Abstract
Numerous instances can be seen in evolution where protein quaternary structures have diverged while the sequences of the building blocks have remained fairly conserved. However, the path through which such divergence has taken place is usually not known. We have designed two synthetic 29-residue α-helical peptides, based on the coiled-coil structural motif, that spontaneously self-assemble into helical nanotubes in vitro. Using electron cryo-microscopy (cryo-EM) with a newly available direct electron detection capability, we can achieve near-atomic resolution of these thin structures. We show how conservative changes of only one or two amino acids results in dramatic changes in quaternary structure, in which the assemblies can be switched between two very different forms. This system provides a framework for understanding how small sequence changes in evolution can translate into very large changes in supramolecular structure, a phenomenon that may have significant implications for the de novo design of synthetic peptide assemblies.
Keywords: electron cryo-microscopy, coiled-coils, helical polymers
Introduction
Native helical assemblies (e.g., filamentous phage capsids (Wang et al., 2006), flagellar filaments (Yonekura et al., 2003), type IV pili (Craig et al., 2006), and the type III secretion system needle (Loquet et al., 2012) encompass diverse functions that would be desirable to emulate in synthetic protein- and peptide-based assemblies, which include dynamic switching, locomotion, controlled release, directional transport, and selective catalysis. However, current limitations in the ability to reliably define the relationship between sequence and supramolecular structure present a significant challenge to the de novo design of novel self-assembled protein architectures, which, in most cases, remains beyond current capabilities of computational structure prediction. This phenomenon is due in part to the fact that, while the tertiary structure of proteins is quite robust with respect to many changes in amino acid sequence (Smith, 1970), the quaternary structure of proteins can be very sensitive to slight variations in sequence. Examples exist such as hemoglobin (Coates, 1975) or bacterial flagellar filaments (Galkin et al., 2008) in which conserved protein folds can assemble into significantly different higher-order forms. Model systems, such as coiled-coil α-helical bundles of the Rop protein, have also shown an unexpected degree of structural plasticity (Amprazi et al., 2014). Since quaternary structures arise from interactions between protomers, these observations raise a significant question regarding the designability of the protein-protein interfaces within biological assemblies (Zhang et al., 2014).
We designed two peptides, Form I and Form II, based on a coiled-coil structural motif, to self-assemble into extended cylindrical assemblies (nanotubes). While coiled-coils have been postulated as potential substrates for the creation of cylindrical assemblies (Walshaw and Woolfson, 2001a), the corresponding formation of synthetic, large-diameter nanotube assemblies has never successfully been accomplished. We demonstrate that these short synthetic peptides can self-assemble into high aspect-ratio fibrillar structures that correspond to helical nanotube assemblies. Using electron cryo-microscopy (cryo-EM) with a newly available direct electron detection capability (Bai et al., 2013; Bammes et al., 2012; Fernandez et al., 2014; Li et al., 2013; Liao et al., 2013; Lu et al., 2014; Wu et al., 2014), we can achieve near-atomic resolution of these thin structures when imaging the equivalent of a single tube that is ~ 17µ m long containing only 80,000 molecules. We show, quite surprisingly, that the structure of these nanotubes can be changed dramatically with only one or two amino acid changes. This peptide system provides direct insight into the designability of extended assemblies based on biologically derived structural motifs, which has potential significance for the de novo design of synthetic peptide-based nanomaterials.
Results
The sequence design of the Form I and Form II peptides (Figure 1) was based on a structural model (Walshaw and Woolfson, 2001a) that derived from a theoretical analysis of the helix-helix packing interactions within bi-faceted coiled-coils (Fig. 2). Antitypic self-association between the two offset hydrophobic faces was proposed to drive self-assembly of the coiled-coil peptides into cylindrical α-helical assemblies in which the individual helices were aligned parallel to the axis of the filament. This structural model was employed to rationalize the interactions of particular α helices within the protein tolC (Calladine et al., 2001; Koronakis et al., 2000), a component of a pump complex associated with multi-drug resistance in gram-negative bacteria.
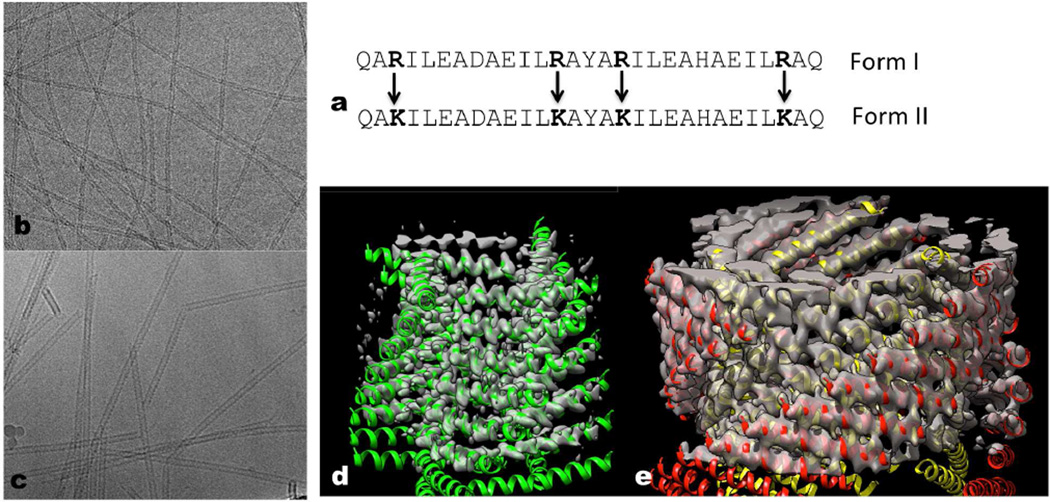
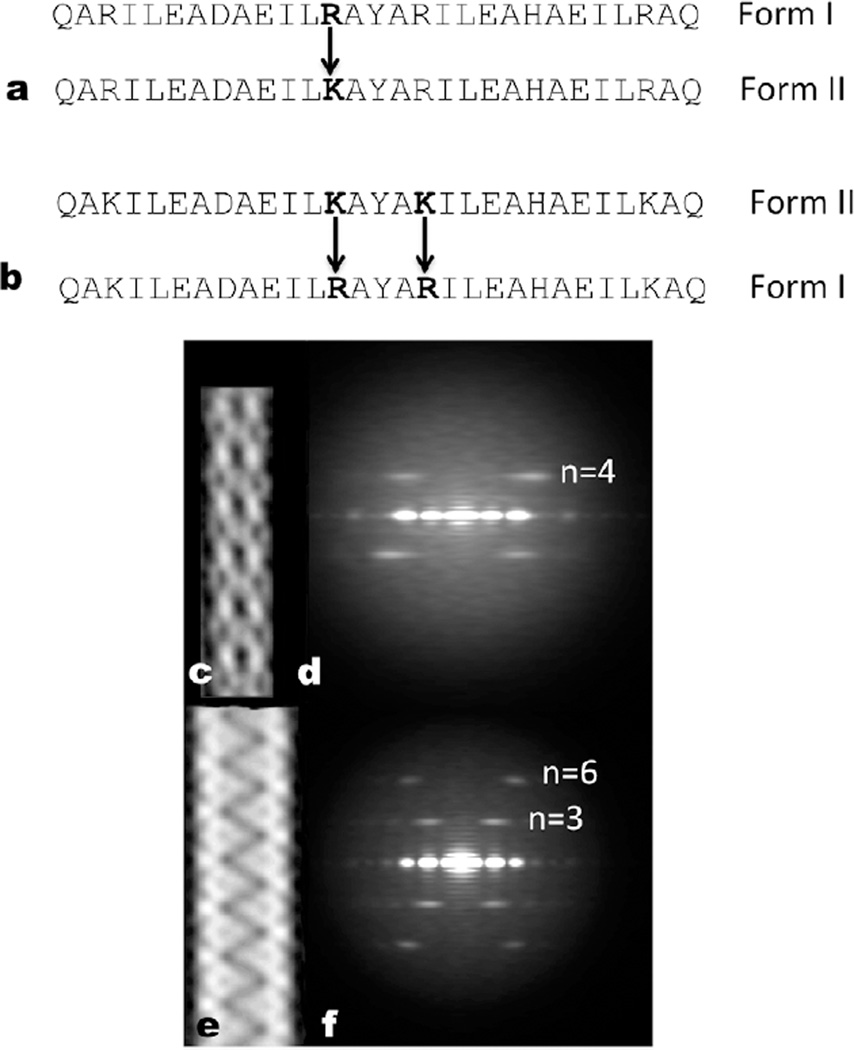
Figure 1. Conservative amino acid changes lead to two different helical forms.
