Abstract
Motivation
Improvements in technology and decreases in price have made de novo bacterial genomic sequencing a reality for many researchers, but it has created a need to evaluate the methods for generating a complete and accurate genome assembly.
Results
We sequenced the GC-rich Caulobacter henricii genome using the Illumina MiSeq, Roche 454, and Pacific Biosciences RS II sequencing systems. To generate a complete genome sequence, we performed assemblies using eight readily available programs and found that builds using the Illumina MiSeq and the Roche 454 data produced accurate yet numerous contigs. SPAdes performed the best followed by PANDAseq. In contrast, the Celera Assembler produced a single genomic contig using the Pacific Biosciences data after error correction with the Illumina MiSeq data. In addition, we duplicated this build using the Pacific Biosciences data with HGAP2.0. The accuracy of these builds was verified by Pulsed Field Gel Electrophoresis of genomic DNA cut with restriction enzymes.
INTRODUCTION
Despite ground breaking advances in the field of prokaryotic biology, there are many unanswered questions left to be studied. Many of these questions require the assembly of high quality bacterial genome sequences. Larger than most viruses but smaller than most eukaryotic genomes, bacterial genomes have been sequenced to understand pathogen-host interactions, to understand the environment specific evolution of species, and also to trace the source of bacterial related disease outbreaks. Since the cost to sequence a genome has dropped dramatically in response to technological advances [12], the number of sequenced bacterial genomes has exploded. For example the Human Microbiome Project, which aims to establish a comprehensive baseline of the microbial diversity at 18 different human body sites, has identified thousands of new microbial strains and has radically increased the number of bacterial genomes that are currently being sequenced [5, 6]. Also, the Wellcome Trust Sanger Institute has recently begun collaborating with Public Health England to complete the sequences of 3,000 bacterial genome strains from PHE's National Collection of Type Cultures (NCTC).
Second- and third-generation genome sequencing technology can now generate high quality, astonishingly fast, high throughput sequencing data. However, there are advantages and disadvantages associated with each individual technology. Things to consider when choosing a technology are read lengths, accuracy, price of sequencing, and the time needed to complete a sequencing run. Pacific Biosciences [22] has developed instrumentation that creates unprecedented read lengths of up to 20,000 bp with a 99.9% accuracy rate. Illumina [21] has technology that can routinely generate 600 billion base pairs (GB) in a single run as well as other iterations that can go from sample to data in as little as 8 hours. A major consideration for all current technologies is cost. In addition to the cost of actual sequence runs, added costs include the cost of sample library preparation. If multiple libraries are used, this cost can be greater than the cost of the sequencing. As such, many researchers have begun to adopt a strategy of sequencing just a single library while relying on deep coverage of the genome to compensate for the lack of multiple libraries.
Typically, whole genome assembly projects have begun by using a combination of two or more short and long read libraries [9]. Short read libraries are often used with paired-end reads generating short fragments less than 800 bp in length. However, if the genome contains repeated sequences that are longer than the read lengths, the sequence data cannot be assembled from short reads. This problem created the need for long fragments that could span the repeated sequences. Pacific Biosciences employs Single Molecule Real Time sequencing (SMRT). Instead of cycles of template amplification, the incorporation of dNTPs by the replicating DNA polymerase is observed in real time. Each nucleotide is attached to a fluorescent dye that is released at the moment of incorporation. The base call is made according to the observed fluorescence of the released dye. This method allows for extremely long read lengths but suffers from less accurate base calling as compared to that of 2nd generation technologies.
Many reviews have been published that assess and compare different strategies for the assembly of genomes and novel metrics have been designed to maximize the quality of the assemblies [18, 19, 16]. These studies demonstrate that there is no one size fits all approach to a quality genome assembly. Each researcher has different needs and queries. Also, as sequencing becomes routine, more researchers with little to no experience in bioinformatics and limited access to assembly experts will be attempting the process of genome assembly. These problems influenced us to compare the efficacy and accuracy of a panel of assembly programs that use input data derived from the GC-rich Caulobacter henricii genome sequenced with the Illumina MiSeq benchtop sequencer, Roche GS FLX 454 sequencer, and the PacBio RS II DNA Sequencing System.
