Summary
How mitochondrial dynamism (fission and fusion) affects mitochondrial quality control is unclear. We uncovered distinct effects on mitophagy of inhibiting Drp1-mediated mitochondrial fission versus mitofusin-mediated mitochondrial fusion. Conditional cardiomyocyte-specific Drp1 ablation evoked mitochondrial enlargement, lethal dilated cardiomyopathy, and cardiomyocyte necrosis. Conditionally ablating cardiomyocyte mitofusins (Mfn) caused mitochondrial fragmentation with eccentric remodeling and no cardiomyocyte dropout. Parallel studies in cultured murine embryonic fibroblasts (MEFs) and in vivo mouse hearts revealed that Mfn1/Mfn2 deletion provoked accumulation of defective mitochondria exhibiting an unfolded protein response, without appropriately increasing mitophagy. Conversely, interrupting mitochondrial fission by Drp1 ablation increased mitophagy and caused a generalized loss of mitochondria. Mitochondrial permeability transition pore (MPTP) opening in Drp1 null mitochondria was associated with mitophagy in MEFs and contributed to cardiomyocyte necrosis and dilated cardiomyopathy in mice. Drp1, MPTP, and cardiomyocyte mitophagy are functionally integrated. Mitochondrial fission and fusion have opposing roles during in vivo cardiac mitochondrial quality control.
Introduction
Mitochondrial fitness is required for normal cell metabolism and to prevent cell damage from mitochondrial-derived reactive oxygen species (ROS). Mitochondrial dynamism is linked to mitochondrial quality control through the process of asymmetric fission, in which mitochondrial fission induced by Dynamin-related protein (Drp)-1 physically segregates dysfunctional mitochondrial components into a depolarized daughter organelle targeted for mitophagy (Twig et al., 2008). Thus, mitochondrial fragmentation and mitophagy often occur concomitantly (Frank et al., 2012; Park et al., 2012; Rambold et al., 2011). Drp1-mediated mitochondrial fission is also pathologically implicated in mitochondrial release of cytochrome c that activates apoptosis signaling (Youle and van der Bliek, 2012) and in post-ischemic myocardial damage (Ong et al., 2010).
Our understanding of the role of mitochondrial fission (and its physiological doppelganger, mitochondrial fusion) in adult cardiac myocytes is confounded by absence of the interconnected mitochondrial networks that characterize most cell types. Given the intrinsically fragmented morphology of cardiomyocyte mitochondria (Dorn, 2013a, b), mitochondrial fission to facilitate “bit-by-bit” mitochondrial autophagy (Yang and Yang, 2013) seems unnecessary. We therefore postulated that mitochondrial morphology does not direct mitochondrial autophagy. In support of this notion, we recently reported that cardiac-specific ablation of the mitochondrial fusion factor mitofusin (Mfn) 2 causes heart failure in mice not by suppressing mitochondrial fusion (Mfn2-deficient mitochondria were larger, not smaller), but because phosphorylated Mfn2 can act as the receptor for parkin on damaged mitochondria (Chen and Dorn, 2013). Here, we interrogated the roles of mitochondrial fission and fusion on mitochondrial fitness using temporally-defined in vivo and in vitro genetic ablation of the pro-fission factor Drp1, or the pro-fusion factors Mfn1 and Mfn2 in combination. Our results uncover distinct effects of inhibiting mitochondrial fission and fusion on mitochondrial autophagy, cell viability, and cardiac remodeling. Rather than just controlling mitochondria network morphometry, our findings reveal that the mitochondrial fission factor Drp1 and fusion factors Mfn1 and Mfn2 interact within an integrated organelle quality control process that orchestrates homeostatic cardiomyocyte mitochondria culling, regeneration, and repair.
Results
Early postnatal cardiomyocyte-specific Drp1 deficiency is lethal
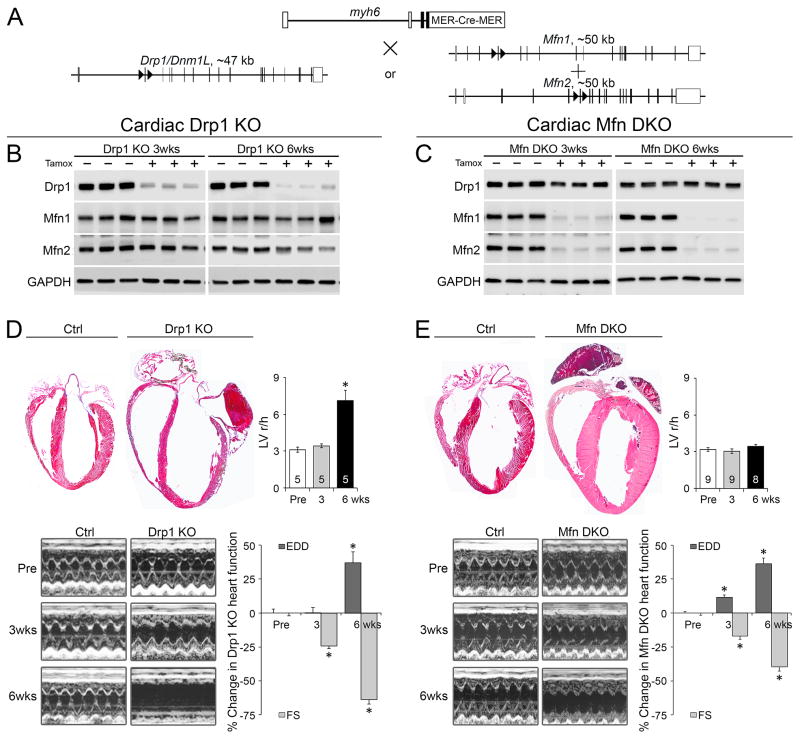
Interrupting mitochondrial fusion by combined ablation of Mfn1 and Mfn2 causes lethal heart failure (Chen et al., 2011; Papanicolaou et al., 2012). Mitochondrial fragmentation is the seminal feature of mitofusin deficiency, but the rapidly lethal cardiomyopathies appear disproportionate to observed organelle dysmorphology, and the cellular basis for cardiac decompensation is still unknown. To gain further insight into the relationship between mitochondrial dysmorphology and mammalian heart function we performed the reciprocal manipulation, genetically ablating Drp1 to interrupt mitochondrial fission in mouse cardiomyocytes. By combining the myh6-Cre transgene with floxed Drp1 alleles (Drp1 fl/fl) (Ishihara et al., 2009) (Supplemental Figure 1a) we deleted Drp1 in cardiomyocytes from the immediate postnatal period (Chen et al., 2012). Immunoblot analysis of myocardium from 4 week old cardiac Drp1 null mice revealed nearly complete absence of Drp1 protein (Supplemental Figure 1b, top). Drp1-deficient hearts were modestly enlarged (Supplemental Figure 1b, middle) and histologically unremarkable (Supplemental Figure 1b, lower). However, whereas myh6-Cre x Drp1 fl/fl mice were born at expected Mendelian ratios, they died by 6 weeks of age (Supplemental Figure 1c), precluding detailed interrogation of causative mechanisms. Accordingly, we abandoned this model in favor of conditional cardiac-specific Drp1 gene ablation in adult mice.
Drp1 deficiency evokes dilated cardiomyopathy; Mfn1/Mfn2 deficiency induces eccentric remodeling
We combined Drp1 fl/fl alleles with a myh6-driven modified estrogen receptor (MER)-Cre transgene to generate mice that could be grown to adulthood prior to cardiac-specific Drp1 deletion using tamoxifen (administered at 8 weeks of age; Figure 1a), as used in our combined cardiac Mfn1/Mfn2 deficiency model (Chen et al., 2011). Importantly, we found no confounding effects of tamoxifen, with or without the myh6-MER-Cre transgene, on cardiac function during the 8 week duration of our studies (Supplemental Figure 2a, 2b).
Figure 1. Conditional deletion of Drp1 or Mfn1/Mfn2 in adult mice evokes different cardiomyopathies.
A. Schematic of tamoxifen-inducible myh6-Cre mediated Drp1 or Mfn1/Mfn2 gene deletion. B-C. Immunobloting of the targeted factors at 3 and 6 weeks after cardiac Drp1 (B) or Mfn1/Mfn2 (C) gene ablation. D–E. Cardiac phenotypes induced by Drp1 (D) or Mfn1/Mfn2 (E) gene deletion. Upper left: Four-chamber view of hearts. Upper right: Ratio of left ventricular (LV) end-diastolic radius (r) to wall thickness (h). Lower left: Serial M-mode echocardiographic analyses. Lower right: Quantitative group data showing % change of LV end-diastolic dimension (EDD) and fractional shortening (FS) (Raw data in Figure S2). Mean ± SEM of numbers of mice indicated in the upper right panel of Fig 1D and 1E, respectively; * = P<0.05 vs identically treated controls. See also Figures S1 and S2.