a, The 29 residue peptide shown has four arginines, and this peptide polymerizes into a single-walled Form I polymer that has circa four subunits per turn. When the four arginines are changed to lysines, the peptides polymerize into a totally different Form II filament with circa three subunits per turn that is double-walled and has two peptides in each asymmetric unit. b–c, Electron cryo-micrographs of the Form I filament (b) (~ 60 Å diameter) and the Form II filament (c) (~ 120 Å diameter). d–e, Three-dimensional reconstructions of the Form I filament (d) and the Form II filament (e) have been fit with atomic models. In the Form II filament (e) there are two peptides in each asymmetric unit, and the outer helices have been colored red while the inner ones are yellow. In the Form I filament (d) each peptide is in an identical environment. The N-terminal region of the Form I helices (d) is melted, and density is not seen for this region at the high threshold used.
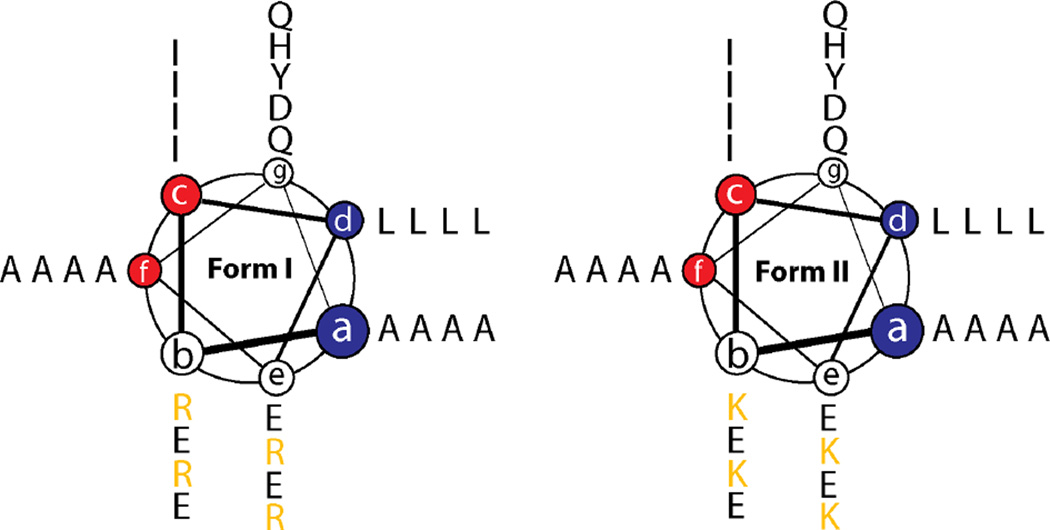
Figure 2. Helical wheel diagram of Form I and Form II peptide sequences in coiled-coil superhelix space.
The two offset hydrophobic faces, (a/d) and (c/f), are highlighted in blue and red, respectively.
The Form I and Form II peptides recapitulate the heptad repeat pattern proposed for tolC, in which the two hydrophobic faces, (a/d) and (c/f), respectively, are offset by two amino acid residues. However, the synthetic sequences diverge from the sequence analysis of tolC in several critical aspects. First, in the structure of tolC, residues at the a- and f-positions of the heptad repeats, which occur at the outer surface of the helix-helix interface, comprise larger side-chain amino acids than those that occur at the inner interface at the c- and d-positions (Calladine et al., 2001). This substitution pattern has the effect of increasing the degree of curvature between interacting helices in the cylindrical assembly, which results in smaller diameter nanotubes that consist of fewer helices than predicted from purely geometric considerations (Walshaw and Woolfson, 2001a). In contrast, the design of Form I and Form II reverses this situation, as smaller alanine residues were encoded at the outer a- and f-positions while larger residues, isoleucine and leucine, respectively, were incorporated at c-and d-positions. This design was selected in order to decrease the curvature of the corresponding cylindrical assemblies, potentially affording larger diameter nanotubes. The substitution of alanine residues at peripheral positions within the heptad repeats has been employed previously in the design of coiled-coil assemblies to promote the formation of higher oligomerization states (Liu et al., 2006; Thomson et al., 2014; Zaccai et al., 2011). In addition, charged residues were introduced at the b- and e- positions of the heptad repeats in order to impart solubility and direct an anti-typic facial interaction between adjacent helices in a parallel orientation. In analogy to tolC, it was anticipated that this mode of interaction would favor formation of closed cylindrical structures based on interactions between α-helices. However, the pattern of charged residues at these positions proved to be critical to the formation of a stable α-helical conformation. Initial designs were prone to adopt a β-strand conformation, which resulted in the formation of amyloid-like assemblies. Negative design was employed in the sequences of Form I and Form II to select against the potential formation of amyloid assemblies through introduction of repulsive Coulombic interactions within alternative conformations having suppositious β-sheet structures. These design considerations placed significant constraints on the resultant peptide sequences, which differed otherwise only in terms of a conservative substitution of four arginines for four lysines at identical positions in Form I versus Form II (Fig. 1a; Fig. 2). The resultant peptides comprised nearly palindromic sequences of 29 amino acids, which complicated the subsequent structural analysis of the resultant nanotubes (see below).
Both peptides self-assemble under mild conditions into high aspect-ratio helical assemblies, which represent the first example of large diameter (≥ 5 nm) tubular assemblies from de novo designed peptides based on coiled-coil structural motifs. Remarkably, the conservative replacement of the four arginines with four lysines leads to a doubling of the diameter of the tubes, as judged by cryo-EM of frozen-hydrated samples (Fig. 1b,c). Using either film or conventional CCD detectors for the EM, we were unable to even determine the helical symmetry of these two forms, let alone generate a three-dimensional reconstruction. However, with a direct electron detector (Bai et al., 2013; Bammes et al., 2012; Fernandez et al., 2014; Li et al., 2013; Liao et al., 2013; Lu et al., 2014; Wu et al., 2014) it was possible to show that the Form I tubes contained a single α-helix in the asymmetric unit (Fig. 1d), while the Form II tubes contained two α-helices in the asymmetric unit (Fig. 1e).
Determination of the helical symmetry proceeded by attempting to index the power spectra (Klug et al., 1958), such as those in Supplementary Movies 1 and 2. Indexing involves assigning a Bessel order to a layer line, and while indexing the power spectrum from an array of atoms all at the same radius is rather simple, many difficulties are present when a helical filament or tube has density extending over a radial range (Egelman, 2010). In fact, for a finite resolution there are frequently ambiguities, such that multiple helical symmetries can all generate the same power spectrum (Egelman, 2010; Egelman, 2014). For both Form I and Form II such a multiplicity of possible solutions existed, and the Iterative Helical Real Space Reconstruction (IHRSR) method (Egelman, 2000) was used for each of the possible symmetries to find one that yielded the correct symmetry. The correct symmetry was apparent in that it showed recognizable secondary structure (α-helices), while incorrect symmetries generated reconstructions that were not interpretable. A complication, however, is that the IHRSR approach can be unstable unless out-of-plane tilt is explicitly treated, and that was done for both forms of the tubes by including reference volumes with out-of-plane tilt. It has been stated previously (Egelman, 2007; Yu and Egelman, 2010) that finding a stable solution in IHRSR is a necessary but not sufficient condition for a helical symmetry to be correct. Ignoring out-of-plane tilt in cryo-EM means that a stable solution is neither necessary nor sufficient for the symmetry used to be correct!
Both forms exhibit the random variability in twist (Fig. 3) which is common in many helical polymers (Egelman et al., 1982), and this helps explain why the symmetry is so difficult to determine in the absence of a direct electron detector since the filaments are far from crystalline. While the reference-based sorting (using models with different pitch) that has been used to generate the histograms in Fig. 3 could lead to an overly broad distribution due to poor signal to noise in the images, out-of-plane tilt, and misclassification errors, the distributions have been confirmed using power spectra generated from the different bins which show that the distributions shown really do arise from the variability in the twist of the long-pitch helices (Supplementary Movies 1 and 2).
