Summary
Background
Eukaryotic cell division is driven by cyclin-dependent kinases (CDKs). Distinct cyclin-CDK complexes are specialized to drive different cell cycle events, though the molecular foundations for these specializations are only partly understood. In budding yeast, the decision to begin a new cell cycle is regulated by three G1 cyclins (Cln1–Cln3). Recent studies revealed that some CDK substrates contain a novel docking motif that is specifically recognized by Cln1 and Cln2, and not by Cln3 or later S- or M-phase cyclins, but the responsible cyclin interface was unknown.
Results
Here, to explore the role of this new docking mechanism in the cell cycle, we first show that it is conserved in a distinct cyclin subtype (Ccn1). Then, we exploit phylogenetic variation to identify cyclin mutations that disrupt docking. These mutations disrupt binding to multiple substrates as well as the ability to use docking sites to promote efficient, multi-site phosphorylation of substrates in vitro. In cells where the Cln2 docking function is blocked, we observed reductions in the polarized morphogenesis of daughter buds and reduced ability to fully phosphorylate the G1/S transcriptional repressor Whi5. Furthermore, disruption of Cln2 docking perturbs the coordination between cell size and division, such that the G1/S transition is delayed.
Conclusions
The findings point to a novel substrate interaction interface on cyclins, with patterns of conservation and divergence that relate to functional distinctions among cyclin subtypes. Furthermore, this docking function helps ensure full phosphorylation of substrates with multiple phosphorylation sites, and this contributes to punctual cell cycle entry.
Introduction
Cyclin-dependent kinases (CDKs) are central regulators of cell division in eukaryotes [1]. The cyclin subunit has a critical role in triggering CDK kinase activity, and plays additional regulatory roles by controlling subcellular localization and substrate selection [2]. While it is possible to construct cells with only a single cyclin-CDK complex [3], eukaryotes invariably have several distinct forms that are specialized for particular tasks. Generally, these cyclin-CDK forms fall into two broad classes: those that drive DNA synthesis and mitosis (in S and M phases), and those that control entry into a new division cycle (in G1 phase). To understand how sequential cell cycle events are properly orchestrated, it is necessary to determine the molecular features of cyclin-CDK forms that impart functional distinctions. For example, how do early forms trigger some events without triggering others that should occur later in S and M phases? One general class of explanation is that early cell cycle events may rely on cyclin-CDK complexes with low activity but strong substrate selectivity [4, 5].
In the budding yeast S. cerevisiae, S and M phases are driven by six B-type cyclins (Clb1–6), whereas cell cycle entry is controlled by three G1 cyclins, Cln1–3 [1, 2]. The G1 phase constitutes a critical period in which cells determine if conditions are appropriate to begin dividing, and this decision is responsive to cues such as nutrient availability, cell size, and inhibitory signals. Ultimately these cues affect the function of Cln1–3, which then drive the CDK phosphorylation events that commit cells to division in a step known as “Start”, followed by the G1 to S transition [6–8]. Key CDK substrates in this period are inhibitors of cell cycle entry such as Whi5, a repressor of G1/S transcription [9, 10], as well as Cdh1 and Sic1, which prevent the expression and activity of Clb cyclins, respectively [1]. Notably, each of these substrates has multiple CDK phosphorylation sites [11–13], which may place unique demands on the cyclin-CDK complex to ensure complete phosphorylation and also dictate the threshold CDK levels required to trigger the regulatory effect [11, 14–16].
Despite some functional overlap among Cln1–3, they have important differences [2, 4], which contribute to a two-stage commitment process: Cln3 plays an early priming role that initiates expression of Cln1 and Cln2, which further enhance their own expression via a positive feedback loop, resulting in a sharp increase in Cln1/2 activity that triggers a decisive entry into the cell cycle [17–19]. Cln3 and Cln1/2 show distinct subcellular distributions [20, 21]. In addition, recent studies show that Cln1/2 cyclins recognize specific docking motifs in select CDK substrates (Figure 1A); these “LP” motifs (enriched in Leu and Pro residues) are not recognized by either Cln3 or Clb1–6, and hence they promote phosphorylation preferentially by Cln1/2-CDK [5, 22]. This mechanism is analogous to recognition of RXL motifs by S-phase cyclins such as yeast Clb5 or mammalian cyclin A [23], but the different motifs are not cross-recognized [5, 22]. Currently, it is unknown what part of the Cln1/2 protein recognizes LP motifs, or why other cyclins do not recognize them. It is also unknown what cell cycle events depend on Cln1/2 docking. To address these issues, in this study we identify and characterize a docking-defective Cln2 mutant. Our findings uncover a novel substrate-docking interface that is conserved among distinct cyclin sub-groups, and demonstrate that this docking function promotes multi-site phosphorylation of substrates and punctual entry into the cell cycle.
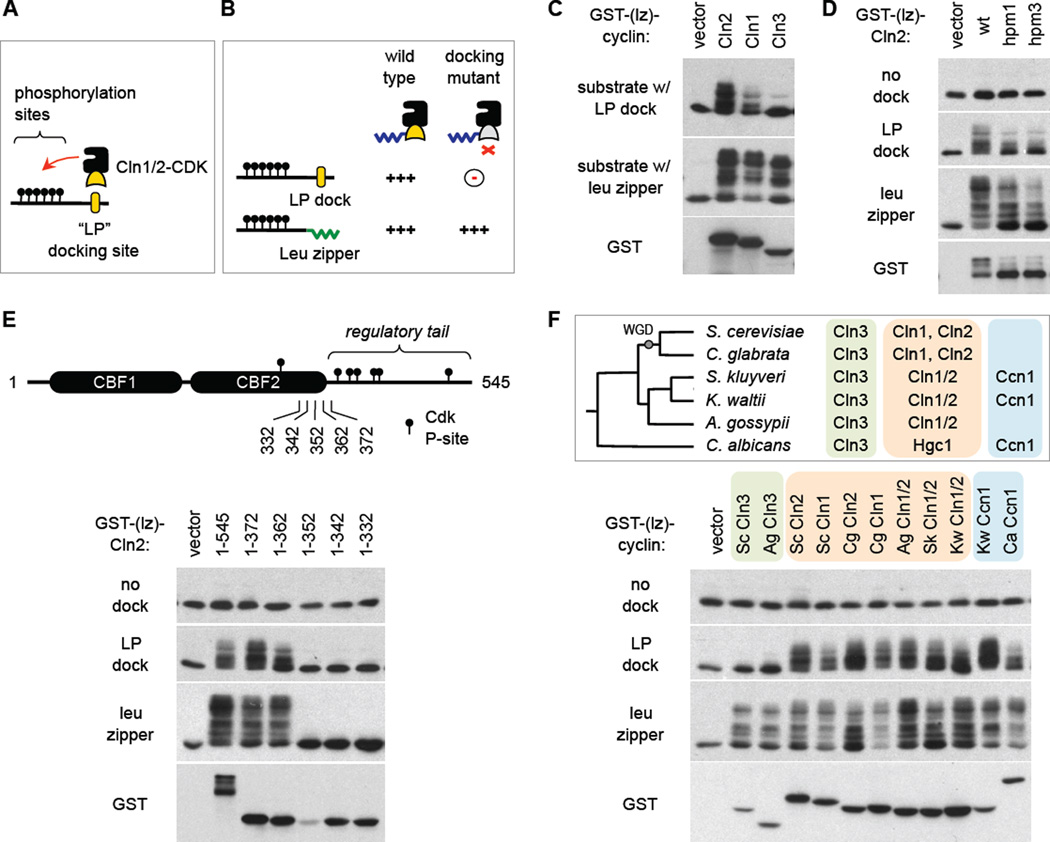
Figure 1. Sequence constraints and evolutionary conservation of docking function.
(A) Interaction of Cln1/2 with “LP” docking sites promotes CDK substrate phosphorylation.
(B) Expected phenotype of a docking-defective Cln1/2 mutant. Defects in docking vs. CDK activity can be discerned by comparing substrates with a docking site vs. a leucine zipper.
(C) Cln3 exemplifies non-docking behavior. Cyclins (with truncated C-termini) were expressed as fusions to GST plus a half leucine zipper (GST-[lz]), along with a Ste5-derived substrate harboring either the partner half leucine zipper or an LP docking site (see Experimental Procedures and Figure S1A). Reduced mobility indicates phosphorylation.