Immunoreactive Drp1 and Mfn1/Mfn2 were virtually undetectable in myocardial homogenates from their respective knockout hearts, without counter-regulation of other dynamics factor(s) (Figures 1b, 1c). Conditional deletion of both Drp1 and Mfn1/Mfn2 induced progressive left ventricular enlargement and a decline in ejection performance (% left ventricular fractional shortening; %LV FS) (Figures 1d, 1e, Supplemental Figure 2c and 2d). Six to seven weeks after Drp1 deletion full-blown heart failure was evident (Figure 1d), but the decline in ejection performance was less severe in Mfn1/Mfn2 double knockout (DKO) hearts (Figure 1e). Indeed, Drp1 deficiency induced ventricular wall thinning and chamber dilatation that increased the ratio of left ventricular in end-diastolic radius to wall thickness (r/h) (Figure 1d); cardiac mass (heart weight corrected for body weight) increased by only 12% 6 weeks after Drp1 deletion (4.63 ± 0.05 mg/g [n=31] vs 5.21 ± 0.11 mg/g [n=23], p<0.001). This is dilated cardiomyopathy. By comparison, combined Mfn1/Mfn2 deletion provoked eccentric ventricular remodeling with ventricular wall thickening with no change in r/h and a >30% increase in cardiac mass 6 weeks after Mfn1/Mfn2 ablation (4.59 ± 0.09 mg/g [n=14] vs 5.98 ± 0.10 mg/g [n=13], p<0.001) (Figure 1e). This is eccentric hypertrophy.
Interrupting mitochondrial fission, but not fusion, induces cardiomyocyte necrosis
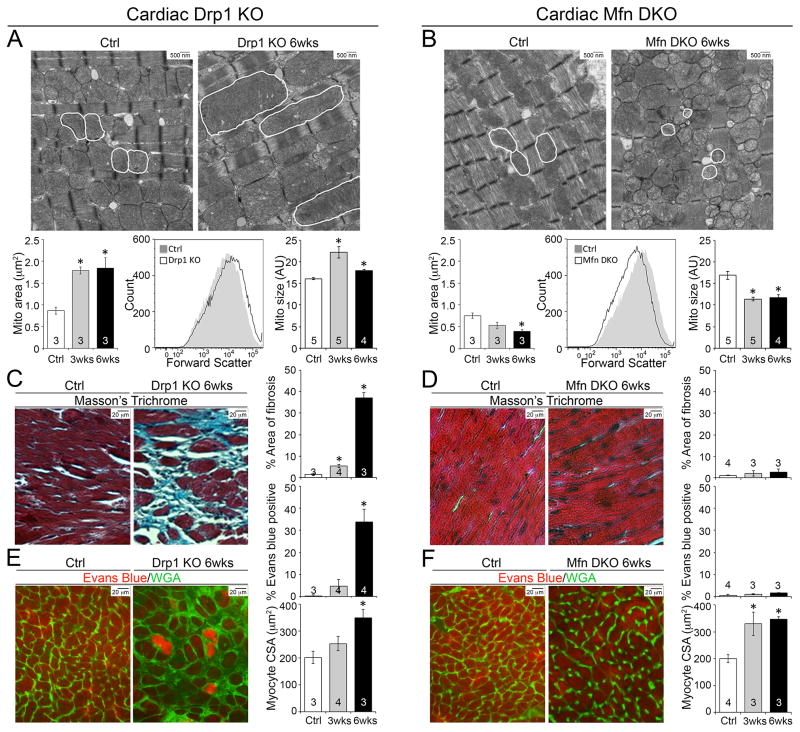
Our expectation was that Drp1 gene ablation would interrupt mitochondrial fission, but deletion of Mfn1 and Mfn2 would suppress fusion. Ultrastructural examination validated these predictions; Drp1 null mitochondria were larger and more elongated, with normal appearing cristae (Figure 2a). By contrast, Mfn1/Mfn2 DKO mitochondria were smaller and exhibited abnormal cristae structures (Figures 2b). Consistent with prior observations (Caffin et al., 2013; Kim, 2013; Papanicolaou et al., 2012), interruption of either mitochondrial fission or fusion was associated with depressed expression of mitochondrial biogenesis genes (Supplemental Figure 3a, 3b).
Figure 2. Inhibition of mitochondrial fission, but not fusion, in adult hearts provokes cardiomyocyte necrosis.
A–B. Mitochondria dysmorphometry evoked by Drp1 (A) or Mfn1/Mfn2 (B) deficiency. Upper: Transmission electron microscopic (TEM) images (5,000x); representative cardiomyocyte mitochondria are outlined in white. Lower left: Mitochondrial area. Lower middle and right: Flow cytometry of mitochondrial size (forward scatter). Representative size distribution curves (black line) are compared to paired controls (grey solid); quantitative data are to the right. C–D. Masson’s trichrome-stained (200x original magnification) Drp1 KO (C) or Mfn DKO (D) hearts. Quantification of blue-staining replacement fibrosis is to the right. E–F. Fluorescence microscopy of Evans blue-perfused Drp1 KO (E) or Mfn DKO (F) hearts. Evans blue dye fluoresces red; green is wheat germ agglutinin staining of cell membranes. Quantitative group data for Evans blue positive cells (%) and cardiomyocyte cross-sectional area (CSA) are to the right. Data are mean ± SEM on indicated numbers of mice; * = P<0.05 vs identically treated controls. See also Figure S3.
We asked why the relatively normal appearing (albeit larger) mitochondria observed after Drp1 ablation were associated with myocardial thinning whereas the “fragmented” and obviously abnormal mitochondria observed after Mfn1/Mfn2 deficiency were not. Because the sine qua non of dilated cardiomyopathy is cardiomyocyte loss with replacement fibrosis (Diwan and Dorn, 2007; Diwan et al., 2008) we examined Drp1- and Mfn1/Mfn2-deficient hearts for cardiomyocyte dropout. Strikingly, Drp1 deficiency provoked a progressive increase in myocardial fibrosis that involved ~40% of the myocardium by 6 weeks (Figure 2c); myocardial fibrosis was not a feature of Mfn1/Mfn2 null hearts (Figure 2d).
Replacement fibrosis is linked to cardiomyocyte death (Diwan et al., 2008). Indeed, TUNEL positive cardiomyocytes were increased in Drp1 null hearts (Supplemental Figure 3c), but does not reliably differentiate between apoptosis and necrosis (Grasl-Kraupp et al., 1995). Previously, mitochondrial fission was associated with increased cardiomyocyte apoptosis (Disatnik et al., 2013; Ong et al., 2010), which appears contrary to our findings. Therefore, we interrogated Drp1 null hearts for evidence of cardiomyocyte necrosis using intravenous infusion of Evans blue dye to assess in vivo sarcolemmal integrity; Evans blue staining of cardiomyocytes was widespread in Drp1 null hearts (Figure 2e). Cardiomyocyte necrosis was confirmed by visualization of focal complement complex activation in Drp1-deficient myocardium (Supplemental Figure 3e). Mfn1/Mfn2 DKO myocardium showed no cardiomyocyte death, consistent with no myocardial fibrosis (Figure 2f, Supplemental Figure 3d and 3f). Modest cardiomyocyte enlargement was observed in both Drp1 null and Mfn1/Mfn2 DKO hearts (Figure 2e, 2f).
Interrupting mitochondrial fusion, but not fission, induces mitochondrial degeneration
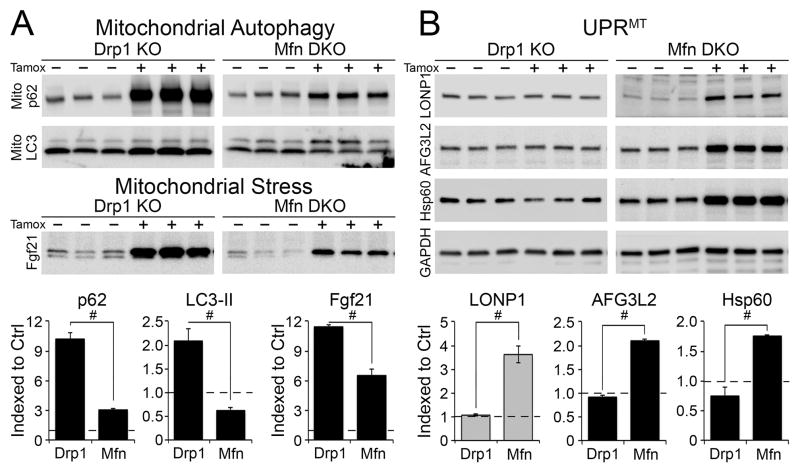
Mitophagic culling of damaged mitochondriais mediated in part by mitochondrial localization of the autophagy chaperone protein p62/SQSTM1, and binding to autophagosomal microtubule associated protein 1 light chain 3 (LC3) (Narendra et al., 2010a). We assayed these two factors specifically in the mitochondrial fractions of our fission- and fusion-defective hearts. Mitochondrial p62 increased ~9-fold in Drp1 null hearts, but was unchanged in Mfn1/Mfn2 DKO hearts. Likewise, mitochondrial LC3-II increased in Drp1 null hearts, but decreased in Mfn1/Mfn2 DKO hearts. Increased mitophagy markers paralleled progression in cardiac phenotype (Figure 3a, Supplemental Figure 4a). Levels of fibroblast growth factor (Fgf) 21, a marker of mitochondria insufficiency (Tyynismaa et al., 2010), increased more rapidly and to a greater extent after Drp1 ablation than after Mfn1/Mfn2 deletion (Figure 3a, Supplemental Figure 4b). Thus, myocardial autophagosome-mitochondria interactions increased after interrupting mitochondrial fission, but not after suppressing mitochondrial fusion. By contrast, levels of two proteases involved in the mitochondrial unfolded protein response (UPRmt), Lon protease (LONP1) and ATPase family gene 3-like 2 (AFG3L2) (Martinelli et al., 2009; Ngo and Davies, 2007), and of the 60 kDa mitochondrial heat shock protein (Hsp60), were normal in Drp1 null hearts, but were increased in Mfn1/Mfn2 DKO hearts (Figure 3b, Supplemental Figure 4c). These results raised the possibility of reciprocal defects in mitochondrial quality control.