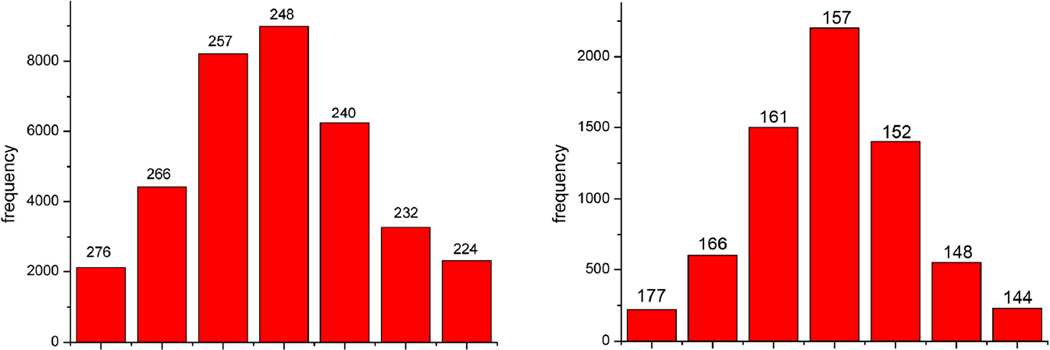
Figure 3. Variability in twist.
The variability in twist found in cryo-EM images can be seen for the Form I filaments (a) and the Form II filaments (b). In (a), continuous four-start helices were used as references for the sorting, with a range of twist from 1.3°/Å to 1.6°/Å for the seven bins shown. In (b), continuous three-start helices were used, with a range of twist from 2.0°/Å to 2.5°/Å. The corresponding pitch of these helices (in Ångstroms) is shown over each bin. The reconstructions were generated by only using images from the central bins of each of these distributions. A test for the reliability of this sorting comes from power spectra (Supplementary Movies 1 and 2), which show that the distributions do reflect the intrinsic variability in pitch.
After sorting by twist (Fig. 3) and using only a subset of the data, we have been able to generate three-dimensional reconstructions (Egelman, 2000) at ~ 3.6 Å resolution for Form I and ~ 5–6 Å resolution for the Form II (Fig. 4). The Form I reconstruction comes from 62,122 overlapping segments, which corresponds to ~ 200,000 unique copies of the peptide. Using a narrower range of twist (Fig, 3a) for the Form I filaments containing ~ 24,000 segments (corresponding to ~ 80,000 unique copies of the peptide) the resolution is not degraded significantly, showing once again (Bai et al., 2013) that a remarkable resolution for cryo-EM can be achieved with a quite limited number of molecules.
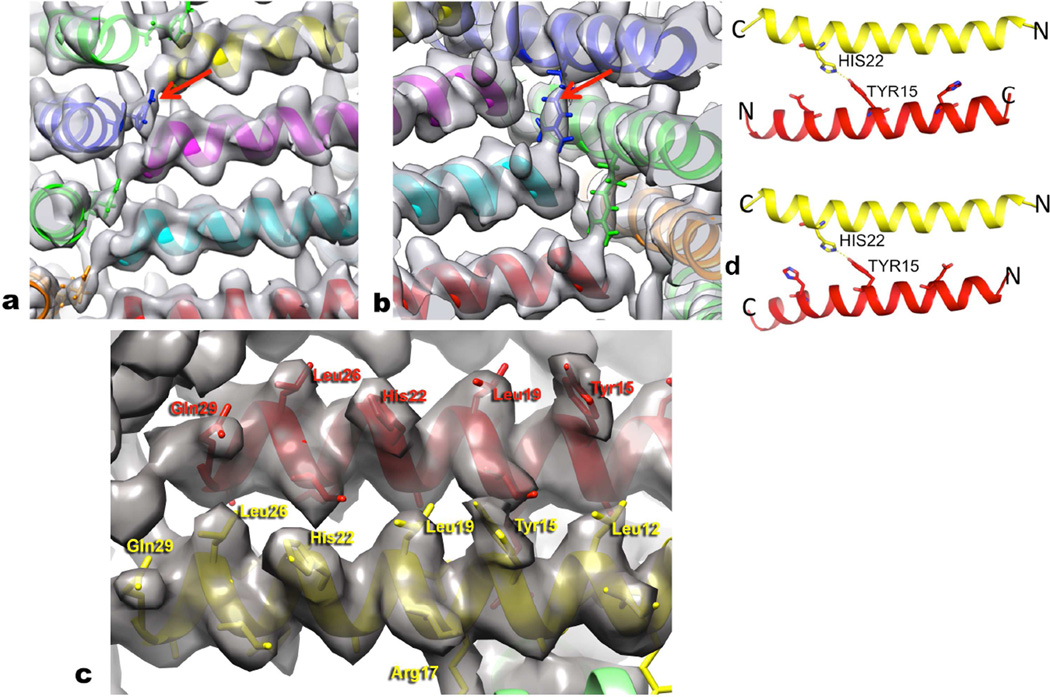
Figure 4. Details of the atomic models are testable.
a, In the Form I filament the single most ordered side chain is Arg13, which is sandwiched between two other subunits at every corner of the square as seen in this view from the outside of the filament. The Arg13 in blue (arrow) can be seen as tightly packed between the subunit in magenta and the one in yellow. b, Arg17 (arrow) provides a C-terminal cap for the α-helix formed by an adjacent peptide in Form I, and stabilizes the inside corners of the square. This view is from the lumen of the filament, and the Arg17 in blue extends to the C-terminus of the cyan helix. c, The quality and resolution of the Form I map can be seen when looking from the outside of the filaments. A number of the larger side chains have been labeled, and density exists to accommodate these residues. d, The lower resolution and nearly-palindromic nature of the sequence leads to ambiguity in the direction of the outer helix in the Form II assembly. In both models, parallel (bottom) and anti-parallel (top), His22 from the inner helix (yellow) interacts with Tyr15 in the outer helix (red). In both models, His22 from both the inner and outer helices is in the space between the two helices.
In Form I, the subunits are arranged along a left-handed one-start helix with ~ 4.1 subunits per turn, generating a nearly square cross-section, with an axial rise per subunit of 2.2 Å. A one-start helix is defined (Klug et al., 1958) as a helix that passes through every subunit, while each of the two strands of a 2-start helix pass through every other subunit, each of the three strands of a three-start helix pass through every third subunit, etc. Thus, the Form I filaments can be defined in terms of four stacks of α-helical subunits, each stack forming a four-start helix. In contrast, in the Form II there are ~ 3.1 subunits per turn with an axial rise per asymmetric unit (containing two helices) of 2.1 Å. The cross-section of the Form II filament is not triangular but circular, due to the fact that the subunits are at a larger radius and exhibit some curvature. The Form II filaments have two layers, an inner one and an outer one, and each layer contains stacks of α-helical subunits forming three-start helices.
The substantially worse resolution for Form II appears to stem from the possibility that the two layers (outer and inner) are not always in perfect register. This was suggested by the fact that iterations attempting to align images only against one layer or the other degenerated rather quickly. This is not surprising, as the packing of helices within the stacks that form each of the two layers is tighter than the packing between the two layers (see below).
The higher resolution of Form I allows us to generate an atomic model with a high degree of confidence (Fig. 4a,b,c), using an approach based upon Rosetta constrained by the cryo-EM map (Loquet et al., 2012). The model has been used to make and test predictions about the role of particular residues. It can be seen (Fig. 4a) that Arg13 makes an important contact with two neighboring subunits at each corner of the square, stabilizing this side chain and allowing us to see the full electron density expected for this residue. Mutating this single arginine to lysine (Fig. 5a) causes a striking switch from Form I to Form II. Arg17 also makes an important interaction in Form I that holds the structure together (Fig. 4b), and making the double mutant K13R, K17R in the peptide that assembles into Form II causes it now to make a Form I filament (Fig. 5b). Thus, guided by our atomic model we see that changing all four arginines to lysines (Fig. 1a), or vice versa, is not needed to switch between these two very different states of the polymer.