(D) Mutations in the hydrophobic patch region of Cln2 (hpm1, 3; see Figure S1B) do not confer specific docking defects but rather a mild general reduction in phosphorylation activity.
(E) Full-length Cln2 (1–545) and truncated forms were tested for substrate phosphorylation (as in C–D). Endpoints are shown relative to regulatory phosphorylation sites (P-sites) and tandem cyclin box folds (CBF1, 2) predicted for Cln2 (see Figure S1C). Lower abundance for 1–352 was reproducible.
(F) Conservation of LP docking in Cln1/2 and Ccn1 cyclins. Top, phylogenetic tree of six yeasts, and their G1 cyclins. WGD, whole genome duplication. The Ccn1 group is related but distinct from Cln3 and Cln1/2 groups. Bottom, cyclins from different yeasts were expressed in S. cerevisiae as GST-(lz) fusions to test substrate phosphorylation. All cyclins had truncated C-termini; see Figure S1C.
Results
Strategic framework for localizing the docking interface in Cln2
Before screening for a docking-defective Cln2 mutant, we needed a strategy to distinguish specific defects in docking from more general defects in CDK activation. Our solution was to compare a CDK substrate harboring a native LP docking site with one that instead uses a leucine zipper to promote cyclin-substrate interaction, expecting that the desired class of mutant would only show defects with the former (Figure 1B). Indeed, this predicted behavior is exemplified by Cln3, which does not recognize LP docking sites (Figure 1C). Hence, in principle we sought a Cln2 mutant that behaves more like Cln3.
Because S-phase cyclins recognize RXL motifs via a hydrophobic patch (HP) on their surface, we wondered if the different makeup of this region in Cln1/2 cyclins allowed recognition of LP rather than RXL motifs. But mutations in this part of Cln2 did not confer a specific defect in LP docking, and instead they caused a general mild reduction in all activity, including auto-phosphorylation of the Cln2 C-terminus (Figure 1D). Hence, we concluded that LP recognition is encoded elsewhere on Cln2. To constrain the candidate sequences, we tested truncations of the Cln2 C-terminus, and found that roughly one-third of the protein was dispensable for LP recognition, whereas truncations that perturb the predicted globular cyclin box fold domains (CBF1, 2) eliminated all activity (Figure 1E). Thus, all positions within the roughly 370-residue core of Cln2 remained candidates.
To evaluate which residues might contribute to docking, we devised a strategy to exploit natural sequence variation. Namely, by expressing G1 cyclins from other yeasts in S. cerevisiae, we found that recognition of LP docking sites was conserved in other Cln1/2 members and lacking among Cln3 members (Figure 1F). Moreover, we tested a distinct class of G1 cyclin, the Ccn1 group, which is absent from S. cerevisiae but present in other yeasts (Figure 1F, top). Remarkably, Ccn1 members were proficient at using an LP docking motif to drive substrate phosphorylation (Figure 1F). This revealed that LP docking exists for a class of cyclin other than Cln1/2, and offered a way to further constrain the possible residues involved in docking.
Identification of a docking defective Cln2 mutant
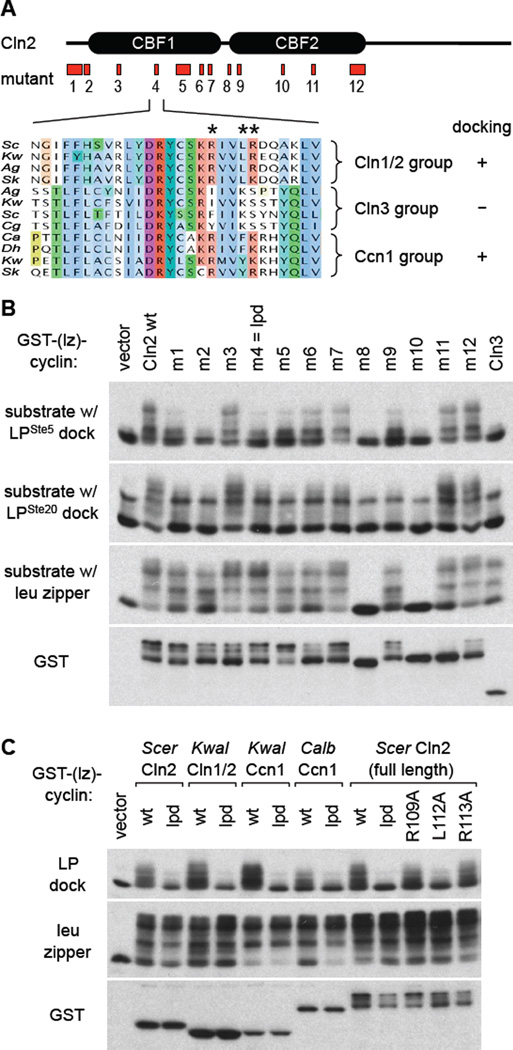
Sequence alignments with multiple members of the Cln1/2, Cln3, and Ccn1 groups revealed that there were many positions where Ccn1 and Cln3 residues were identical (or nearly so), and hence we excluded these from consideration as key residues for LP docking. Then, we scrutinized positions that correlated with docking ability; i.e., similar in Cln1/2 and Ccn1, but different in Cln3. Based on these considerations, twelve Cln2 mutants were designed (Figures 2A, S2).
Figure 2. Identification of a docking-defective Cln2 mutant.
(A) Twelve regions of Cln2 were mutated, emphasizing residues conserved in Cln1/2 and Ccn1 groups but not in Cln3 (see Figure S2A,B). One example (m4/lpd) is illustrated by the sequence alignment, with mutated positions marked by asterisks.
(B) Cln2 mutants were expressed as GST-(lz) fusions to test phosphorylation in vivo of substrates with either of two LP docking sites (from Ste5 or Ste20) or the leucine zipper. Also see Figure S2C.
(C) Left, the lpd mutation was tested in four different cyclins (two Cln1/2 and two Ccn1) for effects on docking-dependent phosphorylation. Right, analysis of single mutations at each of the three residues mutated in the lpd mutant.
These mutants were tested for phosphorylation of substrates with two distinct LP sites or the control leucine zipper (Figures 2B, S2C); we also monitored auto-phosphorylation of the Cln2 C-terminus. Several mutants (m1, m2, m5, m6, m9) showed non-specific reduction in phosphorylation of all substrates, including the leucine zipper control (note the drop in the upper-most bands and the increased proportion of the bottom band), while two mutants (m8, m10) had no detectable activity and no auto-phosphorylation. By contrast, one mutant, Cln2-m4, displayed the desired phenotype, as it showed a specific defect with LP-containing substrates but normal phosphorylation of the control substrate (Figure 2B). Based on these and additional findings to be described below, we hereafter designate this mutant as “lpd” (for “LP docking”). To explore the generality of this phenotype, we compared the analogous mutation in two Cln1/2 members and two Ccn1 members (Figure 2C, left). Each showed the same behavior, in which the lpd mutant was defective at using a native LP dock but fully competent to use the leucine zipper. We also tested each of the single residue changes in the original Cln2-lpd triple mutant, and found that one of them, L112A, was largely responsible for the defect (Figure 2C, right); however, this single mutant was not as defective as the lpd triple mutant, and we observed mild defects with each of the other two (R109A and R113A), and hence we used the original lpd triple mutant for further analyses. Collectively, these findings argue that the lpd mutation disrupts a region of the cyclin with a specific role in utilizing LP docking sites.
