Abstract
Therapeutic resistance remains the principal problem in acute myeloid leukemia (AML). We used area under receiver operator characteristic curves (AUC) to quantify our ability to predict therapeutic resistance in individual patients where AUC=1.0 denotes perfect prediction and AUC=0.5 denotes a coin flip, using data from 4,601 patients with newly diagnosed AML given induction therapy with 3+7 or more intense standard regimens in MRC/NCRI, HOVON, SWOG, and MD Anderson Cancer Center studies. Age, performance status, white blood cell count, secondary disease, cytogenetic risk, and FLT3-ITD/NPM1 mutation status were each independently associated with failure to achieve complete remission despite no early death (“primary refractoriness”). However, the AUC of a bootstrap-corrected multivariable model predicting this outcome was only 0.78, indicating only fair predictive ability. Removal of FLT3-ITD and NPM1 information only slightly decreased the AUC (0.76). Prediction of resistance, defined as primary refractoriness or short relapse-free survival (RFS), was even more difficult. Our ability to forecast resistance based on routinely available pre-treatment covariates provides a rationale for continued randomization between standard and new therapies and supports further examination of genetic and post-treatment data to optimize resistance prediction in AML.
Keywords: Acute myeloid leukemia, area under the receiver operator characteristic curve, prediction, therapeutic resistance
INTRODUCTION
“Resistance”, defined as either failure to achieve initial complete remission (CR) or as relapse from CR, remains the principal problem in adult acute myeloid leukemia (AML).1–3 It is widely appreciated that the likelihood of resistance to therapy differs significantly between individuals; e.g., the complete remission (CR) rate following initial chemotherapy is 80–90% in some patients but below 30% in others.4, 5
The ability to accurately forecast resistance would have considerable significance for the management of AML and the evaluation of new drugs. Specifically, the better our ability to predict resistance to standard therapy, the less need there might be for randomization of patients between such therapy and an investigational therapy. The lack of a need to randomize would address a seldom discussed but very real ethical conundrum: physicians caring for AML patients generally are not enthusiastic about randomizing patients with features associated with resistance (older age, complex cytogenetics, secondary AML, etc.) to standard therapy but are willing to do so because they believe that our ability to predict resistance to standard therapy is only fair and, thus, that randomization is necessary to properly evaluate a new therapy. We therefore set out to test this belief by quantifying our current ability to predict resistance.
Here, it may be important to distinguish between association and prediction. It is well known that clinical, cytogenetic, and molecular characteristics are strongly associated with resistance,1–3 as quantified by odds ratios, hazard ratios, and P-values, the standard measurements of association.6 While valuable, these traditional measurements do not give insight into the degree to which given prognostic factors improve our ability to predict outcome in individual patients. This is the province of measures of prediction such as the area under receiver operating curve (AUC).7 Conceptually, the closer we are to AUCs of 1.0, the less need there might be to randomize patients between standard therapy (i.e. therapy for which the outcome can be predicted) and a novel therapy. Herein, we use AUCs in conjunction with clinical, cytogenetic, and molecular data from adults with newly diagnosed AML treated on trials conducted by the U.K. Medical Research Council/National Cancer Research Institute (MRC/NCRI), the Dutch-Belgian Cooperative Trial Group for Hematology/Oncology and the Swiss Group for Clinical Cancer Research (HOVON/SAKK), the U.S. cooperative group SWOG, and MD Anderson Cancer Center (MDA) to quantify our ability to predict therapeutic resistance using uni- and multiparameter models.
MATERIALS AND METHODS
Study population and treatments
The trials we analyzed primarily enrolled adults with newly diagnosed AML other than acute promyelocytic leukemia, as based on WHO 2008 classification criteria.8 Specifically, we used data from patients who received curative-intent treatment on 6 MRC/NCRI trials from 1988–2010 (AML10, AML11, AML12, AML14, AML15, and AML16), 6 HOVON/SAKK trials from 1987–2008 (HO04, HO04A, HO29, HO42A, HO42, and HO43), or 4 SWOG trials from 1992–2009 (S8931, S93333, S9500, and S0106), or received treatment on various protocols at MDA from 2000–2013. As expected, there were intergroup differences with regard to the type of therapeutic regimens used: for example, while induction treatments generally contained cytarabine and daunorubicin or idarubicin, therapy at MDA more often included cytarabine at >1 g/m2/dose, while in MRC/NCRI typically 10 rather than 7 days of cytarabine at 100–200 mg/m2/dose were used; moreover, HOVON/SAKK post remission therapy often included amsacrine. Institutional review boards of participating institutions approved all protocols, and patients were treated according to the Declaration of Helsinki.
Definitions of outcomes
Early death (“treatment-related mortality” [TRM]) was defined as death within 28 days after initiating therapy9 or study registration, if exact date of initiation of therapy was unknown. Complete remission (CR) was conventionally defined as achievement of a morphologic leukemia-free state (bone marrow blasts <5%, absence of extramedullary disease) and recovery of peripheral blood counts (absolute neutrophil count >1,000/μL and platelet count >100,000/μL).2, 10 Overall survival (OS) was defined as time from initiation of therapy or study registration (if date of initiation of therapy was unknown) to death, with censoring on the day patients were last known to be alive. For patients who achieved CR, relapse-free survival (RFS) was defined as time from achievement of remission until relapse or death from any cause, with patients not known to have relapsed or died being censored at last follow-up.
Rather than settling on a single, arbitrary criterion to define therapeutic resistance, we used several criteria for resistance: (a) failure to attain CR despite surviving at least 28 days from beginning induction therapy (“primary refractory”); (b) primary refractory or RFS ≤3 months; (c) primary refractory or RFS ≤6 months; and (d) primary refractory or RFS ≤12 months.
Statistical analysis
OS and RFS were estimated using the Kaplan-Meier method.11 Chi-squared tests and the Kruskal-Wallis test were used to assess differences between categorical variables and median values of numeric variables across categories, respectively. We used logistic regression analyses to assess the relationship between individual covariates and various working definitions of therapeutic resistance, and then used the AUC to quantify a model’s ability to predict therapeutic resistance; in this approach, an AUC of 1 indicates perfect prediction while an AUC of 0.5 indicates no prediction. It is natural to wonder whether some of the changes in AUC observed, for example from 0.77 to 0.78, are “statistically significant”; however, there is no rule permitting one to say whether a given change in AUC is statistically significant. Rather, this depends on effect size and patient numbers. Given the large number of models we evaluated and the even larger number of pairwise comparisons, we consider it more important to focus on the magnitude of AUC values and changes in these values (rather than on their statistical significance) using the commonly accepted criteria that AUCs of 0.6–0.7, 0.7–0.8, and 0.8–0.9 indicate poor, fair, and good predictive ability, respectively.12–14 An alternative way of interpreting AUC is as the proportion of patients who are correctly ranked. For example, a model with an AUC of 0.85 correctly ranks patients with higher risk has having a larger risk score 85% of the time, so 15% of the time the model incorrectly gives a person with higher risk a lower risk score than a person with lower risk. In this interpretation, the AUC is linear: a model with an AUC of 0.75 is a 25% improvement over a model with an AUC of 0.50. The following pre-treatment covariates were used in the regression modeling: age at study registration, performance status, gender, white blood cell (WBC) count, platelet count, bone marrow blast percentage, disease type (primary vs. secondary), cytogenetic risk, and FLT3-ITD as well as NPM1 mutational status. Missing cytogenetic risk was accounted for as separate category. The relative importance of predictors in the multivariable regression models was evaluated by the value of the partial Wald Chi-squared statistic minus the predictor’s degrees of freedom. Bootstrapping, which has been demonstrated to be a more efficient method for assessing internal model accuracy than cross-validation or splitting data into two groups,15, 16 was used to estimate bias-corrected values of AUC, and all reported AUCs are bootstrap-bias corrected. All analyses were performed using R (http://www.r-project.org).
