplicon-based massive parallel pyrosequencing. J Mol Diagn. 2012;14:623–30. doi: 10.1016/j.jmoldx.2012.05.006. [DOI] [PubMed] [Google Scholar]
- 10.Kim BY, Lee DG, Lee KR, Han SH, Surendran S, Han CW, et al. Identification of BRCA1 and BRCA2 mutations from Korean breast cancer patients using denaturing HPLC. Biochem Biophys Res Commun. 2006;349:604–10. doi: 10.1016/j.bbrc.2006.08.086. [DOI] [PubMed] [Google Scholar]
- 11.Ou J, Wu T, Sijmons R, Ni D, Xu W, Upur H. Prevalence of BRCA1 and BRCA2 germline mutations in breast cancer women of multiple ethnic region in northwest China. J Breast Cancer. 2013;16:50–4. doi: 10.4048/jbc.2013.16.1.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ratajska M, Brozek I, Senkus-Konefka E, Jassem J, Stepnowska M, Palomba G, et al. BRCA1 and BRCA2 point mutations and large rearrangements in breast and ovarian cancer families in Northern Poland. Oncol Rep. 2008;19:263–8. doi: 10.3892/or.19.1.263. [DOI] [PubMed] [Google Scholar]
- 13.Riahi A, Kharrat M, Ghourabi ME, Khomsi F, Gamoudi A, Lariani I, et al. Mutation spectrum and prevalence of BRCA1 and BRCA2 genes in patients with familial and early-onset breast/ovarian cancer from Tunisia. Clin Genet. 2013 doi: 10.1111/cge.12337. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 14.Vuttariello E, Borra M, Calise C, Mauriello E, Greggi S, Vecchione A, et al. A new rapid methodological strategy to assess BRCA mutational status. Mol Biotechnol. 2013;54:954–60. doi: 10.1007/s12033-012-9646-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Struewing JP, Hartge P, Wacholder S, Baker SM, Berlin M, McAdams M, et al. The risk of cancer associated with specific mutations of BRCA1 and BRCA2 among Ashkenazi Jews. N Engl J Med. 1997;336:1401–8. doi: 10.1056/NEJM199705153362001. [DOI] [PubMed] [Google Scholar]
- 16.Roa BB, Boyd AA, Volcik K, Richards CS. Ashkenazi Jewish population frequencies for common mutations in BRCA1 and BRCA2. Nat Genet. 1996;14:185–7. doi: 10.1038/ng1096-185. [DOI] [PubMed] [Google Scholar]
- 17.Tarabeux J, Zeitouni B, Moncoutier V, Tenreiro H, Abidallah K, Lair S, et al. Streamlined ion torrent PGM-based diagnostics: BRCA1 and BRCA2 genes as a model. Eur J Hum Genet. 2014;22:535–41. doi: 10.1038/ejhg.2013.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Doherty E, Marquis-Nicholson R, Love JM, Brookes C, Prosser D, Love DR. Primer design to sequence analysis: A pipeline for a molecular genetic diagnostic laboratory. In: Ivanov O, editor. Applications and Experiences of Quality Control. Rijeka, Croatia: InTech; 2011. pp. 257–72. [Google Scholar]
- 19.National Center for Biotechnology Information. dbSNP. From: www.ncbi.nlm.nih.gov/snp Accessed: Sep 2014.
- 20.National Genetics Reference Laboratory Manchester. SNPCheck. From: www.ngrl.org.uk/Manchester/projects/snpcheck Accessed: Sep 2014.
- 21.Messing J. New M13 vectors for cloning. Methods Enzymol. 1983;101:20–78. doi: 10.1016/0076-6879(83)01005-8. [DOI] [PubMed] [Google Scholar]
- 22.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: Nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–19. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- 23.Human Genome Variation Society (HGVS) Nomenclature for the Description of Sequence Variants. From: www.hgvs.org/mutnomen Accessed: Sep 2014.