To assemble the sequence data, we compared eight assembly programs. Our bacterial data set was generated from a novel GC-rich genome that lacked a reference. These characteristics make it an ideal candidate to test GC bias and the ability of our data sets to produce an accurate and contiguous reference.
METHODS
Genome sequencing
The alphaproteobacterium Caulobacter henricii (ATCC® 15253™) designated CB4 was ordered from the American Type Culture Collection. The bacteria were propagated according to ATCC protocols and genomic DNA was prepared with QIAGEN DNeasy Blood and Tissue Kit [23]. For Pacific Biosciences RS II sequencing, the library prep template for the 10kb protocol was used but the DNA was sheared for 20kb fragments using a Covaris tube and a final 0.4x bead wash for a finished library. The collection protocols for the P4-C2 chemistry were:
-
Protocol: MagBead Standard Seq v2
Movie Time: 120 min
Insert Size (bp): 20000
Stage Start: True
Control: DNA Control 3kb-10kb.
A 250-bp paired end library for Illumina Miseq v2 chemistry and 8-kb paired end library for GS-FLX titanium were prepared, and sequencing was performed according to the manufacturers’ instructions. The 454 and Illumina sequencing processes were performed using the services of EnGenCore LLC (Columbia, SC). The PacBio sequencing was done using the services of University of Washington PacBio Sequencing Services.
The Assemblers
Eight genome assemblers were used in this study:
Celera Assembler 8.0 [15]-http://sourceforge.net/projects/wgs-assembler/files/wgs-assembler/wgs-8.1/
CLC Genomics Workbench 6 (CLC Bio)- http://www.clcbio.com/products/clc-genomics-workbench/
HGAP 2.0 [4]- https://github.com/PacificBiosciences/SMRT-Analysis/wiki/SMRT-Analysis-Release-Notes-v2.1
MaSuRCA v2.1.0 [24]- http://www.genome.umd.edu/masurca.html
Newbler v2.6 [14]- http://454.com/contact-us/software-request.asp
PANDAseq [2]- https://github.com/neufeld/pandaseq/wiki/Installation
DNAStar SeqMan NGen 11.2.1 (DNASTAR, Madison, WI, USA)- http://www.dnastar.com/
SPAdes v2.5.1 [1]- http://bioinf.spbau.ru/spades
As some assemblers can only be used with specific data sets, we ran all assemblers for any data set that was compatible. Our 454 data set was assembled using Newbler. Our Pacbio data set was assembled using HGAP 2.0 and polished using Quiver. The MiSeq data set was assembled using Celera, CLC Genomics, SeqMan, MaSuRCA, and SPAdes. We used one hybrid approach as well. We used the Celera error correction module to error correct our long-read Pacbio data set with the high accuracy short-reads of our MiSeq data set then assembled the error-corrected data set using Celera. This assembly and the HGAP2 assembly both generated the same build which we designated as the reference genome. We used the reference along with the NGA50 contig size (if a contig is misassembled with respect to the reference, it is broken down into smaller pieces) to determine which software produced the best assembly.
All assembly input command lines are supplied in the supplementary information.
Depth of Coverage
We used the 250bp paired-end reads from the C. henricii MiSeq data set which yielded approximately 100X coverage. We used the 600bp reads from the 454 data set which produced approximately 100X coverage. Our Pacbio data set yielded average read lengths of 4289 bp with coverage of approximately 55X.
The Assemblies
We examined various metrics on the performance of each assembler as described in Magoc, et al. [13. All metrics were calculated using the Quality ASsement Tool for genome assembly [10].
Pulsed Field Gel Electrophoresis
Plug genesis and digestion were done as described in Ely and Gerardot [8]. All Pulsed-Field Gels were run at 6 V/cm for 16 hours. Switch times varied from ramped 20-120 seconds; ramped 1-45 seconds; ramped 10-20 seconds. All agarose gels were 1% in SBA buffer and run at a temperature of 14 degrees Celsius.