Figure 3. Induction of mitophagy in Drp1 null hearts and the mitochondrial unfolded protein response (UPRmt) in Mfn1/Mfn2 DKO hearts.
A. Immunoblot analysis of autophagy markers p62 and LC3-II in mitochondrial fractions (top) and the mitochondrial stress marker Fgf21 in cardiac homogenates (bottom). B. Immunoblot analysis of UPRmt markers LONP1, AFG3L2, and Hsp60 in cardiac homogenates. Each lane is a different mouse heart. p62 is Sequestosome1; LC3-II is microtubule associated protein 1 light chain 3, processed form; Fgf21 is fibroblast growth factor 21; LONP1 is Lon peptidase 1; AFG3L2 is ATPase family member 3-like 2; Hsp60 is heat shock protein chaperone 60; GAPDH is loading control. Quantitative data comparing Drp1 KO to Mfn1/Mfn2 DKO are at the bottom; black columns are 6 weeks, and grey columns 3 weeks, after tamoxifen. Horizontal dashed line represents normal value from Cre-negative controls. Data are mean ± SEM of 4 hearts per group; # = P<0.05 vs identically treated Drp1 KO hearts. See also Figure S4.
Conditional ablation of Drp1 or Mfn1 and Mfn2 in MEFs recapitulates mitochondrial phenotypes from knockout hearts
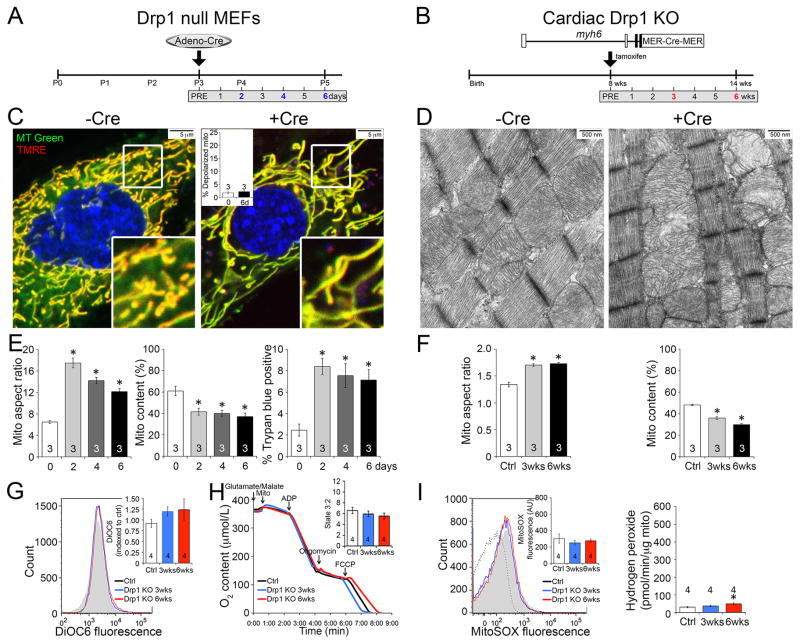
It is not currently possible to directly measure mitochondrial autophagy in vivo. Thus, we developed in vitro systems wherein Drp1 or Mfn1 and Mfn2 were conditionally manipulated using adenoviral-encoded Cre (adeno-Cre) and their respective floxed genes in cultured murine embryonic fibroblasts (MEFs) (Figures 4a) and interrogated the time-dependent consequences of acutely interrupting mitochondrial fission or fusion.
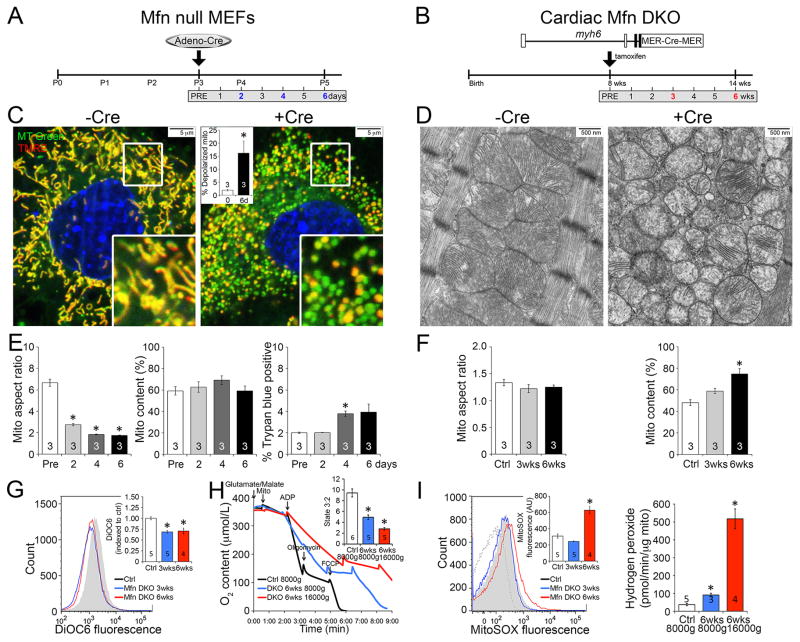
Figure 4. Conditional ablation of Drp1 in cultured MEFs recapitulates mitochondrial phenotypes from conditional Drp1 KO hearts.
A–B. Schematic depiction of time course experimental design in conditional Drp1 KO MEFs (Drp1fl/fl MEFs treated with adeno-Cre) and Drp1 KO adult mice, respectively. C–D. Mitochondria of MEFs co-stained with MitoTracker Green and TMRE (C) and hearts by TEM (10,000x) (D). Red TMRE stains fully polarized mitochondria that appear yellow in merged images; inset is quantitative group data. E. Quantitative mitochondrial aspect ratio (left), mitochondrial content (middle), and cell membrane permeability (right) in Drp1 KO MEFs. F. Quantitative data for mitochondrial aspect ratio (left) and mitochondrial content (right) in Drp1 KO hearts based on TEM images. G. Flow cytometry of isolated cardiac mitochondrial transmembrane electrical potential measured with DiOC6; inset is group data. H. Cardiac mitochondrial respiration; inset is quantitative data for state 3 (ADP stimulated)/state 2. I. Mitochondrial ROS production: Left, O2− measured with MitoSOX Red; Right, H2O2 measured with Amplex Red. Figure 4G-4I: white bar (black line) is ctrl, blue is 3 wks Drp1 KO, and red is 6 wks Drp1 KO. Data are mean ± SEM of indicated number of experiments; * = P<0.05 vs Cre negative controls. See also Figure S5.
The consequences of Cre-mediated conditional ablation of Drp1 on mitochondria in cultured MEFs and in vivo mouse hearts are shown in Figure 4. Confocal fluorescence studies of Drp1-deficient MEFs stained with MitoTracker Green showed mitochondrial hyper-fusion (increased organelle aspect ratio) with a loss of mitochondrial content within two days (Figures 4c, 4e); we found the same correlation between increased mitochondrial aspect ratio and decreased mitochondrial abundance in our Drp1-deficient hearts (Figures 4d, 4f, Supplemental Figure 5a, 5b and 5e). Consistent with the loss of sarcolemmal integrity in Drp1-deficient hearts (see Figure 2e), we observed increased MEF cell membrane permeability after Drp1 ablation (Figure 4e, right panel).
To assess the effects of Drp1 ablation on mitochondrial fitness we assayed fluorescence of the mitochondrial electrochemical potential (Δψm) sensitive dye, tetramethylrhodamine ethyl ester (TMRE). Δψm was not adversely impacted by mitochondrial elongation induced by Drp1 deletion in either MEFs (Figure 4c), or in Drp1 null hearts using flow cytometric analysis of another Δψm sensitive dye, 3,3′-dihexyloxacarbocyanine iodide (DiOC6) (Figure 4g). Normal respiratory function (Figure 4h) and absence of increased ROS production (Figure 4i) in isolated Drp1-deficient cardiac mitochondria further support a high fitness level in the retained mitochondria.
