Summary
Brown adipose tissue (BAT) possesses the inherent ability to dissipate metabolic energy as heat through uncoupled mitochondrial respiration. An essential component of the mitochondrial electron transport chain is coenzyme Q (CoQ). While cells mostly synthesize CoQ endogenously, exogenous supplementation with CoQ has been successful as a therapy for patients with CoQ deficiency. However, which tissues depend on exogenous CoQ uptake as well as the mechanism by which CoQ is taken up by cells and the role of this process in BAT function is not well understood. Here we report that the scavenger receptor CD36 drives the uptake of CoQ by BAT and is required for normal BAT function. BAT from mice lacking CD36 displays CoQ deficiency, impaired CoQ uptake, hypertrophy, altered lipid metabolism, mitochondrial dysfunction, and defective non-shivering thermogenesis. Together, these data reveal an important new role for the systemic transport of CoQ to BAT and its function in thermogenesis.
Introduction
Classical brown adipose tissue (BAT) is a unique type of adipose tissue that is composed of adipocytes with multilocular lipid droplets and a large amount of mitochondria, making it a highly metabolically active organ that is responsible for nonshivering thermogenesis both in neonate and adult humans (Aherne and Hull, 1966; Cypess et al., 2009; Heaton, 1972; van Marken Lichtenbelt et al., 2009; Virtanen et al., 2009). The unabated growth of the obesity epidemic and associated metabolic diseases such as Type 2 diabetes reflects our current lack of efficient strategies for intervention and treatment of metabolic diseases (Zimmet et al., 2001). Because BAT possesses the inherent ability to dissipate metabolic energy as heat through uncoupled mitochondrial respiration, expanding BAT or enhancing its respiratory activity could be a strategy for therapeutic intervention.
In order to generate a high metabolic rate for heat production, brown adipocytes contain a large amount of mitochondria densely packed with cristae (Sell et al., 2004). These cells possess an unparalleled ability to generate heat due to the dissociation of electron transport chain respiration from ATP production through uncoupling protein 1 (UCP1) (Argyropoulos and Harper, 2002). Located in the inner mitochondrial membrane, UCP1 provides an alternative route of entry for protons, allowing them to bypass ATP synthase and in turn dissipate energy to generate heat (Argyropoulos and Harper, 2002). Brown adipocyte mitochondria also display an elevated capacity for substrate utilization, relying predominantly on the β-oxidation of long chain fatty acids (LCFA) (Sell et al., 2004). LCFA can be taken up from the circulation via fatty acid transport proteins (FATPs) or can be generated from endocytosed lipoproteins through a process mediated in part by the scavenger receptor CD36 (Anderson and Stahl, 2013; Coburn et al., 2001; Kuniyasu et al., 2002; Zeng et al., 2003).
The scavenger receptor (SR) family of transmembrane glycoproteins mediates the binding and uptake of a broad range of ligands in a variety of tissues (Greaves and Gordon, 2009). The SR family is comprised of several classes, SR-A through H (Pluddemann et al., 2007). SR class B receptors are unique from other classes of SRs in that they have two transmembrane domains, an extracellular ligand binding loop, and two short intracellular tails (Pluddemann et al., 2007). A defining member of the SR class B receptors is CD36, an 88kDa scavenger receptor with multiple binding pockets and hydrophobic segments (Su and Abumrad, 2009). CD36 has been shown to be required in a variety of tissues for the uptake of several hydrophobic molecules, including LCFA and the carotenoid lycopene (Harmon and Abumrad, 1993; Moussa et al., 2011). Originally identified as fatty acid translocase (FAT), CD36 is also an established receptor for compounds of different chemical nature, such as oxidized lipoproteins, thrombospondin, and collagen (Nergiz-Unal et al., 2011). CD36 is structurally most similar to SR-BI, which has been shown to mediate the selective uptake of cholesteryl esters from HDL (Acton et al., 1996; Calvo et al., 1995; Calvo et al., 1998). More recently, it was demonstrated that CD36 is required for the endocytosis of lipoproteins by both macrophages and BAT (Bartelt et al., 2011; Febbraio et al., 2000).
The lipid coenzyme Q (CoQ; also known as ubiquinone) is an essential component of the mitochondrial electron transport chain, functioning as a transporter of electrons from several electron carriers (including complex I, complex II, glycerol-3-phosphate dehydrogenase (GPDH) and the electron transferring flavoprotein of fatty acid β-oxidation) to complex III as well as an antioxidant (Crane, 2001). CoQ consists of a quinone ring and an isoprenoid side chain of varying length that is derived from the same mevalonate pathway as cholesterol (Bentinger et al., 2010). The longer the isoprenoid side chain, the more hydrophobic the CoQ molecule (Bentinger et al., 2010). Mice have predominantly CoQ with 9 isoprenoid units (CoQ9), whereas humans have mostly CoQ10 (Sohet and Delzenne, 2012). A large portion of CoQ is found in the inner mitochondrial membrane, but a smaller portion can also be found in the membranes of other organelles and the cytosol, suggesting it may have other functions in addition to transporting electrons in the mitochondria (Bhagavan and Chopra, 2006).
Low CoQ levels are associated with cardiomyopathies, aging, and statin-induced myopathies (Bentinger et al., 2010). There are also several genetic mutations directly affecting proteins involved in the synthesis of CoQ, resulting in primary CoQ deficiencies (Quinzii et al., 2008). Therefore, increasing CoQ levels could be therapeutically beneficial for a variety of metabolic diseases. Many tissues endogenously synthesize CoQ, and the contribution of dietary CoQ is thought to be comparatively small (Bhagavan and Chopra, 2006). However, exogenous uptake of CoQ from the circulation can be significant in situations where CoQ levels are already low, such as in genetic deficiencies, myopathies, and aging (Artuch et al., 2009). Exactly how tissues take up CoQ and which tissues primarily depend on exogenous CoQ has not been well studied (Padilla-Lopez et al., 2009), posing a considerable challenge to the therapeutic use of CoQ in metabolic diseases, aging, and mitochondrial function.
We speculated that, given the ability of CD36 and related receptors to mediate the uptake of hydrophobic molecules in a variety of tissues, CD36 may be required in BAT for the uptake of similar molecules, such as CoQ. Here we report that CD36 is required for CoQ uptake in BAT and the maintenance of normal CoQ levels, and thus non-shivering thermogenesis and BAT function. The ability of CD36 deficient BAT (Cd36−/−) to take up CoQ was greatly impaired, and analysis of mitochondrial CoQ levels showed that loss of CD36 caused a BAT-specific decrease in CoQ9 and CoQ10. While BAT fatty acid uptake rates were unchanged, the ability of Cd36−/− BAT to take up another lipid, CoQ, was greatly impaired. This CD36-dependent CoQ deficiency impaired processes linked to classical CoQ function, such as oxidative damage and reduced respiration. Together, these data demonstrate a unique role for exogenous CoQ provided by CD36 in BAT mitochondrial function and reveal an unexpected dependence of BAT homeostasis on the cellular levels of CoQ.