RESULTS
Characterization of study population
Our dataset included 4,955 predominantly adults with newly diagnosed AML treated on MRC/NCRI, HOVON/SAKK, or SWOG protocols or given treatment at MDA and in whom pre-treatment information (age, gender, performance status, WBC and platelet count, bone marrow blast percentage, secondary AML, FLT3-ITD and NPM1 mutational status) and outcome data were available. To better assess resistance following initial chemotherapy, we excluded patients who died within 28 days of initiation of induction chemotherapy (n=335) or were lost to follow-up before this time (n=19), leaving 4,601 patients (median age: 52 years) from our 4 treatment sites (MRC/NCRI [n=2,615], HOVON/SAKK n=1,134], SWOG [n=443], and MDA [n=409]). Patients’ baseline characteristics are presented in Table 1. As summarized in Table 2, a CR to the initial 1–2 courses of induction chemotherapy was achieved in 3,613 (79%) of patients, whereas 988 (21%) were “primary refractory”. 1,318/4,552 patients (29%) with sufficient follow-up time were either primary refractory or had a RFS of 3 months or less after CR achievement, 1,784/4,503 patients (40%) with sufficient follow-up time were either primary refractory or had a RFS of 6 months or less after CR achievement, and 2,547/4,457 patients (57%) with sufficient follow-up time were primary refractory or had a RFS of 12 months or less after CR achievement.
TABLE 1.
Baseline characteristics of study population
| Parameter | MRC/NCRI n = 2,615 |
HOVON/SAKK n = 1,134 |
SWOG n = 443 |
MDA n = 409 |
All n = 4,601 |
|---|---|---|---|---|---|
| Age [years], median (range) | 53 (16–90) | 48 (15–77) | 51 (19–84) | 54 (18–82) | 52 (15–90) |
| Patients aged ≥60 years, n (%) | 981 (38) | 95 (8) | 90 (21) | 108 (26) | 1,274 (28) |
| Male Gender, n (%) | 1,407 (54) | 588 (52) | 234 (53) | 213 (52) | 2,442 (53) |
| Secondary AML1, n (%) | 332 (13) | 100 (9) | 19 (4) | 208 (51) | 659 (14) |
| Karyotype1, n (%) | |||||
| Favorable | 259 (10) | 142 (13) | 66 (15) | 50 (12) | 517 (11) |
| Intermediate | 1,447 (55) | 682 (60) | 242 (55) | 197 (48) | 2,568 (56) |
| Unfavorable | 275 (11) | 275 (24) | 90 (20) | 152 (37) | 792 (17) |
| Missing | 634 (24) | 35 (3) | 45 (10) | 10 (2) | 724 (16) |
| NPM1 Mutation1, n (%) | |||||
| Yes | 828 (32) | 325 (29) | 125 (28) | 89 (22) | 1,367 (30) |
| No | 1,787 (68) | 809 (71) | 318 (72) | 320 (78) | 3,234 (70) |
| FLT3-ITD Mutation1, n (%) | |||||
| Yes | 582 (22) | 279 (25) | 98 (22) | 77 (19) | 1,036 (23) |
| No | 2,033 (78) | 855 (75) | 345 (78) | 332 (81) | 3,565 (77) |
| NPM1, FLT3-ITD Mutation1, n (%) | |||||
| NPM1−, FLT3-ITD− | 1,553 (59) | 692 (61) | 268 (60) | 279 (68) | 2,792 (61) |
| NPM1−, FLT3-ITD+ | 234 (9) | 117 (10) | 50 (11) | 41 (10) | 442 (10) |
| NPM1+, FLT3-ITD− | 480 (18) | 163 (14) | 77 (17) | 53 (13) | 773 (17) |
| NPM1+, FLT3-ITD+ | 348 (13) | 162 (14) | 48 (11) | 36 (9) | 594 (13) |
| WBC1 [x103/μL], median (range) | 15 (0–559) | 19 (1–510) | 14 (0–370) | 8 (1–340) | 15 (0–559) |
| Platelets1 [x103/μL], median (range) | 56 (2–983) | 66 (3–998) | 54 (2–9,300) | 41 (3–1,069) | 58 (2–9,300) |
| Bone marrow blasts1, median% (range) | 70 (0–100) | 60 (0–98) | 67 (3–100) | 47 (6–96) | 65 (0–100) |
| Performance status1, n (%) | |||||
| 0–1 | 2,081 (80) | 1,051 (93) | 368 (83) | 374 (91) | 3,874 (84) |
| 2 | 534 (20) | 83 (7) | 76 (17) | 35 (9) | 727 (16) |
At diagnosis.
Abbreviations: WBC, white blood cells.
TABLE 2.
Treatment outcomes in study population
| Parameter | MRC/NCRI n = 2,615 |
HOVON/SAKK n = 1,134 |
SWOG n = 443 |
MDA n = 409 |
All n = 4,601 |
|---|---|---|---|---|---|
| Response to initial 1–2 courses of induction chemotherapy | |||||
| CR, n (%) | 2,074 (79) | 954 (84) | 303 (68) | 282 (69) | 3,613 (79) |
| Failure to achieve CR, n (%) | 541 (21) | 180 (16) | 140 (32) | 127 (31) | 988 (21) |
| OS [months], median (95% CI) | 18 (18–20) | 24 (20–33) | 27 (23–42) | 18 (16–24) | 20 (19–22) |
| RFS [months], median (95% CI) | 13 (13–15) | 18 (15–26) | 17 (14–27) | 16 (13–26) | 15 (14–16) |
| Patients evaluable at 3 months, n (%) | 2,573 (98) | 1,134 (100) | 443 (100) | 402 (98) | 4,552 (99) |
| Failure to achieve CR or to have RFS ≤3 months, n (%) | 720 (28) | 283 (25) | 159 (36) | 156 (39) | 1,318 (29) |
| Patients evaluable at 6 months, n (%) | 2,544 (97) | 1,130 (99) | 441 (100) | 388 (95) | 4,503 (98) |
| Failure to achieve CR or to have RFS ≤6 months, n (%) | 1,004 (39) | 396 (35) | 192 (44) | 192 (49) | 1,784 (40) |
| Patients evaluable at 12 months, n (%) | 2,523 (96) | 1,124 (99) | 439 (99) | 371 (91) | 4,457 (97) |
| Failure to achieve CR or to have RFS ≤12 months, n (%) | 1,471 (58) | 580 (52) | 259 (59) | 237 (64) | 2,547 (57) |
Abbreviations: CR, complete remission; OS, overall survival; RFS, relapse-free survival.
Prediction of failure to achieve CR with initial induction chemotherapy
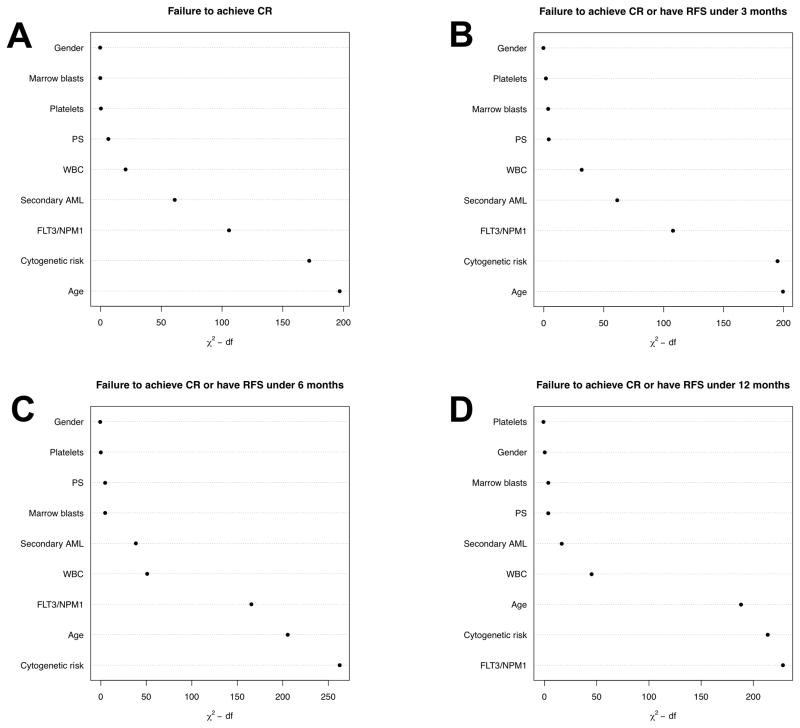
Patients with primary refractory AML are widely considered those exhibiting the highest degree of resistance. Not surprisingly, but serving as a data reliability check, age (P<0.001), performance status (P=0.006), WBC (P<0.001), secondary disease (P<0.001), cytogenetic risk (favorable or intermediate vs. adverse; P<0.001 for each comparison), and NPM1 mutation as well as FLT3-ITD status (NPM1neg/FLT3-ITDneg or NPM1neg/FLT3-ITDpos vs. NPM1pos/FLT3-ITDpos; P<0.005 for each comparison) were independently associated with being primary refractory to induction chemotherapy when considering all patients (Table 3). As shown in Figure 1A, age was the most important individual covariate in predicting failure to achieve CR followed by cytogenetic risk and NPM1 and FLT3/ITD status, as indicated by values of the partial Wald Chi-squared statistic. Despite the strong associations between such covariates and primary refractory AML, AUCs validated by bootstrap-adjustment suggested that the predictive ability was less impressive. Specifically, in univariate models, AUC values were 0.68, 0.66, and 0.60 for age, cytogenetic risk, and NPM1 and FLT3-ITD mutational status, A bootstrap-corrected multivariable model incorporating all covariates (age, performance status, WBC, platelet count, bone marrow blast percentage, gender, disease type [primary vs. secondary], cytogenetic risk, and FLT3-ITD as well as NPM1 mutational status) yielded an AUC of 0.78. Despite their strong associations with primary refractory AML, removal of FLT3-ITD and NPM1 from the model, while having a statistically significant effect on AUC, only decreased the AUC slightly (from 0.78 to 0.76; see Supplemental Figure 1). AUCs varied considerably among the 4 treatment sites: from 0.82/0.81 (with/without FLT3-ITD and NPM1 mutation information) to 0.69/0.66.