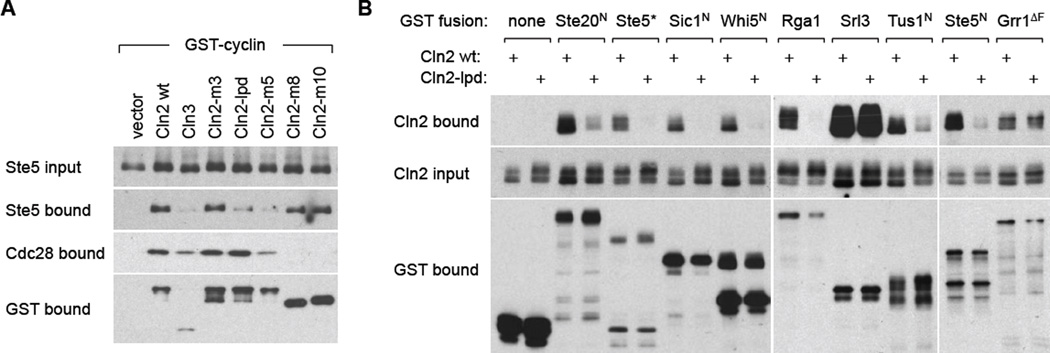
Cln2 docking function is required for interaction with multiple substrates
To verify that Cln2-lpd was defective at docking interactions, we assayed substrate binding. First, we compared several of the initial Cln2 mutants, expressed as GST fusions, for their ability to co-precipitate Ste5, a substrate with an LP motif [22]. As predicted for a docking mutant, Cln2-lpd showed reduced binding to Ste5 but normal binding to its partner CDK molecule, Cdc28 (Figure 3A). The specific nature of this phenotype was reinforced by comparison to other mutants: (i) Cln2-m3 showed no binding defects, consistent with its full activity in phosphorylation assays; (ii) Cln2-m5 showed reduced binding to both Ste5 and Cdc28, suggesting a non-specific defect that agrees with its reduced activity against all substrates; and, (iii) Cln2-m8 and Cln2-m10 were defective at binding Cdc28 but bound Ste5 normally, which agrees with their inactivity in phosphorylation assays and shows that binding Cdc28 is not required to bind substrates.
Figure 3. Docking-defective Cln2 mutant shows reduced binding to multiple partners.
(A) Cells co-expressed a galactose-inducible GST-cyclin (or vector) with V5-tagged Ste5. After galactose induction, GST fusions and co-bound proteins were captured with glutathione-Sepharose. Bound and input proteins were analyzed by immunoblots.
(B) Cells expressed V5-tagged Cln2 (wt or lpd) and galactose-inducible GST fusions to full-length proteins or N-terminal fragments. Ste5* is a hybrid fragment [22, 29], in which the Cln2-docking site in the Ste20 N-terminus is replaced with one from Ste5. Grr1ΔF lacks its F-box, to prevent it from driving degradation of Cln2 [25]. Also see Figure S3.
We also conducted reciprocal assays in which GST-substrate fusions were used to co-precipitate V5-tagged Cln2 (Figures 3B, S3A), and found that Cln2-lpd showed substantially reduced binding to most substrates tested (i.e., Ste20, Ste5, Sic1, Whi5, Rga1, Tus1). An exception was Srl3, which binds Cln2 especially strongly; this might indicate a distinct mode of binding (e.g., involving the CDK) or that the reduction in binding is too mild to register in this assay. As a further control, we tested binding to Grr1, an F-box protein that promotes Cln2 ubiquitination; this binding was unaffected by the lpd mutation (Figure 3B), consistent with the fact that Grr1 recognizes the phosphorylated C-terminus of Cln2 rather than the globular CBF domains [24, 25]. Finally, we also assayed substrate binding by one of the Ccn1 family members (Figure S3B). This cyclin bound only a subset of the substrates that bind Cln2, indicating some divergence in docking motif recognition (to be pursued separately), but each was disrupted by the Ccn1-lpd mutation.
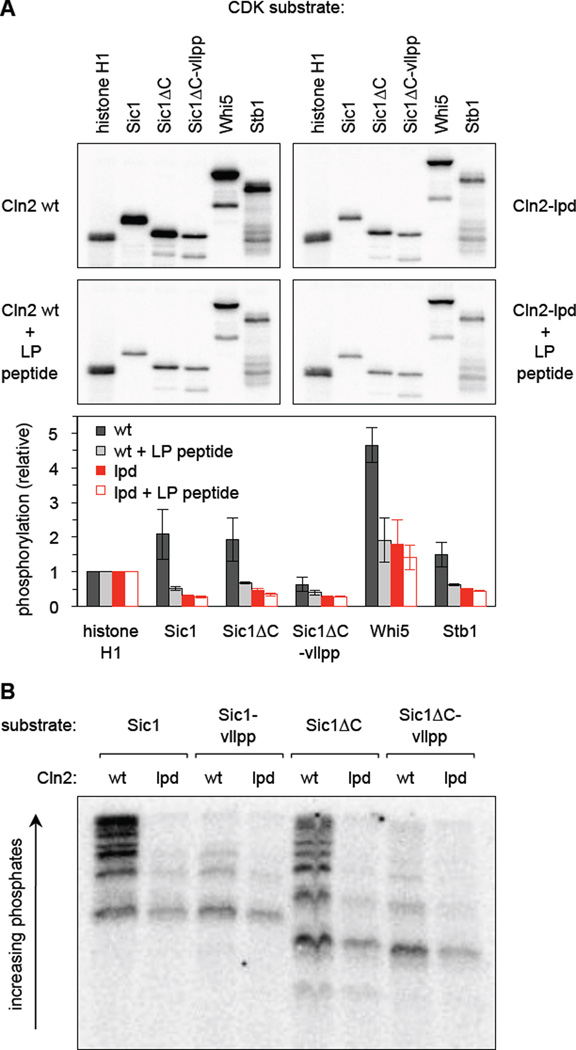
In vitro kinase assays confirm a specific defect in docking function
To probe the biochemical phenotypes in vitro, we purified wt and lpd versions of the Cln2-Cdc28 complex from yeast cells (Figure S4A). Cln2-lpd co-purified with normal amounts of Cdc28 and, when using histone H1 as a generic substrate, wt and lpd complexes showed indistinguishable kinase activity. In contrast, with other substrates the Cln2-lpd complex showed defects that indicated a specific failure in LP docking function (Figure 4A, S4B). Namely, for several substrates (Sic1, Sic1ΔC, Whi5, Stb1), the activity of the Cln2-lpd complex was reduced to a level comparable to that seen when the wt complex was inhibited with a competitor LP peptide (Figure 4A). The competitor peptide had negligible effect on the Cln2-lpd complex, indicating that its reduced activity specifically reflects an inability to recognize the LP docking site to enhance phosphorylation. Similarly, when the LP docking site on Sic1 was mutated (vllpp), this reduced phosphorylation by the wt complex but did not affect the lpd complex (Figure 4A, S4B).
Figure 4. Docking promotes multi-site phosphorylation of substrates in vitro.
(A) Cln2-Cdc28 complexes containing either wt or lpd Cln2 were purified from yeast cells and assayed for substrate phosphorylation in vitro, both with and without a competitor LP peptide. Sic1ΔC is residues 1–215, and vllpp denotes mutation of its LP docking site. Top, total 32P incorporation into substrates. Bottom, 32P incorporation was quantified from two reaction times (8 and 16 min.), and then normalized to the results with histone H1 for each of the 4 conditions; bars show the mean ± range. Also see Figure S4B.
(B) The indicated Sic1 substrates were phosphorylated by Cln2-Cdc28 (wt or lpd), and the products were separated on a Phos-tag gel to assess the multiplicity of phosphorylation. Also see Figures S4B, S4C.
To further assess the Cln2-lpd defect, the products of these kinase reactions were analyzed on Phos-tag gels, which can resolve substrate isoforms differing in the number of phosphates added, and do so especially clearly for the substrate Sic1 [14, 15]. Notably, the Cln2-lpd complex could not generate the most highly phosphorylated products, and instead yielded products modified on few sites, similar to when the LP site in the substrate was mutated (vllpp; Figure 4B, S4C). Other substrates were also less highly modified (Figure S4B), though resolution of the intermediates was poorer. Collectively, these in vitro findings show that the Cln2-lpd mutant has a specific defect in utilizing LP docking sites to drive extensive, multi-site phosphorylation.
Cln2-substrate docking helps coordinate the G1/S transition with cell size
To assess how docking contributes to the cellular functions of Cln2, we first asked if Cln2-lpd supports cell division when expressed from its native promoter. Indeed, it permitted growth when provided as the only G1 cyclin (Figure S5A), as well as under conditions where CLN3 was not sufficient (i.e., cln1Δ cln2Δ pcl1Δ pcl2Δ; Figure S5B). These results confirm that Cln2-lpd remains generally active, and indicate that docking is not the only function that discriminates Cln1/2 from Cln3.
Because G1 phase is a period of growth before division, defects in the control of cell cycle entry can affect cell size [7, 26]. When we measured cell volumes in asynchronous cultures (using cln1Δ strains to eliminate redundancy between Cln1 and Cln2), cln2-lpd cultures were shifted toward mildly larger cells (Figure 5A, top), consistent with a known role for Cln1/2 in setting critical size [19, 27, 28]. Remarkably, this difference was amplified in cells lacking the CDK inhibitor protein Far1 (Figures 5A, S5C). We were primed to consider such an effect by our recent finding that Far1 competitively interferes with Cln2-substrate docking [29]; this raised the possibility that mutations in the docking interface of Cln2 could impair interactions with both substrates and Far1, so that the resulting defects in positive functions of Cln2 are partly counteracted by reduced inhibition from Far1 (see Discussion). Hence, to avoid differential inhibition, further analysis used far1Δ strains.