Results
CD36 drives CoQ uptake by BAT
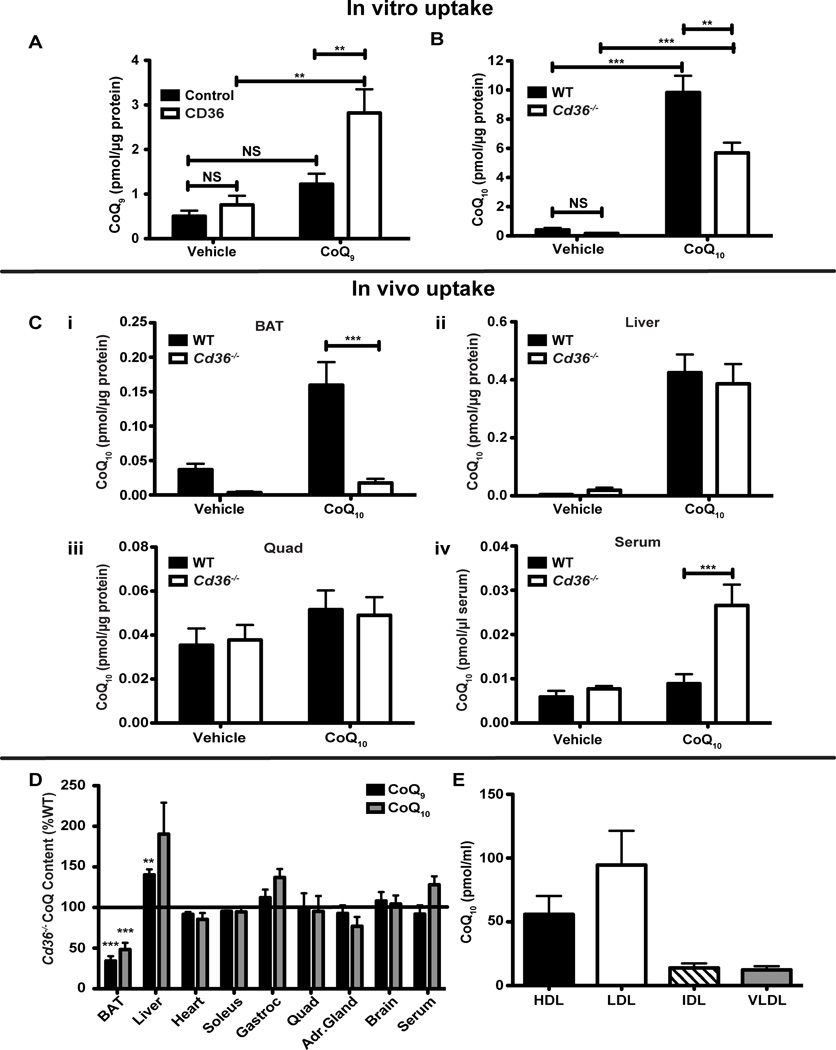
CD36 has been shown to enhance uptake of long-chain fatty acids (LCFA) in the heart, skeletal muscle, and white adipose tissue (Abumrad et al., 1993; Coburn et al., 2000; Van Nieuwenhoven et al., 1995). Besides LCFAs, CD36 and related receptors have been shown to play a role in the cellular uptake of a variety of other hydrophobic molecules, including the selective uptake of cholesterol esters from HDL by SR-B1 (Acton et al., 1996) and carotenoids such as lycopenes by CD36 (Moussa et al., 2011). Since both lycopenes and CoQ are isoprenoids, we wanted to determine if CD36 can enhance cellular uptake of CoQ, particularly in the context of a mitochondrial-rich tissue. CoQ was administered to cells and animals via Intralipid, which is an emulsion of soy bean oil, phospholipids, and glycerin (Taskinen et al., 1983). We compared the uptake of solvent- and detergent-solubilized CoQ to Intralipid and found that receptor-mediated uptake was only observed with Intralipid (data not shown). When we treated HEK293 cells overexpressing CD36 with CoQ, the CD36-expressing cells took up 57% more CoQ9 than control cells (Figure 1A). Conversely, mature brown adipocytes from Cd36−/− BAT took up 42% less CoQ10 than WT cells, as quantified by HPLC (Figure 1B and S1A). It is important to note that CoQ10 was used as a substrate for uptake, as endogenous levels are low in murine cells and differed only slightly between WT and Cd36−/− mature brown adipocytes (Figure 1B). However, total CoQ levels, which are dominated by CoQ9, were significantly suppressed in Cd36−/− mature brown adipocytes (WT 1.55 ± 0.18 pmol total CoQ/µg protein vs. Cd36−/− 0.96 ± 0.11 pmol total CoQ/µg protein, *p=0.015). Taken together, these results suggest a role for CD36 in CoQ uptake, particularly by BAT.
Figure 1. CD36 is required for CoQ transport.
(A) HEK293 cells stably expressing CD36 or parent vector (control) were treated with CoQ9 for 24 h (n=5–6 per group). CoQ levels were measured by HPLC and normalized to µg of protein. (B) Primary mature brown adipocytes were isolated from WT and Cd36−/− BAT and treated with either vehicle or CoQ10 for 3 h (n=5). (Ci–iv) WT and Cd36−/− mice were injected intraperitoneally with either vehicle (Intralipid) or CoQ10. After 24 h tissues were isolated and CoQ10 levels were measured by HPLC and normalized to µg of protein or µl of serum (n=8–9). (D) BAT, liver, heart, soleus, gastrocnemius, quadriceps, adrenal gland, brain, and serum CoQ9 and CoQ10 levels in Cd36−/− relative to WT (100%) were measured by HPLC and normalized to µg of protein (n=3–6). A complete table with the absolute CoQ values is included in Supplementary Table 1. (E) CoQ10 levels were measured in lipoprotein fractions from human serum by HPLC and normalized to ml of fraction (n=6). *p<0.05; **p<0.005; ***p<0.0005. Error bars, SEM. See also Figure S1 and Table S1.
To determine if CD36 enhances CoQ uptake in vivo, we administered a CoQ-containing Intralipid mix to WT and Cd36−/− mice by intraperitoneal injection and then measured CoQ levels by HPLC. Twenty-four hours post-injection, Cd36−/− BAT had taken up significantly less CoQ10 than WT mice (WT 0.159 ± 0.034 pmol CoQ10/µg protein vs. Cd36−/− 0.018 ± 0.006 pmol CoQ10/µg protein) (Figure 1Ci). Conversely, uptake of exogenous CoQ10 was unaffected in the liver, which took up significant amounts of injected CoQ, and skeletal muscle, which did not take up appreciable amounts of exogenous CoQ (Figure 1Cii–iii). Interestingly, while there was no difference in serum CoQ levels between WT and Cd36−/− mice at baseline, Cd36−/− mice had significantly more CoQ10 in their serum following injection of CoQ10 (WT 0.009 ± 0.002 pmol CoQ10/µl serum vs. Cd36−/− 0.026 ± 0.005 pmol CoQ10/µl serum) (Figure 1Civ).
To determine if CoQ levels are perturbed in other tissues from Cd36−/− mice at baseline, we measured CoQ9 and CoQ10 in BAT, liver, heart, soleus, gastrocnemius, quadriceps, adrenal gland, brain, and serum. While CoQ10 is the predominant species in humans, the mouse CoQ pool is composed of approximately 90% CoQ9 and 10% CoQ10 (Lass et al., 1997). Interestingly, both CoQ9 and CoQ10 levels in Cd36−/− BAT were significantly decreased by approximately 40–50% (Figure 1D). This finding was confirmed by mass spectrometry, which showed significantly decreased levels of both CoQ9 and CoQ10 in Cd36−/− BAT (64% and 80% of WT, respectively) (Figure S1B–C). In contrast, CoQ levels in the heart, skeletal muscle, adrenal glands, brain and serum from Cd36−/− mice were normal, while levels in the liver were slightly increased (Figure 1D and Table S1).
These loss- and gain-of-function studies in vitro and in vivo suggest that CD36 facilitates direct cellular uptake of CoQ in BAT. Given that CD36 can facilitate uptake of lipoprotein particles in BAT (Bartelt et al., 2011), we wanted to see in which lipoprotein fraction CoQ is present. CoQ is most abundant in the LDL fraction (Figure 1E), but absent from the non-lipoprotein fraction (data not shown), suggesting that it may be transported through the circulation and presented to the cell as a lipoprotein particle component (Tomasetti et al., 1999).