TABLE 3.
Multivariate logistic regression models for failure to achieve CR, with inclusion of NPM1 and FLT3-ITD mutational information
| Parameter | MRC/NCRI n = 2,615 |
HOVON/SAKK n = 1,134 |
SWOG n = 443 |
MDA n = 409 |
All n = 4,601 |
|---|---|---|---|---|---|
|
| |||||
| Age (per 1 year) | 1.06 (1.05–1.07), P<0.001 | 1.01 (1.00–1.02), P=0.15 | 1.02 (1.00–1.03), P=0.04 | 1.03 (1.01–1.05), P=0.003 | 1.04 (1.04–1.05), P<0.001 |
|
| |||||
| Performance status | |||||
| 0–1 | 1.00 (reference) | 1.00 (reference) | 1.00 (reference) | 1.00 (reference) | 1.00 (reference) |
| 2–4 | 1.64 (1.22–2.20), P=0.001 | 1.52 (0.81–2.89), P=0.20 | 1.10 (0.62–1.96), P=0.75 | 0.89 (0.39–2.04), P=0.79 | 1.36 (1.08–1.70), P=0.006 |
|
| |||||
| WBC (per 10,000/μL) | 1.32 (1.07–1.62), P=0.009 | 2.08 (1.51–2.86), P<0.001 | 1.69 (1.06–2.70), P=0.026 | 1.89 (0.88–4.05), P=0.10 | 1.42 (1.23–1.65), P<0.001 |
|
| |||||
| Platelets (per 10,000/μL) | 1.15 (1.02–1.30), P=0.025 | 1.35 (1.16–1.58), P<0.001 | 0.97 (0.90–1.05), P=0.47 | 1.19 (0.88–1.61), P=0.25 | 1.02 (0.99–1.05), P=0.25 |
|
| |||||
| BM blasts (per 10%) | 1.01 (0.97–1.06), P=0.68 | 0.96 (0.89–1.03), P=0.24 | 0.97 (0.88–1.07), P=0.53 | 0.95 (0.85–1.05), P=0.31 | 0.99 (0.95–1.02), P=0.36 |
|
| |||||
| Gender | |||||
| Female | 1.00 (reference) | 1.00 (reference) | 1.00 (reference) | 1.00 (reference) | 1.00 (reference) |
| Male | 1.06 (0.84–1.33), P=0.63 | 1.09 (0.77–1.55), P=0.63 | 0.82 (0.53–1.27), P=0.38 | 1.37 (0.86–2.20), P=0.18 | 1.07 (0.92–1.26), P=0.38 |
|
| |||||
| Type of AML | |||||
| De novo (primary) | 1.00 (reference) | 1.00 (reference) | 1.00 (reference) | 1.00 (reference) | 1.00 (reference) |
| Secondary | 2.95 (2.24–3.88), P<0.001 | 1.51 (0.90–2.55), P=0.12 | 2.50 (0.85–7.33), P=0.094 | 1.55 (0.96–2.49), P=0.071 | 2.19 (1.80–2.65), P<0.001 |
|
| |||||
| Cytogenetics | |||||
| Favorable | 0.13 (0.05–0.33), P<0.001 | 0.36 (0.16–0.79), P<0.001 | 0.26 (0.11–0.62), P=0.002 | 0.16 (0.05–0.56), P=0.0039 | 0.23 (0.15–0.36), P<0.001 |
| Intermediate | 1.00 (reference) | 1.00 (reference) | 1.00 (reference) | 1.00 (reference) | 1.00 (reference) |
| Unfavorable | 3.66 (2.66–5.06), P<0.001 | 2.49 (1.68–3.70), P<0.001 | 2.02 (1.16–3.51), P=0.013 | 1.93 (1.15–3.25), P=0.013 | 2.75 (2.26–3.35), P<0.001 |
|
| |||||
| NPM1, FLT3-ITD Status | |||||
| NPM1−, FLT3-ITD− | 1.00 (reference) | 1.00 (reference) | 1.00 (reference) | 1.00 (reference) | 1.00 (reference) |
| NPM1−, FLT3-ITD+ | 1.14 (0.78–1.67), P=0.49 | 2.57 (1.60–4.16), P<0.001 | 1.22 (0.62–2.40), P=0.56 | 0.84 (0.38–1.87), P=0.67 | 1.45 (1.13–1.86), P=0.005 |
| NPM1+, FLT3−ITD− | 0.30 (0.20–0.43), P<0.001 | 0.18 (0.08–0.44), P<0.001 | 0.41 (0.21, 0.80), P=0.009 | 0.25 (0.10–0.63), P=0.004 | 0.31 (0.23–0.41), P<0.001 |
| NPM1+, FLT3−ITD+ | 0.48 (0.32–0.71), P<0.001 | 0.40 (0.20–0.79), P=0.004 | 0.59 (0.27–1.26), P=0.17 | 0.59 (0.23–1.51), P=0.27 | 0.47 (0.35–0.63), P<0.001 |
|
| |||||
| Bootstrap-corrected AUC | 0.82 | 0.77 | 0.69 | 0.74 | 0.78 |
Data is presented as odds ratio (95% CI). The model also contained a covariate “missing cytogenetics”.
Abbreviations: BM, bone marrow; CR, complete remission; WBC, white blood cells.
Figure 1. Prediction of Therapeutic Resistance.
Importance of individual covariates to predict (A) failure to achieve CR with initial 1–2 courses of induction chemotherapy, (B) failure to achieve CR with initial 1–2 courses of induction chemotherapy or RFS ≤3 months, (C) failure to achieve CR with initial 1–2 courses of induction chemotherapy or RFS ≤6 months, and (D) failure to achieve CR with initial 1–2 courses of induction chemotherapy or RFS ≤12 months, using χ2 values.