Figure 5. Only one or two amino acid changes are needed for dramatic rearrangements.
a, The R13K mutation is all that is necessary to make a Form I polymer (c,d) into a Form II polymer (e,f). b, The K13R,K17R double mutation turns a Form IIfilament (e,f) into a Form I filament (c,d). c,e, “Reference-free” averages (Penczek et al., 1992) from images of negatively-stained filaments of the K13R,K17R double mutation (c) and the R13K mutation (e). d,f, Averaged power spectra from negatively stained filaments of the K13R,K17R double mutation (d) and the R13K mutation (f). The layer line Bessel orders are indicated, and it can be seen that the only layer line visible in (d) arises from the four-start helix, while in (f) the first visible layer line arises from the three-start helix and the next layer line is the second order of that (n=6).
While the lower resolution of Form II does not allow the same confidence in generating a full atomic model, various features of the model that was built appear to be quite reliable. The packing between helices within the inner layer that are separated by one turn of the helix (those lying on top of each other in the same stack) are very similar to the packing between helices in the outer layer, and both are similar to the packing in the stacks that constitute the Form I single layered tubes. In contrast, a new interface appears in Form II that is weaker, as evidenced by the increased spacing between helices across the inner and outer layers in comparison to the stacking of helices within either of the two layers. In addition, there are no contacts between adjacent helices along the one-start helix in the outer layer, while in the inner layer such contacts (formed by the termini of the helices) are much weaker than in the more compact Form I polymer. Models suggest that the inner layer of the Form II assembly is stabilized by a pair of hydrogen bonds between Gln1 in one subunit and Gln29 in an adjacent subunit; these interactions appear weaker than the more extensive hydrogen bonding network of the Form I polymer (involving Glu10, Arg13, Arg17, Glu20, and Gln29).
In support of these results, the structural models that we generated appear to fit experimental data that have been acquired from other modes of structural analysis and to account for the observed properties of the assemblies. The structural models for the Form I and Form II assemblies were employed to generate theoretical scattering curves that compared well to experimental data acquired using synchrotron SAXS measurements (Supplementary Figure 6). This correspondence strongly suggests that the tubular structures persist in dilute aqueous solution and that the models are representative of the structure of the respective assemblies.
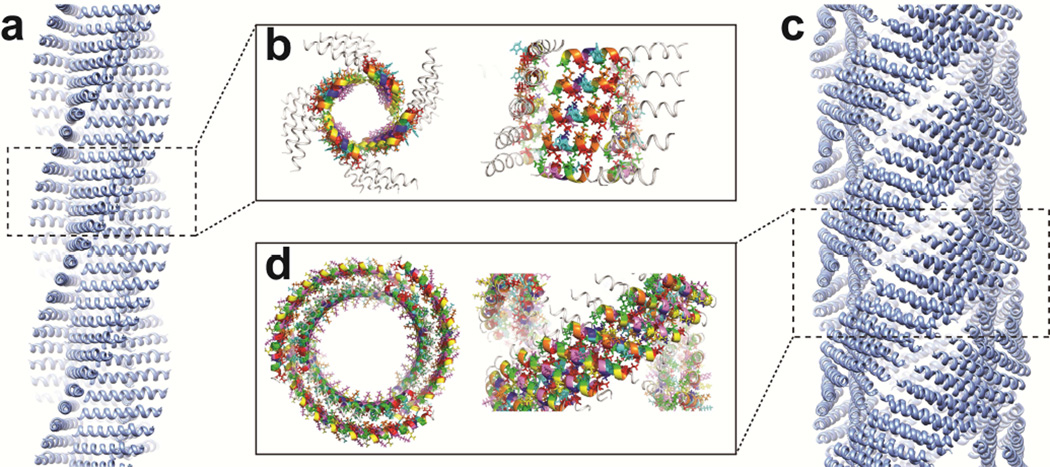
While the sequence design of the Form I and II peptides embodied concepts deduced from the structural analysis of the tolC complex, the structures of the corresponding assemblies deviated significantly from the structures of native α-helical barrels as well as from each other. However, commonalities were observed among the different structures especially at lower levels of structural hierarchy. SOCKET analysis (Walshaw and Woolfson, 2001b) of the structural models for the Form I and Form II assemblies revealed the presence of knobs-into-holes (KIH) interactions between adjacent subunits within the stacks of helices (Fig. 6). The assignment of heptad register is consistent with the proposed anti-typic interaction between the offset (a/d)-and (c/f)-interfaces. At the default packing cut-off of 7 Å, type 4 knobs-into-holes interactions were observed across a limited extent of the helix-helix interface, which became slightly more extended using a more relaxed packing cut-off of 7.4 Å. Structural analysis of the tolC interface indicated a similar relaxation of the KIH interactions at the helical interfaces that defined the structure of the α-helical barrel (Calladine et al., 2001; Walshaw and Woolfson, 2001a). In addition, prior analysis of packing within α-helical barrels suggested that the structures should distort to some degree from idealized coiled-coil structures due to the need to simultaneously interact across two interfaces. One of the structural consequences of this distortion is an alternating pattern of long and short distances between β-carbons at the helix-helix interfaces, especially those comprising the smaller side-chain amino acids (Calladine et al., 2001). This alternating pattern of long/short distances was observed between the methyl groups of successive alanine residues along helical interfaces within the stacks of the Form I and Form II assemblies.
Figure 6. SOCKET analysis of the knobs-into-holes (KIH) packing for the Form I and Form II assemblies.
a, Atomic model of Form I filament for SOCKET analysis. b, Top (left) and side (right) view of consecutive KIH interactions (colored segments) at C-terminal ordered region of Form I helices. c, Atomic model of Form II filament for SOCKET analysis. d, Top (left) and side (right) view of consecutive KIH interactions (colored segments) in Form II filaments. KIH packing is limited to helices within the same wall (inner or outer). No helix-helix KIH packing is identified across the interface between inner and outer walls. The default packing cut-off of 7 Å was employed in the SOCKET analysis (http://coiledcoils.chm.bris.ac.uk/socket/server.html).
Despite similarities in the local interfacial packing interactions, the structures of the Form I and Form II assemblies did not form closed cylindrical structures of the type that was anticipated in the original design of the peptide sequence and was previously observed in tolC and other naturally occurring α-helical barrels (Koronakis et al., 2000; Olia et al., 2011; Sun et al., 2014). In the original sequence design, the steric demand at the inner/outer interface was reversed for the synthetic peptides with respect to the α-helical barrel of tolC. The resulting decrease in curvature at the helix-helix interface of Form I and Form II resulted in helical assemblies that could not form discrete closed cylindrical structures. The formation of the nanotubes from Form I and Form II required coalescence of the respective helical stacks into the 4-start and 3-start helical assemblies, respectively. This process was mediated through relatively weak, non-covalent interactions that involved the participation of a small number of amino acid residues. In retrospect, such weak interactions would have been difficult to predict a priori and the emergence of stable nanotube structures from coalescence of the stacks was both fortuitous and fortunate. The difference in structure between the Form I and Form II assemblies can be attributed solely to adventitious differences in local interactions that arise due to the presence of arginine versus lysine in the respective peptide sequences. The fact that mutagenesis of the residues at positions 13 and 17 cause the structure of the assemblies to flip between the two forms underscores the observation that local interactions, propagated on a large scale through repetition within the helical structures, profoundly influence the structure of the resultant assemblies.
The behavior of these peptide systems may have significant implications with respect to the designability of higher order assemblies of synthetic peptides. The effect of amino acid substitutions in the peptide sequences is best considered in terms of their relative influence on the structural hierarchy. Within coiled-coil layers, the knobs-into-holes packing, which determines the helix-helix interactions within a stack, appears to be conserved in the Form I and Form II assemblies, which suggests that these interactions may be designable, i.e., relatively robust in sequence space (Zhang et al., 2014). The helix-helix interactions can be understood in terms of the design principles proposed from the structural analysis of tolC, even though global design features such as the interfacial curvature may be varied. In contrast, the presence and nature of the higher order interactions between stacks may be difficult to predict and control. In support of this hypothesis, we have observed that the introduction of conservative mutations within the helix-helix interface in the sequences of Form I and Form II usually abrogated self-assembly of the nanotubes but not the formation of thin fibrils of similar dimensions to the stacks that comprise the assemblies. In fact, at least for the Form II assemblies, these thin fibrils appear initially in TEM images prior to nanotube assembly and may be intermediates on the pathway to the nanotubes. Thin fibrils are observed in EM images to emerge from the ends or from localized unwinding of the Form II nanotube assemblies. STEM measurements on these fibrils permitted experimental determination of the mass per length value (718 ± 61 Dalton/Å), which compared well with the value of 719 Dalton/Å calculated based on the helix-helix spacing within the double-layered stacks of the Form II assemblies (Supplementary Figure 7).