We used the same approach to better understand the consequences of ablating Mfn1 and Mfn2. Mfn1/Mfn2 double floxed MEFs were generated and the genes combinatorially deleted using adeno-Cre (Figure 5a). Conditional ablation of Mfn1 and Mfn2 decreased mitochondrial aspect ratio, reflecting so-called fragmentation induced by unopposed fission, with no change in mitochondrial content (Figures 5c, 5e). These findings mirrored those induced by conditional Mfn1/Mfn2 deletion in mouse hearts, except that the aspect ratio did not decrease (because cardiac mitochondria are already round) (Figures 5d, 5f, Supplemental Figure 5c, 5d and 5e). Similar to Mfn1/Mfn2 DKO hearts, cell membrane integrity was not impaired by conditional ablation of Mfn1 and Mfn2 in MEFs (Figure 5e, right panel). Nevertheless, dissipation of mitochondrial Δψm was clearly evident in the fragmented mitochondria of MEFs lacking both mitofusins (loss of red TMRE fluorescence producing green organelles on the merged image in Figure 5c). In Mfn1/Mfn2 DKO hearts the same phenomenon was revealed by a leftward shift in the DiOC6 fluorescence distribution curve of isolated cardiac mitochondria (Figure 5g), pointing to widespread mitochondrial dysfunction in fusion-defective organelles. Respiratory impairment (Figure 5h) and ROS production (Figure 5i) were more severe in the most fragmented mitochondria of Mfn1/Mfn2 DKO hearts. Taken together, the results depicted in Figures 4 and 5 show that conditional interruption of mitochondrial fission or fusion have parallel effects in MEFs and mouse hearts: mitochondrial fitness is maintained after ablating Drp1, but is severely compromised by combined mitofusin deficiency.
Figure 5. Conditional ablation of Mfn1/Mfn2 in cultured MEFs recapitulates mitochondrial phenotypes from Mfn DKO heart.
A–F, exactly as described for Figure 4 except using Mfn1/Mfn2 floxed allele MEFs and mice. G–I: white bar (black line) is ctrl, blue is 3 wks Mfn DKO (16,000g fragmented mitochondria) or 6 wks Mfn DKO (8,000g normal mitochondria), and red is 6 wks Mfn DKO (16,000g fragmented mitochondria). See also Figure S5.
Conditional ablation of Drp1 but not Mfn1 and Mfn2 stimulates MPTP-dependent mitochondrial autophagy in MEFs
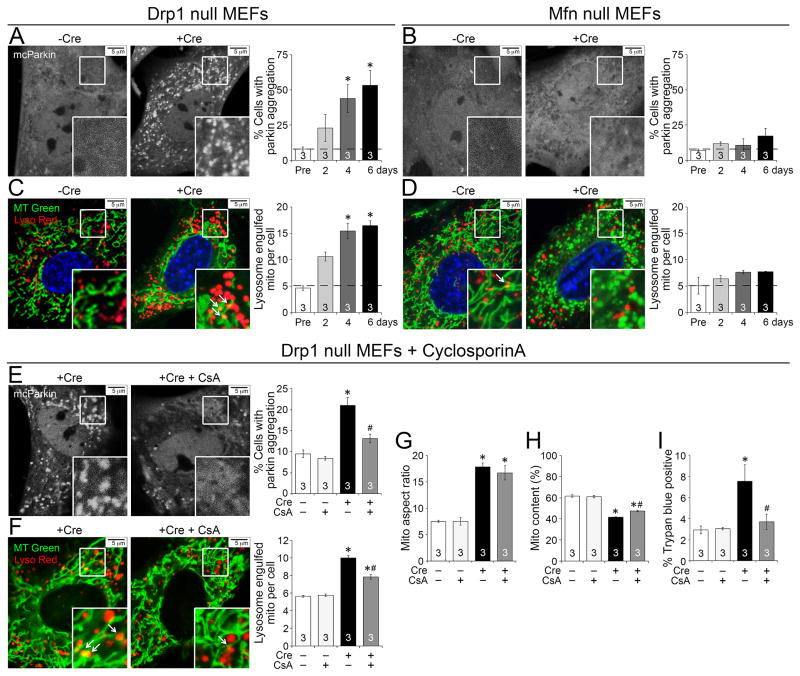
The above findings indicate conservation of cellular mechanisms in our in vivo and in vitro models. Accordingly, we used the conditional MEF ablation system to ask if the increase in mitochondrial autophagy markers in Drp1 null hearts increased parkin-mediated mitophagy and transfer to lysosomes (Youle and van der Bliek, 2012). In control MEFs, fluorescent mcherry-parkin (mcParkin) is homogenously distributed throughout the cytosol (Figures 6a, 6b, −Cre), but mcParkin spontaneously cluistered at cell mitochondria in MEFs after Drp1 gene deletion (Figure 6a, +Cre) (Narendra et al., 2008). Concomitantly, mitochondrial engulfment into lysosomes increased (Figure 6c). Moreover, we observed mitochondria inside cardiomyocyte autophagosomes in Drp1 null hearts (Supplemental Figure 6). By contrast, mcParkin clumping and lysosomal incorporation of mitochondria were rare in Mfn1/Mfn2-deficient MEFs (Figures 6b, 6d).
Figure 6. Conditional ablation of Drp1 in MEFs increases MPTP-dependent mitochondrial autophagy.
A–B. Parkin aggregation in conditional Drp1 KO (A) or Mfn DKO (B) MEFs infected with adeno-mcherry-parkin (mcParkin) before and 6 days after adeno-Cre mediated gene ablation. Quantitative group data are to the right. C–D. Confocal co-localization of lysosomes (LysoTracker Red) and mitochondria (MitoTracker Green) in conditional Drp1 KO (C) or Mfn DKO (D) MEFs before and 6 days after adeno-Cre mediated gene ablation. Mitochondria engulfed in lysosomes appear yellow (arrows). E–F. Parkin aggregation (E) and co-localization of lysosome and mitochondria (F) in conditional Drp1 KO MEFs in the absence or presence of 2 μM cyclosporinA (CsA) at 2 days after adeno-Cre. Quantitative data are to the right. G–H. Group data for mitochondria aspect ratio (G) and content (H) of Drp1 KO MEFs in the absence or presence of CsA. I. Cell membrane permeability assessed by Trypan blue exclusion in Drp1 KO MEFs in the absence or presence of CsA. Horizontal dashed line represents normal value from Cre-negative controls. Data are mean ± SEM of indicated number of experiments; * = P<0.05 vs identically treated Cre-negative controls; # = P<0.05 vs conditional Drp1 KO MEFs (no CsA). See also Figure S6.
Mitophagy in cultured cardiac myocytes can be stimulated by opening of mitochondrial permeability transition pores (MPTP) (Carreira et al., 2010). We assessed the consequences of inhibiting MPTP opening with cyclosporin A on Drp1-deficient MEFs; both mcParkin aggregation and mitochondrial engulfment by lysosomes were suppressed (Figure 6e, 6f). Although Drp1-deficient MEF mitochondria are still hyper-fused (Figure 6g), the loss of organelles and compromised membrane integrity were improved (Figure 6h, 6i). Thus, genetic interdiction of mitochondrial fission accelerates MPTP-dependent mitophagy and reduces overall mitochondrial content, but the mitochondria that are not culled are normal.