We performed several additional analyses of prediction of primary refractory AML. First, because information on the mutational status of FLT3 and NPM1 is currently thought to be clinically most helpful to guide decision-making in patients with cytogenetically normal AML, we restricted our analysis to patients with normal karyotypes at diagnosis. Here, the multivariable models with or without NPM1 and FLT3-ITD information for the combined dataset yielded AUCs of 0.75 and 0.72, respectively, with AUCs for the individual treatment sites of 0.72-0.81 (with inclusion of FLT3-ITD and NPM1) and 0.63–0.80 (without inclusion of FLT3-ITD and NPM1). Thus, these AUC values were not higher than the values in the unrestricted dataset presented above, and only in 1 of the 4 individual cohorts did the inclusion of FLT3-ITD and NPM1 mutational information improve the prediction performance of the models compared to models incorporating all cytogenetic groups. In the U.S., selection of induction chemotherapy for newly diagnosed AML is commonly made before cytogenetic and mutational information is available. For a second additional analysis aimed at evaluating AUCs under these circumstances, we built multivariable models without inclusion of molecular or cytogenetic information. The accuracy of such models was significantly lower, as indicated by the AUC obtained in the entire patient cohort of 0.70, with AUCs for the individual 4 treatment sites ranging from 0.59 to 0.78. Third, we examined whether predictive accuracy was improved if we focused on relatively homogeneous subsets. However, an analysis limited to our 3,942 patients with de novo AML yielded bootstrap-corrected AUCs of only 0.75 and 0.73 for multivariable models with or without NPM1 and FLT3-ITD information, i.e. lower than in our entire study cohort (0.78 and 0.76). Similarly restricting attention separately to patients aged 60 years or younger (n=3,425) and those older than 60 years (n=1,176), AUCs again were not higher than those in the entire study cohort (0.76/0.72 [younger patients] and 0.73/0.73 [older patients] for multivariable models with or without NPM1 and FLT3-ITD information, respectively). Finally, because recent studies indicated that many patients can achieve CR with an identical second course of chemotherapy after a first course failed,17 we restricted our analysis to patients declared primary refractory only after 2 courses of chemotherapy. Information on receipt of 1 vs. 2 cycles of induction therapy was only available in the SWOG cohort, in which 62 patients failed the first course of induction chemotherapy and did not receive re-induction on protocol. In a subset analysis on the remaining 381 patients, bootstrap-corrected AUCs were 0.66 and 0.64 for multivariable models with or without NPM1 and FLT3-ITD information, i.e. values that were lower than in the entire SWOG cohort (0.69 and 0.66, respectively).
Prediction of failure to achieve CR or to have short RFS
To assess whether the results were similar when we used other criteria for resistance, we performed additional analyses in which we considered resistance not only as primary refractory AML but also as an RFS of no more than 3, 6, or 12 months from date of first CR. As shown in Figures 1A–D, the relative importance of individual covariates changed slightly when the criterion for resistance was altered, with FLT3-ITD and NPM1 mutational status and cytogenetic risk gradually replacing age as the most important individual covariates. As the time over which prediction was attempted increased, predictive ability decreased. Specifically, when analyzing the entire study cohort, the AUCs for models predicting primary refractory disease or to have RFS of ≤3, ≤6, or ≤12 months were 0.75/0.74 (with/without inclusion of FLT3-ITD and NPM1 mutational data; Table 4 and Supplemental Table 2), 0.76/0.73 (Table 5 and Supplemental Table 3), and 0.75/0.71 (Table 6 and Supplemental Table 4), respectively, vs. 0.78/0.76 when considering primary refractory AML alone.
TABLE 4.
Multivariate logistic regression models for failure to achieve CR or RFS ≤3 months, with inclusion of NPM1 and FLT3-ITD mutational information
| Parameter | MRC/NCRI n = 2,573 |
HOVON/SAKK n = 1,134 |
SWOG n = 443 |
MDA n = 402 |
All n = 4,552 |
|---|---|---|---|---|---|
|
| |||||
| Age (per 1 year) | 1.05 (1.04–1.06), P<0.001 | 1.02 (1.00–1.03), P=0.006 | 1.02 (1.01–1.04), P=0.01 | 1.02 (1.01–1.05), P=0.0069 | 1.04 (1.03–1.04), P<0.001 |
|
| |||||
| Performance status | |||||
| 0–1 | 1.00 (reference) | 1.00 (reference) | 1.00 (reference) | 1.00 (reference) | 1.00 (reference) |
| 2–4 | 1.38 (1.07–1.79), P=0.014 | 0.87 (0.49–1.57), P=0.65 | 1.22 (0.69–2.17), P=0.49 | 2.37 (1.04–5.41), P=0.039 | 1.26 (1.03–1.54), P=0.023 |
|
| |||||
| WBC (per 10,000/μL) | 1.42 (1.19–1.70), P<0.001 | 1.90 (1.44–2.52), P<0.001 | 1.53 (0.96–2.43), P=0.076 | 2.23 (0.99–5.06), P=0.054 | 1.49 (1.30–1.70), P<0.001 |
|
| |||||
| Platelets (per 10,000/μL) | 1.14 (1.02–1.28), P=0.023 | 1.19 (1.03–1.38), P=0.016 | 1.01 (0.98–1.04), P=0.56 | 1.17 (0.87–1.56), P=0.31 | 1.04 (0.99–1.09), P=0.096 |
|
| |||||
| BM blasts (per 10%) | 0.97 (0.94–1.01), P=0.21 | 0.95 (0.90–1.01), P=0.12 | 0.98 (0.89–1.08), P=0.72 | 0.97 (0.88–1.08), P=0.61 | 0.97 (0.94–1.00), P=0.030 |
|
| |||||
| Gender | |||||
| Female | 1.00 (reference) | 1.00 (reference) | 1.00 (reference) | 1.00 (reference) | 1.00 (reference) |
| Male | 1.01 (0.83–1.24), P=0.91 | 1.09 (0.81–1.46), P=0.58 | 0.87 (0.56–1.33), P=0.51 | 1.50 (0.95–2.36), P=0.079 | 1.06 (0.92–1.23), P=0.39 |
|
| |||||
| Type of AML | |||||
| De novo (primary) | 1.00 (reference) | 1.00 (reference) | 1.00 (reference) | 1.00 (reference) | 1.00 (reference) |
| Secondary | 2.81 (2.14–3.68), P<0.001 | 1.65 (1.04–2.61), P=0.033 | 2.43 (0.79–7.51), P=0.12 | 1.45 (0.92–2.30), P=0.11 | 2.14 (1.77–2.58), P<0.001 |
|
| |||||
| Cytogenetics | |||||
| Favorable | 0.35 (0.21–0.57), P<0.001 | 0.76 (0.45–1.30), P=0.32 | 0.25 (0.11–0.59), P=0.002 | 0.16 (0.05–0.48), P=0.001 | 0.39 (0.29–0.54), P<0.001 |
| Intermediate | 1.00 (reference) | 1.00 (reference) | 1.00 (reference) | 1.00 (reference) | 1.00 (reference) |
| Unfavorable | 3.50 (2.58–4.75), P<0.001 | 2.74 (1.93–3.90), P<0.001 | 2.47 (1.41–4.32), P=0.002 | 1.91 (1.15–3.17), P=0.012 | 2.90 (2.40–3.49), P<0.001 |
|
| |||||
| NPM1, FLT3-ITD Status | |||||
| NPM1-, FLT3-ITD− | 1.00 (reference) | 1.00 (reference) | 1.00 (reference) | 1.00 (reference) | 1.00 (reference) |
| NPM1-, FLT3-ITD+ | 1.23 (0.87–1.73), P=0.24 | 3.10 (1.99–4.82), P<0.001 | 1.90 (0.87–3.74), P=0.06 | 1.31 (0.62–2.77), P=0.48 | 1.74 (1.38–2.20), P<0.001 |
| NPM1+, FLT3-ITD− | 0.38 (0.28–0.52), P<0.001 | 0.32 (0.17–0.60), P<0.001 | 0.40 (0.20–0.77), P=0.006 | 0.25 (0.11–0.69), P=0.002 | 0.37 (0.29–0.47), P<0.001 |
| NPM1+, FLT3-ITD+ | 0.69 (0.50–0.97), P=0.030 | 0.93 (0.57–1.52), P=0.77 | 0.77 (0.37–1.59), P=0.48 | 0.64 (0.27–1.52), P=0.31 | 0.74 (0.58–0.94), P=0.012 |
|
| |||||
| Bootstrap-corrected AUC | 0.78 | 0.73 | 0.73 | 0.74 | 0.75 |
Data is presented as odds ratio (95% CI). The model also contained a covariate “missing cytogenetics”.
Abbreviations: BM, bone marrow; CR, complete remission; WBC, white blood cells.
TABLE 5.