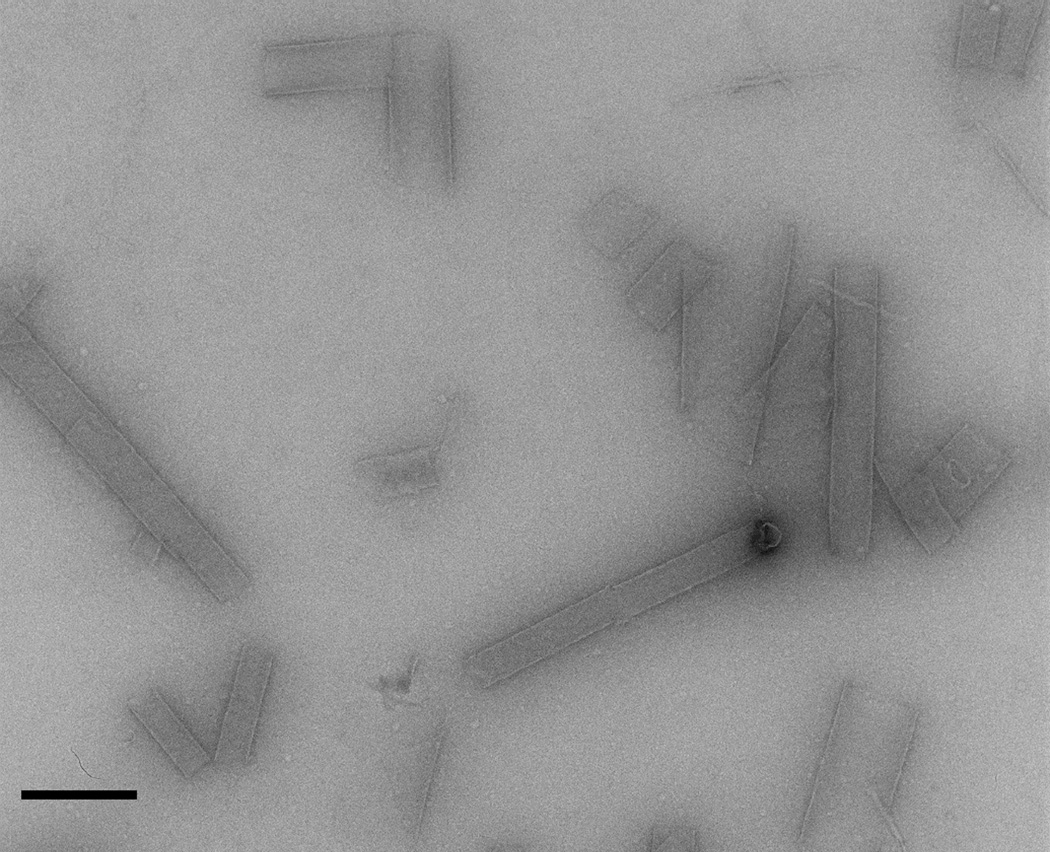
One interesting exception to the aforementioned mutagenesis studies occurs when the weak interface between the inner and outer layers of the Form II assemblies is altered. While ambiguity exists regarding the relative orientation of the inner versus outer helices, due to the lower resolution of the Form II reconstruction and the fact that the peptide sequence is nearly palindromic, in either the parallel or anti-parallel arrangement His22 from the inner helix forms a hydrogen bond with Tyr15 from the outer helix (Fig. 4d). In addition, in both models His22 from both the outer and inner helices would be in the space between the inner and outer helices (Fig. 4d). Mutagenesis of His22 to Asp eliminates the assembly of the nanotubes, and instead favors the formation of very large but variable diameter tubes (Fig. 7). This observation provides further evidence that single amino acid changes in peptide sequence can result in dramatically different polymorphs of the resultant higher-order assemblies. However, once again, these structural differences are observed at higher levels of structural hierarchy and involve weaker interactions, while the local interactions between helices appear conserved.
Figure 7.
- QAKILEADAEILKAYAKILEAHAEILKAQ
- QAKILEADAEILKAYAKILEADAEILKAQ
Our results suggest that simple amino acid substitutions within the sequences of self-assembling peptides may strongly influence the energy landscape that defines the formation of different higher order structures. These structural alterations are manifested as a consequence of the additive effect of short-range interactions in local structural space. The helical symmetry of the assemblies permits these interactions to be propagated over large length-scales, which can potentially reinforce the higher order interactions that underlie nanotube formation. Native biological assemblies display a progression of structural hierarchy that is ultimately encoded at the molecular level, often resulting in exquisite structural order over multiple length-scales. A dramatic example of which is the D-periodic banded structure of fibrillar collagen (Prockop and Fertala, 1998). Similarly, directional non-covalent interactions (hydrogen bonds, salt bridges, metal coordination) have been engineered into synthetic peptide-based assemblies (Lai et al., 2012) to introduce crystalline order (Lanci et al., 2012; Ogihara et al., 2001; Papapostolou et al., 2007; Sharp et al., 2012), or to switch between structurally ordered polymorphs (Brodin et al., 2012; Jiang et al., 2014a; Jiang et al., 2014b). Similar approaches may be envisioned to direct the formation of specific helical protein assemblies. Indeed, polar interactions involving amino acid side-chains have been observed at the helix-helix interfaces within cylindrical structures resulting from self-assembly of tolC (Calladine et al., 2001) and the ΦX-H-protein (Sun et al., 2014). These directional interactions may not only exert an influence on the thermodynamic stability of the assembly, but also select for specific oligomerization states. Simple peptide model systems such as the one that we have described represent an attractive test bed to address these design considerations, especially since the high-resolution structures of Form I and Form II provide a contextual framework through which further modifications can be evaluated using cryo-EM structural analysis.
Conclusion
The structural characterization of supramolecular assemblies at atomic resolution has historically presented a significant technical problem, which has hindered the development of an understanding of the principles that govern the interactions between the subunits that stabilize the assemblies. Crystallography has been one of the main tools for generating atomic detail of such assemblies, but unless a helix contains exactly two, three, four or six subunits per turn it cannot be crystallized with every subunit in an equivalent environment. Recent improvements in imaging hardware, reconstruction algorithms, and computational methods of structural refinement can provide rapid access to near-atomic resolution structures of synthetic helical assemblies. However, the structural analysis of the Form I and Form II assemblies highlights a critical challenge in the de novo design of structurally defined supramolecular materials, i.e., the structural plasticity of interactions at protein interfaces. In contrast to the robustness of protein tertiary structure, quaternary structure is surprisingly labile with respect to mutagenesis of residues located at the protein-protein interface. The interaction between helices stacked on top of each other (forming the walls of the tubes) resembles the packing of coiled-coil motifs as was expected from the design. However, the interactions between the ends of the peptides for Form I and Form II were significantly different to the degree that the structures would have been difficult to predict a priori, highlighted by the fact that structurally conservative amino acid substitutions can completely alter the packing of the assembly. Moreover, the Form I and Form II assemblies can be interconverted through a small subset of amino acid residues. The prediction of global structural effects that result from hierarchical propagation of weak local interactions presents a considerable challenge to current methods of computational analysis and design. Although limited structural data were available to inform the design of the Form I and Form II peptides, nonetheless novel helical assemblies were observed with lateral dimensions similar to native filamentous structures in which certain structural aspects of the initial design were manifested. As greater numbers of atomic-level structures become available for supramolecular assemblies from high-resolution cryo-EM analysis, sufficient information may become available to more reliably design novel protein-based nanomaterials of defined structure and controllable function.