Inhibition of the mitochondrial permeability transition alleviates cardiomyocyte death and mitochondrial loss induced by cardiac Drp1 deficiency
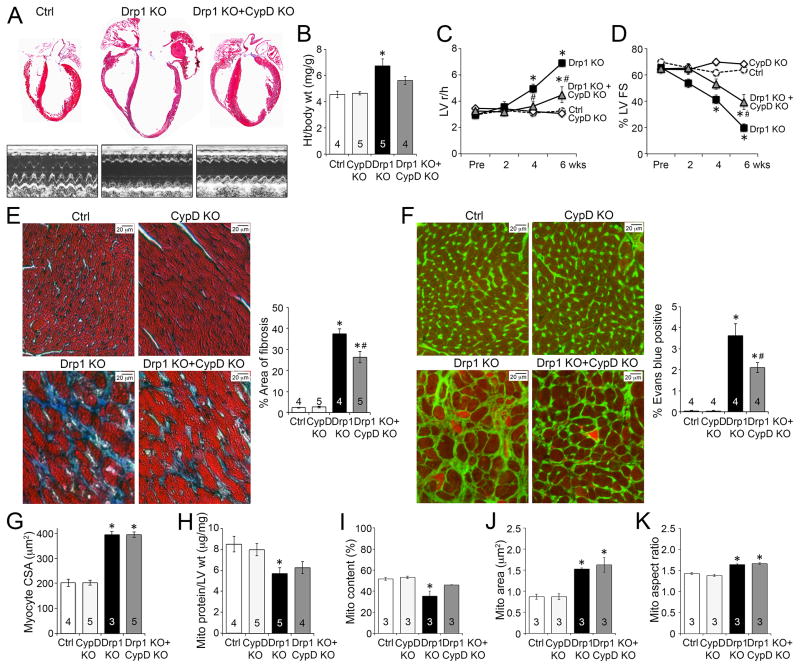
We examined the role of MPTP activation in the dilated cardiomyopathy of Drp1-deficient hearts by concomitantly deleting Drp1 and the ppif gene (ppif encodes the MPTP regulatory protein, cyclophilin D) (Baines et al., 2005). Because the genetic background differed slightly in the ppif crosses, we performed new sets of conditional cardiac Drp1 deletion experiments as matched littermate controls. As in our initial studies (see Figure 1), conditional Drp1 ablation provoked a progressive dilated cardiomyopathy over 6 weeks (Figures 7a–d). By itself, ablation of ppif had no effect on any measured cardiac endpoint (Figure 7). However, ppif ablation diminished cardiac enlargement (Figures 7a, 7b), protected against adverse ventricular remodeling (r/h; Figure 7c), and improved contractile dysfunction (Figure 7d) in cardiac Drp1 null mice. At the cellular level, ppif ablation in Drp1 null hearts reduced myocardial fibrosis and cardiomyocyte necrosis, without affecting cardiomyocyte size (Figure 7e, 7f, 7g). Likewise, mitochondrial content was improved (Figure 7h, 7i), but the characteristic elongated mitochondrial morphology was not affected (Figure 7j, 7k) by ppif ablation in Drp1 null hearts. We have previously shown that ppif ablation does not protect against cardiomyopathy in Mfn2 insufficiency, consistent with absence of cardiomyocyte necrosis in that model (Song et al., 2014).
Figure 7. Inhibition of MPTP opening improves the cardiomyopathy provoked by conditional Drp1 ablation.
A. Representative four-chamber view and echocardiographic images of hearts 6weeks after Drp1 gene deletion in the presence or absence of MPTP regulatory protein, cyclophilin D (CypD). B. Ratio of heart weight to body weight. C–D. Echocardiographic LV r/h (C) and %FS (D). E. Masson’s trichrome-staining of myocardial fibrosis; group data are to the right. F. Fluorescence microscopy of Evans blue-perfused hearts co-stained with wheat germ agglutinin; quantitative group data are to the right. G. Group quantitative data for cardiomyocyte cross-sectional area (CSA). H. Ratio of mitochondrial protein to LV weight. I–K. Group data for myocardial mitochondrial content (I), individual mitochondrial area (J), and mitochondrial aspect ratio (K) derived from TEM images. Data are mean ± SEM; * = P<0.05 vs identically treated controls (Drp1 f/f ± CypD KO); # = P<0.05 vs Drp1 KO.
Discussion
These comparative studies of mitochondrial fission- and fusion-defective hearts and cells show that Drp1-mediated mitochondrial fission is essential to: 1. properly target mitochondrial autophagy and 2. restrain MPTP-mediated cell necrosis. As we uncovered similar effects in cultured fibroblasts and in vivo mouse hearts, these may be general functions of Drp1 in mitochondrial quality control. The pathophysiological consequences of Drp1 deficiency in adult mouse hearts were striking, with progressive chamber enlargement and functional decompensation leading to fulminant and ultimately lethal heart failure. Evidence for widespread cardiomyocyte necrosis induced by Drp1 ablation, but not by Mfn1/Mfn2 ablation, revealed MPTP activation in fission-defective, but not fusion-defective, cells. Indeed, inhibiting MPTP favorably impacted the cellular and cardiac phenotypes evoked by Drp1 ablation, without affecting the underlying mitochondrial elongation. Thus, mitochondrial morphology per se does not appear to be the driving force behind cardiac dysfunction and cardiomyocyte death. Rather, hyper-fused mitochondria in Drp1 deficiency were associated with opening of MPTP (Szabadkai et al., 2004), which can both activate mitophagy (Carreira et al., 2010) and induce cell necrosis (Baines et al., 2005). Additionally, these studies support the proposed roles of mitochondrial fission in quality control and cell death pathways (Twig et al., 2008; Whelan et al., 2012). We believe these findings warrant a re-evaluation of the assumption that mitochondrial fusion is inherently beneficial and mitochondrial fission is inherently detrimental (Ong et al., 2010), and that mitochondrial fission is a pre-requisite for mitophagy (Twig et al., 2008).
By comparing conditional Drp1 null hearts and MEFs to conditional Mfn1/Mfn2 DKO hearts and cells in a unique side-by-side multi-platform comparison, we generated insights into the role of both fusion and fission. We previously showed that cardiomyopathy develops after combined Mfn1/Mfn2 deletion in adult hearts, but the underlying mechanism was not identified (Chen et al., 2011). Our objective in that study was simply to determine if mitochondrial fusion occurs in adult hearts, and the only abnormalities we described were mitochondrial fragmentation and impaired respiration in isolated cardiomyocytes; the respiratory defect in cardiomyocytes, but not in isolated mitochondria, was unexplained (Chen et al., 2011). Here, we have resolved that paradox: conventional differential centrifugation techniques we originally used to isolate cardiac mitochondria (Frezza et al., 2007) did not efficiently capture the fragmented mitochondria produced by mitofusin ablation. Using a modified protocol we demonstrate here that the fragmented Mfn1/Mfn2-deficient mitochondria do indeed have a severe respiratory defect. Additionally, we newly demonstrate increased mitochondrial ROS production, loss of Δψm, and stimulation of the mitochondrial UPR in mitofusin-deficient mitochodria, all tied to a mitophagy defect.
An advantage of our conditional in vivo and in vitro systems is that the consequences of the genetic manipulation can be followed over time. Opportunistic compensation by functionally related factors that might be upregulated in response to gene ablation is also minimized by this approach. Our findings in hearts were largely recapitulated in MEFs, demonstrating that our findings are not relevant exclusively to cardiomyocytes. Thus, loss of mitochondrial fission is linked to enhanced MPTP opening, stimulation of mitophagy, and cell death by necrosis. By comparison, loss of mitochondrial fusion suppressed the normal mitophagic response to mitochondrial damage, likely explaining the associated accumulation of toxic mitochondria.
Mitophagy helps to maintain the overall health of the cellular mitochondrial pool by selectively identifying and eliminating damaged or dysfunctional organelles. Mitophagy was first mechanistically connected with mitochondrial fission by Shirihai’s group, who observed that asymmetric fission of modestly damaged (i.e. partially depolarized) mitochondria produced one healthy (hyperpolarized) daughter to join the functional pool, and one depolarized daughter specifically targeted for mitophagic elimination (Twig et al., 2008) (Supplemental Figure 7a). According to this schema, mitochondrial fission normally precedes mitophagy. Here, we discovered what happens when fission is suppressed. Without the ability to segregate the healthy and corrupted components of a moderately damaged organelle through asymmetric fission, the mitochondrion continues to degrade until it reaches the damage threshold activating mitophagy, which eliminates the senescent parent organelle without producing a healthy daughter to be restored to the cellular pool. In Drp1-deficient hearts wherein mitophagy is increased and mitochondria are depleted, the organelles that remain are not markedly dysfunctional (Supplemental Figure 7b). As a consequence, there is little evidence for accumulation of abnormal mitochondrial proteins (the UPRmt) until death from heart failure is imminent. Instead, Fgf21 is increased in a manner that recapitulates metabolic (or in this case mitochondrial) insufficiency (Tyynismaa et al., 2010).
Mitochondrial morphology can affect the spatiotemporal properties of mitochondrial Ca2+ signaling. Thus, intra-mitochondrial Ca2+ waves that stimulate MPTP opening (Baumgartner et al., 2009) diffuse more efficiently in fused mitochondria, a process that is interrupted by Drp1-mediated mitochondrial fission (Szabadkai et al., 2004). In our Drp1-deficient hearts and MEFs, inhibition of Drp1-mediated fission produced elongated mitochondria with increased MPTP opening (Szabadkai et al., 2004) (Supplemental Figure 7b). Thus, pharmacological MPTP inhibition in Drp1 null MEFs prevented parkin aggregation, decreased lysosomal engulfment of mitochondria, and increased mitochondrial content, while genetic MPTP ablation in Drp1 null hearts alleviated both the cardiomyopathy and accompanying cardiomyocyte necrosis.
Mitochondrial fusion is also connected with mitophagy. Youle’s group first described how loss of the normal highly polarized electrochemical gradient across mitochondrial inner membranes (mitochondrial depolarization) stabilizes the kinase PINK1, which recruits the E3 ubiquitin ligase parkin (Narendra et al., 2010b). These two Parkinson’s disease factors constitute a critical regulatory switch enabling selective elimination of damaged mitochondria. Recently, we discovered that, in hearts, Mfn2 acts as the mitochondrial receptor for parkin when it is phosphorylated by PINK, mechanistically linking fusion with mitophagy (Chen and Dorn, 2013). Potent induction of the UPRmt just 3 weeks after Mfn1/Mfn2 ablation, when cardiac abnormalities are modest and mitochondrial mass is increased, provides additional support for a contributory role of impaired mitochondrial quality control in this phenotype (Supplemental Figure 7c).