Multivariate logistic regression models for failure to achieve CR or RFS ≤6 months, with inclusion of NPM1 and FLT3-ITD mutational information
| Parameter | MRC/NCRI n = 2,544 |
HOVON/SAKK n = 1,130 |
SWOG n = 441 |
MDA n = 388 |
All n = 4,503 |
|---|---|---|---|---|---|
|
| |||||
| Age (per 1 year) | 1.04 (1.03–1.05), P<0.001 | 1.02 (1.01–1.03), P=0.022 | 1.03 (1.01–1.05), P<0.001 | 1.02 (1.00–1.04), P=0.024 | 1.04 (1.03–1.04), P<0.001 |
|
| |||||
| Performance status | |||||
| 0–1 | 1.00 (reference) | 1.00 (reference) | 1.00 (reference) | 1.00 (reference) | 1.00 (reference) |
| 2–4 | 1.24 (0.98–1.57), P=0.069 | 0.90 (0.53–1.54), P=0.70 | 1.45 (0.82–2.59), P=0.20 | 2.95 (1.18–7.33), P=0.02 | 1.26 (1.04–1.52), P=0.016 |
|
| |||||
| WBC (per 10,000/μL) | 1.59 (1.34–1.88), P<0.001 | 1.96 (1.49–2.57), P<0.001 | 1.59 (0.97–2.60), P=0.065 | 5.20 (1.79–15.1), P=0.0025 | 1.64 (1.43–1.88), P<0.001 |
|
| |||||
| Platelets (per 10,000/μL) | 1.12 (1.00–1.26), P=0.046 | 1.03 (0.90–1.19), P=0.65 | 1.01 (0.97–1.04), P=0.74 | 1.11 (0.82–1.50), P=0.49 | 1.02 (0.98–1.06), P=0.30 |
|
| |||||
| BM blasts (per 10%) | 0.97 (0.93–1.01), P=0.099 | 0.96 (0.91–1.02), P=0.17 | 1.01 (0.92–1.12), P=0.77 | 0.90 (0.81–1.00), P=0.054 | 0.97 (0.94–0.99), P=0.016 |
|
| |||||
| Gender | |||||
| Female | 1.00 (reference) | 1.00 (reference) | 1.00 (reference) | 1.00 (reference) | 1.00 (reference) |
| Male | 0.97 (0.81–1.17), P=0.79 | 1.14 (0.86–1.49), P=0.36 | 0.88 (0.57–1.35), P=0.55 | 1.33 (0.85–2.10), P=0.21 | 1.04 (0.91–1.19), P=0.56 |
|
| |||||
| Type of AML | |||||
| De novo (primary) | 1.00 (reference) | 1.00 (reference) | 1.00 (reference) | 1.00 (reference) | 1.00 (reference) |
| Secondary | 2.36 (1.78–3.13), P<0.001 | 1.33 (0.85–2.11), P=0.22 | 3.01 (0.83–10.96), P=0.094 | 1.33 (0.84–2.10), P=0.23 | 1.85 (1.53–2.25), P<0.001 |
|
| |||||
| Cytogenetics | |||||
| Favorable | 0.33 (0.21–0.48), P<0.001 | 0.67 (0.41–1.10), P=0.12 | 0.21 (0.09–0.49), P<0.001 | 0.10 (0.03–0.30), P<0.001 | 0.34 (0.26–0.45), P<0.001 |
| Intermediate | 1.00 (reference) | 1.00 (reference) | 1.00 (reference) | 1.00 (reference) | 1.00 (reference) |
| Unfavorable | 4.27 (3.12–5.84), P<0.001 | 3.52 (2.50–4.92), P<0.001 | 2.52 (1.41–4.48), P=0.002 | 1.85 (1.10–3.10), P=0.02 | 3.22 (2.67–3.88), P<0.001 |
|
| |||||
| NPM1, FLT3-ITD Status | |||||
| NPM1−, FLT3-ITD− | 1.00 (reference) | 1.00 (reference) | 1.00 (reference) | 1.00 (reference) | 1.00 (reference) |
| NPM1−, FLT3-ITD+ | 1.74 (1.27–2.40) P<0.001 | 3.71 (2.37–5.79), P<0.001 | 2.20 (1.08–4.49), P=0.030 | 1.42 (0.65–3.13), P=0.38 | 2.17 (1.73–2.73), P<0.001 |
| NPM1+, FLT3-ITD− | 0.33 (0.25–0.44), P<0.001 | 0.35 (0.21–0.60), P<0.001 | 0.36 (0.19–0.68), P=0.002 | 0.39 (0.19–0.80), P=0.011 | 0.35 (0.29–0.44), P<0.001 |
| NPM1+, FLT3-ITD+ | 0.71 (0.53–0.95), P=0.022 | 1.66 (1.08–2.54), P=0.020 | 1.00 (0.49–2.04), P=0.99 | 0.86 (0.36–2.03), P=0.73 | 0.90 (0.73–1.11), P=0.36 |
|
| |||||
| Bootstrap-corrected AUC | 0.78 | 0.74 | 0.75 | 0.75 | 0.76 |
Data is presented as odds ratio (95% CI). The model also contained a covariate “missing cytogenetics”.
Abbreviations: BM, bone marrow; CR, complete remission; WBC, white blood cells.
TABLE 6.
Multivariate logistic regression models for failure to achieve CR or RFS ≤12 months, with inclusion of NPM1 and FLT3-ITD mutational information
| Parameter | MRC/NCRI n = 2,523 |
HOVON/SAKK n = 1,124 |
SWOG n = 439 |
MDA n = 371 |
All n = 4,457 |
|---|---|---|---|---|---|
|
| |||||
| Age (per 1 year) | 1.04 (1.03–1.05), P<0.001 | 1.01 (0.99–1.02), P=0.15 | 1.03 (1.01–1.04), P<0.001 | 1.03 (1.01–1.05), P=0.0052 | 1.03 (1.03–1.04), P<0.001 |
|
| |||||
| Performance status | |||||
| 0–1 | 1.00 (reference) | 1.00 (reference) | 1.00 (reference) | 1.00 (reference) | 1.00 (reference) |
| 2–4 | 1.25 (1.00–1.57), P=0.053 | 0.74 (0.44–1.23), P=0.24 | 1.36 (0.76–2.45), P=0.31 | 1.96 (0.70–5.51), P=0.20 | 1.22 (1.02–1.47), P=0.030 |
|
| |||||
| WBC (per 10,000/μL) | 1.50 (1.25–1.79), P<0.001 | 1.97 (1.50–2.60), P<0.001 | 2.52 (1.33–4.78), P=0.0045 | 9.45 (2.05–43.5), P=0.0039 | 1.64 (1.42–1.89), P<0.001 |
|
| |||||
| Platelets (per 10,000/μL) | 1.01 (0.90–1.13), P=0.85 | 1.09 (0.94–1.25), P=0.25 | 0.99 (0.96–1.03), P=0.72 | 1.01 (0.74–1.37), P=0.95 | 1.00 (0.97–1.03), P=0.93 |
|
| |||||
| BM blasts (per 10%) | 0.96 (0.93–1.00), P=0.044 | 0.96 (0.91–1.01), P=0.11 | 1.02 (0.93–1.12), P=0.71 | 0.93 (0.83–1.04), P=0.20 | 0.97 (0.94–0.99), P=0.031 |
|
| |||||
| Gender | |||||
| Female | 1.00 (reference) | 1.00 (reference) | 1.00 (reference) | 1.00 (reference) | 1.00 (reference) |
| Male | 1.09 (0.91–1.31), P=0.36 | 1.04 (0.80–1.34), P=0.79 | 0.88 (0.57–1.35), P=0.56 | 1.37 (0.83–2.26), P=0.21 | 1.08 (0.94–1.23), P=0.26 |
|
| |||||
| Type of AML | |||||
| De novo (primary) | 1.00 (reference) | 1.00 (reference) | 1.00 (reference) | 1.00 (reference) | 1.00 (reference) |
| Secondary | 1.94 (1.40–2.69), P<0.001 | 1.37 (0.86–2.19), P=0.18 | 2.17 (0.55–8.58), P=0.27 | 1.07 (0.65–1.77), P=0.79 | 1.56 (1.27–1.92), P<0.001 |
|
| |||||
| Cytogenetics | |||||
| Favorable | 0.42 (0.31–0.57), P<0.001 | 0.63 (0.41–0.97), P=0.037 | 0.41 (0.21–0.79), P=0.009 | 0.09 (0.03–0.24), P<0.001 | 0.43 (0.34–0.54), P<0.001 |
| Intermediate | 1.00 (reference) | 1.00 (reference) | 1.00 (reference) | 1.00 (reference) | 1.00 (reference) |
| Unfavorable | 4.21 (2.87–6.17), P<0.001 | 3.22 (2.29–4.53), P<0.001 | 2.09 (1.13–3.87), P=0.018 | 1.88 (1.05–3.36), P=0.034 | 2.94 (2.39–3.61), P<0.001 |
|
| |||||
| NPM1, FLT3-ITD Status | |||||
| NPM1−, FLT3-ITD− | 1.00 (reference) | 1.00 (reference) | 1.00 (reference) | 1.00 (reference) | 1.00 (reference) |
| NPM1−, FLT3-ITD+ | 2.48 (1.74–3.53), P<0.001 | 3.01 (1.87–4.83), P<0.001 | 2.46 (1.08–5.59), P=0.032 | 1.59 (0.63–4.01), P=0.32 | 2.49 (1.93–3.21), P<0.001 |
| NPM1+, FLT3-ITD− | 0.31 (0.24–0.40), P<0.001 | 0.39 (0.26–0.69), P<0.001 | 0.30 (0.16–0.55), P<0.001 | 0.22 (0.10–0.47), P<0.001 | 0.32 (0.27–0.39), P<0.001 |
| NPM1+, FLT3-ITD+ | 0.85 (0.65–1.13), P=0.27 | 1.46 (0.97–2.20), P=0.071 | 1.40 (0.52–3.14), P=0.42 | 1.60 (0.58–4.41), P=0.36 | 1.01 (0.82–1.25), P=0.64 |
|
| |||||
| Bootstrap-corrected AUC | 0.77 | 0.72 | 0.74 | 0.76 | 0.75 |
Data is presented as odds ratio (95% CI). The model also contained a covariate “missing cytogenetics”.