Experimental Procedures
Peptide Synthesis
Peptides were either purchased from GenScript USA, Inc. (Piscataway, NJ) or synthesized in-house. In the latter case, peptides were prepared as the capped (N-acetyl, C-amide) derivatives using microwave-assisted synthesis on a CEM Liberty solid-phase peptide synthesis instrument using PAL-PEG-PS resin from Applied Biosystems, Inc. (Foster City, CA). Standard Fmoc protection chemistry was employed with coupling cycles based on HBTU/DIEA-mediated activation protocols and base-induced deprotection (20% piperidine in DMF with 0.1 M HOBt) of the Fmoc group. The peptides were purified via RP-HPLC on a C18 column with a gradient of water–acetonitrile (0.1% trifluoroacetic acid). The purity was assessed to be above 95% by analytical HPLC (Supp. Fig. 1). Peptide mass was confirmed using MALDI mass spectrometry (Supp. Fig. 2). The peptides were lyophilized, sealed, and stored at −20° C.
For analytical measurements, peptides were dissolved in acetate buffer (10 mM, pH 4.0). Peptide concentration was determined spectrophotometrically from measurement of the absorbance at 280 nm (A280). For peptides containing Tyr, Trp or Cys residues, the peptide concentration can be calculated from equation (1):
| (1) |
in which c is the concentration of peptide in mg/mL, and nY, nW, and nC are the numbers of tyrosine, tryptophan and cysteine residues, respectively, in the peptide sequence (Gill and von Hippel, 1989). Since the peptide derivatives contain only a single tyrosine residue per molecule, then c = MW×A280/1280. To eliminate error in determination of absorbance that could arise as a result of UV light scattering due to peptide self-assembly, aqueous solutions of peptide were mixed with 6 M guanidinium chloride in 1:9 v/v ratio and seated at room temperature for 2 h to completely denature the sample prior to performing the absorbance measurements. Samples for EM were heated to 95° C in acetate buffer (10 mM, pH 4.0) and slowly cooled to 4° C to promote assembly of the nanotubes.
EM and Image Analysis
Samples (2.5 µL) were applied to lacey carbon grids and vitrified in a Vitrobot Mark IV (FEI, Inc.). They were imaged in a Titan Krios (FEI, Inc.) at 300 keV using a K2 Summit direct electron detector (Gatan, Inc.), with a magnification yielding 1.02 Å/pixel. Images were collected in the movie mode, with a total exposure of 50 e−/Å2. The full integrated exposure was used for determination of the contrast transfer function (CTF) using CTFFIND3 (Mindell and Grigorieff, 2003) as well as for boxing filaments using the e2helixboxer routine in EMAN2 (Tang et al., 2007). The defocus range used was from 1.4 to 4.4 µ. Overlapping segments that were 400 px long were then cut from these filaments, using a shift of 7 px (98% overlap) that was possible due to the small axial rise of the subunits (~ 2 Å). The SPIDER software package (Frank et al., 1996) was used for most image processing steps, including the implementation of the IHRSR algorithm (Egelman, 2000). For both the Form I and the Form II filaments the correct helical symmetry was only able to be determined by a brute-force trial-and-error approach, as the information contained in the power spectra (Sup. Movies 1 and 2) led to a multiplicity of possible solutions, only one of which would be correct. Only one solution for each of the two forms generated recognizable α-helices, establishing that this was the correct solution.
For the Form I filaments, a total of 113,581 segments were extracted from 260 micrographs. After sorting by the pitch of the 4-start helices (Fig. 3a), 24,111 segments were used for a three-dimensional reconstruction. Initially, frames totaling a dose of 30 e−/Å2 were employed in the IHRSR procedure. For the last cycle, however, the alignment parameters (shifts and Euler angle assignments) from the higher dose frames were applied to initial frames with a dose of 10 e−/Å2 which yielded a significant improvement in the reconstruction due to minimizing both movement and radiation damage. Attempts to directly apply motion correction to the frames (Li et al., 2013) failed due to the weak scattering of the filaments and the absence of carbon in many images. Due to the non-specific binding of unpolymerized peptides to the hydrophobic outer surface of the filaments, a mask was imposed (Fig. 4c) to remove some of this peripheral noise (seen in Fig. 1d) which only becomes evident at lower thresholds. In contrast, the lumen of the tubes contained no noise at any of the thresholds used. A small improvement in the map was obtained by using a broader range of twist (Fig. 3a) containing 62,122 segments, showing that the improvements in signal-to-noise from using more segments was offset by the greater heterogeneity of the larger set.
A similar approach was used for the Form II filaments, but because of the greater disorder, an additional search was made to optimize the box length. Using lengths of 192, 288 and 400 pixels for all procedures, starting with sorting by the pitch of the 3-start helices (Fig. 3b), it was found that the best resolution was obtained with the 288 pixel long boxes. A total of 26,896 segments (each 288 pixels) were used for the final reconstruction.
We suggest that the appearance of the map and comparison with the model is the best measure of resolution, as it provides a reality-based standard (Fig. 4c). In contrast, the standard Fourier Shell Correlation (FSC) is at best a measure of internal consistency, and does not necessarily show the true resolution of a map. That is why one can impose the wrong symmetry on a helical structure and obtain an FSC curve that is completely meaningless (Wu et al., 2014; Xu et al., 2014). Using the FSC measure for helical filaments is further complicated by the fact that the helical filaments must be masked at the top and bottom to prevent large artifacts from arising due to the discontinuity in density at the ends of the filament. Nevertheless, Supplementary Fig. 3 shows a standard FSC curve for the Form I filaments. If one uses the FSC=0.143 criterion (Rosenthal and Henderson, 2003) after dividing the data set into two halves and using a common alignment, then an overly-optimistic resolution of 3.3 Å is obtained. This FSC has actually been lowered by the peripheral density (seen in Fig. 1d) on the outside of the filament. Masking this density can produce even “better” FSC curves, but these all depend upon the details of the mask that is used.
For the negatively stained samples, 2% uranyl acetate was used for staining. Grids were imaged on a Tecnai T12 microscope at an accelerating voltage of 120 keV and recorded on film. Micrographs were scanned with a Nikon Coolscan 9000 densitometer with a raster of 4.2 Å/px.
Atomic Models
To construct models of the Form I assembly, a single polyalanine helix was first docked into the density. Helical assembly parameters were determined from the experimental data, and a symmetric polyalanine assembly was constructed following these parameters. Since the placement of N- and C-termini of the models were ambiguous from the data, 14 different threadings of the designed sequence onto this polyalanine helix were considered: seven with the N-termini inward, and seven with the N-termini outward. Following previous work, particles were split into two sets, and independent reconstructions of each set were constructed. Each of these fourteen models was refined against one of the reconstructions using Rosetta’s cryoEM refinement protocol (DiMaio et al., 2013); all refinement was done in the context of the helical assembly. Refined models were then evaluated against the reconstruction not used in the fitting process, assessing agreement against this independent set in high-resolution shells (10-4 Å). Looking at the average model-map agreement of five models refined for each threading gives a clear signal for one particular threading (Supp. Fig. 4).
A similar strategy was used for model determination of the Form II assembly. As the asymmetric unit contains two helices, and the lower resolution of the data made helix placement ambiguous, additional threadings were considered. For each helix, 22 different models were generated: 11 threadings in both N-terminal to C-terminal directions. Then, all 484 combinations of these helices were refined (in the helical assembly) using the same protocol as Form I. Evaluating models of the inner and outer layers separately, we see a clear signal for the configuration of the inner layer (Supp. Fig. 5). The helix-helix interactions of the inner layer are identical to that of the Form I, but with additional interactions between GLN1 and GLN29 between adjacent helices stabilizing the assembly. The configuration of the outer layer – due to relatively lower local resolution and the nearly palindromic nature of the sequence – was ambiguous under our analysis (Fig. 4d, Supp. Fig. 5b). Assessing agreement of model and map to independent reconstructions identifies one particular configuration of helices in the outer layer, but whether helices in this layer run parallel or antiparallel to helices in the inner layer is unclear (Supp. Fig. 5c). In both models of the Form II assembly, the helix-helix interactions within the outer layer are similar to those of the inner layer, but shifted by two turns.