Alterations of lipid metabolism in Cd36−/− BAT
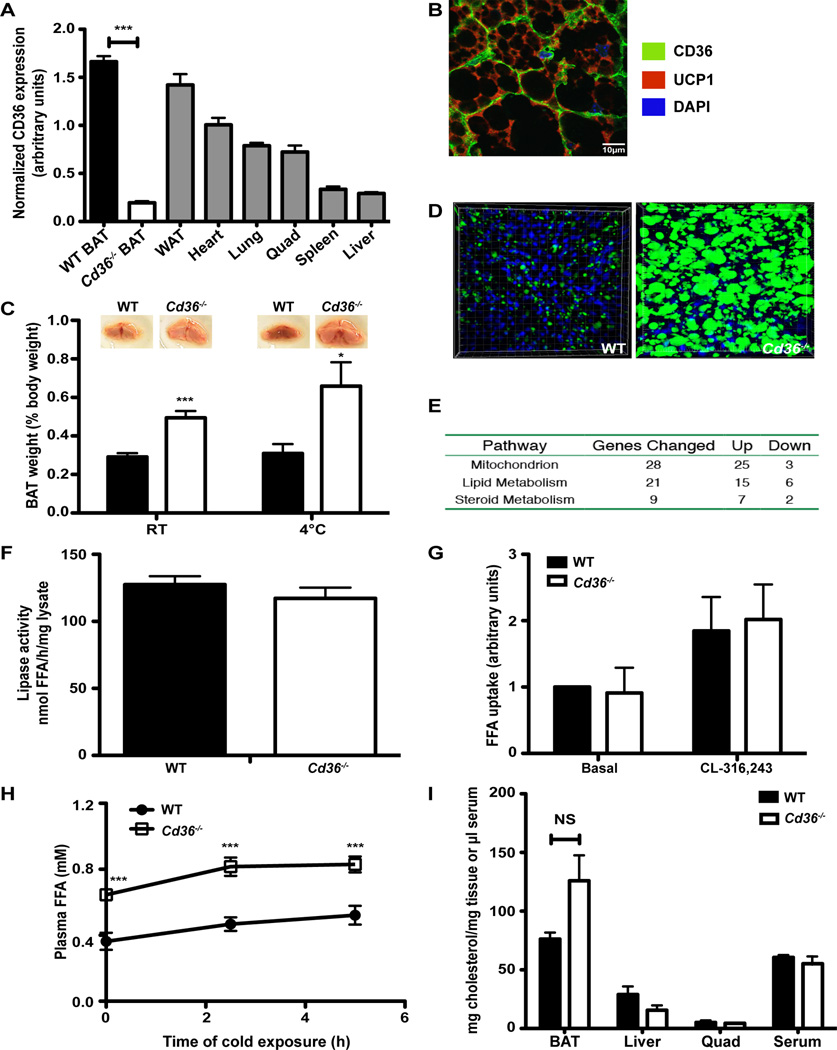
CD36 is most abundant in adipose tissue, heart, lung, and muscle, with less expression in spleen and liver (Figure 2A; Figure S2A–B). In addition, there is no effect of browning in WAT by CL-316,243 on the expression of Cd36 in subcutaneous and visceral WAT depots (Figure S2C). To elucidate the role of CD36 in BAT, we took advantage of mice lacking CD36 (Cd36−/−) (Febbraio et al., 1999). The absence of CD36 in the BAT of these mice was confirmed by Western blot (Figure 2A and Figure S2A). To identify specifically where CD36 is expressed in BAT, we performed an immunolocalization study for CD36 and UCP1. Interestingly, CD36 was found to be located exclusively on the plasma membrane in BAT, supporting a role for CD36 in the uptake of hydrophobic molecules from the circulation (Figure 2B).
Figure 2. Pathologic TAG storage in Cd36−/− BAT.
(A) Western blot for CD36 and β-tubulin was performed on lysates from WT and Cd36−/− BAT, WAT, heart, lung, quadriceps muscle, spleen, and liver (n=5). Data is presented in arbitrary units as a ratio of CD36 normalized to tubulin. (B) Co-localization of CD36 (green), UCP1 (red), and DAPI (blue) in a WT BAT cryosection. Scale bar represents 10µm. (C) Images of BAT from WT and Cd36−/− mice and their weights before and after cold exposure (n=3–6). (D) Representative images of 3-D reconstructions from cold-exposed mouse BAT sections stained with the fluorescently-labeled TAG probe BODIPY 493/503 (green) and DAPI (blue). (E) Genome-wide expression analysis was performed on RNA from WT and Cd36−/− BAT (n=3 per group). A summary table of misregulated genes identified using a p≤0.05 threshold is presented. A complete list of misregulated genes is included in Supplementary Tables 2 and 3. (F) Lipase activity (expressed as nmol of FFA released/hour/mg protein of tissue lysate) from WT and Cd36−/− BAT chunks (n=4). (G) Maximal BODIPY 510/512-labeled FFA uptake capacity of untreated and CL-316,243-treated primary mature brown adipocytes from WT and Cd36−/− mice (n=3). (H) WT and Cd36−/− fasting plasma FFA levels at 0, 2.5 and 5 h of cold exposure (n=4). (I) Cholesterol was extracted from BAT, liver, quadriceps, and serum and normalized to either mg of tissue or µl of serum (n=3). *p<0.05; **p<0.005; ***p<0.0005. Error bars, SEM. See also Figures S2 and S3 and Tables S2 and S3.
Given that CD36 is robustly expressed in BAT and that Cd36−/− mice have decreased CoQ levels in BAT, we wanted to determine if mice lacking CD36 had normal BAT morphology and function. An appreciable difference in the morphology of Cd36−/− BAT compared to wild type (WT) BAT was immediately noticed by direct observation (Figure 2C). BAT lacking CD36 was significantly larger and paler in color compared to BAT from WT mice at room temperature (Figure 2C). Following cold exposure, the characteristic browning of BAT was absent in Cd36−/− mice while the weight difference remained (Figure 2C). Three-dimensional reconstruction of confocal microscopy scans of BAT sections stained for neutral lipids with BODIPY confirmed that the marked hypertrophy in Cd36−/− BAT was mainly due to increased lipid droplet accumulation (Figure 2D) and lipid droplet volume (median volume: 18.5 µm3 WT vs. 205 µm3 Cd36−/−). These results show that BAT lacking CD36 displays whitening and increased lipid droplet size.
To determine if the accumulation of lipids in Cd36−/− BAT was due to defective lipolysis, we first measured lipase activity in mature brown adipocytes from WT and Cd36−/− mice. No significant difference in lipase activity was observed between WT and Cd36−/− BAT (Figure 2F). The lipolysis rate in white adipose tissue (WAT) was unchanged (Figure S3A), indicating that the release of fatty acids from WAT into the circulation was unaffected in Cd36−/− mice. However, the lipolysis rate in both the basal state and after stimulation by forskolin/IBMX was significantly reduced in Cd36−/− BAT (Figure S3B).
We speculated that the defects observed in Cd36−/− BAT morphology could arise from alterations in lipid trafficking. We found that LCFA uptake rates of isolated primary mature brown adipocytes from Cd36−/− mice were comparable to WT and responded normally to stimulation by the β3-adrenergic receptor agonist CL-316,243 (WT 1.84 ± 0.51 fold change over untreated vs. Cd36−/− 2.02 ± 0.53 fold change over untreated) (Figure 2G), while Lpl expression in Cd36−/− BAT was elevated by 1.48-fold compared to WT BAT (*p=0.007) (Table S3), arguing against defective free fatty acid (FFA) uptake or production. To determine if there was a defect in circulating FFA, we measured FFA levels in the serum of WT and Cd36−/− mice. Consistent with previous reports (Coburn et al., 2000; Febbraio et al., 1999), circulating FFA levels were actually elevated 2-fold in Cd36−/− mice both before and after cold exposure (Figure 2H). Cholesterol levels were unchanged between WT and Cd36−/− mice in BAT, liver, quad, and serum (Figure 2I).
Impaired mitochondrial substrate utilization in Cd36−/− BAT
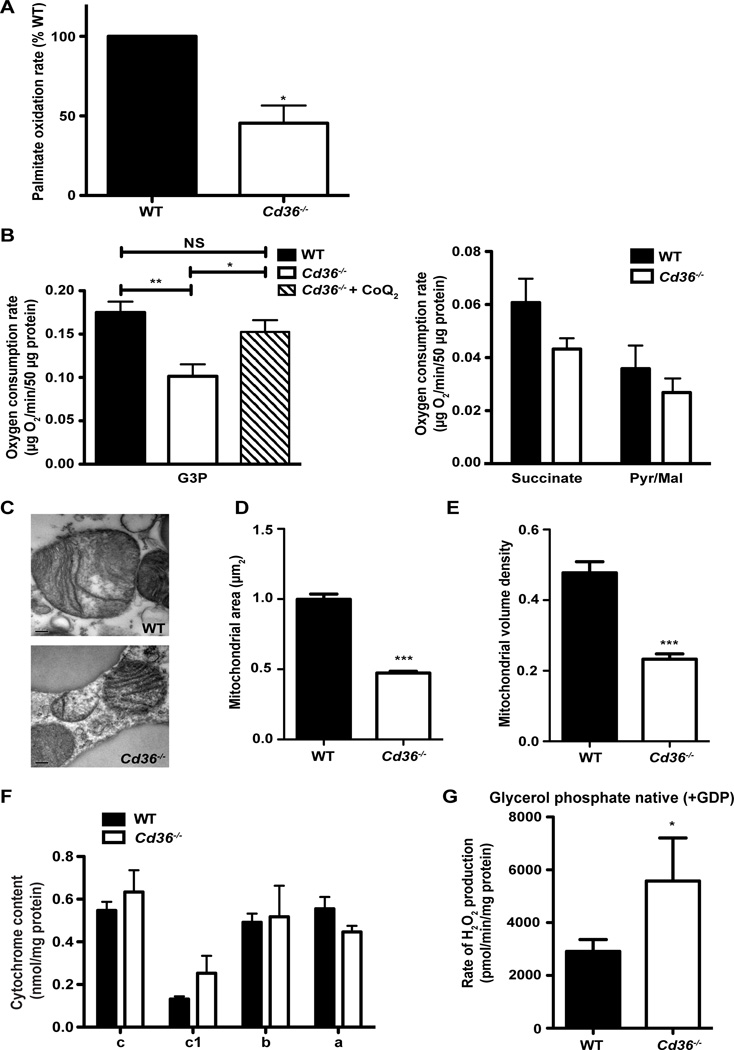
To determine if there are bioenergetic consequences as a result of BAT CoQ deficiency, we assessed LCFA β-oxidation in intact mature brown adipocytes using [14C]-palmitic acid. Despite normal fatty acid uptake potential (Figure 2G), production of 14CO2 by Cd36−/− brown adipocytes was reduced by approximately 50% compared to WT brown adipocytes, demonstrating that substrate utilization is decreased in Cd36−/− BAT and suggesting a potential mitochondrial defect (Figure 3A).