Abbreviations: BM, bone marrow; CR, complete remission; WBC, white blood cells.
Inclusion of patients who incurred TRM
Although our previous data suggest that 28 days is a reasonable criterion for distinguishing patients who fail therapy because of TRM,9 it is plausible that patients who die between days 14 and 28 had sufficient AML present at death that they would have been considered resistant had they lived beyond day 28. Hence, exclusion of patients who died before day 28 might affect conclusions. We therefore performed additional analyses in which we included the 7% of our patients (354/4955) who died before day 28 or were lost to follow-up before that time. As summarized in Table 7, the conclusions were essentially unaffected by inclusion of these patients, with AUCs for models predicting primary refractory disease or to have RFS of ≤3, ≤6, or ≤12 months being 0.77, 0.75, 0.76, and 0.75, respectively, when mutational data on FLT3-ITD and NPM1 were included.
TABLE 7.
Multivariate logistic regression models for failure to achieve CR or to have short RFS, with inclusion of patients who experienced TRM
| Parameter | Failure to achieve CR n = 4,955 |
Failure to achieve CR or to have RFS ≤ 3 months n = 4,897 |
Failure to achieve CR or to have RFS ≤ 6 months n = 4,848 |
Failure to achieve CR or to have RFS ≤ 12 months n = 4,802 |
|
|---|---|---|---|---|---|
|
| |||||
| Age (per 1 year) | 1.05 (1.04–1.05), P<0.001 | 1.04 (1.04–1.05), P<0.001 | 1.04 (1.04–1.04), P<0.001 | 1.04 (1.03–1.04), P<0.001 | |
|
| |||||
| Performance status | |||||
| 0–1 | 1.00 (reference) | 1.00 (reference) | 1.00 (reference) | 1.00 (reference) | |
| 2–4 | 1.75 (1.45–2.11), P<0.001 | 1.57 (1.31–1.87), P<0.001 | 1.48 (1.25–1.76), P<0.001 | 1.38 (1.16–1.65), P<0.001 | |
|
| |||||
| WBC (per 10,000/μL) | 1.61 (1.42–1.84), P<0.001 | 1.64 (1.45–1.86), P<0.001 | 1.77 (1.56–2.02), P<0.001 | 1.76 (1.53–2.02), P<0.001 | |
|
| |||||
| Platelets (per 10,000/μL) | 1.01 (0.98–1.04), P=0.53 | 1.03 (0.99–1.07), P=0.14 | 1.01 (0.98–1.05), P=0.44 | 1.00 (0.97–1.03), P=0.90 | |
|
| |||||
| BM blasts (per 10%) | 1.01 (0.98–1.04), P=0.54 | 0.99 (0.96–1.01), P=0.36 | 0.98 (0.95–1.01), P=0.14 | 0.98 (0.95–1.00), P=0.11 | |
|
| |||||
| Gender | |||||
| Female | 1.00 (reference) | 1.00 (reference) | 1.00 (reference) | 1.00 (reference) | |
| Male | 1.12 (0.97–1.29), P=0.12 | 1.10 (0.96–1.25), P=0.17 | 1.06 (0.93–1.21), P=0.37 | 1.09 (0.95–1.24), P=0.21 | |
|
| |||||
| Type of AML | |||||
| De novo (primary) | 1.00 (reference) | 1.00 (reference) | 1.00 (reference) | 1.00 (reference) | |
| Secondary | 2.02 (1.69–2.43), P<0.001 | 2.04 (1.70–2.44), P<0.001 | 1.81 (1.50–2.18), P<0.001 | 1.56 (1.27–1.91), P<0.001 | |
|
| |||||
| Cytogenetics | |||||
| Favorable | 0.34 (0.24–0.47), P<0.001 | 0.46 (0.35–0.61), P<0.001 | 0.39 (0.30–0.50), P<0.001 | 0.45 (0.36–0.55), P<0.001 | |
| Intermediate | 1.00 (reference) | 1.00 (reference) | 1.00 (reference) | 1.00 (reference) | |
| Unfavorable | 2.73 (2.27–3.28), P<0.001 | 2.89 (2.42–3.46), P<0.001 | 3.24 (2.70–3.89), P<0.001 | 3.00 (2.45–3.68), P<0.001 | |
|
| |||||
| NPM1, FLT3-ITD Status | |||||
| NPM1−, FLT3-ITD− | 1.00 (reference) | 1.00 (reference) | 1.00 (reference) | 1.00 (reference) | |
| NPM1−, FLT3-ITD+ | 1.22 (0.97–1.55), P=0.092 | 1.50 (1.20–1.88), P<0.001 | 1.94 (1.55–2.42), P<0.001 | 2.32 (1.80–2.99), P<0.001 | |
| NPM1+, FLT3-ITD− | 0.41 (0.33–0.51), P<0.001 | 0.44 (0.36–0.54), P<0.001 | 0.40 (0.33–0.48), P<0.001 | 0.34 (0.28–0.41), P<0.001 | |
| NPM1+, FLT3-ITD+ | 0.51 (0.40–0.65), P<0.001 | 0.71 (0.57–0.88), P=0.0023 | 0.85 (0.69–1.04), P=0.12 | 0.95 (0.77–1.17), P=0.65 | |
|
| |||||
| Bootstrap-corrected AUC | 0.77 | 0.75 | 0.76 | 0.75 | |
Data is presented as odds ratio (95% CI). The model also contained a covariate “missing cytogenetics”.
Abbreviations: BM, bone marrow; CR, complete remission; WBC, white blood cells.
AUCs in specific cytogenetic risk groups
One might suspect that the ability to predict resistance is higher in patients with high-risk or, conversely, favorable cytogenetics. To test this possibility, we performed separate analyses of resistance in favorable-risk, intermediate-risk, and unfavorable-risk cytogenetic groups. However, there was no evidence that AUCs differed in different cytogenetic risk groups regardless of whether FLT3-ITD and NPM1 data were included. Even in the unfavorable-risk group AUCs remained relatively low for models predicting primary refractory disease or to have RFS of ≤3, ≤6, or ≤12 months (0.64–0.65).
DISCUSSION
AML patients fail to be cured primarily because of resistance to therapy. The dominant role of resistance has been noted previously for patients during induction18 as well as while in remission.19 Analyses from our patient cohort are consistent with these previous data in that only 7% of patients experienced TRM and 16% died while in CR. Accurate methods to predict resistance to standard therapy are needed if physicians are to reliably advise AML patients. Specifically, if the ability to predict resistance was sufficiently high, it would be difficult to recommend standard therapy to some patients and similarly difficult to recommend investigational therapies to others. Experience suggests that these recommendations are commonly made; however, they are made in the absence of quantitative measurements of predictive ability. While perhaps not surprisingly confirming that age, performance status, WBC, secondary disease, cytogenetic risk, and NPM1/FLT3-ITD mutational status are each strongly and independently associated (P<0.001) with resistance, our results suggest that our ability to predict resistance is much less impressive. Indeed, using standard criteria relating AUC to predictive ability, this ability is only “fair” and is little improved by inclusion of data on aberrations in FLT3 and NPM1; the same was true if attention was restricted to more homogeneous subsets such as younger patients or those with de novo AML. This finding, denoting only a fair ability to predict CR, is reminiscent of a recent study by Krug et al., who observed similar AUCs (0.72 and 0.68) with multivariable models in their study cohort, although their definition of primary refractory AML were slightly different.20 While some clinicians might understandably feel that even fair predictive ability justifies a recommendation that some patients receive standard therapy and others investigational therapy, others might feel that the relatively low AUCs for resistance support randomization between standard and investigational therapies. Thus this study provides a rationale for continued randomization as the best means to accurately evaluate the comparative efficacies of new and standard therapies.