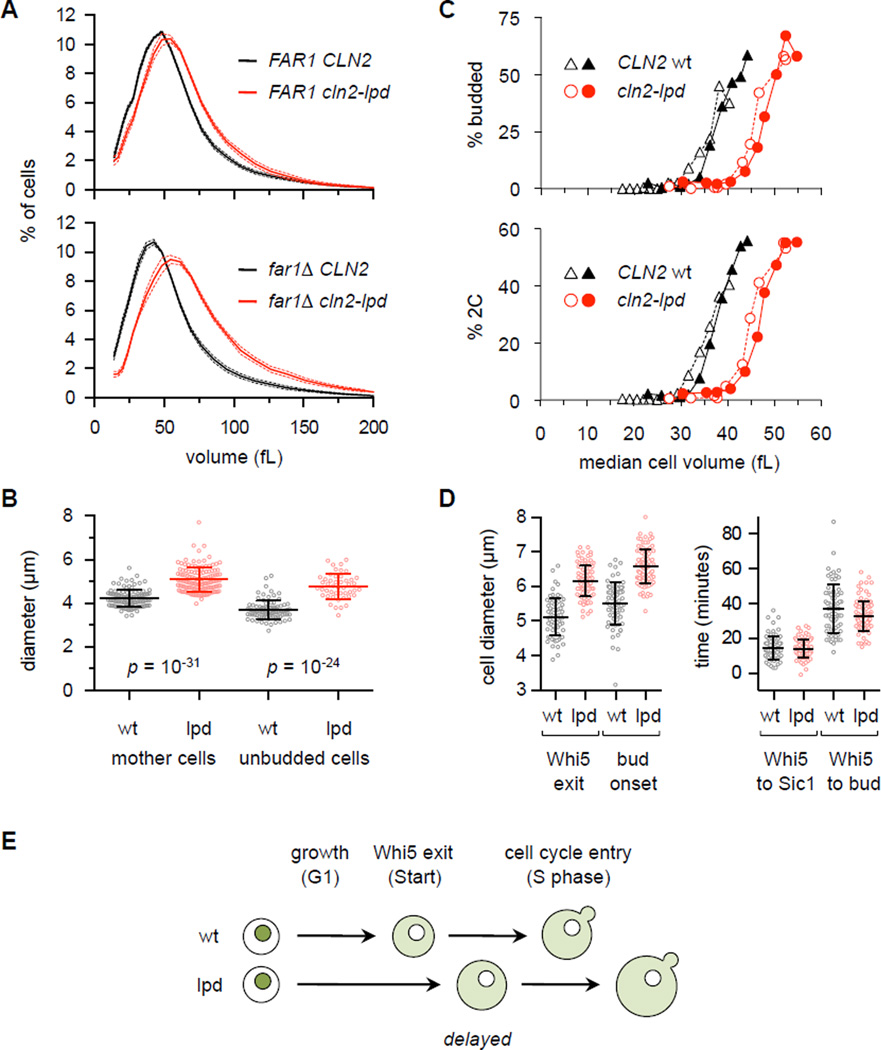
Figure 5. Cln2-lpd alters the critical cell size for cell cycle entry.
(A) Cell volume is increased in cln2-lpd strains. Note that all strains are cln1Δ. Solid and dashed lines show mean ± SEM (n = 6, in YPD). Also see Figure S5C.
(B) Increased diameters of mother and unbudded cells in cln2-lpd strains compared to CLN2-wt, in far1Δ cln1Δ background. Cells were grown in SC/raffinose. Lines denote mean ± SD. Also see Figure S5D.
(C) Small G1 daughter cells (in far1Δ cln1Δ background), grown in SC/raffinose, were isolated by centrifugal elutriation and inoculated in fresh YPD medium (note, cln2-lpd cells were born larger than wt cells); samples were collected at 15-min. intervals to assay cell volume, budding, and DNA replication. For each strain, we assayed two elutriator fractions (denoted by open and closed symbols) with distinct starting median cell volumes (17 and 23 fL for CLN2 wt, or 27 and 30 fL for cln2-lpd), to confirm that the phenotypes relate to size rather than incubation time.
(D) Time-lapse microscopy was used to monitor the diameter of cells at the times of Whi5 nuclear exit and bud emergence (left), as well as the time intervals separating Whi5 exit from Sic1 degradation and budding (right). Only the first G1 of newly born daughter cells was monitored. Lines show mean ± SD.
(E) Summary of phenotypes. In cln2-lpd cells, Whi5 nuclear exit is delayed and Start occurs at a larger cell size. After the delayed Start, subsequent events (DNA replication, bud emergence) occur with relatively normal timing.
In the microscope, mother cells and unbudded cells in cln2-lpd cultures had significantly larger diameters compared to wt (Figures 5B, S5D), suggesting that they did not begin dividing until reaching a larger size. Indeed, when G1 cells were isolated by centrifugal elutriation (Figure 5C), the cln2-lpd cells delayed budding and DNA synthesis until they reached a volume roughly one-third greater than wt (i.e., ~ 40 vs. 30 fL). Thus, disruption of Cln2 docking skews the calibration between cell size and cell cycle entry. To pinpoint the defect, we conducted time-lapse microscopy with fluorescent forms of two key G1/S regulators, the transcriptional repressor Whi5 and the CDK inhibitor Sic1. In cln2-lpd cells, nuclear exit of Whi5 was delayed, whereas Sic1 degradation and budding occurred with relatively normal timing thereafter (Figure 5D). Thus, the predominant effect of Cln2-lpd is to delay Start (Figure 5E), which coincides with Whi5 exit and the onset of G1/S transcription [19, 30]. This delay did not cause an accumulation of G1 cells in asynchronous cultures (Figure S5E), likely because cells that bud at a larger size also produce larger daughters (i.e., with a larger birth size), and hence the time from birth to budding remains comparable. Size is also regulated by nutrients such as carbon source [7, 26], but the cln2-lpd cells still shifted to smaller sizes in poor carbon media and were always larger than wt regardless of nutrients (Figure S5F). Therefore, nutrient regulation remains intact, but the eventual execution of Start is delayed in cln2-lpd cells.
Docking promotes Whi5 phosphorylation and bud polarization
To analyze Whi5 phosphorylation in vivo, we first eliminated delays in achieving critical cell size by synchronizing cells with a prolonged G1 arrest in mating pheromone. Cells arrested in G1 continue to grow [31], and so upon release can begin cell cycle entry without further growth delay [32], similar to large mother cells [33]. To further equalize initial conditions, we used cells with a separate PMET3-CLN2 construct [19, 27] that was expressed only during initial propagation and then repressed during the experiment.
Using these conditions, we monitored Whi5 gel mobility (Figure 6A), which reflects CDK phosphorylation [9, 10, 12]. V5-tagged Whi5 was resolved into three species: one in G1 arrested cells, plus two higher forms that appeared after release (Figure 6Ai–ii). In CLN2-wt cells the highest form eventually became the predominant species, whereas this never occurred in cln2-lpd cells and instead the middle form was predominant. This pattern suggests that Cln2-lpd cannot drive Whi5 phosphorylation as extensively as wt, in agreement with the in vitro assays. GFP-tagged Whi5 was resolved into only two species (Figure 6Aiii), but again we observed less of the top form in cln2-lpd cells. This reduced phosphorylation of Whi5 was associated with mildly reduced G1/S transcription (Figure 6B) and budding (Figure S6A), whereas DNA synthesis was unaffected (Figure S6A), consistent with Cln1/2 serving a more unique role in bud emergence whereas Clb5/6 expression drives DNA synthesis [27, 32, 34]. Strikingly, in the absence of Cln2 (i.e., empty vector), phosphorylation of Whi5 by Cln3 was barely (if at all) detectable (Figure 6Aii–iii), despite prior suggestions that Cln3 triggers its initial inactivation [10, 19]. This implies that Cln3-CDK phosphorylates Whi5 at most only weakly, whereas Cln1/2-CDK drives more complete phosphorylation (see Discussion). Altogether, the findings indicate that Cln2-lpd is primarily deficient at driving full modification of substrates.