- 24.Mardis ER. The impact of next-generation sequencing technology on genetics. Trends Genet. 2008;24:133–41. doi: 10.1016/j.tig.2007.12.007. [DOI] [PubMed] [Google Scholar]
- 25.De Leeneer K, De Schrijver J, Clement L, Baetens M, Lefever S, De Keulenaer S, et al. Practical tools to implement massive parallel pyrosequencing of PCR products in next generation molecular diagnostics. PLoS One. 2011;6:e25531. doi: 10.1371/journal.pone.0025531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ewing B, Green P. Base-calling of automated sequencer traces using phred: II - Error probabilities. Genome Res. 1998;8:186–94. doi: 10.1101/gr.8.3.175. [DOI] [PubMed] [Google Scholar]
- 27.Ellard S, Charlton R, Lindsay H, Camm N, Watson C, Abbs S, et al. Clinical Molecular Genetics Society Practice guidelines for Targeted Next Generation Sequencing Analysis and Interpretation. From: www.cmgsweb.shared.hosting.zen.co.uk/BPGs/BPG%20for%20targeted%20next%20generation%20sequencing%20final.pdf Accessed: Feb 2014.
- 28.Craig DW, Pearson JV, Szelinger S, Sekar A, Redman M, Corneveaux JJ, et al. Identification of genetic variants using barcoded multiplexed sequencing. Nat Methods. 2008;5:887–93. doi: 10.1038/nmeth.1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gargis AS, Kalman L, Berry MW, Bick DP, Dimmock DP, Hambuch T, et al. Assuring the quality of next-generation sequencing in clinical laboratory practice. Nat Biotechnol. 2012;30:1033–6. doi: 10.1038/nbt.2403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Balzer S, Malde K, Lanzén A, Sharma A, Jonassen I. Characteristics of 454 pyrosequencing data: Enabling realistic simulation with flowsim. Bioinformatics. 2010;26:i420–5. doi: 10.1093/bioinformatics/btq365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Balzer S, Malde K, Jonassen I. Systematic exploration of error sources in pyrosequencing flowgram data. Bioinformatics. 2011;27:i304–9. doi: 10.1093/bioinformatics/btr251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Berkeley Drosophila Genome Project (BDGP) Splice Site Prediction by Neural Network. From: www.fruitfly.org/seq_tools/splice.html Accessed: Sep 2014.
- 33.Reese MG, Eeckman FH, Kulp D, Haussler D. Improved splice site detection in Genie. J Comput Biol. 1997;4:311–23. doi: 10.1145/267521.267766. [DOI] [PubMed] [Google Scholar]
- 34.National Human Genome Research Institute; National Institutes of Health. Breast Cancer Information Core: An open access on-line breast cancer mutation data base. From: www.research.nhgri.nih.gov/bic/ Accessed: Sep 2014.
- 35.BIOBASE Biological Databases HGMD® Human Gene Mutation Database. From: www.biobase-international.com/product/hgmd Accessed: Sep 2014.
- 36.Alansari A, Al-Rawahi S, Ba-Omar T, Al-Nabhani M, Date A. The identification of Pompe disease mutations in archival tissues and development of a rapid molecular-based test. Sultan Qaboos Univ Med J. 2013;13:502–9. doi: 10.12816/0003308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ellard S, Charlton R, Lindsay H, Camm N, Watson C, Abbs S, et al. Clinical Molecular Genetics Society Practice guidelines for Targeted Next Generation Sequencing Analysis and Interpretation. From: www.cmgsweb.shared.hosting.zen.co.uk/BPGs/BPG%20for%20targeted%20next%20generation%20sequencing%20final.pdf Accessed: Feb 2014.
- 38.Jones MA, Bhide S, Chin E, Ng BG, Rhodenizer D, Zhang VW, et al. Targeted polymerase chain reaction-based enrichment and next generation sequencing for diagnostic testing of congenital disorders of glycosylation. Genet Med. 2011;13:921–32. doi: 10.1097/GIM.0b013e318226fbf2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mattocks CJ, Morris MA, Matthijs G, Swinnen E, Corveleyn A, Dequeker E, et al. A standardized framework for the validation and verification of clinical molecular genetic tests. Eur J Hum Genet. 2010;18:1276–88. doi: 10.1038/ejhg.2010.101. [DOI] [PMC free article] [PubMed] [Google Scholar]