Resistance is undoubtedly difficult to predict because AML is an extraordinarily complex and heterogeneous disease, whose diversity is only very partially captured by clinical parameters, cytogenetic data, and information about aberrations in NPM1 and FLT3, the covariates we examined. Mutations in many additional genes may carry prognostic significance,21–25 the degree of which may depend on the co-existence of other alterations;26 it would therefore be interesting to assess the degree to which AUCs could be improved by incorporating data from additional genetic or molecular analyses or pre-treatment assays that test AML signaling pathways functionally. Moreover, different AML subclones may exhibit differential chemotherapy sensitivities, with resistance reflecting emergence of subclones whose importance cannot be determined at initial presentation.27 Emergence of these clones will likely make it more difficult to forecast resistance as the period over which the forecast is attempted increases. Such increasing difficulty was seen as we moved from predicting primary resistance to predicting primary resistance and RFS. In contrast, our previous efforts to predict death within 28 days of beginning chemotherapy were more successful.9 Integration of post-treatment data, e.g. early disease clearance in the case of primary refractory AML or achievement of CR vs. CRp/CRi and assessment of minimal residual disease,28–33 in the case of RFS may improve prediction of resistance.
While our data are derived from well-annotated patients treated with contemporary AML regimens, several limitations need to be acknowledged. First, our results may only apply to newly diagnosed AML treated with “3+7” or more intense regimens and may be influenced by allogeneic transplantation, about which we have only limited information. Second, rather than “harmonizing” cytogenetic risk categories across our study cohort, we used the cooperative study group’s separate classification; however, recoding of the SWOG patient cohort to the refined MRC/NCRI classification34 only slightly changed the AUC of a multivariable prediction model (change from 0.68 to 0.70) and additional categorization of patients with monosomal karyotype35 as separate risk entity did not further improve the AUC (AUC=0.70). Third, we did not distinguish between relapse and death in CR as events contributing to short RFS, a criterion for resistance in some of our analyses. While patients who die in CR are not necessarily “resistant”, previous studies indicated that patients who die in CR are much more similar to patients who relapse than those who remain alive in CR.19 And fourth, we did not use an independent dataset to obtain an unbiased estimate of the AUC of any of the models presented in this paper. However, the goal of this paper was not to generate one predictive model; rather, our aim was to investigate how the predictive ability of a model changes as cytogenetics and molecular information is incorporated into the model, and to assess whether there were similar trends across separate cohorts. For this purpose, validation via bootstrapping15, 16 provides a useful approach to assess the internal model accuracy. Of note, experience suggests that models do not perform as well in independent datasets as in the dataset used to derive them. Thus, the use of another dataset is unlikely to change our fundamental conclusion (i.e. that the usefulness of current prognostic factors for guiding treatment decisions is very likely overestimated), which was supported by separate analyses of data from SWOG, MDA, HOVON, and MRC.
In conclusion, our ability to predict therapeutic resistance based on routinely available clinical covariates, even with inclusion of commonly used molecular data on FLT3 and NPM1, is relatively limited. This finding appears to have significant clinical consequences. Not infrequently, multivariate analysis is used to examine whether, after accounting for covariates such as cytogenetics, NPM1 and FLT3 status, a new treatment is superior to an older one. However if the predictive ability of these covariates is limited as our results indicate, reliance on historical controls to assess the new treatment is problematic. Thus, our results emphasize the continued importance of randomized treatment assignment for the testing of new drugs in AML.
Supplementary Material
Acknowledgments
Research reported in this publication was supported by grants from the National Cancer Institute/National Institutes of Health (NCI/NIH; R21-CA182010 to R.B.W. and M.O., and R01-CA090998-09 to M.O.). SWOG trials were supported in part by the following PHS Cooperative Agreement grant numbers awarded by the NCI/NIH: U10-CA032102, U10-CA038926, and U10-CA105409. R.B.W. is a Leukemia & Lymphoma Society Scholar in Clinical Research.
Footnotes
Presented in part at the 55th Annual Meeting of the American Society of Hematology, December 7–10, 2013, New Orleans, LA
CONFLICT OF INTEREST
The authors declare no competing financial interests.
Supplementary information is available at Leukemia’s website.
References
- 1.Estey E, Döhner H. Acute myeloid leukaemia. Lancet. 2006;368(9550):1894–1907. doi: 10.1016/S0140-6736(06)69780-8. [DOI] [PubMed] [Google Scholar]
- 2.Döhner H, Estey EH, Amadori S, Appelbaum FR, Büchner T, Burnett AK, et al. Diagnosis and management of acute myeloid leukemia in adults: recommendations from an international expert panel, on behalf of the European LeukemiaNet. Blood. 2010;115(3):453–474. doi: 10.1182/blood-2009-07-235358. [DOI] [PubMed] [Google Scholar]
- 3.Ferrara F, Schiffer CA. Acute myeloid leukaemia in adults. Lancet. 2013;381(9865):484–495. doi: 10.1016/S0140-6736(12)61727-9. [DOI] [PubMed] [Google Scholar]
- 4.Burnett AK, Russell NH, Hills RK, Hunter AE, Kjeldsen L, Yin J, et al. Optimization of chemotherapy for younger patients with acute myeloid leukemia: results of the medical research council AML15 trial. J Clin Oncol. 2013;31(27):3360–3368. doi: 10.1200/JCO.2012.47.4874. [DOI] [PubMed] [Google Scholar]
- 5.Kayser S, Zucknick M, Döhner K, Krauter J, Köhne CH, Horst HA, et al. Monosomal karyotype in adult acute myeloid leukemia: prognostic impact and outcome after different treatment strategies. Blood. 2012;119(2):551–558. doi: 10.1182/blood-2011-07-367508. [DOI] [PubMed] [Google Scholar]
- 6.Harrell FE., Jr . Regression modeling strategies: with applications to linear models, logistic regression, and survival analysis. Springer; New York, NY: 2001. [Google Scholar]
- 7.Pepe MS. The statistical evaluation of medical tests for classification and prediction. Oxford University Press; New York, NY: 2004. [Google Scholar]
- 8.Swerdlow SH, Campo E, Harris NL, Jaffe ES, Pileri SA, Stein H, et al. WHO Classification of Tumours of Haematopoietic and Lymphoid Tissue. 4. IARC; Lyon, France: 2008. [Google Scholar]
- 9.Walter RB, Othus M, Borthakur G, Ravandi F, Cortes JE, Pierce SA, et al. Prediction of early death after induction therapy for newly diagnosed acute myeloid leukemia with pretreatment risk scores: a novel paradigm for treatment assignment. J Clin Oncol. 2011;29(33):4417–4423. doi: 10.1200/JCO.2011.35.7525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cheson BD, Bennett JM, Kopecky KJ, Buchner T, Willman CL, Estey EH, et al. Revised recommendations of the International Working Group for Diagnosis, Standardization of Response Criteria, Treatment Outcomes, and Reporting Standards for Therapeutic Trials in Acute Myeloid Leukemia. J Clin Oncol. 2003;21(24):4642–4649. doi: 10.1200/JCO.2003.04.036. [DOI] [PubMed] [Google Scholar]
- 11.Kaplan EL, Meier P. Nonparametric estimation from incomplete observations. J Am Stat Assoc. 1958;53:457–481. [Google Scholar]