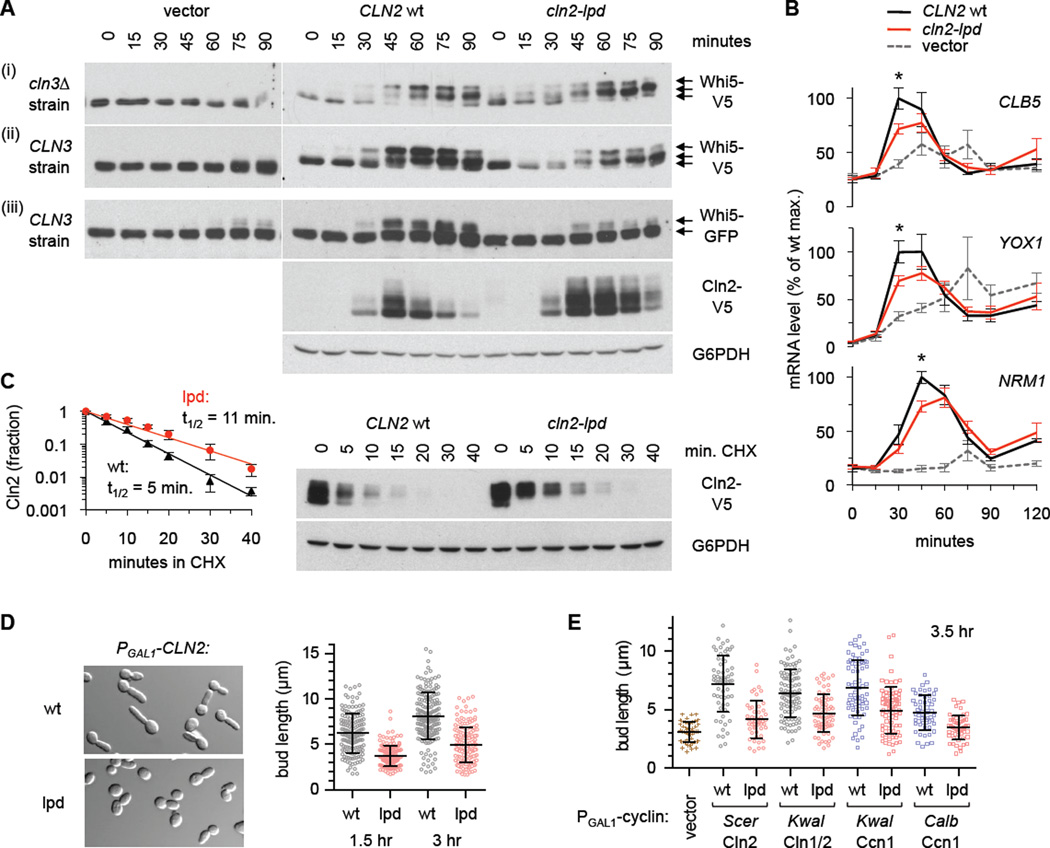
Figure 6. Whi5 phosphorylation and bud polarization depend on Cln2 docking.
(A) CLN3 cln1Δ cln2Δ PMET3-CLN2 and cln3Δ cln1Δ cln2Δ PMET3-CLN2 cells harbored a CLN2 plasmid (wt or lpd; native promoter) or empty vector. Cells were arrested in G1 phase with α factor (plus methionine to repress PMET3-CLN2), and then released; aliquots were harvested at times shown. Whi5 phosphorylation was monitored by immunoblotting for Whi5-V5 (i, ii) or Whi5-GFP (iii); Cln2-V5 levels and a loading control (G6PDH) are shown for set (iii).
(B) mRNA levels were measured, in the cln3Δ background, using conditions as in panel (A). Graphs plot mean ± SEM (n = 5), and asterisks indicate p < 0.05 (t-test) for wt vs. lpd.
(C) The Cln2-lpd protein shows reduced turnover. Cells harboring a CLN2-V5 plasmid (wt or lpd; as in panel Aiii; growing in +Met medium), were treated with 50 µg/mL cycloheximide (CHX). Cln2-V5 and G6PDH levels were measured at times indicated. The graph plots the fraction of protein remaining (mean ± SD; n = 4) with exponential trendlines; half-lives (t1/2) were calculated by fitting to an exponential decay.
(D) Cells harboring a PGAL1-CLN2 plasmid (wt or lpd) were induced with galactose for 1.5 or 3 hr. Images are from 1.5 hr. Plots show individual bud lengths with mean ± SD (n > 150); p = 10−29 (1.5 hr) or 10−33 (3 hr) by two-tailed t-test.
(E) Cells harboring PGAL1-CLN2 or PGAL1-CCN1 plasmids (as in Figure 2C) were induced with galactose for 3.5 hr. Plots show bud lengths with mean ± SD (n > 50); for all pair-wise comparisons of wt versus lpd, p < 10−4 (two-tailed t-test). Also see Figure S6B.
Interestingly, in these synchronous cultures, the Cln2-lpd protein was initially expressed on schedule but then persisted long after the wt protein declined (Figure 6Aiii), suggesting that it might have a reduced turnover rate. Indeed, in a cycloheximide chase assay, the half-life was roughly doubled for the mutant protein (Figure 6C). Hence, Cln2 turnover might involve docking with specific partner proteins (see Discussion). This stabilization is also noteworthy because the increased protein levels or duration in the mutant might partially counteract its defects.
Finally, we analyzed how docking contributes to cell polarization. Early in the cell cycle, Cln1 and Cln2 promote highly polarized apical growth of new buds, which then shifts to an isotropic pattern as Cln1/2 levels decline and Clb1–6 cyclins take their place [35]. If Cln2 is expressed continuously from a foreign promoter (e.g., PGAL1), it can drive incessant apical growth and hyperpolarized buds [35]. We found that this hyperpolarized growth was greatly diminished for the docking mutant, Cln2-lpd (Figure 6D). Furthermore, the hyperpolarized phenotype was also observed upon expression of other members of the Cln1/2 and Ccn1 subgroups (Figures 6E, S6B), and in each case this was disrupted by the lpd mutation. Thus, docking helps each of these cyclins drive directionally persistent growth, which may be of particular importance in fungi that form hyphal filaments.
Discussion
We have identified a mutant form of the yeast cyclin Cln2 that is deficient in recognition of LP docking sites on CDK substrates. The detection of key residues was aided by the discovery that LP docking is conserved in both Cln1/2 and Ccn1 groups, and this conservation attests to a selectively advantageous function. Cln3 and Cln1/2 likely diverged from each other after they split from the Ccn1 group (N. Buchler, personal communication), implying that LP docking existed in the prior common ancestor but was then lost in Cln3. Indeed, most non-yeast fungi (dikarya) harbor only one G1 cyclin, and initial studies suggest that they can recognize LP motifs (S. B. and P.M.P., unpublished observations). Still, LP docking may not be identical in each case, as Ccn1 bound only some partners of Cln1/2 (Figure 3D). Thus, evolution of distinct cyclin sub-types likely included both loss of docking and divergence of motif preferences.