We observed evidence of decreased mitochondrial biogenesis in both loss of mitochondrial fission and loss of mitochondrial fusion. While the specific processes by which mitochondria communicate with the nucleus to activate biogenic gene expression have proven elusive, our results and prior studies are noteworthy because they show that an intact fission-fusion cycle is important for normal biogenesis (Caffin et al., 2013; Kim, 2013; Papanicolaou et al., 2012). This is consistent with biogenesis functioning as a pipeline for replacement components to be incorporated into pre-existing mitochondria as part of the normal renewal mechanism (as when the healthy daughter organelle of a fission event is re-integrating into the generally healthy mitochondrial pool), rather than as a means by which mitochondria are generated de-novo. Indeed, we have previously suggested that there are no de-novo mitochondria (Chen et al., 2013). Instead, as endosymbionts, mitochondria retain central features of the life cycles shared with their protobacterial ancestors who invaded unicellular organisms over a billion years ago (Gray et al., 1999; Yang et al., 1985). Mitochondria accumulate and synthesize protein and lipid components (which we call biogenesis), they exchange genetic information through fusion, and parent mitochondria replicate by undergoing fission into daughter organelles. We envision that mitochondrial homeostasis depends upon balanced organelle fission that segregates normal and impaired components into daughter organelles, mitophagy that selectively removes damaged organelles, and biogenesis and fusion that function in concert to repair, re-grow, and reintroduce healthy daughter mitochondria into the general cellular pool.
In our studies we tried to minimize potential opportunistic compensation for deleted mitochondrial fission and fusion factors by employing temporally-controlled in vivo and in vitro systems, and assessing the consequences over time. This experimental detail distinguishes our work from the recent report by Kageyama, et al (Kageyama Y, 2014), which used a version of the Drp1 cardiac knockout model we elected not to use because of many confounding factors (see Supplemental Figure 1). Although it remains to be formally shown that increased mitophagy is the proximal cause of the cardiomyopathy induced by conditional cardiac Drp1 deficiency, our data offer a possible explanation for why mitochondrial dynamics factors are highly expressed in adult cardiac myocytes, wherein structural mitochondrial remodeling is so rare that it has never been directly observed (Dorn, 2013a). Cardiomyocyte mitochondria are normally static until, through damage or senescence, they are unable to maintain a normal Δψm and partially depolarize. Mitochondrial depolarization is a potent stimulus for Drp1-mediated asymmetric organelle fission that separates and segregates damaged from healthy mitochondrial components (Twig et al., 2008). Since mitochondrial dysfunction is a continuum, but the decision to mitophagically eliminate a damaged organelle is binary, preventing the segregation of healthy from damaged mitochondrial components by interrupting Drp1-mediated fission has consequences that evolve over time. Initially, healthy daughter mitochondria are not generated to renew the mitochondrial pool, and fully depolarized daughter mitochondria are not generated to be disposed of via mitophagy. Since mitochondrial fusion is uninterrupted, damaged mitochondria are able to fuse with their normal counterparts, accelerating cell-wide mitochondrial dysfunction (Bhandari et al., 2014). Later, as the phenotype progresses, an increasing proportion of mitochondria reach the threshold for mitophagy activation and are eliminated. The ultimate effect of removing mitochondria that cannot be repaired would therefore be mitochondrial depletion, as observed. In the reciprocal condition, where mitochondrial fusion is suppressed by combined Mfn1/Mfn2 deletion, two concomitant events appear to interrupt normal mitochondrial quality control mechanisms and result in accumulation of toxic, ROS-producing mitochondria. First, Mfn2 ablation impairs mitophagy signaling independent of mitochondrial fusion (Chen and Dorn, 2013), preventing the normal elimination of damaged mitochondria. Second, the organelle fusion renewal mechanism for re-introduction of the healthy daughter mitochondria produced by asymmetric organelle fission cannot occur in the absence of mitofusins.
In summary, we propose that mitochondrial fission performs a critical triage function by which healthy and damaged components are segregated; fission may also regulate mitochondrial morphology to protect against MPTP-mediated cell death. On the other hand, we conceive of mitochondrial fusion as a mechanism for mitochondrial repair and regeneration. When fission-mediated triage and fusion-mediated repair are intact, impaired organelles are properly targeted for elimination, and healthy organelles are renewed. When fission-mediated organelle triage is interrupted however, fusion changes from a regenerative process to a contaminating one.
Experimental Procedures
More detailed methods are described in the Supplemental Experimental Procedures.
Mouse Generation and Phenotypic Analyses
Drp1loxp/loxp (Ishihara et al., 2009), Mfn1loxp/loxp and Mfn2loxp/loxp (Chen et al., 2003; Chen et al., 2007), and ppif null (Baines et al., 2005) mice have been described. Drp1loxp/loxp mice were crossed onto the myh6-nuclear-directed Cre transgenic line (Oka et al., 2006) for cardiomyocyte-specific gene deletion after birth. For conditional cardiac ablation, floxed mouse lines were crossed onto the myh6-MER-Cre-Mer mice; gene recombination was induced by intraperitoneal (i.p.) administration of tamoxifen (Sohal et al., 2001). Weekly M-mode echocardiography was performed on unsedated mice (Chen et al., 2011); controls were sex- and age-matched Drp1loxp/loxp± ppif−/− littermates, Mfn1loxp/loxp+Mfn2loxp/loxp littermates and myh6-MER-Cre-Mer mice with or without tamoxifen administration. All experimental procedures were approved by the Washington University Institutional Animal Care and Use Committee.
Western Blotting
Myocardial homogenates and mitochondrial fractions were prepared and protein expression levels were measured by standard western blotting (Chen et al., 2010).
Histological Studies
Hematoxylin-eosin stain, Masson’s trichrome stain, TUNEL assay, and immunofluorescent staining were performed on sections from formalin-fixed, paraffin-embedded tissues. Mice received 0.01 ml/g 1% Evans blue dye solution (in PBS) i.p. ~24 h prior to tissue sampling (Hamer et al., 2002). Evans blue positive cardiomyocytes fluoresced in red.
Antibodies
For western blotting, primary antibodies against Drp1 (1:1000, ab56788), Mfn1 (1:500, ab57602), Mfn2 (1:1000, ab56889), GAPDH (1:3000, ab8245), p62 (1:1000, ab56416), LC3 I/II (1:1000, ab128025), Fgf21 (1:1000, ab171941), LONP1 (1:500, ab103809), Hsp60 (1:2000, ab46798), and OXPHOS (a premixed cocktail of antibodies against CI-NDUFB8, CII-SDHB, CIII-UQCRC2, CIV-MTCO1 and CV-ATP5A, 1:500, MS604) were from Abcam. Primary antibody against AFG3L2 (1:500, sc-84687) was from Santa Cruz Biotechnology. Horseradish peroxidase (HRP) linked secondary antibodies anti-mouse IgG (1:3000, cs7076) and anti-rabbit IgG (1:3000, cs7074) were from Cell Signaling Technology.
For immunohistochemistry, primary antibodies against C5b-9 (1:200, ab55811) and heavy chain cardiac Myosin (1:200, ab15) were from Abcam. Alexa Fluor conjugated secondary antibodies anti-mouse IgG (1:400, A-11029) and anti-rabbit IgG (1:400, A-11035) were from Invitrogen.
Transmission Electron Microscopy
Ultrastructural examination of osmium tetroxide/uranyl acetate stained mouse heart thin sections (90 nm) used a Jeol electron microscope (JEM-1400) at 1,500x - 30,000x direct magnifications (JEOL, Tokyo, Japan). Mitochondrial content was measured as the areas taken by mitochondria compared to those of the cardiomyocytes using ImageJ.
Isolation of Cardiac Mitochondria
Isolation of cardiac mitochondria used a modification of the standard published protocol (Frezza et al., 2007). Briefly, mouse hearts were collected, minced, and incubated with trypsin before homogenization with a glass/teflon Potter Elvehjem homogenizer. Heart homogenates were centrifuged at 800 g x 10 min at 4 °C and the supernatant collected and centrifuged at 8,000 g x 10 min at 4 °C; both the pellet and the supernatant were collected. The pellet was again washed and centrifuged at 8,000 g x 10 min at 4 °C to obtain normal-size mitochondria, which were resuspended for analyses. The 8,000 g supernatant was centrifuged at 16,000 g x 10 min at 4 °C to pellet smaller fragmented mitochondria.
Mitochondrial Respiration
A micro Clark-type electrode was used in a closed, magnetically stirred glass chamber as previously described (Chen et al., 2011). Non-stimulated (-ADP) respiration and stimulated (+ADP) respiration were measured, indicated as state 2 and state 3, respectively. Following ADP depletion, oligomycin was added to check proton leak. Carbonyl cyanide 4-(trifluoromethoxy) phenylhydrazone (FCCP) was added to measure maximal uncoupled respiration.