Figure 3. Impaired substrate utilization and mitochondrial dysfunction in Cd36−/− BAT.
(A) Production of 14CO2 from 14C-[Palmitic acid] in Cd36−/− primary mature brown adipocytes relative to WT (100%) and normalized for mg of protein (n=3). (B) WT and Cd36−/− BAT mitochondrial respiration rates using glycerol-3-phosphate (G3P) with and without CoQ2, succinate, and pyruvate/malate (Pyr/Mal). Oxygen consumption rates (OCR) were measured and presented as µg O2/min/50 µg protein (n=6–9). (C) Representative electron microscopy (EM) images from WT and Cd36−/− BAT. 43,000× magnification. Scale bar represents 0.2µm. (D) Quantification of the mitochondrial area from WT and Cd36−/− BAT EM images (n=4). (E) Quantification of the mitochondrial volume density from WT and Cd36−/− BAT EM images (n=4). (F) Cytochrome content was measured in isolated mitochondria from WT and Cd36−/− BAT and normalized to mg of mitochondrial protein (n=3). (G) The rate of H2O2 production was measured in WT and Cd36−/− BAT mitochondria and presented as pmol/min/mg protein (n=4). *p<0.05; ***p<0.0005. Error bars, SEM.
A series of Clark electrode respiratory tests with isolated BAT mitochondria using a variety of substrates was performed. In isolated mitochondria, electrons from pyruvate and malate enter complex I, while electrons from succinate and glycerol-3-phosphate (G3P) bypass complex I and are transferred directly to CoQ by complex II or GPDH respectively (Crane, 2001). The basal rates of respiration were decreased in BAT mitochondria from Cd36−/− mice (although not statistically significant for succinate and pyruvate/malate) (Figure 3B). Furthermore, the defect in respiration with G3P could be rescued with the addition of CoQ2, suggesting that the cause of impaired respiration is due to the decreased levels of CoQ (Figure 3B). Electron microscopy revealed significantly smaller mitochondria in Cd36−/− BAT compared to WT (Figure 3C). Mitochondria in Cd36−/− BAT were 53% smaller in area compared to WT (Figure 3D). Furthermore, the mitochondrial volume density in Cd36−/− BAT was 51% that of WT (Figure 3E). Taken together these results demonstrate impaired mitochondrial function in Cd36−/− BAT due to decreased CoQ levels.
Genome-wide expression analysis in BAT from WT and Cd36−/− mice showed changes in the expression of several functional families, including mitochondrial related genes, but no misregulation of genes encoding electron transport chain proteins (Figure 2E; Tables S2 and S3). To determine if cytochrome levels within the mitochondrial electron transport chain were affected, we measured the content of cytochromes c, c1, b, and a, but saw no differences in their levels in Cd36−/− BAT mitochondria (Figure 3F).
Low CoQ levels are associated with increased reactive oxygen species (ROS) production and oxidative damage (Murphy, 2009). To determine if Cd36−/− BAT mitochondria are undergoing oxidative stress, we measured H2O2 production. Cd36−/− BAT mitochondria had a significantly higher rate of H2O2 production (Figure 3G), indicating that there is more ROS production and possibly oxidative damage in the mitochondria of Cd36−/− BAT.
Cd36−/− mice are cold intolerant in spite of increased shivering
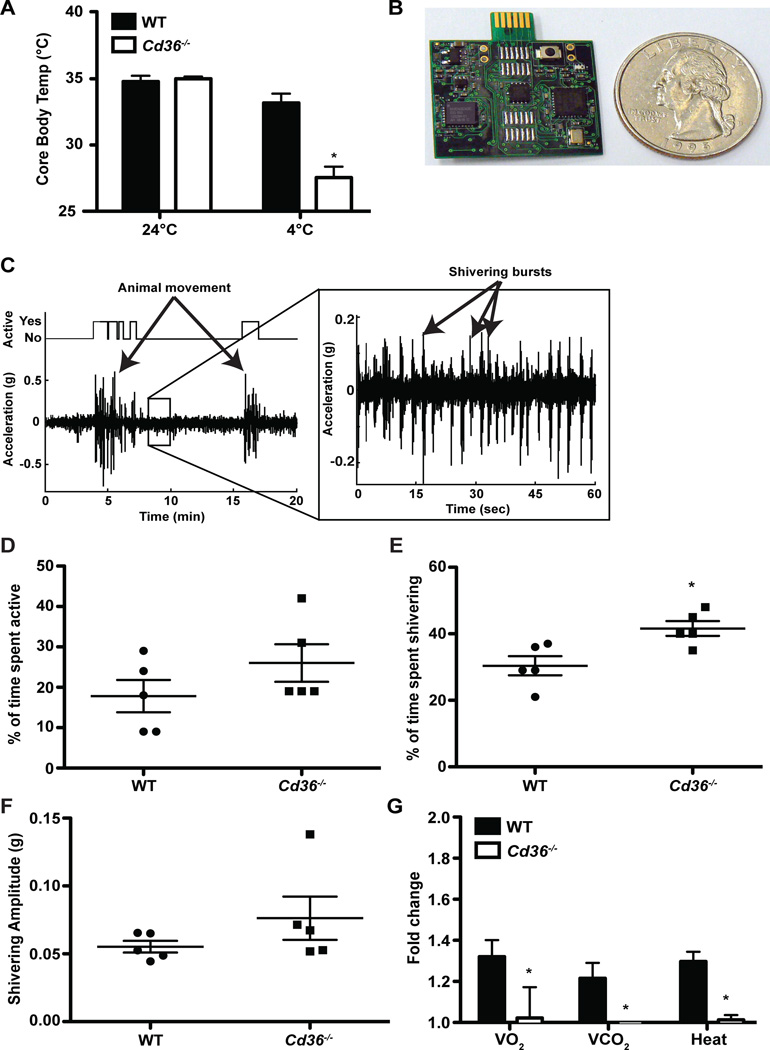
To determine if the observed altered lipid metabolism and decrease in CoQ levels affected BAT function, we exposed WT and Cd36−/− mice to 4°C for five hours and then measured their core body temperature. While there was no difference in body temperature under normal conditions, we did detect a prominent thermogenic defect in Cd36−/− mice compared to WT mice housed at 4°C (Figure 4A). Since CD36 is also expressed in muscle, we wanted to exclude a contribution of defects from shivering thermogenesis (Bastie et al., 2004; Van Nieuwenhoven et al., 1995). Therefore, we assessed shivering thermogenesis utilizing an innovative wireless accelerometer (Figure 4B) that can be used to detect both animal movement as well as shivering (Figure 4C). We found that neither overall activity (Figure 4D), nor shivering amplitude (Figure 4F) were impaired, yet shivering duration was increased (Figure 4E). These results indicate that Cd36−/− mice display a defect in non-shivering thermogenesis and a compensatory but insufficient upregulation of shivering thermogenesis in response to cold exposure. This hypothesis was further supported by the finding that, while resting metabolic rates were comparable between WT and Cd36−/− mice, the characteristic increase in VO2, VCO2, and heat production in response to a single injection of the β3-adrenergic receptor agonist CL-316,243 was absent in Cd36−/− mice (Figure 4G). This suggests that altered lipid metabolism and CoQ deficiency in Cd36−/− BAT has functional consequences.