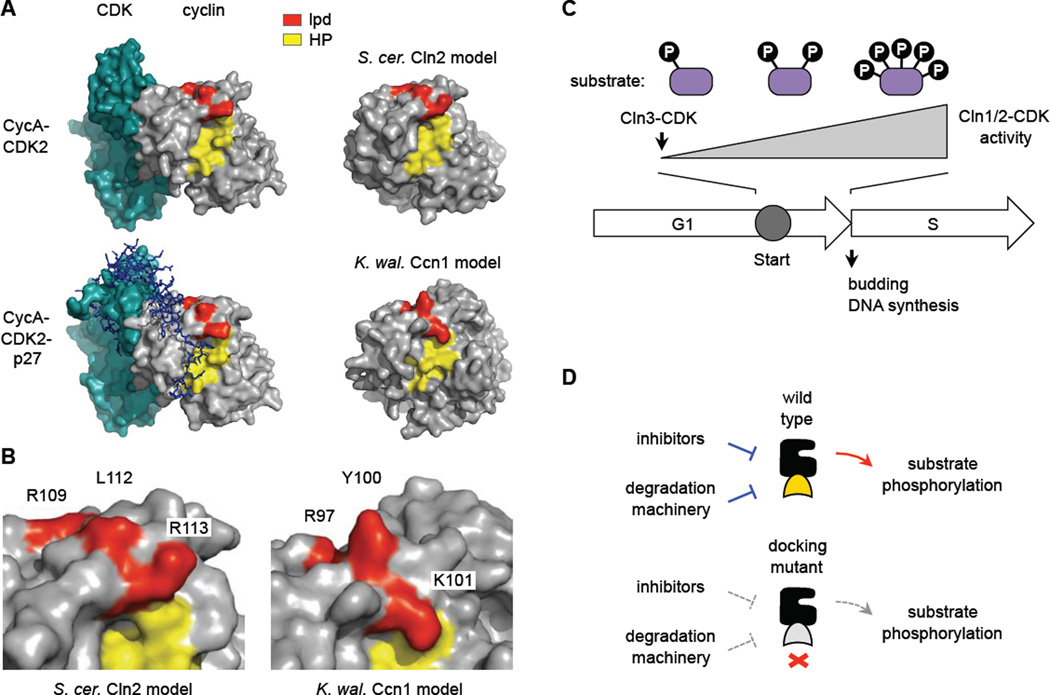
A structural comparison (Figure 7A, B) suggests that the region required for LP docking is close but separated from the hydrophobic patch (HP) region that allows S-phase cyclins to bind RXL motifs [36–38]. The lpd residues begin at a ridge bordering the HP cleft and then proceed along the edge of an adjacent plateau. The non-polar Leu112 residue that is most critical in Cln2 is predicted to be solvent-exposed, and thus is well suited to interact favorably with Leu/Pro-rich LP motifs. Interestingly, although RXL peptides from CDK substrates do not encroach upon the lpd plateau [39, 40], the inhibitor protein p27, which binds cyclin A-CDK2 over a broad interface [41], does make contacts in this region as it traverses from cyclin to CDK (Figure 7A). Hence, LP docking and CDK inhibition could be evolutionarily related.
Figure 7. Docking interfaces and models.
(A) Structures of a mammalian cyclin A-CDK2 complex, with and without the inhibitor protein p27 (PDB IDs: 1H26, 1JSU), compared to models for S. cerevisiae Cln2 and K. waltii Ccn1 generated by the I-TASSER algorithm [56]. The hydrophobic patch (HP, yellow) and residues altered in the lpd mutants (red) are highlighted.
(B) Close-up of the boundary between HP and lpd regions in the predicted models, with mutated residues labeled.
(C) General model. Cln3-CDK initiates expression of CLN1/2 (and then Cln1/2-CDK further increase this expression). As Cln1/2-CDK activity accumulates, the efficiency with which it fully phosphorylates multi-site substrates may affect the duration of Start and the G1/S transition.
(D) Disruption of Cln2 docking may simultaneously disrupt both positive output and negative regulation. See text for discussion.
In vitro, the docking function of Cln2 was required for multi-site phosphorylation of substrates, which involves processive catalysis and the Cks1 subunit of the cyclin-CDK-Cks1 complex [15, 42]. Thus, in vivo, events that require extensive substrate phosphorylation may be especially dependent on docking, and this could underlie some functional distinctions among G1 cyclins. For example, although Cln3 is thought to initiate inactivation of Whi5, Cln1/2 substantially accelerates its chromatin dissociation and nuclear exit [10, 19]. Likewise, we saw that Cln3 is not as proficient as Cln2 at driving extensive Whi5 modification in vivo, and the superior activity of Cln2 depends at least in part on docking. Whi5 might be phosphorylated via a two-stage relay, first by Cln3 and then more completely by Cln1/2; this would be analogous to Rb phosphorylation by cyclins D and E in animal cells [43], and to the sequential phosphorylation of Sic1 by Cln1/2 and Clb5 in yeast [14, 16]. Yet, while our results clearly reveal phosphorylation of Whi5 by Cln1/2, there is scant (if any) evidence that Cln3 can do so, and hence this remains an important issue for future studies.
Although substrate docking by Cln2 helps cells initiate division at the proper size, Cln1/2 are not expected to be involved in the size sensing mechanism per se, but rather in the robust execution of molecular events that drive cell cycle entry once the sensing mechanism is satisfied [7, 26]. The commitment point at Start comprises a brief interval in which Cln1/2-CDK activity rapidly accumulates (Figure 7C), and many key regulators of the G1/S transition are proteins with multiple CDK sites, including not only Whi5 but also Sic1 and Cdh1 [11, 13]. Accordingly, the docking function of Cln1/2 likely contributes to a rapid and decisive transition by ensuring that full phosphorylation of substrates occurs promptly once cyclins arise. The need for docking might be reduced in G1 arrest/release experiments because cells grow beyond the critical size by the time of release, and this can alter the CDK threshold required to pass Start [32, 33].
The contribution of LP docking to polarized morphogenesis likely relates to Cln2 substrates involved in cell polarization such as regulators of Rho GTPases (reviewed in [44, 45]), some examples of which (i.e., Rga1, Tus1) showed reduced binding to Cln2-lpd. Relatedly, G1 cyclins drive persistent hyphal tip growth in filamentous fungi [45–48]. Substrate docking by Cln1/2 and Ccn1 could help maintain high levels of phosphorylation in a local region or even co-localize the cyclin with substrates at sites of polarized growth.
In addition to its positive role, the docking interface on Cln2 might promote regulation by antagonists (Figure 7D). First, the Cln2-lpd protein showed reduced turnover, suggesting that the docking interface might engage a degradation factor, such as Cdc48 [49]. Second, enhanced phenotypic differences between wt and cln2-lpd alleles in far1Δ cells suggest that Far1 might inhibit Cln2-wt and Cln2-lpd differentially. Far1 blocks Cln2-substrate docking [29], perhaps by engaging the docking interface; indeed, in a recently developed in vitro assay for Far1 inhibition (E.V. and M.L., in preparation), the Cln2-lpd mutation increased the Ki of Far1 by roughly ten-fold. Competition among substrates and regulators for a common interface may provide a simple mechanism to integrate multiple signals, and there are precedents in cyclin-CDK complexes [36, 40, 41] and MAP kinases [50].
In conclusion, our findings uncover a novel substrate-docking interface, conserved among distinct G1 cyclin sub-groups, that contributes to efficient, multi-site phosphorylation of CDK substrates and to the punctual entry into the cell cycle. Future studies of LP docking could shed light on cyclin evolution and the role of CDKs in polarized growth, plus illuminate how robust CDK phosphorylation influences cell-to-cell variability of Start [28, 33] and the coherence of distinct events at the G1/S transition [19].
Experimental Procedures
Yeast Methods
Standard procedures were used for growth and genetic manipulation of yeast [51, 52]. Cells were grown at 30°C in yeast extract/peptone medium with 2% glucose (YPD) or galactose (YPGal), or in synthetic (SC) medium with 2% glucose and/or raffinose. Strains and plasmids are listed in the Supplemental Information.
In Vivo Phosphorylation Assays
As described previously [22, 29], cells harboring PGAL1-GST-cyclin constructs and HA-tagged CDK substrates were induced with 2% galactose (for 2.5 hr) to drive cyclin expression; as before [29], most cyclins were truncated to remove destabilizing C-termini (see Figure S1C), thus allowing comparable expression. All substrates were based on the Ste5 N-terminus (1–260), with or without docking sites: (i) the “LP dock” was from Ste5 (261–315 or 261–337), except where noted as being from Ste20 (80–115); (ii) “no dock” substrates had either no added sequence or a mutated Ste5 LP motif; (iii) leucine zipper substrates had the E34(N) sequence (see Figure S1A). Substrate phosphorylation was assessed by SDS-PAGE and immunoblotting of whole cell lysates (see Supplemental Information).
Protein Binding Assays
GST co-precipitation assays were performed as described [29]. Briefly, 10 mL cultures were treated with 2% galactose (1.5 hr) to express GST fusion proteins. Extracts were prepared by glass bead lysis, and GST fusions and co-bound proteins were collected on glutathione-sepharose.
In Vitro Kinase Assays
Cln2-Cdc28-Cks1 complexes were purified from yeast, and substrates (except histone H1) were purified from bacteria. Reactions contained kinase and substrate with or without competitor LP peptide. For detailed methods, see the Supplemental Information.
Synchronous Cultures
Small G1 cells were purified by centrifugal elutriation as described previously [53]. Initial cultures were grown in SC/raffinose, and elutriated cells were resuspended in YPD. For G1 arrest/release, PMET3-CLN2 cells harboring CLN2 plasmids were arrested in +Met medium containing α factor, and released in +Met medium without α factor; see Supplemental Information for details.