Determination of Mitochondrial H2O2 Production
H2O2 production of isolated cardiac mitochondria was determined using Amplex Red hydrogen peroxide/peroxidase assay kit (Song et al., 2014).
Flow Cytometric Analyses of Isolated Mitochondria
Isolated cardiac mitochondria were stained with 200 nM MitoTracker Green, 2.5 μM MitoSOX red, or 100 nM 3,3′-dihexyloxacarbocyanine iodide (DiOC6) at room temperature for 20 min and washed twice with PBS. Flow cytometric analyses of mitochondrial size (forward scatter, FSC), mitochondrial superoxide level (MitoSOX red signal intensity detected by PE channel), or mitochondrial membrane potential (DiOC6 signal intensity detected by FITC channel) were performed on a BD LSR II Flow Cytometer (Becton Dickinson, San Jose, CA). Data are shown as histograms for, and as bar graph of average signal intensity of, 50,000 ungated events.
Cell Culture
Drp1 KO MEFs and Mfn1/Mfn2 DKO MEFs were generated from Drp1loxp/loxp and Mfn1loxp/loxp+Mfn2loxp/loxp embryos, respectively. MEFs were maintained in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum, 1x non-essential amino acids, 2 mM L-glutamine, 100 U/ml penicillin and 100 μg/ml streptomycin.
Adenovirus Infection and Cyclosporin A Treatment
Cre adenovirus was from Vector Biolabs. Adeno-mcherry parkin was a gift from Dr. Åsa Gustafsson, University of California San Diego. MEFs were passaged every 2 days and plated on 25 mm glass round coverslips at a density of 1.0×105 cells per well in 6-well plates. Adeno-Cre was added after cell attaching at a multiplicity of infection (MOI) of 100 at day 0 and kept in the medium for 48 hours; adeno-mcherry-parkin (MOI of 20) and/or cyclosporin A (CsA, 2 μM) were added at 48 hours prior to imaging.
Confocal Microscopy
Live cell imaging used a Nikon Ti confocal microscope equipped with a 60x 1.3NA oil immersion objective. For visualization of mitochondria and measurement of mitochondrial membrane potential, MEFs were stained with 200 nM MitoTracker Green, 200 nM of tetramethylrhodamine, ethyl ester (TMRE) and 10 μg/ml Hoechst at 37 °C for 30 min. Fluorescence was excited with a 405 nm laser diode (Hoechst), a 488 nm Argon laser (MitoTracker Green) and a 543 nm HeNe laser (TMRE). For detection of parkin aggregation, MEFs were stained with MitoTracker Green and Hoechst; fluorescence for mcherry-parkin was excited with a 543 nm HeNe laser. For assessment of lysosomal engulfed mitochondria, MEFs were stained with 50 nM LysoTracker Red, MitoTracker Green and Hoechst; fluorescence for LysoTracker Red was excited with a 543 nm HeNe laser.
Image Analysis
Mitochondrial aspect ratio (the ratio of length/width) and content (% of mitochondrial area compared to whole-cell area) was quantified using ImageJ. Mitochondrial depolarization was calculated as % of the area of green mitochondria compared to that of all the mitochondria visualized on MitoTracker Green and TMRE merged images; data are presented as green ÷ (green + yellow mitochondria) × 100%. Parkin aggregation was calculated as % of cells with clumping mcherry-parkin compared to all the cells. Lysosomal engulfment of mitochondria was calculated by counting the number of co-localized lysosomes and mitochondria per cell detected by confocal co-localization of LysoTracker Red and MitoTracker Green.
RNA Expression Analysis
RNA expression analysis was performed using Taqman qRT-PCR master mix with predesigned primer/probes sets.
Statistical Analysis
Data represent mean ± SEM of representative experiments, unless otherwise stated. Statistical comparisons used unpaired Student’s t-test or one-way ANOVA with Tukey’s corrections, as appropriate. P < 0.05 was considered statistically significant.
Supplementary Material
Acknowledgments
Supported by NIH HL59888 (GWD), an American Heart Association predoctoral fellowship award 14PRE18970093 (MS), and by Telethon GGP06254A, GPP10005B (LS). TEM was performed with assistance of Marilyn Levy, Department of Cell Biology, Washington University.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Baines CP, Kaiser RA, Purcell NH, Blair NS, Osinska H, Hambleton MA, Brunskill EW, Sayen MR, Gottlieb RA, Dorn GW, 2nd, et al. Loss of cyclophilin D reveals a critical role for mitochondrial permeability transition in cell death. Nature. 2005;434:658–662. doi: 10.1038/nature03434. [DOI] [PubMed] [Google Scholar]
- Baumgartner HK, Gerasimenko JV, Thorne C, Ferdek P, Pozzan T, Tepikin AV, Petersen OH, Sutton R, Watson AJ, Gerasimenko OV. Calcium elevation in mitochondria is the main Ca2+ requirement for mitochondrial permeability transition pore (mPTP) opening. J Biol Chem. 2009;284:20796–20803. doi: 10.1074/jbc.M109.025353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhandari P, Song M, Chen Y, Burelle Y, Dorn GW., 2nd Mitochondrial contagion induced by Parkin deficiency in Drosophila hearts and its containment by suppressing mitofusin. Circ Res. 2014;114:257–265. doi: 10.1161/CIRCRESAHA.114.302734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caffin F, Prola A, Piquereau J, Novotova M, David DJ, Garnier A, Fortin D, Alavi MV, Veksler V, Ventura-Clapier R, et al. Altered skeletal muscle mitochondrial biogenesis but improved endurance capacity in trained OPA1-deficient mice. J Physiol. 2013;591:6017–6037. doi: 10.1113/jphysiol.2013.263079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carreira RS, Lee Y, Ghochani M, Gustafsson AB, Gottlieb RA. Cyclophilin D is required for mitochondrial removal by autophagy in cardiac cells. Autophagy. 2010;6:462–472. doi: 10.4161/auto.6.4.11553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Detmer SA, Ewald AJ, Griffin EE, Fraser SE, Chan DC. Mitofusins Mfn1 and Mfn2 coordinately regulate mitochondrial fusion and are essential for embryonic development. J Cell Biol. 2003;160:189–200. doi: 10.1083/jcb.200211046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, McCaffery JM, Chan DC. Mitochondrial fusion protects against neurodegeneration in the cerebellum. Cell. 2007;130:548–562. doi: 10.1016/j.cell.2007.06.026. [DOI] [PubMed] [Google Scholar]
- Chen Y, Csordas G, Jowdy C, Schneider TG, Csordas N, Wang W, Liu Y, Kohlhaas M, Meiser M, Bergem S, et al. Mitofusin 2-containing mitochondrial-reticular microdomains direct rapid cardiomyocyte bioenergetic responses via interorganelle Ca2+ crosstalk. Circ Res. 2012;111:863–875. doi: 10.1161/CIRCRESAHA.112.266585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Dorn GW., 2nd PINK1-phosphorylated mitofusin 2 is a Parkin receptor for culling damaged mitochondria. Science. 2013;340:471–475. doi: 10.1126/science.1231031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Lewis W, Diwan A, Cheng EH, Matkovich SJ, Dorn GW., 2nd Dual autonomous mitochondrial cell death pathways are activated by Nix/BNip3L and induce cardiomyopathy. Proc Natl Acad Sci USA. 2010;107:9035–9042. doi: 10.1073/pnas.0914013107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Liu Y, Dorn GW., 2nd Mitochondrial fusion is essential for organelle function and cardiac homeostasis. Circ Res. 2011;109:1327–1331. doi: 10.1161/CIRCRESAHA.111.258723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Sparks M, Bhandari P, Matkovich SJ, Dorn GW., 2nd Mitochondrial Genome Linearization Is a Causative Factor for Cardiomyopathy in Mice and Drosophila. Antioxid Redox Signal. 2014;21:1949–1959. doi: 10.1089/ars.2013.5432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Disatnik MH, Ferreira JC, Campos JC, Gomes KS, Dourado PM, Qi X, Mochly-Rosen D. Acute inhibition of excessive mitochondrial fission after myocardial infarction prevents long-term cardiac dysfunction. J Am Heart Assoc. 2013;2:e000461. doi: 10.1161/JAHA.113.000461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diwan A, Dorn GW., 2nd Decompensation of cardiac hypertrophy: cellular mechanisms and novel therapeutic targets. Physiology (Bethesda) 2007;22:56–64. doi: 10.1152/physiol.00033.2006. [DOI] [PubMed] [Google Scholar]