RESULTS
Data
The first data set was produced by the Roche GS FLX 454 system using standard FLX chemistry to sequence the genome of Caulobacter henricii strain CB4. The second data set consists of 2x250bp Illumina MiSeq paired-end reads that was obtained using Reagent Kit v2. The third data set was generated using the P4-C2 chemistry of the PacBio RS II system. The data sets were post processed using BLASTn in order to discard contaminating and plasmid sequences.
Generation of the Reference Genome
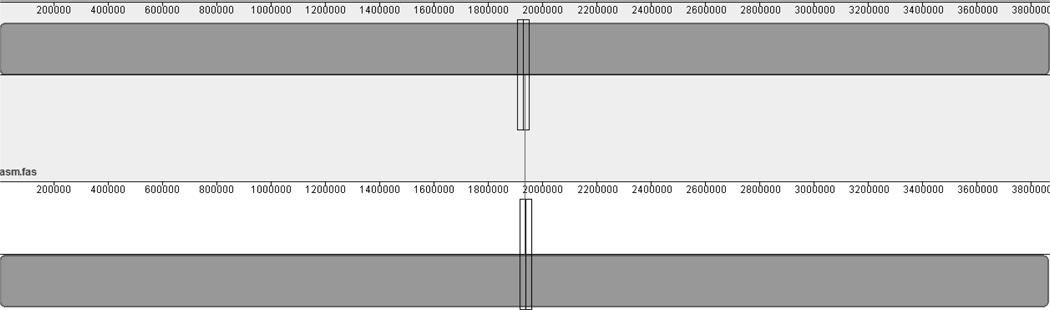
One of our main goals in this research was to generate a finished genome with no gaps. We were able to accomplish this goal using two different methods. As described in Koren, [11] we used an approach that utilized the short, high-accuracy sequences of MiSeq to correct the error inherent in the long, single-molecule sequence reads generated by the Pacbio RS II using different modules found in the Celera Assembler 8.0. The corrected “hybrid” PBcR (PacBio corrected Reads) were then assembled de novo into 2 contigs consisting of a 3,870,958 bp contig and a 100,699 bp plasmid (3,971,657 total bp). We also used the HGAP 2.0 assembler that self-corrected the PacBio long-reads to create a draft assembly. This draft was then polished with Quiver to generate a more highly accurate consensus sequence. It produced 2 contigs, the first being 3,868,732 bp and a 97,894 bp plasmid (3,966,626 total bp). The plasmid sequences in all builds were easily identified through comparison with the reference and BLASTn and were subsequently removed in all downstream analyses. We used Mauve [7] to compare the two assemblies and discovered that at 99.99999879% similarity, they were virtually identical to each other (Figure 1). We determined the extra base pairs from the PBcR were simply repeats of the ends of the genome reinforcing its circular nature.
Figure 1.
Mauve visualization of two assemblies. The top bar represents the PBcR assembly. The bottom bar represents the HGAP 2.0 assembly. The colors represent identical DNA sequence that is shared between the two builds.