Flow Cytometry, Budding, and Cell Size Measurements
As described previously [54], DNA content was measured by flow cytometry using Sytox Green, and budding status was assayed using formaldehyde-fixed cells (200 cells were counted per condition). Cell volume was measured using a Beckman Coulter Multisizer 3 (Beckman Coulter). Cell diameters and bud lengths were measured microscopically (see Supplemental Information).
mRNA Analysis
RNA was prepared as described previously [54]. Synthesis of cDNA and quantitative real-time PCR was performed as described in the Supplemental Information.
Time-Lapse Microscopy
Cells in SC/glucose medium were entrapped in a CellASIC microfluidic device as described [30]. Images were acquired every 3 min., and multiple fields were followed simultaneously. Fluorescence data were analyzed via automated algorithms [55] to determine midpoint times of Whi5-mCherry nuclear exit and Sic1-GFP degradation. Phase-contrast images were inspected manually to determine the time of bud emergence and the cell diameters at Whi5 exit and budding onset.
Supplementary Material
Highlights.
Recognition of LP docking sites is conserved between Cln1/2 and Ccn1 cyclins
A novel docking interface on the cyclin surface allows LP motif recognition
Docking by Cln2 allows efficient, multi-site phosphorylation of CDK substrates
Cln2-substrate docking promotes bud polarization and timely cell cycle entry
Acknowledgements
We thank R. Baker, A. Gladfelter, O. Rando, and C. Specht for fungal DNA samples, J. Benanti and J. Skotheim for strains and plasmids, D. McCollum, N. Rhind, and E. Torres for use of lab equipment, and N. Buchler and J. Skotheim for discussions. This work was supported by grants from the National Institutes of Health (GM57769) to P.M.P, and from the Estonian Science Agency (IUT2-21) to M.L.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions
S.B. and P.M.P. designed the project. S.B. performed most experiments, with assistance from M.J.W. and B.G. In vitro kinase assays and time-lapse microscopy were performed by E.V. in the lab of M.L. P.M.P. analyzed data and prepared the manuscript.
References
- 1.Morgan DO. The Cell Cycle: Principles of Control. London: New Science Press; 2007. [Google Scholar]
- 2.Bloom J, Cross FR. Multiple levels of cyclin specificity in cell-cycle control. Nat Rev Mol Cell Biol. 2007;8:149–160. doi: 10.1038/nrm2105. [DOI] [PubMed] [Google Scholar]
- 3.Coudreuse D, Nurse P. Driving the cell cycle with a minimal CDK control network. Nature. 2010;468:1074–1079. doi: 10.1038/nature09543. [DOI] [PubMed] [Google Scholar]
- 4.Levine K, Huang K, Cross FR. Saccharomyces cerevisiae G1 cyclins differ in their intrinsic functional specificities. Mol Cell Biol. 1996;16:6794–6803. doi: 10.1128/mcb.16.12.6794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kõivomägi M, Valk E, Venta R, Iofik A, Lepiku M, Morgan DO, Loog M. Dynamics of Cdk1 Substrate Specificity during the Cell Cycle. Mol Cell. 2011;42:610–623. doi: 10.1016/j.molcel.2011.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cross FR. Starting the cell cycle: what's the point? Curr Opin Cell Biol. 1995;7:790–797. doi: 10.1016/0955-0674(95)80062-x. [DOI] [PubMed] [Google Scholar]
- 7.Jorgensen P, Tyers M. How cells coordinate growth and division. Curr Biol. 2004;14:R1014–R1027. doi: 10.1016/j.cub.2004.11.027. [DOI] [PubMed] [Google Scholar]
- 8.Johnson A, Skotheim JM. Start and the restriction point. Curr Opin Cell Biol. 2013;25:717–723. doi: 10.1016/j.ceb.2013.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Costanzo M, Nishikawa JL, Tang X, Millman JS, Schub O, Breitkreuz K, Dewar D, Rupes I, Andrews B, Tyers M. CDK activity antagonizes Whi5, an inhibitor of G1/S transcription in yeast. Cell. 2004;117:899–913. doi: 10.1016/j.cell.2004.05.024. [DOI] [PubMed] [Google Scholar]
- 10.de Bruin RA, McDonald WH, Kalashnikova TI, Yates J, 3rd, Wittenberg C. Cln3 activates G1-specific transcription via phosphorylation of the SBF bound repressor Whi5. Cell. 2004;117:887–898. doi: 10.1016/j.cell.2004.05.025. [DOI] [PubMed] [Google Scholar]
- 11.Nash P, Tang X, Orlicky S, Chen Q, Gertler FB, Mendenhall MD, Sicheri F, Pawson T, Tyers M. Multisite phosphorylation of a CDK inhibitor sets a threshold for the onset of DNA replication. Nature. 2001;414:514–521. doi: 10.1038/35107009. [DOI] [PubMed] [Google Scholar]
- 12.Wagner MV, Smolka MB, de Bruin RA, Zhou H, Wittenberg C, Dowdy SF. Whi5 regulation by site specific CDK-phosphorylation in Saccharomyces cerevisiae. PLoS One. 2009;4:e4300. doi: 10.1371/journal.pone.0004300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zachariae W, Schwab M, Nasmyth K, Seufert W. Control of cyclin ubiquitination by CDK-regulated binding of Hct1 to the anaphase promoting complex. Science. 1998;282:1721–1724. doi: 10.1126/science.282.5394.1721. [DOI] [PubMed] [Google Scholar]
- 14.Kõivomägi M, Valk E, Venta R, Iofik A, Lepiku M, Balog ER, Rubin SM, Morgan DO, Loog M. Cascades of multisite phosphorylation control Sic1 destruction at the onset of S phase. Nature. 2011;480:128–131. doi: 10.1038/nature10560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kõivomägi M, Ord M, Iofik A, Valk E, Venta R, Faustova I, Kivi R, Balog ER, Rubin SM, Loog M. Multisite phosphorylation networks as signal processors for Cdk1. Nat Struct Mol Biol. 2013;20:1415–1424. doi: 10.1038/nsmb.2706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang X, Lau KY, Sevim V, Tang C. Design principles of the yeast G1/S switch. PLoS Biol. 2013;11:e1001673. doi: 10.1371/journal.pbio.1001673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cross FR, Tinkelenberg AH. A potential positive feedback loop controlling CLN1 and CLN2 gene expression at the start of the yeast cell cycle. Cell. 1991;65:875–883. doi: 10.1016/0092-8674(91)90394-e. [DOI] [PubMed] [Google Scholar]
- 18.Dirick L, Nasmyth K. Positive feedback in the activation of G1 cyclins in yeast. Nature. 1991;351:754–757. doi: 10.1038/351754a0. [DOI] [PubMed] [Google Scholar]
- 19.Skotheim JM, Di Talia S, Siggia ED, Cross FR. Positive feedback of G1 cyclins ensures coherent cell cycle entry. Nature. 2008;454:291–296. doi: 10.1038/nature07118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miller ME, Cross FR. Distinct subcellular localization patterns contribute to functional specificity of the Cln2 and Cln3 cyclins of Saccharomyces cerevisiae. Mol Cell Biol. 2000;20:542–555. doi: 10.1128/mcb.20.2.542-555.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Edgington NP, Futcher B. Relationship between the function and the location of G1 cyclins in S. cerevisiae. J Cell Sci. 2001;114:4599–4611. doi: 10.1242/jcs.114.24.4599. [DOI] [PubMed] [Google Scholar]
- 22.Bhaduri S, Pryciak PM. Cyclin-specific docking motifs promote phosphorylation of yeast signaling proteins by G1/S Cdk complexes. Curr Biol. 2011;21:1615–1623. doi: 10.1016/j.cub.2011.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Archambault V, Buchler NE, Wilmes GM, Jacobson MD, Cross FR. Two-faced cyclins with eyes on the targets. Cell Cycle. 2005;4:125–130. doi: 10.4161/cc.4.1.1402. [DOI] [PubMed] [Google Scholar]