- Diwan A, Wansapura J, Syed FM, Matkovich SJ, Lorenz JN, Dorn GW., 2nd Nix-mediated apoptosis links myocardial fibrosis, cardiac remodeling, and hypertrophy decompensation. Circulation. 2008;117:396–404. doi: 10.1161/CIRCULATIONAHA.107.727073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorn GW., 2nd Mitochondrial dynamics in heart disease. Biochim Biophys Acta. 2013a;1833:233–241. doi: 10.1016/j.bbamcr.2012.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorn GW., 2nd Mitochondrial dynamism and cardiac fate. Circ J. 2013b;77:1370–1379. doi: 10.1253/circj.cj-13-0453. [DOI] [PubMed] [Google Scholar]
- Frank M, Duvezin-Caubet S, Koob S, Occhipinti A, Jagasia R, Petcherski A, Ruonala MO, Priault M, Salin B, Reichert AS. Mitophagy is triggered by mild oxidative stress in a mitochondrial fission dependent manner. Biochim Biophys Acta. 2012;1823:2297–2310. doi: 10.1016/j.bbamcr.2012.08.007. [DOI] [PubMed] [Google Scholar]
- Frezza C, Cipolat S, Scorrano L. Organelle isolation: functional mitochondria from mouse liver, muscle and cultured fibroblasts. Nat Protoc. 2007;2:287–295. doi: 10.1038/nprot.2006.478. [DOI] [PubMed] [Google Scholar]
- Grasl-Kraupp B, Ruttkay-Nedecky B, Koudelka H, Bukowska K, Bursch W, Schulte-Hermann R. In situ detection of fragmented DNA (TUNEL assay) fails to discriminate among apoptosis, necrosis, and autolytic cell death: a cautionary note. Hepatology. 1995;21:1465–1468. doi: 10.1002/hep.1840210534. [DOI] [PubMed] [Google Scholar]
- Gray MW, Burger G, Lang BF. Mitochondrial evolution. Science. 1999;283:1476–1481. doi: 10.1126/science.283.5407.1476. [DOI] [PubMed] [Google Scholar]
- Hamer PW, McGeachie JM, Davies MJ, Grounds MD. Evans Blue Dye as an in vivo marker of myofibre damage: optimising parameters for detecting initial myofibre membrane permeability. J Anat. 2002;200:69–79. doi: 10.1046/j.0021-8782.2001.00008.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishihara N, Nomura M, Jofuku A, Kato H, Suzuki SO, Masuda K, Otera H, Nakanishi Y, Nonaka I, Goto Y, et al. Mitochondrial fission factor Drp1 is essential for embryonic development and synapse formation in mice. Nat Cell Biol. 2009;11:958–966. doi: 10.1038/ncb1907. [DOI] [PubMed] [Google Scholar]
- Kageyama Y, Hoshijima M, Seo K, Bedja D, Sysa-Shah P, Andrabi SA, Chen W, Höke A, Dawsn VL, Dawson TM, et al. Parkin-independent mitophagy requires Drp1 and maintains the integrity of mammalian heart and brain. EMBO J. 2014;e2:01488658. doi: 10.15252/embj.201488658. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim B, Kim JS, Yoon Y, Santiago MD, Brown MD, Park JY. Inhibition of Drp1-dependent mitochondrial division impairs myogenic differentiation. Am J Physiol Regul Integr Comp Physiol. 2013;305:R927–938. doi: 10.1152/ajpregu.00502.2012. [DOI] [PubMed] [Google Scholar]
- Martinelli P, La Mattina V, Bernacchia A, Magnoni R, Cerri F, Cox G, Quattrini A, Casari G, Rugarli EI. Genetic interaction between the m-AAA protease isoenzymes reveals novel roles in cerebellar degeneration. Hum Mol Genet. 2009;18:2001–2013. doi: 10.1093/hmg/ddp124. [DOI] [PubMed] [Google Scholar]
- Narendra D, Kane LA, Hauser DN, Fearnley IM, Youle RJ. p62/SQSTM1 is required for Parkin-induced mitochondrial clustering but not mitophagy; VDAC1 is dispensable for both. Autophagy. 2010a;6:1090–1106. doi: 10.4161/auto.6.8.13426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narendra D, Tanaka A, Suen DF, Youle RJ. Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J Cell Biol. 2008;183:795–803. doi: 10.1083/jcb.200809125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narendra DP, Jin SM, Tanaka A, Suen DF, Gautier CA, Shen J, Cookson MR, Youle RJ. PINK1 is selectively stabilized on impaired mitochondria to activate Parkin. PLoS Biol. 2010b;8:e1000298. doi: 10.1371/journal.pbio.1000298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngo JK, Davies KJ. Importance of the lon protease in mitochondrial maintenance and the significance of declining lon in aging. Ann N Y Acad Sci. 2007;1119:78–87. doi: 10.1196/annals.1404.015. [DOI] [PubMed] [Google Scholar]
- Oka T, Maillet M, Watt AJ, Schwartz RJ, Aronow BJ, Duncan SA, Molkentin JD. Cardiac-specific deletion of Gata4 reveals its requirement for hypertrophy, compensation, and myocyte viability. Circ Res. 2006;98:837–845. doi: 10.1161/01.RES.0000215985.18538.c4. [DOI] [PubMed] [Google Scholar]
- Ong SB, Subrayan S, Lim SY, Yellon DM, Davidson SM, Hausenloy DJ. Inhibiting mitochondrial fission protects the heart against ischemia/reperfusion injury. Circulation. 2010;121:2012–2022. doi: 10.1161/CIRCULATIONAHA.109.906610. [DOI] [PubMed] [Google Scholar]
- Papanicolaou KN, Kikuchi R, Ngoh GA, Coughlan KA, Dominguez I, Stanley WC, Walsh K. Mitofusins 1 and 2 are essential for postnatal metabolic remodeling in heart. Circ Res. 2012;111:1012–1026. doi: 10.1161/CIRCRESAHA.112.274142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SJ, Shin JH, Kim ES, Jo YK, Kim JH, Hwang JJ, Kim JC, Cho DH. Mitochondrial fragmentation caused by phenanthroline promotes mitophagy. FEBS Lett. 2012;586:4303–4310. doi: 10.1016/j.febslet.2012.10.035. [DOI] [PubMed] [Google Scholar]
- Rambold AS, Kostelecky B, Elia N, Lippincott-Schwartz J. Tubular network formation protects mitochondria from autophagosomal degradation during nutrient starvation. Proc Natl Acad Sci USA. 2011;108:10190–10195. doi: 10.1073/pnas.1107402108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohal DS, Nghiem M, Crackower MA, Witt SA, Kimball TR, Tymitz KM, Penninger JM, Molkentin JD. Temporally regulated and tissue-specific gene manipulations in the adult and embryonic heart using a tamoxifen-inducible Cre protein. Circ Res. 2001;89:20–25. doi: 10.1161/hh1301.092687. [DOI] [PubMed] [Google Scholar]
- Song M, Chen Y, Gong G, Murphy E, Rabinovitch PS, Dorn GW., 2nd Super-suppression of mitochondrial reactive oxygen species signaling impairs compensatory autophagy in primary mitophagic cardiomyopathy. Circ Res. 2014;115:348–353. doi: 10.1161/CIRCRESAHA.115.304384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabadkai G, Simoni AM, Chami M, Wieckowski MR, Youle RJ, Rizzuto R. Drp-1-dependent division of the mitochondrial network blocks intraorganellar Ca2+ waves and protects against Ca2+-mediated apoptosis. Mol Cell. 2004;16:59–68. doi: 10.1016/j.molcel.2004.09.026. [DOI] [PubMed] [Google Scholar]
- Twig G, Elorza A, Molina AJ, Mohamed H, Wikstrom JD, Walzer G, Stiles L, Haigh SE, Katz S, Las G, et al. Fission and selective fusion govern mitochondrial segregation and elimination by autophagy. EMBO J. 2008;27:433–446. doi: 10.1038/sj.emboj.7601963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyynismaa H, Carroll CJ, Raimundo N, Ahola-Erkkila S, Wenz T, Ruhanen H, Guse K, Hemminki A, Peltola-Mjosund KE, Tulkki V, et al. Mitochondrial myopathy induces a starvation-like response. Hum Mol Genet. 2010;19:3948–3958. doi: 10.1093/hmg/ddq310. [DOI] [PubMed] [Google Scholar]
- Whelan RS, Konstantinidis K, Wei AC, Chen Y, Reyna DE, Jha S, Yang Y, Calvert JW, Lindsten T, Thompson CB, et al. Bax regulates primary necrosis through mitochondrial dynamics. Proc Natl Acad Sci USA. 2012;109:6566–6571. doi: 10.1073/pnas.1201608109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang D, Oyaizu Y, Oyaizu H, Olsen GJ, Woese CR. Mitochondrial origins. Proc Natl Acad Sci US A. 1985;82:4443–4447. doi: 10.1073/pnas.82.13.4443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang JY, Yang WY. Bit-by-bit autophagic removal of parkin-labelled mitochondria. Nat Commun. 2013;4:2428. doi: 10.1038/ncomms3428. [DOI] [PubMed] [Google Scholar]
- Youle RJ, van der Bliek AM. Mitochondrial fission, fusion, and stress. Science. 2012;337:1062–1065. doi: 10.1126/science.1219855. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.