Figure 4. Defective non-shivering thermogenesis in Cd36−/− BAT.
(A) Mouse core body temperature measured rectally before and after 5 h of cold exposure (n=6). (B) The wireless accelerometer used in this study was built at UC Berkeley and is approximately the size of a coin (a US quarter). Shivering and movement were monitored by attaching the device shown to the mouse back. (C) Accelerograph showing millisecond-resolution monitoring of movement and shivering in a WT mouse. The large bursts of acceleration indicate animal movement. A zoomed-in accelerograph displays regular shivering spikes in a cold-exposed WT mouse. Motor activity (D), time spent shivering (E), and shivering amplitude (F) of fasted mice exposed to 4°C for 2 h (n=5). (G) Oxygen consumption and carbon dioxide and heat production measurements 3 h after intraperitoneal administration of the β3-adrenergic receptor agonist CL-316,243 (n=3). Data is presented as fold change compared to before CL injection. *p<0.05. Error bars, SEM.
Discussion
The unique ability of mitochondria-rich BAT to generate heat and regulate non-shivering thermogenesis, coupled with its recent identification in adult humans, has driven research into the therapeutic use of this tissue for metabolic diseases. The presence of CD36 in BAT and its known role as a transporter of lipids in other tissues prompted us to examine whether CD36 could function as a transporter of the highly hydrophobic mitochondrial electron carrier CoQ in BAT. Here we show that CD36 drives the uptake of CoQ by BAT and is required for normal BAT function. The findings presented in this study demonstrate a unique relationship between CD36 and CoQ in brown fat and identify CD36 and CoQ as new factors that may limit the metabolic activity of BAT.
Efficient import of CoQ specifically into BAT by CD36 is an interesting finding. A number of studies have indicated the importance of CD36 and other class B scavenger receptors in the uptake of hydrophobic isoprenoids, such as lycopene, lutein, and β-carotene (Borel et al., 2011; Borel et al., 2013; Harmon and Abumrad, 1993; Kiefer et al., 2002; Moussa et al., 2011), whose transport in aqueous environments is reliant on specific protein carriers such as CD36 (Kuzuyama and Seto, 2012; Reboul and Borel, 2011). In addition, genetic variants within CD36 are associated with varying blood levels of carotenoids (Borel et al., 2011; Borel et al., 2013). Carotenoids are synthesized from isoprenoids, which are highly hydrophobic precursors whose transport in aqueous environments is reliant on specific protein carriers such as CD36 (Kuzuyama and Seto, 2012; Reboul and Borel, 2011). CoQ is another molecule with a hydrophobic isoprenoid side chain whose cellular uptake may be facilitated by transport proteins (Bentinger et al., 2010; Padilla-Lopez et al., 2009). Our finding that Cd36−/− BAT had a smaller CoQ pool than WT BAT suggested that CD36 may be required for efficient CoQ uptake or catabolism by BAT.
Loss- and gain-of-function studies in vitro and in vivo demonstrate that, indeed, CD36 facilitates direct cellular uptake of CoQ, and show a clear BAT-autonomous CoQ uptake defect. While the precise mechanism of CoQ uptake by CD36 in BAT remains unclear at this point, uptake of lipoprotein particles by CD36, long believed to be restricted to liver and macrophages, was elegantly shown recently to also occur in BAT (Bartelt et al., 2011). In other tissues, CD36 has been shown to take up cholesteryl esters from HDL, as well as oxidized LDL, through endocytosis (Acton et al., 1996; Calvo et al., 1998). Furthermore, CoQ is most abundant in the LDL fraction (Figure 1E), suggesting that it may be transported through the circulation and presented to the cell as a lipoprotein particle component (Tomasetti et al., 1999). Therefore, it is possible that CD36 mediates the uptake of lipoproteins containing CoQ through endocytosis. Alternatively, CD36 alone or in conjunction with other proteins may mediate the selective uptake of CoQ from bound lipoproteins.
It is important to note that CoQ deficiency and impaired uptake were found only in BAT lacking CD36 and not in several other tissues tested. In both WT and Cd36−/− mice the liver, which has very little CD36, was still able to take up CoQ while the quadriceps muscle, which has moderate levels of CD36, did not take up exogenous CoQ (Figure 1Cii–iii). This pattern of tissue CoQ uptake is very similar to the pattern of CD36-dependent lipoprotein uptake by Cd36−/− tissues which, like our CoQ uptake, falls into 3 categories: CD36-dependent uptake (i.e. BAT), CD36-independent uptake (i.e. liver), and low overall uptake, as exemplified by skeletal muscle (Bartelt et al., 2011). These findings demonstrate a tissue-specific role for CD36 not only in lipoprotein uptake but also CoQ uptake.
Overall, BAT is a major sink for exogenous CoQ. In spite of its size, BAT had the highest CoQ uptake rate of all the tissues examined, removing 5 times as much circulating CoQ as the 16 times larger liver. Moreover, the quadriceps muscle takes up 250 times less CoQ than BAT. Remarkably, Cd36−/− mice had three times more CoQ10 in their serum compared to WT mice, yet they had eight times less CoQ10 in their BAT, suggesting that CoQ accumulated in the circulation instead of being taken up by BAT in Cd36−/− mice. These data suggest that, out of the tissues examined, BAT is a major destination for circulating CoQ and that CD36 plays a role in the clearance of CoQ from the serum.
While we cannot exclude a contribution of CoQ de novo synthesis to the CD36-dependent CoQ deficiency, it is most likely that CoQ synthesis is weakly, if at all, affected. Gene expression analysis by microarray identified a multitude of genes involved in pathways leading to CoQ synthesis that were up- rather than downregulated in Cd36−/− BAT (Tables S2 and S3). This could be interpreted as a possible compensatory response to the decrease in the pools of CoQ in Cd36−/− BAT. For example, HMG-CoA synthase, which catalyzes an early step in the mevalonate synthesis pathway, was upregulated (Tables S2 and S3). This pathway generates both cholesterol and CoQ, and reduction of HMG-CoA synthase, a common precursor in the synthesis of these two molecules, suggested deficiencies in the levels of mevalonate or its derivatives cholesterol and CoQ (Bentinger et al., 2010). However, the level of cholesterol was unchanged between WT and Cd36−/− BAT (Figure 2I), indicating that the early components of the CoQ synthesis pathway were not affected in Cd36−/− BAT. The unchanged cholesterol levels could also hint at a facilitative rather than endocytic uptake mechanism, although further studies are required to address the point. Furthermore, while all tissues synthesize CoQ endogenously at varying rates (Thelin et al., 1992), the biosynthesis of CoQ in BAT is very low, yet its levels are quite high, suggesting that CoQ must be taken up from the circulation (Sekhar et al., 1990). The low CoQ synthesis rate in BAT coupled with our findings that cholesterol levels are unaffected in Cd36−/− BAT suggest that loss of CD36 most likely does not affect CoQ synthesis.
Mice lacking CD36 also displayed defective non-shivering thermogenesis. Unfortunately we were unable to test if exogenous CoQ would rescue the thermogenic defect in Cd36−/− mice, as CoQ10 supplementation by a single IP injection failed to raise total CoQ levels in Cd36−/− BAT to that of baseline WT levels (Figure S4). This is most likely due to the fact that CD36 is required for the uptake of sufficient CoQ by BAT.
The data presented in this study identify a mechanism for the regulation of BAT function and thermogenesis via CD36-mediated CoQ uptake. We have shown a clear requirement for CD36 in BAT function and CoQ homeostasis. Identifying the precise mechanism of CoQ uptake by CD36 will aid in the enhancement of CoQ supplementation therapies for patients with CoQ deficiencies as well as a way to stimulate brown fat activity for the treatment of obesity and associated metabolic disorders.