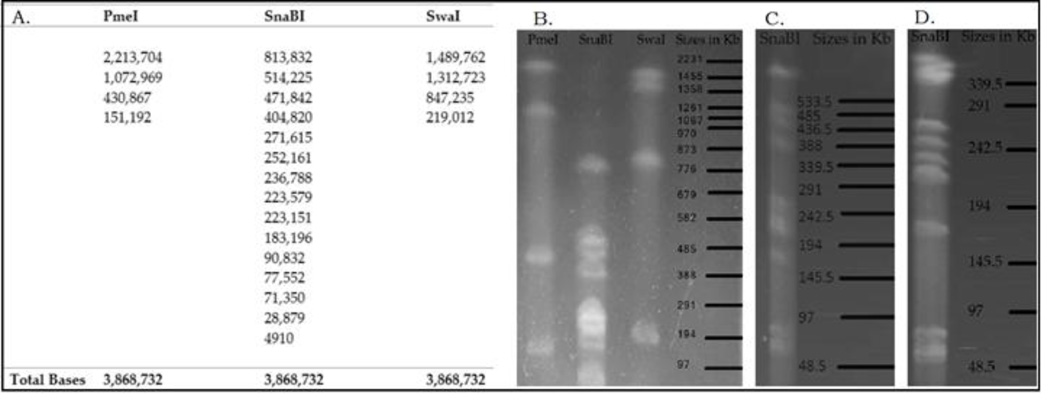
We tested the accuracy of each build by downloading the consensus of each assembly into the Webcutter 2.0 program [3] and generating a theoretical digest using the SnaBI enzyme which cut the genomic sequence 15 times (Supplemental 1). This digest was predicted to produce moderate to large fragments of the genomic DNA that could be easily identified via Pulsed Field Gel Electrophoreses (PFGE) (Figure 2). We also predicted the PmeI and SwaI digestion patterns which both cut the genome 4 times and would confirm the legitimacy of the assembly in conjunction with the SnaBI data (Figure 2).
Figure 2.
A. Predicted and actual fragment sizes of HGAP2 build after enzymatic digestion, B. Switch times ramped from 20-120 seconds. C. Switch times ramped from 1-45 seconds. D. Switch times ramped from 10-20 seconds. The black lines indicate the positions and sizes of the fragments generated by a lambda DNA reference ladder.
When the C. henricii DNA was digested with each of the restriction enzymes and the resulting fragments were resolved by PFGE, there was a one to one correspondence between the bands observed on the gel and those predicted from the assembled nucleotide sequence (Figure 2). These data indicate that these genome assemblies matched the organization of the actual C. henricii chromosome. We decided to use the HGAP 2.0 assembly as our reference based on the fact that it used the default settings in SMRT Analysis 2.1 to generate this build and would be easier to duplicate as opposed to the PBcR assembly which used many steps to achieve the final output. A previous study also showed that PacBio consensus accuracy always exceeded that of the second-generation sequencing data and consistently matched or exceeded the quality of both short-read and hybrid assemblies [11]. However, we found that at both “ends” of the HGAP2 assembly there were fragmented protein reading frames due to missing bases.
Comparison of Assemblies
Using the HGAP 2.0 build as a reference, we computed the NGA50 (corrected N50) sizes of our assemblies. NGA50 values convey more information about a build because the program breaks the misassembled contigs at perceived misjoins to provide a superior gauge of assembly quality. If an assembler incorrectly merges two contigs, then this results in a larger N50 size. Since N50 is often used to determine how well an assembler performed, these incorrect builds appear to be better than they actually are. Using the 454 data set of the C. henricii CB4 genome, Newbler generated 69 contigs with a N50 and NGA50 value of 128 Kb and a genome fraction of 99.721% (Table 1). The combined length of all 69 contigs was 3,950,077 bp.
Table 1.
Comparison of assembler builds using the HGAP2 assembly as the reference. NGA50 values are boxed in. The data were generated with QUAST.