Synchrotron SAXS/WAXS Measurements
Synchrotron SAXS/WAXS measurements were performed at the 12- ID-B beamline of the Advanced Photon Source at Argonne National Laboratory. A SAXS/WAXS simultaneous setup was utilized, and the sample-to-detector distances were set such that the overall scattering momentum transfer q range was achieved from 0.003 to 2.4 Å−1, where q = 4πsinθ/λ, 2θ denoting the scattering angle and λ the X-ray wavelength. The wavelength was set at 1.033 Å during the measurements. Scattered X-ray intensities were measured using a Pilatus 2 M (DECTRIS Ltd.) detector for SAXS and Pilatus 300K for WAXS. SAXS/WAXS measurements were performed on aqueous solutions of the Form I and Form II peptide assemblies at concentrations of 4 mg/mL (circa 1.2 mM) in acetate buffer (10 mM, pH 4.0) at 25 °C. A flow cell equipped with a quartz capillary (1.5 mm diameter) was used to prevent radiation damage. Twenty images were collected for each sample and buffer. The 2-D scattering images were converted to 1-D SAXS curves through azimuthally averaging after solid angle correction and then normalizing with the intensity of the transmitted X-ray beam using the software package at beamline 12ID-B. The 1-D curves of the samples were averaged and subtracted with the background measured from the corresponding buffers. The simulated SAXS curves were calculated using program CRYSOL Svergun et al., 1995).
Scanning Transmission Electron Microscopy
STEM data were acquired at Brookhaven National Laboratory (BNL) using methods described previously (Xu et al., 2013). Specimen quality and mass calibration were checked by detailed comparison of the image to the known structure of tobacco mosaic virus (TMV). Mass measurements were performed off-line with customized software (PCMass, available at ftp.stem.bnl. gov).
Supplementary Material
Highlights.
Nanotubes self-assemble from a 29-residue peptide that contains coiled-coil motifs.
Two very different forms can be made using only a single amino acid substitution.
Cryo-EM has been used to achieve near-atomic resolution of one state.
This provides insights into the lability of quaternary structure in protein evolution.
Acknowledgments
We thank Z. Hong Zhou and Peng Ge for access to the Titan Krios at UCLA, and Chris Booth Gatan, Inc.) for the use of the K2 Summit direct electron detector. We thank Dr. Xiaobing Zuo and Dr. Joseph Wall for assistance in acquisition of the synchrotron SAXS and STEM mass-per-length data, respectively. The synchrotron SAXS/WAXS measurements benefited from the use of the APS funded by the US DOE Office of Basic Energy Sciences, Division of Material Sciences, under contract W-31-109-Eng-38. This work was supported by NIH EB001567 (E.H.E) and DOE-BES DE-ER15377 (V.P.C.). Models and volumes for Form I have been deposited with IDs xxxx.PDB and EMDB-yyyy, respectively. For Form II, the models and volumes are zzzz.PDB and EMDB-wwww, respectively.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Amprazi M, Kotsifaki D, Providaki M, Kapetaniou EG, Fellas G, Kyriazidis I, Perez J, Kokkinidis M. Structural plasticity of 4-alpha-helical bundles exemplified by the puzzle-like molecular assembly of the Rop protein. Proc Natl Acad Sci U S A. 2014;111:11049–11054. doi: 10.1073/pnas.1322065111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai XC, Fernandez IS, McMullan G, Scheres SH. Ribosome structures to near-atomic resolution from thirty thousand cryo-EM particles. eLife. 2013;2:e00461. doi: 10.7554/eLife.00461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bammes BE, Rochat RH, Jakana J, Chen DH, Chiu W. Direct electron detection yields cryo-EM reconstructions at resolutions beyond 3/4 Nyquist frequency. Journal of Structural Biology. 2012;177:589–601. doi: 10.1016/j.jsb.2012.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodin JD, Ambroggio XI, Tang C, Parent KN, Baker TS, Tezcan FA. Metal-directed, chemically tunable assembly of one-, two- and three-dimensional crystalline protein arrays. Nature chemistry. 2012;4:375–382. doi: 10.1038/nchem.1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calladine CR, Sharff A, Luisi B. How to untwist an alpha-helix: structural principles of an alpha-helical barrel. J Mol Biol. 2001;305:603–618. doi: 10.1006/jmbi.2000.4320. [DOI] [PubMed] [Google Scholar]
- Coates ML. Hemoglobin function in the vertebrates: an evolutionary model. Journal of molecular evolution. 1975;6:285–307. doi: 10.1007/BF01794636. [DOI] [PubMed] [Google Scholar]
- Craig L, Volkmann N, Arvai AS, Pique ME, Yeager M, Egelman EH, Tainer JA. Type IV pilus structure by cryo-electron microscopy and crystallography: implications for pilus assembly and functions. Molecular Cell. 2006;23:651–662. doi: 10.1016/j.molcel.2006.07.004. [DOI] [PubMed] [Google Scholar]
- DiMaio F, Zhang J, Chiu W, Baker D. Cryo-EM model validation using independent map reconstructions. Protein Science. 2013;22:865–868. doi: 10.1002/pro.2267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egelman EH. A robust algorithm for the reconstruction of helical filaments using single-particle methods. Ultramicroscopy. 2000;85:225–234. doi: 10.1016/s0304-3991(00)00062-0. [DOI] [PubMed] [Google Scholar]
- Egelman EH. The iterative helical real space reconstruction method: Surmounting the problems posed by real polymers. Journal of Structural Biology. 2007;157:83–94. doi: 10.1016/j.jsb.2006.05.015. [DOI] [PubMed] [Google Scholar]
- Egelman EH. Reconstruction of helical filaments and tubes. Methods in Enzymology. 2010;482:167–183. doi: 10.1016/S0076-6879(10)82006-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egelman EH. Helical Ambiguities. eLife. 2014 doi: 10.7554/eLife.04969. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egelman EH, Francis N, DeRosier DJ. F-actin is a helix with a random variable twist. Nature. 1982;298:131–135. doi: 10.1038/298131a0. [DOI] [PubMed] [Google Scholar]
- Fernandez IS, Bai XC, Murshudov G, Scheres SH, Ramakrishnan V. Initiation of translation by cricket paralysis virus IRES requires its translocation in the ribosome. Cell. 2014;157:823–831. doi: 10.1016/j.cell.2014.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank J, Radermacher M, Penczek P, Zhu J, Li Y, Ladjadj M, Leith A. SPIDER and WEB: Processing and visualization of images in 3D electron microscopy and related fields. Journal of Structural Biology. 1996;116:190–199. doi: 10.1006/jsbi.1996.0030. [DOI] [PubMed] [Google Scholar]
- Galkin VE, Yu X, Bielnicki J, Heuser J, Ewing CP, Guerry P, Egelman EH. Divergence of quaternary structures among bacterial flagellar filaments. Science. 2008;320:382–385. doi: 10.1126/science.1155307. [DOI] [PubMed] [Google Scholar]
- Gill SC, von Hippel PH. Calculation of protein extinction coefficients from amino acid sequence data. Anal Biochem. 1989;182:319–326. doi: 10.1016/0003-2697(89)90602-7. [DOI] [PubMed] [Google Scholar]
- Jiang T, Xu C, Liu Y, Liu Z, Wall JS, Zuo X, Lian T, Salaita K, Ni C, Pochan D, et al. Structurally defined nanoscale sheets from self-assembly of collagen-mimetic peptides. Journal of the American Chemical Society. 2014a;136:4300–4308. doi: 10.1021/ja412867z. [DOI] [PubMed] [Google Scholar]
- Jiang T, Xu C, Zuo X, Conticello VP. Structurally homogeneous nanosheets from self-assembly of a collagen-mimetic peptide. Angewandte Chemie. 2014b;53:8367–8371. doi: 10.1002/anie.201403780. [DOI] [PubMed] [Google Scholar]
- Klug A, Crick FH, Wyckoff HW. Diffraction by helical structures. Acta Crystallographica. 1958;11:199–213. [Google Scholar]