Experimental Procedures
Animal Experiments
All animal procedures were approved by the University of California Berkeley Animal Care and Use Committee. Cd36−/− mice on a C57BL/6J background were kindly provided by Dr. Maria Febbraio (Febbraio et al., 1999). Control C57BL/6J mice (referred to as WT in this paper) were purchased from the Jackson Laboratory. All animal experiments were conducted in 8–20 week-old male mice housed under standard conditions. They were given free access to water and rodent chow (Harlan Teklad #2018). For cold exposure studies, singly caged mice were housed at 4°C for 5 h. Core body temperature was measured rectally using a lubricated thermistor probe (Yellow Springs Instruments) at 30 min intervals. Shivering was determined by attaching a wireless accelerometer (weight=1.5g) to the mouse back and quantified with millisecond resolution in three dimensions for 2 h. For serum analysis, retro-orbital blood was collected from anesthetized mice prior to and following 2.5 and 5 h cold exposure. Serum samples were analyzed for non-esterified fatty acids and glucose using colorimetric kits according to the manufacturer’s protocol (Wako Chemicals).
CoQ Uptake Assays
For cellular uptake of CoQ, cells were incubated with 10µM CoQ9 or CoQ10. A 1mM CoQ stock solution was generated by dissolving CoQ in 100% ethanol and heating for 5 min to 65°C. 10µL of 1mM CoQ was then added to 90µL Intralipid (Sigma #I141) and heated to 65°C for 5 min. This solution was then mixed with 1ml pre-warmed complete media and immediately added to cells. Cells were incubated at 37°C with 10µM CoQ for up to 6 h, at which time the media was removed, cells were washed three times with PBS, and harvested for CoQ extraction. For in vivo uptake of CoQ, mice were injected intraperitoneally with 200µL of either a commercial CoQ10 solution (LiQSorb, Tishcon Corp) or a CoQ10/Intralipid solution. To make this solution, a 29mM CoQ10 stock solution was generated by dissolving CoQ10 (Sigma #C9538) in 100% ethanol and heating for 15 min to 65°C. 20µL of 29mM CoQ10 was then added to 180µL Intralipid (Sigma #I141) and heated to 65°C for 15 min. 200µl of this 1.45mM CoQ10/Intralipid was injected intraperitoneally per mouse. Serum and tissues were collected 24 h following injection for CoQ extraction.
Coenzyme Q Extraction and Measurement by HPLC
CoQ levels were determined as previously described (Podda et al., 1999). In brief, whole tissue, isolated mitochondria, or cells were homogenized in 1ml PBS containing 0.5mg/ml of the antioxidant butylated hydroxytoluene (BHT). 1ml of 0.1M SDS was added to samples, followed by vortexing and sonication. An aliquot (50µl) was removed for determining protein concentration by BCA assay, and CoQ4 was then added as an internal standard. 2ml of 100% ethanol was added, followed by vortexing and sonication. 2ml of hexane was then added and the samples were vortexed for 90 seconds and centrifuged at 1000g for 3 min. The upper hexane layer was dried under gentle nitrogen stream in a 37°C water bath. The dried remnants were then resuspended in 160µl of 100% ethanol and briefly heated at 65°C. Quantification by HPLC was determined using the Waters HPLC-UV detection system. 100µl of the resuspension was injected and run on a C18-ODS Hypersil reverse phase column (Thermo Scientific #30105-254630). A gradient was used consisting of solution A (80% methanol/20% water) and solution B (100% ethanol) with a flow rate of 1ml/min for 40 min: Min 0–16, 39% A and 61% B; over min 16–18 change to 100% B; min 18–28 100% B; over min 28–30 change to 39% A and 61% B; min 30–40, 39% A and 61% B. CoQ9 peak appears around 24 min and CoQ10 peak appears around 25 min. A standard curve was generated using commercial CoQ9 and CoQ10 (Sigma) to determine absolute amounts.
Lipolysis and Lipase Activity
Lipase activity was performed as previously described (Ahmadian et al., 2009). For lipolysis rates, 5mg of BAT tissue chunks was incubated in serum-free DMEM with and without 200µM IBMX and 10µM forskolin (Sigma). Lipolysis rate was determined at 15 min intervals as the amount of glycerol released using the free glycerol reagent from Sigma (#F6428) according to the manufacturer’s recommendations.
Long Chain Fatty Acid Uptake
Fatty acid uptake assays were performed as previously described (Wu et al., 2006) using primary mature brown adipocytes at 37°C in 1× HBSS containing 0.1% fatty acid-free BSA and BODIPY 500/510 C1, C12-labeled fatty acid (Invitrogen #D3823). The reaction was stopped at 0.5, 1, and 5 minutes by the addition of ice-cold PBS containing 0.1% BSA and 500µM Phloretin (Sigma). Single-cell fluorescence was determined at 510nm by fluorescence activated cell sorting (FACS) using the FACScalibur instrument (BD Biosciences).
Palmitic Acid Oxidation
Palmitic acid oxidation assays were performed as previously described (Ahmadian et al., 2009). Primary mature brown adipocytes were incubated at 37°C for 1 hour in sealed vials containing modified Krebs buffer plus 1% BSA, 5mM glucose, and 5µM [14C]-Palmitate (50mCi/mmol, PerkinElmer). The reaction was stopped with 5N H2SO4 and the 14CO2 produced was trapped in filter papers soaked in 200µL 2-phenethylamine (Sigma). The filter papers were transferred to scintillation vials containing 5mL scintillation liquid (Ecoscint XR, National Diagnostics) and the radioactivity was measured in a LS 6000 scintillation counter (Beckman Coulter).
Mitochondrial Isolation, Respiration, and Cytochrome Content
Mitochondria were purified as previously described (Parker et al., 2009). All steps were carried out at 4°C. Tissues were chopped with scissors in STE buffer (250mM sucrose, 5mM Tris and 2mM EGTA, pH 7.4) plus 1% BSA, homogenized manually (3 strokes with a loose-fit and 10 strokes with a tight-fit dounce homogenizer) and centrifuged at 8500g for 10 min. For BAT, the top fat layer was removed and the tube walls were cleaned of fat. The pellet was resuspended in STE with 1% BSA and centrifuged at 400g for 10 min. The supernatant was placed in a new tube and centrifuged at 8500g. The mitochondrial pellet was washed twice in BSA-free STE and resuspended in BSA-free STE. Protein concentration was determined by the BCA method. Mitochondrial respiration rates were recorded in a Clark-type electrode (Strathkelvin Instruments, UK) as previously described (Oelkrug et al., 2010). 50µg of mitochondria were incubated at 37°C under agitation in a final volume of 100µL in buffer containing 50mM KCl, 10mM TES, 1mM KH2PO4 and 2mM MgCl2, 1mM EGTA, 0.46mM CaCl2, 0.4% BSA, and 1µg/mL oligomycin (Sigma), pH 7.4. 10mM glycerol-3-phosphate (G3P), 5mM pyruvate, 5mM malate, or 4mM succinate were used as substrates. 20µM CoQ2 was used in rescue experiments. Cytochrome difference spectra were determined by dithionite reduced minus air oxidized scans on a DW-2 Olis dual-beam spectrophotometer. Isolated mitochondria (~0.5–1 mg/mL) were incubated in SHE medium. Concentrations of cytochromes c, c1, b, and a were calculated by simultaneous equation solving according to previously published studies (Schneider et al., 1980; Williams, 1964). The wavelength pairs and extinction coefficients for each cytochrome were: cytochrome c 550–535 nm, 25.1 mM−1 cm−1; cytochrome c1 554–540 nm, 24.1 mM−1 cm−1; cytochrome b 563–577 nm, 23.2 mM−1 cm−1; cytochrome a 605–630 nm, 13.1 mM−1 cm−1.
Statistical Analysis
Differences between two groups were determined using unpaired t test. Differences between three or more groups were analyzed by one-way ANOVA followed by Tukey post-hoc test for multiple comparisons. Power calculations were performed using online software (StatisticalSolutions.net) with a desired sample size of 10 on data sets with an n=3. Data is presented as the mean value, and error bars represent SEM. Asterisks indicate significant comparisons and specific p-values are listed in each figure legend.
Supplementary Material
Acknowledgements
This study was supported by NIH grants R01DK089202 and R01DK066336 to A.S. We thank Xiaoyue Zhao for the gene arrays statistical analysis, Harris Ingle for assistance with the laser microscopy, Kris Pister for the wireless accelerometer, Katie Wojnoonski and Ronald Krauss for providing the human serum lipoprotein fractions, Hei Sook Sul for critical reading of this manuscript, and Jessica Archambault and Amy Henkin for maintenance of mouse colonies, genotyping, and experimental assistance.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Supplemental Information
Supplemental information includes Supplemental Experimental Procedures, four figures, and three tables and can be found with this article online.