- 24.Berset C, Griac P, Tempel R, La Rue J, Wittenberg C, Lanker S. Transferable domain in the G(1) cyclin Cln2 sufficient to switch degradation of Sic1 from the E3 ubiquitin ligase SCF(Cdc4) to SCF(Grr1) Mol Cell Biol. 2002;22:4463–4476. doi: 10.1128/MCB.22.13.4463-4476.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Landry BD, Doyle JP, Toczyski DP, Benanti JA. F-box protein specificity for g1 cyclins is dictated by subcellular localization. PLoS Genet. 2012;8:e1002851. doi: 10.1371/journal.pgen.1002851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Turner JJ, Ewald JC, Skotheim JM. Cell size control in yeast. Curr Biol. 2012;22:R350–R359. doi: 10.1016/j.cub.2012.02.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dirick L, Bohm T, Nasmyth K. Roles and regulation of Cln-Cdc28 kinases at the start of the cell cycle of Saccharomyces cerevisiae. EMBO J. 1995;14:4803–4813. doi: 10.1002/j.1460-2075.1995.tb00162.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ferrezuelo F, Colomina N, Palmisano A, Gari E, Gallego C, Csikasz-Nagy A, Aldea M. The critical size is set at a single-cell level by growth rate to attain homeostasis and adaptation. Nat Commun. 2012;3:1012. doi: 10.1038/ncomms2015. [DOI] [PubMed] [Google Scholar]
- 29.Pope PA, Bhaduri S, Pryciak PM. Regulation of cyclin-substrate docking by a G1 arrest signaling pathway and the Cdk inhibitor Far1. Curr Biol. 2014;24:1390–1396. doi: 10.1016/j.cub.2014.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Doncic A, Falleur-Fettig M, Skotheim JM. Distinct interactions select and maintain a specific cell fate. Mol Cell. 2011;43:528–539. doi: 10.1016/j.molcel.2011.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Goranov AI, Cook M, Ricicova M, Ben-Ari G, Gonzalez C, Hansen C, Tyers M, Amon A. The rate of cell growth is governed by cell cycle stage. Genes Dev. 2009;23:1408–1422. doi: 10.1101/gad.1777309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schwob E, Nasmyth K. CLB5 and CLB6, a new pair of B cyclins involved in DNA replication in Saccharomyces cerevisiae. Genes Dev. 1993;7:1160–1175. doi: 10.1101/gad.7.7a.1160. [DOI] [PubMed] [Google Scholar]
- 33.Di Talia S, Skotheim JM, Bean JM, Siggia ED, Cross FR. The effects of molecular noise and size control on variability in the budding yeast cell cycle. Nature. 2007;448:947–951. doi: 10.1038/nature06072. [DOI] [PubMed] [Google Scholar]
- 34.Moffat J, Andrews B. Late-G1 cyclin-CDK activity is essential for control of cell morphogenesis in budding yeast. Nat Cell Biol. 2004;6:59–66. doi: 10.1038/ncb1078. [DOI] [PubMed] [Google Scholar]
- 35.Lew DJ, Reed SI. Morphogenesis in the yeast cell cycle: regulation by Cdc28 and cyclins. J Cell Biol. 1993;120:1305–1320. doi: 10.1083/jcb.120.6.1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schulman BA, Lindstrom DL, Harlow E. Substrate recruitment to cyclin-dependent kinase 2 by a multipurpose docking site on cyclin A. Proc Natl Acad Sci U S A. 1998;95:10453–10458. doi: 10.1073/pnas.95.18.10453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wilmes GM, Archambault V, Austin RJ, Jacobson MD, Bell SP, Cross FR. Interaction of the S-phase cyclin Clb5 with an "RXL" docking sequence in the initiator protein Orc6 provides an origin-localized replication control switch. Genes Dev. 2004;18:981–991. doi: 10.1101/gad.1202304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Loog M, Morgan DO. Cyclin specificity in the phosphorylation of cyclin-dependent kinase substrates. Nature. 2005;434:104–108. doi: 10.1038/nature03329. [DOI] [PubMed] [Google Scholar]
- 39.Cheng KY, Noble ME, Skamnaki V, Brown NR, Lowe ED, Kontogiannis L, Shen K, Cole PA, Siligardi G, Johnson LN. The role of the phospho-CDK2/cyclin A recruitment site in substrate recognition. J Biol Chem. 2006;281:23167–23179. doi: 10.1074/jbc.M600480200. [DOI] [PubMed] [Google Scholar]
- 40.Lowe ED, Tews I, Cheng KY, Brown NR, Gul S, Noble ME, Gamblin SJ, Johnson LN. Specificity determinants of recruitment peptides bound to phospho-CDK2/cyclin A. Biochemistry. 2002;41:15625–15634. doi: 10.1021/bi0268910. [DOI] [PubMed] [Google Scholar]
- 41.Russo AA, Jeffrey PD, Patten AK, Massague J, Pavletich NP. Crystal structure of the p27Kip1 cyclin-dependent-kinase inhibitor bound to the cyclin A-Cdk2 complex. Nature. 1996;382:325–331. doi: 10.1038/382325a0. [DOI] [PubMed] [Google Scholar]
- 42.McGrath DA, Balog ER, Koivomagi M, Lucena R, Mai MV, Hirschi A, Kellogg DR, Loog M, Rubin SM. Cks confers specificity to phosphorylation-dependent CDK signaling pathways. Nat Struct Mol Biol. 2013;20:1407–1414. doi: 10.1038/nsmb.2707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Narasimha AM, Kaulich M, Shapiro GS, Choi YJ, Sicinski P, Dowdy SF. Cyclin D activates the Rb tumor suppressor by mono-phosphorylation. Elife. 2014:e02872. doi: 10.7554/eLife.02872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Enserink JM, Kolodner RD. An overview of Cdk1-controlled targets and processes. Cell Div. 2010;5:11. doi: 10.1186/1747-1028-5-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang Y. CDKs and the yeast-hyphal decision. Curr Opin Microbiol. 2009;12:644–649. doi: 10.1016/j.mib.2009.09.002. [DOI] [PubMed] [Google Scholar]
- 46.Hungerbuehler AK, Philippsen P, Gladfelter AS. Limited functional redundancy and oscillation of cyclins in multinucleated Ashbya gossypii fungal cells. Eukaryot Cell. 2007;6:473–486. doi: 10.1128/EC.00273-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zheng X, Wang Y. Hgc1, a novel hypha-specific G1 cyclin-related protein regulates Candida albicans hyphal morphogenesis. EMBO J. 2004;23:1845–1856. doi: 10.1038/sj.emboj.7600195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Loeb JD, Sepulveda-Becerra M, Hazan I, Liu H. A G1 cyclin is necessary for maintenance of filamentous growth in Candida albicans. Mol Cell Biol. 1999;19:4019–4027. doi: 10.1128/mcb.19.6.4019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Archambault V, Chang EJ, Drapkin BJ, Cross FR, Chait BT, Rout MP. Targeted proteomic study of the cyclin-Cdk module. Mol Cell. 2004;14:699–711. doi: 10.1016/j.molcel.2004.05.025. [DOI] [PubMed] [Google Scholar]
- 50.Remenyi A, Good MC, Bhattacharyya RP, Lim WA. The role of docking interactions in mediating signaling input, output, and discrimination in the yeast MAPK network. Mol Cell. 2005;20:951–962. doi: 10.1016/j.molcel.2005.10.030. [DOI] [PubMed] [Google Scholar]
- 51.Rothstein R. Targeting, disruption, replacement, and allele rescue: integrative DNA transformation in yeast. Methods Enzymol. 1991;194:281–301. doi: 10.1016/0076-6879(91)94022-5. [DOI] [PubMed] [Google Scholar]
- 52.Sherman F. Getting started with yeast. Methods Enzymol. 2002;350:3–41. doi: 10.1016/s0076-6879(02)50954-x. [DOI] [PubMed] [Google Scholar]
- 53.Strickfaden SC, Winters MJ, Ben-Ari G, Lamson RE, Tyers M, Pryciak PM. A mechanism for cell-cycle regulation of MAP kinase signaling in a yeast differentiation pathway. Cell. 2007;128:519–531. doi: 10.1016/j.cell.2006.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pope PA, Pryciak PM. Functional overlap among distinct G1/S inhibitory pathways allows robust G1 arrest by yeast mating pheromones. Mol Biol Cell. 2013;24:3675–3688. doi: 10.1091/mbc.E13-07-0373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Doncic A, Eser U, Atay O, Skotheim JM. An algorithm to automate yeast segmentation and tracking. PLoS One. 2013;8:e57970. doi: 10.1371/journal.pone.0057970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Roy A, Kucukural A, Zhang Y. I-TASSER: a unified platform for automated protein structure and function prediction. Nat Protoc. 2010;5:725–738. doi: 10.1038/nprot.2010.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.