- 12.Muller MP, Tomlinson G, Marrie TJ, Tang P, McGeer A, Low DE, et al. Can routine laboratory tests discriminate between severe acute respiratory syndrome and other causes of community-acquired pneumonia? Clin Infect Dis. 2005;40(8):1079–1086. doi: 10.1086/428577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ishihara R, Yamada T, Iishi H, Kato M, Yamamoto S, Yamamoto S, et al. Quantitative analysis of the color change after iodine staining for diagnosing esophageal high-grade intraepithelial neoplasia and invasive cancer. Gastrointest Endosc. 2009;69(2):213–218. doi: 10.1016/j.gie.2008.04.052. [DOI] [PubMed] [Google Scholar]
- 14.Roelen CA, van Rhenen W, Groothoff JW, van der Klink JJ, Bultmann U, Heymans MW. The development and validation of two prediction models to identify employees at risk of high sickness absence. Eur J Public Health. 2013;23(1):128–133. doi: 10.1093/eurpub/cks036. [DOI] [PubMed] [Google Scholar]
- 15.Harrell FE, Jr, Lee KL, Mark DB. Multivariable prognostic models: issues in developing models, evaluating assumptions and adequacy, and measuring and reducing errors. Stat Med. 1996;15(4):361–387. doi: 10.1002/(SICI)1097-0258(19960229)15:4<361::AID-SIM168>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 16.Harrell FE, Jr, Margolis PA, Gove S, Mason KE, Mulholland EK, Lehmann D, et al. Development of a clinical prediction model for an ordinal outcome: the World Health Organization Multicentre Study of Clinical Signs and Etiological agents of Pneumonia, Sepsis and Meningitis in Young Infants. WHO/ARI Young Infant Multicentre Study Group. Stat Med. 1998;17(8):909–944. doi: 10.1002/(sici)1097-0258(19980430)17:8<909::aid-sim753>3.0.co;2-o. [DOI] [PubMed] [Google Scholar]
- 17.Othus M, Appelbaum FR, Petersdorf S, Erba HP, Estey EH. Evaluation of which patients get a second course of 3+7 on cooperative group trials for newly diagnosed acute myeloid leukemia: a report from SWOG [abstract] Blood. 2013;122(21):3925. [Google Scholar]
- 18.Appelbaum FR, Gundacker H, Head DR, Slovak ML, Willman CL, Godwin JE, et al. Age and acute myeloid leukemia. Blood. 2006;107(9):3481–3485. doi: 10.1182/blood-2005-09-3724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yanada M, Garcia-Manero G, Borthakur G, Ravandi F, Kantarjian H, Estey E. Relapse and death during first remission in acute myeloid leukemia. Haematologica. 2008;93(4):633–634. doi: 10.3324/haematol.12366. [DOI] [PubMed] [Google Scholar]
- 20.Krug U, Röllig C, Koschmieder A, Heinecke A, Sauerland MC, Schaich M, et al. Complete remission and early death after intensive chemotherapy in patients aged 60 years or older with acute myeloid leukaemia: a web-based application for prediction of outcomes. Lancet. 2010;376(9757):2000–2008. doi: 10.1016/S0140-6736(10)62105-8. [DOI] [PubMed] [Google Scholar]
- 21.Taskesen E, Bullinger L, Corbacioglu A, Sanders MA, Erpelinck CA, Wouters BJ, et al. Prognostic impact, concurrent genetic mutations, and gene expression features of AML with CEBPA mutations in a cohort of 1182 cytogenetically normal AML patients: further evidence for CEBPA double mutant AML as a distinctive disease entity. Blood. 2011;117(8):2469–2475. doi: 10.1182/blood-2010-09-307280. [DOI] [PubMed] [Google Scholar]
- 22.Ley TJ, Ding L, Walter MJ, McLellan MD, Lamprecht T, Larson DE, et al. DNMT3A mutations in acute myeloid leukemia. N Engl J Med. 2010;363(25):2424–2433. doi: 10.1056/NEJMoa1005143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Paschka P, Schlenk RF, Gaidzik VI, Habdank M, Kronke J, Bullinger L, et al. IDH1 and IDH2 mutations are frequent genetic alterations in acute myeloid leukemia and confer adverse prognosis in cytogenetically normal acute myeloid leukemia with NPM1 mutation without FLT3 internal tandem duplication. J Clin Oncol. 2010;28(22):3636–3643. doi: 10.1200/JCO.2010.28.3762. [DOI] [PubMed] [Google Scholar]
- 24.Metzeler KH, Becker H, Maharry K, Radmacher MD, Kohlschmidt J, Mrozek K, et al. ASXL1 mutations identify a high-risk subgroup of older patients with primary cytogenetically normal AML within the ELN Favorable genetic category. Blood. 2011;118(26):6920–6929. doi: 10.1182/blood-2011-08-368225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rücker FG, Schlenk RF, Bullinger L, Kayser S, Teleanu V, Kett H, et al. TP53 alterations in acute myeloid leukemia with complex karyotype correlate with specific copy number alterations, monosomal karyotype, and dismal outcome. Blood. 2012;119(9):2114–2121. doi: 10.1182/blood-2011-08-375758. [DOI] [PubMed] [Google Scholar]
- 26.Patel JP, Gonen M, Figueroa ME, Fernandez H, Sun Z, Racevskis J, et al. Prognostic relevance of integrated genetic profiling in acute myeloid leukemia. N Engl J Med. 2012;366(12):1079–1089. doi: 10.1056/NEJMoa1112304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Walter MJ, Shen D, Ding L, Shao J, Koboldt DC, Chen K, et al. Clonal architecture of secondary acute myeloid leukemia. N Engl J Med. 2012;366(12):1090–1098. doi: 10.1056/NEJMoa1106968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Haferlach T, Kern W, Schoch C, Schnittger S, Sauerland MC, Heinecke A, et al. A new prognostic score for patients with acute myeloid leukemia based on cytogenetics and early blast clearance in trials of the German AML Cooperative Group. Haematologica. 2004;89(4):408–418. [PubMed] [Google Scholar]
- 29.Elliott MA, Litzow MR, Letendre LL, Wolf RC, Hanson CA, Tefferi A, et al. Early peripheral blood blast clearance during induction chemotherapy for acute myeloid leukemia predicts superior relapse-free survival. Blood. 2007;110(13):4172–4174. doi: 10.1182/blood-2007-07-104091. [DOI] [PubMed] [Google Scholar]
- 30.Loken MR, Alonzo TA, Pardo L, Gerbing RB, Raimondi SC, Hirsch BA, et al. Residual disease detected by multidimensional flow cytometry signifies high relapse risk in patients with de novo acute myeloid leukemia: a report from Children’s Oncology Group. Blood. 2012;120(8):1581–1588. doi: 10.1182/blood-2012-02-408336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lacombe F, Arnoulet C, Maynadié M, Lippert E, Luquet I, Pigneux A, et al. Early clearance of peripheral blasts measured by flow cytometry during the first week of AML induction therapy as a new independent prognostic factor: a GOELAMS study. Leukemia. 2009;23(2):350–357. doi: 10.1038/leu.2008.296. [DOI] [PubMed] [Google Scholar]
- 32.Freeman SD, Virgo P, Couzens S, Grimwade D, Russell N, Hills RK, et al. Prognostic relevance of treatment response measured by flow cytometric residual disease detection in older patients with acute myeloid leukemia. J Clin Oncol. 2013;31(32):4123–4131. doi: 10.1200/JCO.2013.49.1753. [DOI] [PubMed] [Google Scholar]
- 33.Terwijn M, van Putten WL, Kelder A, van der Velden VH, Brooimans RA, Pabst T, et al. High Prognostic Impact of Flow Cytometric Minimal Residual Disease Detection in Acute Myeloid Leukemia: Data From the HOVON/SAKK AML 42A Study. J Clin Oncol. 2013;31(31):3889–3897. doi: 10.1200/JCO.2012.45.9628. [DOI] [PubMed] [Google Scholar]
- 34.Grimwade D, Hills RK, Moorman AV, Walker H, Chatters S, Goldstone AH, et al. Refinement of cytogenetic classification in acute myeloid leukemia: determination of prognostic significance of rare recurring chromosomal abnormalities among 5876 younger adult patients treated in the United Kingdom Medical Research Council trials. Blood. 2010;116(3):354–365. doi: 10.1182/blood-2009-11-254441. [DOI] [PubMed] [Google Scholar]
- 35.Breems DA, Van Putten WL, De Greef GE, Van Zelderen-Bhola SL, Gerssen-Schoorl KB, Mellink CH, et al. Monosomal karyotype in acute myeloid leukemia: a better indicator of poor prognosis than a complex karyotype. J Clin Oncol. 2008;26(29):4791–4797. doi: 10.1200/JCO.2008.16.0259. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.