Author Contributions
C.M.A., M.K., J.W., S.V., R.L.S.G., C.L.Q., R.N., M.J., D.I.B., B.N., C.H., A.N.V., M.J.P., D.Y., K.C., A.Y., and P.V. designed experiments and collected, analyzed, and interpreted data. A.S. conceived and designed the study and analyzed data. M.F. provided the Cd36−/− mice. D.N., J.N., and M.D.B. designed experiments and analyzed data. C.M.A., M.K., and A.S. were involved in drafting or critically revising the manuscript. All authors approved the final version of the manuscript.
The authors declare no competing financial interests.
References
- Abumrad NA, el-Maghrabi MR, Amri EZ, Lopez E, Grimaldi PA. Cloning of a rat adipocyte membrane protein implicated in binding or transport of long-chain fatty acids that is induced during preadipocyte differentiation. Homology with human CD36. J Biol Chem. 1993;268:17665–17668. [PubMed] [Google Scholar]
- Acton S, Rigotti A, Landschulz KT, Xu S, Hobbs HH, Krieger M. Identification of scavenger receptor SR-BI as a high density lipoprotein receptor. Science. 1996;271:518–520. doi: 10.1126/science.271.5248.518. [DOI] [PubMed] [Google Scholar]
- Aherne W, Hull D. Brown adipose tissue and heat production in the newborn infant. J Pathol Bacteriol. 1966;91:223–234. doi: 10.1002/path.1700910126. [DOI] [PubMed] [Google Scholar]
- Ahmadian M, Duncan RE, Varady KA, Frasson D, Hellerstein MK, Birkenfeld AL, Samuel VT, Shulman GI, Wang Y, Kang C, et al. Adipose overexpression of desnutrin promotes fatty acid use and attenuates diet-induced obesity. Diabetes. 2009;58:855–866. doi: 10.2337/db08-1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson CM, Stahl A. SLC27 fatty acid transport proteins. Mol Aspects Med. 2013;34:516–528. doi: 10.1016/j.mam.2012.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Argyropoulos G, Harper ME. Uncoupling proteins and thermoregulation. J Appl Physiol. 2002;92:2187–2198. doi: 10.1152/japplphysiol.00994.2001. [DOI] [PubMed] [Google Scholar]
- Artuch R, Salviati L, Jackson S, Hirano M, Navas P. Coenzyme Q10 deficiencies in neuromuscular diseases. Adv Exp Med Biol. 2009;652:117–128. doi: 10.1007/978-90-481-2813-6_8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartelt A, Bruns OT, Reimer R, Hohenberg H, Ittrich H, Peldschus K, Kaul MG, Tromsdorf UI, Weller H, Waurisch C, et al. Brown adipose tissue activity controls triglyceride clearance. Nat Med. 2011;17:200–205. doi: 10.1038/nm.2297. [DOI] [PubMed] [Google Scholar]
- Bastie CC, Hajri T, Drover VA, Grimaldi PA, Abumrad NA. CD36 in myocytes channels fatty acids to a lipase-accessible triglyceride pool that is related to cell lipid and insulin responsiveness. Diabetes. 2004;53:2209–2216. doi: 10.2337/diabetes.53.9.2209. [DOI] [PubMed] [Google Scholar]
- Bentinger M, Tekle M, Dallner G. Coenzyme Q--biosynthesis and functions. Biochem Biophys Res Commun. 2010;396:74–79. doi: 10.1016/j.bbrc.2010.02.147. [DOI] [PubMed] [Google Scholar]
- Bhagavan HN, Chopra RK. Coenzyme Q10: absorption, tissue uptake, metabolism and pharmacokinetics. Free Radic Res. 2006;40:445–453. doi: 10.1080/10715760600617843. [DOI] [PubMed] [Google Scholar]
- Borel P, de Edelenyi FS, Vincent-Baudry S, Malezet-Desmoulin C, Margotat A, Lyan B, Gorrand JM, Meunier N, Drouault-Holowacz S, Bieuvelet S. Genetic variants in BCMO1 and CD36 are associated with plasma lutein concentrations and macular pigment optical density in humans. Ann Med. 2011;43:47–59. doi: 10.3109/07853890.2010.531757. [DOI] [PubMed] [Google Scholar]
- Borel P, Lietz G, Goncalves A, Szabo de Edelenyi F, Lecompte S, Curtis P, Goumidi L, Caslake MJ, Miles EA, Packard C, et al. CD36 and SR-BI are involved in cellular uptake of provitamin A carotenoids by Caco-2 and HEK cells, and some of their genetic variants are associated with plasma concentrations of these micronutrients in humans. J Nutr. 2013;143:448–456. doi: 10.3945/jn.112.172734. [DOI] [PubMed] [Google Scholar]
- Calvo D, Dopazo J, Vega MA. The CD36, CLA-1 (CD36L1), and LIMPII (CD36L2) gene family: cellular distribution, chromosomal location, and genetic evolution. Genomics. 1995;25:100–106. doi: 10.1016/0888-7543(95)80114-2. [DOI] [PubMed] [Google Scholar]
- Calvo D, Gomez-Coronado D, Suarez Y, Lasuncion MA, Vega MA. Human CD36 is a high affinity receptor for the native lipoproteins HDL, LDL, and VLDL. J Lipid Res. 1998;39:777–788. [PubMed] [Google Scholar]
- Coburn CT, Hajri T, Ibrahimi A, Abumrad NA. Role of CD36 in membrane transport and utilization of long-chain fatty acids by different tissues. J Mol Neurosci. 2001;16:117–121. doi: 10.1385/JMN:16:2-3:117. discussion 151-117. [DOI] [PubMed] [Google Scholar]
- Coburn CT, Knapp FF, Jr, Febbraio M, Beets AL, Silverstein RL, Abumrad NA. Defective uptake and utilization of long chain fatty acids in muscle and adipose tissues of CD36 knockout mice. J Biol Chem. 2000;275:32523–32529. doi: 10.1074/jbc.M003826200. [DOI] [PubMed] [Google Scholar]
- Crane FL. Biochemical functions of coenzyme Q10. J Am Coll Nutr. 2001;20:591–598. doi: 10.1080/07315724.2001.10719063. [DOI] [PubMed] [Google Scholar]
- Cypess AM, Lehman S, Williams G, Tal I, Rodman D, Goldfine AB, Kuo FC, Palmer EL, Tseng YH, Doria A, et al. Identification and importance of brown adipose tissue in adult humans. N Engl J Med. 2009;360:1509–1517. doi: 10.1056/NEJMoa0810780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Febbraio M, Abumrad NA, Hajjar DP, Sharma K, Cheng W, Pearce SF, Silverstein RL. A null mutation in murine CD36 reveals an important role in fatty acid and lipoprotein metabolism. J Biol Chem. 1999;274:19055–19062. doi: 10.1074/jbc.274.27.19055. [DOI] [PubMed] [Google Scholar]
- Febbraio M, Podrez EA, Smith JD, Hajjar DP, Hazen SL, Hoff HF, Sharma K, Silverstein RL. Targeted disruption of the class B scavenger receptor CD36 protects against atherosclerotic lesion development in mice. J Clin Invest. 2000;105:1049–1056. doi: 10.1172/JCI9259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greaves DR, Gordon S. The macrophage scavenger receptor at 30 years of age: current knowledge and future challenges. J Lipid Res. 2009;50(Suppl):S282–S286. doi: 10.1194/jlr.R800066-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmon CM, Abumrad NA. Binding of sulfosuccinimidyl fatty acids to adipocyte membrane proteins: isolation and amino-terminal sequence of an 88-kD protein implicated in transport of long-chain fatty acids. J Membr Biol. 1993;133:43–49. doi: 10.1007/BF00231876. [DOI] [PubMed] [Google Scholar]
- Heaton JM. The distribution of brown adipose tissue in the human. J Anat. 1972;112:35–39. [PMC free article] [PubMed] [Google Scholar]
- Kiefer C, Sumser E, Wernet MF, Von Lintig J. A class B scavenger receptor mediates the cellular uptake of carotenoids in Drosophila. Proc Natl Acad Sci U S A. 2002;99:10581–10586. doi: 10.1073/pnas.162182899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuniyasu A, Hayashi S, Nakayama H. Adipocytes recognize and degrade oxidized low density lipoprotein through CD36. Biochem Biophys Res Commun. 2002;295:319–323. doi: 10.1016/s0006-291x(02)00666-6. [DOI] [PubMed] [Google Scholar]