| Statistics without reference | Celera | CLCGenomics | DNAStar | SPAdes | MaSuRCA | NewMer | PANDAseq | PBcR | HGAP2 |
|---|---|---|---|---|---|---|---|---|---|
| # contigs | 210 | 119 | 78 | 28 | 60 | 69 | 27 | 1 | 1 |
| Largest contig | 96 335 | 228 321 | 512 281 | 1 717 07 | 501 495 | 414 950 | 683 332 | 3 870 958 | 3 868 732 |
| Total length | 3 885 508 | 3 872 940 | 3 954 246 | 3 875 49 | 3 931 679 | 3 950 077 | 3 954 266 | 3 870 958 | 3 868 732 |
| N50 | 31 035 | 68 799 | 311 910 | 849 521 | 283 078 | 128 030 | 349 035 | 3 870 958 | 3 868 732 |
| Misassemblies | |||||||||
| # misassemblies | 0 | 2 | 1 | 1 | 3 | 1 | 1 | 1 | 0 |
| Misassembled contigs length | 0 | 148 706 | 428 086 | 1 717 07 | 632 172 | 211 829 | 523 614 | 3 870 958 | 0 |
| Mismatches | |||||||||
| # mismatches per 100 kbp | 0.65 | 3.61 | 0.93 | 0.91 | 25.08 | 0.41 | 0.23 | 0.36 | 0 |
| # indels per 100 kbp | 0.52 | 1.27 | 0.91 | 0.91 | 1.65 | 1.74 | 0.91 | 0.85 | 0 |
| # N's per 100 kbp | 0.03 | 0 | 0.13 | 0 | 0 | 0.99 | 0 | 0 | 0 |
| Genome statistics | |||||||||
| Genome fraction (%) | 99.393 | 99.704 | 99.836 | 99.834 | 99.981 | 99.721 | 99.729 | 100 | 100 |
| Duplication ratio | 1.01 | 1.002 | 1.003 | 1 | 1.017 | 1.006 | 1 | 1.002 | 1 |
| NGA50 | 31 035 | 66 099 | 262 235 | 720 050 | 217 552 | 127 991 | 349 035 | 2 754 062 | 3 868 732 |
| Predicted genes | |||||||||
| # predicted genes (unique) | 3843 | 3743 | 3750 | 3696 | 3720 | 3745 | 3764 | 3639 | 3643 |
| # predicted genes (>= 0 bp) | 3843 | 3743 | 3783 | 3697 | 3744 | 3745 | 3764 | 3643 | 3644 |
| # predicted genes (>= 300 bp) | 3427 | 3370 | 3411 | 3342 | 3380 | 3389 | 3405 | 3314 | 3313 |
| # predicted genes (>= 1500 bp) | 536 | 552 | 558 | 562 | 561 | 574 | 575 | 558 | 557 |
| # predicted genes (>= 3000 bp) | 40 | 43 | 46 | 46 | 44 | 45 | 48 | 47 | 46 |
Using the MiSeq data set, SPAdes generated the assembly with the highest N50 and NGA50 scores of 849 Kb and 720 Kb respectively. PANDAseq was next with a N50 and NGA50 value of 349 Kb. It also generated the build with the fewest contigs while DNAStar produced the build with the largest total genome length at 3,954,246 bp. All assemblies displayed genome fraction percentages of over 99.4 with the MaSuRCA build reaching 99.981%.
DISCUSSION
We compared the efficacy and accuracy of a panel of assemblers on a 66% GC bacterial genome consisting of input data derived from next generation sequencing technologies. In terms of 2nd generation data, the SPAdes assembler generated the largest contig sizes in terms of N50 and NGA50 as compared to the other assemblers (Table 1). The build generated from PANDAseq created the second best results. As expected, all the assemblies were improved when the data was mapped to the reference genome. The MaSuRCA assembly generated only one gap when aligned to the reference and a 99.981% genome fraction (Table 1); SPAdes generated two gaps while DNASTAR produced five gaps.
In terms of assembly errors, the Celera Assembler produced none and no assembler had more than three. However, the Celera Assembler performed the poorest in terms of the number of contigs and N50 scores (Table 1). This is unsurprising as Celera utilizes the Overlap-Layout-Consensus method of contig generation which favors long-read input.
The number of misjoined contigs did not greatly reduce the NGA50 values but we did find false detection of errors in some builds. The C. henricii genome is circular and in some instances, a contig started at the end or beginning of the reference and “wrapped around” to the other end of the reference. This resulted in that contig falsely being labeled as misjoined. Such was the case with the PBcR and SPAdes assembly (Supplemental 2).