- Koronakis V, Sharff A, Koronakis E, Luisi B, Hughes C. Crystal structure of the bacterial membrane protein TolC central to multidrug efflux and protein export. Nature. 2000;405:914–919. doi: 10.1038/35016007. [DOI] [PubMed] [Google Scholar]
- Lai YT, King NP, Yeates TO. Principles for designing ordered protein assemblies. Trends in cell biology. 2012;22:653–661. doi: 10.1016/j.tcb.2012.08.004. [DOI] [PubMed] [Google Scholar]
- Lanci CJ, MacDermaid CM, Kang SG, Acharya R, North B, Yang X, Qiu XJ, DeGrado WF, Saven JG. Computational design of a protein crystal. Proc Natl Acad Sci U S A. 2012;109:7304–7309. doi: 10.1073/pnas.1112595109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Mooney P, Zheng S, Booth CR, Braunfeld MB, Gubbens S, Agard DA, Cheng Y. Electron counting and beam-induced motion correction enable near-atomic-resolution single-particle cryo-EM. NatMethods. 2013;10:584–590. doi: 10.1038/nmeth.2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao M, Cao E, Julius D, Cheng Y. Structure of the TRPV1 ion channel determined by electron cryo-microscopy. Nature. 2013;504:107–112. doi: 10.1038/nature12822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Zheng Q, Deng Y, Cheng CS, Kallenbach NR, Lu M. A seven-helix coiled coil. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:15457–15462. doi: 10.1073/pnas.0604871103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loquet A, Sgourakis NG, Gupta R, Giller K, Riedel D, Goosmann C, Griesinger C, Kolbe M, Baker D, Becker S, et al. Atomic model of the type III secretion system needle. Nature. 2012;486:276–279. doi: 10.1038/nature11079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu A, Magupalli VG, Ruan J, Yin Q, Atianand MK, Vos MR, Schroder GF, Fitzgerald KA, Wu H, Egelman EH. Unified Polymerization Mechanism for the Assembly of ASC-Dependent Inflammasomes. Cell. 2014;156:1193–1206. doi: 10.1016/j.cell.2014.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mindell JA, Grigorieff N. Accurate determination of local defocus and specimen tilt in electron microscopy. Journal of Structural Biology. 2003;142:334–347. doi: 10.1016/s1047-8477(03)00069-8. [DOI] [PubMed] [Google Scholar]
- Ogihara NL, Ghirlanda G, Bryson JW, Gingery M, DeGrado WF, Eisenberg D. Design of three-dimensional domain-swapped dimers and fibrous oligomers. Proc Natl Acad Sci U S A. 2001;98:1404–1409. doi: 10.1073/pnas.98.4.1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olia AS, Prevelige PE, Jr, Johnson JE, Cingolani G. Three-dimensional structure of a viral genome-delivery portal vertex. Nat Struct Mol Biol. 2011;18:597–603. doi: 10.1038/nsmb.2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papapostolou D, Smith AM, Atkins ED, Oliver SJ, Ryadnov MG, Serpell LC, Woolfson DN. Engineering nanoscale order into a designed protein fiber. Proc Natl Acad Sci U S A. 2007;104:10853–10858. doi: 10.1073/pnas.0700801104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penczek PA, Radermacher M, Frank J. Three-dimensional reconstruction of single particles embedded in ice. Ultramicroscopy. 1992;40:33–53. [PubMed] [Google Scholar]
- Prockop DJ, Fertala A. The collagen fibril: the almost crystalline structure. J Struct Biol. 1998;122:111–118. doi: 10.1006/jsbi.1998.3976. [DOI] [PubMed] [Google Scholar]
- Rosenthal PB, Henderson R. Optimal Determination of Particle Orientation, Absolute Hand, and Contrast Loss in Single-particle Electron Cryomicroscopy. Journal of Molecular Biology. 2003;333:721–745. doi: 10.1016/j.jmb.2003.07.013. [DOI] [PubMed] [Google Scholar]
- Sharp TH, Bruning M, Mantell J, Sessions RB, Thomson AR, Zaccai NR, Brady RL, Verkade P, Woolfson DN. Cryo-transmission electron microscopy structure a gigadalton peptide fiber of de novo design. Proc Natl Acad Sci U S A. 2012;109:13266–13271. doi: 10.1073/pnas.1118622109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JM. Natural selection and the concept of a protein space. Nature. 1970;225:563–564. doi: 10.1038/225563a0. [DOI] [PubMed] [Google Scholar]
- Sun L, Young LN, Zhang X, Boudko SP, Fokine A, Zbornik E, Roznowski AP, Molineux IJ, Rossmann MG, Fane BA. Icosahedral bacteriophage PhiX174 forms a tail for DNA transport during infection. Nature. 2014;505:432–435. doi: 10.1038/nature12816. [DOI] [PubMed] [Google Scholar]
- Svergun D, Barberato C, Koch MHJ. CRYSOL– a Program to Evaluate X-ray Solution Scattering of Biological Macromolecules from Atomic Coordinates. Journal of Applied Crystallography. 1995;28:768–773. [Google Scholar]
- Tang G, Peng L, Baldwin PR, Mann DS, Jiang W, Rees I, Ludtke SJ. EMAN2: an extensible image processing suite for electron microscopy. Journal of Structural Biology. 2007;157:38–46. doi: 10.1016/j.jsb.2006.05.009. [DOI] [PubMed] [Google Scholar]
- Thomson AR, Wood CW, Burton AJ, Bartlett GJ, Sessions RB, Brady RL, Woolfson DN. Computational design of water-soluble alpha-helical barrels. Science. 2014;346:485–488. doi: 10.1126/science.1257452. [DOI] [PubMed] [Google Scholar]
- Walshaw J, Woolfson DN. Open-and-shut cases in coiled-coil assembly: alpha-sheets and alpha-cylinders. Protein Science. 2001a;10:668–673. doi: 10.1110/ps.36901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walshaw J, Woolfson DN. Socket: a program for identifying and analysing coiled-coil motifs within protein structures. J Mol Biol. 2001b;307:1427–1450. doi: 10.1006/jmbi.2001.4545. [DOI] [PubMed] [Google Scholar]
- Wang YA, Yu X, Overman S, Tsuboi M, Thomas GJ, Jr, Egelman EH. The structure of a filamentous bacteriophage. Journal of Molecular Biology. 2006;361:209–215. doi: 10.1016/j.jmb.2006.06.027. [DOI] [PubMed] [Google Scholar]
- Wu B, Peisley A, Tetrault D, Li Z, Egelman EH, Magor KE, Walz T, Penczek PA, Hur S. Molecular Imprinting as a Signal-Activation Mechanism of the Viral RNA Sensor RIG-I. Mol Cell. 2014;55:511–523. doi: 10.1016/j.molcel.2014.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C, Liu R, Mehta AK, Guerrero-Ferreira RC, Wright ER, Dunin-Horkawicz S, Morris K, Serpell LC, Zuo X, Wall JS, et al. Rational design of helical nanotubes from self-assembly of coiled-coil lock washers. Journal of the American Chemical Society. 2013;135:15565–15578. doi: 10.1021/ja4074529. [DOI] [PubMed] [Google Scholar]
- Xu H, He X, Zheng H, Huang LJ, Hou F, Yu Z, de la Cruz MJ, Borkowski B, Zhang X, Chen ZJ, et al. Structural basis for the prion-like MAVS filaments in antiviral innate immunity. eLife. 2014;3:e01489. doi: 10.7554/eLife.01489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonekura K, Maki-Yonekura S, Namba K. Complete atomic model of the bacterial flagellar filament by electron cryomicroscopy. Nature. 2003;424:643–650. doi: 10.1038/nature01830. [DOI] [PubMed] [Google Scholar]
- Yu X, Egelman EH. Helical filaments of human Dmc1 protein on single-stranded DNA: a cautionary tale. Journal of Molecular Biology. 2010;401:544–551. doi: 10.1016/j.jmb.2010.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaccai NR, Chi B, Thomson AR, Boyle AL, Bartlett GJ, Bruning M, Linden N, Sessions RB, Booth PJ, Brady RL, et al. A de novo peptide hexamer with a mutable channel. Nature chemical biology. 2011;7:935–941. doi: 10.1038/nchembio.692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Zheng F, Grigoryan G. Design and designability of protein-based assemblies. Curr Opin Struct Biol. 2014;27:79–86. doi: 10.1016/j.sbi.2014.05.009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.