- Kuzuyama T, Seto H. Two distinct pathways for essential metabolic precursors for isoprenoid biosynthesis. Proc Jpn Acad Ser B Phys Biol Sci. 2012;88:41–52. doi: 10.2183/pjab.88.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lass A, Agarwal S, Sohal RS. Mitochondrial ubiquinone homologues, superoxide radical generation, and longevity in different mammalian species. J Biol Chem. 1997;272:19199–19204. doi: 10.1074/jbc.272.31.19199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moussa M, Gouranton E, Gleize B, Yazidi CE, Niot I, Besnard P, Borel P, Landrier JF. CD36 is involved in lycopene and lutein uptake by adipocytes and adipose tissue cultures. Mol Nutr Food Res. 2011;55:578–584. doi: 10.1002/mnfr.201000399. [DOI] [PubMed] [Google Scholar]
- Murphy MP. How mitochondria produce reactive oxygen species. Biochem J. 2009;417:1–13. doi: 10.1042/BJ20081386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nergiz-Unal R, Rademakers T, Cosemans JM, Heemskerk JW. CD36 as a multiple-ligand signaling receptor in atherothrombosis. Cardiovasc Hematol Agents Med Chem. 2011;9:42–55. doi: 10.2174/187152511794182855. [DOI] [PubMed] [Google Scholar]
- Oelkrug R, Kutschke M, Meyer CW, Heldmaier G, Jastroch M. Uncoupling protein 1 decreases superoxide production in brown adipose tissue mitochondria. J Biol Chem. 2010;285:21961–21968. doi: 10.1074/jbc.M110.122861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padilla-Lopez S, Jimenez-Hidalgo M, Martin-Montalvo A, Clarke CF, Navas P, Santos-Ocana C. Genetic evidence for the requirement of the endocytic pathway in the uptake of coenzyme Q6 in Saccharomyces cerevisiae. Biochim Biophys Acta. 2009;1788:1238–1248. doi: 10.1016/j.bbamem.2009.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker N, Crichton PG, Vidal-Puig AJ, Brand MD. Uncoupling protein-1 (UCP1) contributes to the basal proton conductance of brown adipose tissue mitochondria. J Bioenerg Biomembr. 2009;41:335–342. doi: 10.1007/s10863-009-9232-8. [DOI] [PubMed] [Google Scholar]
- Pluddemann A, Neyen C, Gordon S. Macrophage scavenger receptors and host-derived ligands. Methods. 2007;43:207–217. doi: 10.1016/j.ymeth.2007.06.004. [DOI] [PubMed] [Google Scholar]
- Podda M, Weber C, Traber MG, Milbradt R, Packer L. Sensitive high-performance liquid chromatography techniques for simultaneous determination of tocopherols, tocotrienols, ubiquinols, and ubiquinones in biological samples. Methods Enzymol. 1999;299:330–341. doi: 10.1016/s0076-6879(99)99033-x. [DOI] [PubMed] [Google Scholar]
- Quinzii CM, Lopez LC, Naini A, DiMauro S, Hirano M. Human CoQ10 deficiencies. Biofactors. 2008;32:113–118. doi: 10.1002/biof.5520320113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reboul E, Borel P. Proteins involved in uptake, intracellular transport and basolateral secretion of fat-soluble vitamins and carotenoids by mammalian enterocytes. Prog Lipid Res. 2011;50:388–402. doi: 10.1016/j.plipres.2011.07.001. [DOI] [PubMed] [Google Scholar]
- Schneider H, Lemasters JJ, Hochli M, Hackenbrock CR. Fusion of liposomes with mitochondrial inner membranes. Proc Natl Acad Sci U S A. 1980;77:442–446. doi: 10.1073/pnas.77.1.442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekhar BS, Kurup CK, Ramasarma T. Microsomal redox systems in brown adipose tissue: high lipid peroxidation, low cholesterol biosynthesis and no detectable cytochrome P-450. Mol Cell Biochem. 1990;92:147–157. doi: 10.1007/BF00218132. [DOI] [PubMed] [Google Scholar]
- Sell H, Deshaies Y, Richard D. The brown adipocyte: update on its metabolic role. Int J Biochem Cell Biol. 2004;36:2098–2104. doi: 10.1016/j.biocel.2004.04.003. [DOI] [PubMed] [Google Scholar]
- Sohet FM, Delzenne NM. Is there a place for coenzyme Q in the management of metabolic disorders associated with obesity? Nutr Rev. 2012;70:631–641. doi: 10.1111/j.1753-4887.2012.00526.x. [DOI] [PubMed] [Google Scholar]
- Su X, Abumrad NA. Cellular fatty acid uptake: a pathway under construction. Trends Endocrinol Metab. 2009;20:72–77. doi: 10.1016/j.tem.2008.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taskinen MR, Nikkila EA, Kuusi T, Tulikoura I. Changes of high density lipoprotein subfraction concentration and composition by Intralipid in vivo and by lipolysis of Intralipid in vitro. Arteriosclerosis (Dallas, Tex) 1983;3:607–615. doi: 10.1161/01.atv.3.6.607. [DOI] [PubMed] [Google Scholar]
- Thelin A, Schedin S, Dallner G. Half-life of ubiquinone-9 in rat tissues. FEBS Lett. 1992;313:118–120. doi: 10.1016/0014-5793(92)81425-l. [DOI] [PubMed] [Google Scholar]
- Tomasetti M, Alleva R, Solenghi MD, Littarru GP. Distribution of antioxidants among blood components and lipoproteins: significance of lipids/CoQ10 ratio as a possible marker of increased risk for atherosclerosis. Biofactors. 1999;9:231–240. doi: 10.1002/biof.5520090218. [DOI] [PubMed] [Google Scholar]
- van Marken Lichtenbelt WD, Vanhommerig JW, Smulders NM, Drossaerts JM, Kemerink GJ, Bouvy ND, Schrauwen P, Teule GJ. Cold-activated brown adipose tissue in healthy men. N Engl J Med. 2009;360:1500–1508. doi: 10.1056/NEJMoa0808718. [DOI] [PubMed] [Google Scholar]
- Van Nieuwenhoven FA, Verstijnen CP, Abumrad NA, Willemsen PH, Van Eys GJ, Van der Vusse GJ, Glatz JF. Putative membrane fatty acid translocase and cytoplasmic fatty acid-binding protein are co-expressed in rat heart and skeletal muscles. Biochem Biophys Res Commun. 1995;207:747–752. doi: 10.1006/bbrc.1995.1250. [DOI] [PubMed] [Google Scholar]
- Virtanen KA, Lidell ME, Orava J, Heglind M, Westergren R, Niemi T, Taittonen M, Laine J, Savisto NJ, Enerback S, et al. Functional brown adipose tissue in healthy adults. N Engl J Med. 2009;360:1518–1525. doi: 10.1056/NEJMoa0808949. [DOI] [PubMed] [Google Scholar]
- Williams JN., Jr A method for the simultaneous quantitative estimation of cytochromes a, b, c1, and c in mitochondria. Arch Biochem Biophys. 1964;107:537–543. doi: 10.1016/0003-9861(64)90313-3. [DOI] [PubMed] [Google Scholar]
- Wu Q, Kazantzis M, Doege H, Ortegon AM, Tsang B, Falcon A, Stahl A. Fatty acid transport protein 1 is required for nonshivering thermogenesis in brown adipose tissue. Diabetes. 2006;55:3229–3237. doi: 10.2337/db06-0749. [DOI] [PubMed] [Google Scholar]
- Zeng Y, Tao N, Chung KN, Heuser JE, Lublin DM. Endocytosis of oxidized low density lipoprotein through scavenger receptor CD36 utilizes a lipid raft pathway that does not require caveolin-1. J Biol Chem. 2003;278:45931–45936. doi: 10.1074/jbc.M307722200. [DOI] [PubMed] [Google Scholar]
- Zimmet P, Alberti KG, Shaw J. Global and societal implications of the diabetes epidemic. Nature. 2001;414:782–787. doi: 10.1038/414782a. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.