With an average genomic GC content of 65.72% and some genomic regions reaching 80% GC, the C. crescentus CB4 genome was a good choice to test the performance of assemblies where GC bias was a problem. Interestingly, each build covers at least 99% of the reference suggesting that the reason for multiple contigs and unfinished assemblies was not incomplete coverage from GC bias, but the ability of the algorithms to process and reconcile repetitive regions. The SPAdes build, for example, produced twenty-eight contigs after assembly but two contigs when aligned to the reference. We analyzed the ends of contigs that should have aligned and discovered that they contained sequences that were repeated at the ends of multiple contigs found in the assembly. Since there were multiple ways these contigs could be assembled, they were unable to be assembled further.
These issues were addressed in the builds of the 3rd generation PacBio RS II data. With a mean read length of 4,289 bp and maximum read lengths approaching 20,000 bp, these repetitive regions could easily be resolved. The weakness of this technology has traditionally been its low accuracy but this problem has been addressed with assemblers such as Celera that includes steps to error correct these reads with high accuracy short reads. Recently, PacBio developed the HGAP 2.0 assembler that self-corrects these errors and further polishes the consensus with Quiver to produce a result that is equal to the Celera correction method. Thus the short read data is no longer necessary since we achieved a complete and accurate (QV 39=99.987% accuracy) genome assembly using only PacBio data.
Overall, we conclude that the latest genome assemblers can produce very good yet incomplete de novo assemblies using single, deep coverage, short-read libraries of 2nd generation sequencers. However, these assemblers are limited by repetitive regions that can be difficult to resolve with the short-reads of these libraries. This result verifies the findings that repeated sequence in the genome induces complexity and poses the greatest challenge to all assembly algorithms [17]. Therefore, consistent with [22, 20], we suggest that the simplest and most effective way to produce a de novo GC-rich bacterial genome assembly is with PacBio RS II long-reads using HGAP 2.0 assembler to self-correct the reads. Further, we were able to complete a reference genome by using only one SMRT cell. This negates the need for multiple libraries and decreases the cost of a sequencing project. We duplicated this method with the novel bacterium Brevundimonas DS20. This strain is a relative of the Caulobacter genus and also has a high GC content. Some caveats to this method are that the DNA sent off for sequencing must be extremely pure with no contaminants. If this condition is not met then the sequencing run will likely end in error at worst or gives a highly fragmented library at best. Neither of these scenarios will result in the completion of sequence assembly.
Supplementary Material
ACKNOWLEDGEMENTS
This work was funded in part by a fellowship from The Southern Region Educational Board (SREB) to DS and NIH grant GM076277 to BE. We would like to thank Nicole Rapicavoli at Pacific Biosciences for her assistance with the HGAP2 assembly, Alexey Gurevich and Anton Korobeynikov at the Algorithmic Biology Lab, St. Petersburg, Russia for their support with the SPAdes and QUAST programs, and special thanks to Nathan Elger and Paul Sagona who are a part of the Research Cyberinfrastructure at The University of South Carolina.
Footnotes
Supplementary information: Supplemental data are available at Current Microbiology online.
REFERENCES
- 1.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bartram AK, Lynch Michael DJ, Stearns Jennifer C, Moreno-Hagelsieb Gabriel. Generation of Multimillion-Sequence 16S rRNA Gene Libraries from Complex Microbial Communities by Assembling Paired-End Illumina Reads. APPLIED AND ENVIRONMENTAL MICROBIOLOGY. 2011;77:3846–3852. doi: 10.1128/AEM.02772-10. a.J.D.N. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.bio.biomedicine.gu.se/cutter2/.
- 4.Chin CS, Alexander DH, Marks P, Klammer AA, Drake J, Heiner C, Clum A, Copeland A, Huddleston J, Eichler EE, et al. Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat Methods. 2013;10:563–569. doi: 10.1038/nmeth.2474. [DOI] [PubMed] [Google Scholar]
- 5.Consortium T.H.M.P. A framework for human microbiome research. Nature. 2012;486:215–221. doi: 10.1038/nature11209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Consortium T.H.M.P. Structure, function and diversity of the healthy human microbiome. Nature. 2012;486:207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Darling AE, Mau B, Perna NT. progressiveMauve: multiple genome alignment with gene gain, loss and rearrangement. PLoS One. 2010;5:e11147. doi: 10.1371/journal.pone.0011147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ely B, Gerardot CJ. Use of pulsed-field-gradient gel electrophoresis to construct a physical map of the Caulobacter crescentus genome. Gene. 1988;68:323–333. doi: 10.1016/0378-1119(88)90035-2. [DOI] [PubMed] [Google Scholar]
- 9.Fleischmann RD, Adams MD, White O, Clayton RA, Kirkness EF, Kerlavage AR, Bult CJ, Tomb JF, Dougherty BA, Merrick JM, et al. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science. 1995;269:496–512. doi: 10.1126/science.7542800. [DOI] [PubMed] [Google Scholar]
- 10.Gurevich A, Saveliev V, Vyahhi N, Tesler G. QUAST: quality assessment tool for genome assemblies. Bioinformatics. 2013;29:1072–1075. doi: 10.1093/bioinformatics/btt086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Koren S, Harhay GP, Smith TP, Bono JL, Harhay DM, McVey SD, Radune D, Bergman NH, Phillippy AM. Reducing assembly complexity of microbial genomes with single-molecule sequencing. Genome Biology. 2013;14:R101. doi: 10.1186/gb-2013-14-9-r101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jackman SD, Birol I. Assembling genomes using short-read sequencing technology. Genome Biol. 2010;11:202. doi: 10.1186/gb-2010-11-1-202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Magoc T, Pabinger S, Canzar S, Liu X, Su Q, Puiu D, Tallon LJ, Salzberg SL. GAGE-B: an evaluation of genome assemblers for bacterial organisms. Bioinformatics. 2013;29:1718–1725. doi: 10.1093/bioinformatics/btt273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Margulies M, Egholm M, Altman WE, Attiya S, Bader JS, Bemben LA, Berka J, Braverman MS, Chen YJ, Chen Z, et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437:376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Myers EW, Sutton GG, Delcher AL, Dew IM, Fasulo DP, Flanigan MJ, Kravitz SA, Mobarry CM, Reinert KH, Remington KA, et al. A whole-genome assembly of Drosophila. Science. 2000;287:2196–2204. doi: 10.1126/science.287.5461.2196. [DOI] [PubMed] [Google Scholar]
- 16.Narzisi G, Mishra B. Comparing de novo genome assembly: the long and short of it. PLoS One. 2011;6:e19175. doi: 10.1371/journal.pone.0019175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Phillippy AM, Schatz MC, Pop M. Genome assembly forensics: finding the elusive mis-assembly. Genome Biol. 2008;9:R55. doi: 10.1186/gb-2008-9-3-r55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Quail MA, Smith M, Coupland P, Otto TD, Harris SR, Connor TR, Bertoni A, Swerdlow HP, Gu Y. A tale of three next generation sequencing platforms: comparison of Ion Torrent, Pacific Biosciences and Illumina MiSeq sequencers. BMC Genomics. 2012;13:341. doi: 10.1186/1471-2164-13-341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schatz MC, Phillippy AM, Sommer DD, Delcher AL, Puiu D, Narzisi G, Salzberg SL, Pop M. Hawkeye and AMOS: visualizing and assessing the quality of genome assemblies. Brief Bioinform. 2011;14:213–224. doi: 10.1093/bib/bbr074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shin SC, Ahn do H, Kim SJ, Lee H, Oh TJ, Lee JE, Park H. Advantages of Single-Molecule Real-Time Sequencing in High-GC Content Genomes. PLoS One. 2013;8:e68824. doi: 10.1371/journal.pone.0068824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.www.illumina.com
- 22.www.pacificbiosciences.com
- 23.www.qiagen.com
- 24.Zimin AV, Marcais G, Puiu D, Roberts M, Salzberg SL, Yorke JA. The MaSuRCA genome assembler. Bioinformatics. 2013;29:2669–2677. doi: 10.1093/bioinformatics/btt476. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.