Summary
In most eukaryotes, centromeres are defined epigenetically by presence of the histone H3 variant CENP-A [1-3]. CENP-A containing chromatin recruits the constitutive centromere-associated network (CCAN) of proteins, which in turn directs assembly of the outer kinetochore to form microtubule attachments and ensure chromosome segregation fidelity [4-6]. While the mechanisms that load CENP-A at centromeres are being elucidated, the functions of its divergent N-terminal tail remain enigmatic [7-12]. Here, we employ the well-studied fission yeast centromere [13-16] to investigate the function of the CENP-A (Cnp1) N-tail. We show that alteration of the N-tail did not affect Cnp1 loading at centromeres, outer kinetochore formation, or spindle checkpoint signaling, but nevertheless elevated chromosome loss. N-Tail mutants exhibited synthetic lethality with an altered centromeric DNA sequence, with rare survivors harboring chromosomal fusions in which the altered centromere was epigenetically inactivated. Elevated centromere inactivation was also observed for N-tail mutants with unaltered centromeric DNA sequences. N-tail mutants specifically reduced localization of the CCAN proteins Cnp20/CENP-T and Mis6/CENP-I, but not Cnp3/CENP-C. Overexpression of Cnp20/CENP-T suppressed defects in an N-tail mutant, suggesting a link between reduced CENP-T recruitment and the observed centromere inactivation phenotype. Thus, the Cnp1 N-tail promotes epigenetic stability of centromeres in fission yeast, at least in part via recruitment of the CENP-T branch of the CCAN.
Keywords: centromere, CENP-A, CenH3, histone variant, kinetochore, CENP-T, CCAN, mitosis, chromosome segregation, aneuploidy
Results & Discussion
Cnp1 N-Tail Variants Support Viability but Exhibit Elevated Chromosome Missegregation
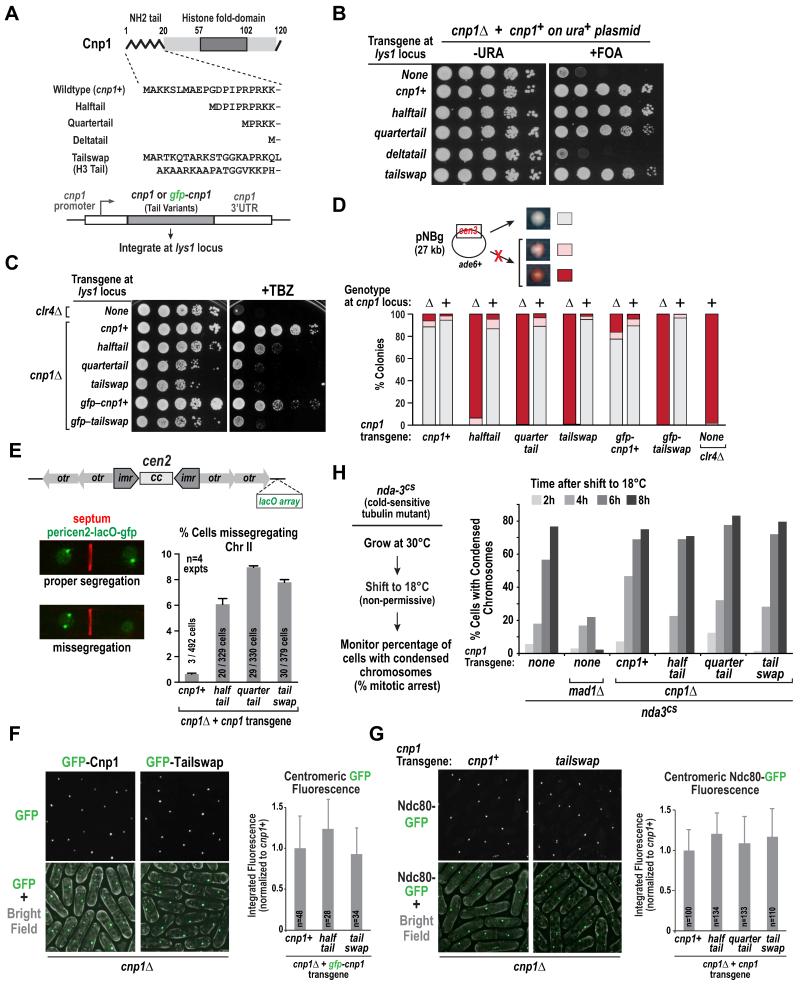
To investigate the function of the 20 amino acid Cnp1 N-tail, we generated Halftail, Quartertail and Deltatail variants, and a Tailswap variant in which the Cnp1 N-tail was replaced by the N-tail of histone H3 (Fig. 1A). All tested transgenes (either untagged or with GFP coding sequence inserted immediately after the start codon) were inserted in single copy at the lys1 locus (Fig. 1A). We first assessed the ability of these variants to rescue lethality of cnp1Δ cells observed following 5-Fluoroorotic acid (FOA)-mediated removal of plasmid-encoded cnp1+ (Fig. 1B). The cnp1+, halftail, quartertail and tailswap transgenes rescued inviability, whereas deltatail did not (Fig. 1B). Immunoblotting indicated that Deltatail was not expressed well (Fig. S1A); consistent with this, overexpression of Deltatail rescued inviability of cnp1Δ and cnp1ts (Fig. S1B,C). Thus, the N-tail of Cnp1 is dispensable for viability of a cnp1Δ cell population.
Figure 1. Cnp1 N-tail variants support viability, centromere loading, and outer kinetochore formation yet exhibit elevated chromosome missegregation.
A. Schematic of the Cnp1 N-tail variants used in this study. All transgenes were integrated at the lys1 locus.
B. Plasmid shuffle assay, employing 5-fluorooritic acid (5-FOA) to select against plasmid-encoded wild-type Cnp1, to analyze rescue of cnp1Δ cells by indicated transgenes. A ten-fold dilution series is shown for each condition.
C. Sensitivity of cnp1Δ cells expressing indicated Cnp1 N-tail variants to the spindle poison thiabendazole (TBZ). Growth was assayed at 30°C using 10-fold serial dilutions plated on rich YES medium with (right) or without (left) 10 μg/ml TBZ. clr4Δ serves as a TBZ-sensitive control.
D. Minichromosome (pNBg) maintenance assay in wildtype and cnp1Δ (indicated as + or Δ, respectively) cells expressing Cnp1 N-tail variants. clr4Δ serves as a control with elevated missegregation. Cell with a minichromosome generate white colonies; minichromosome loss results in red or sectored colonies. >900 colonies were scored per condition.
E. Segregation of Chr II marked with a pericentromeric LacO array labeled with LacI-GFP (pericen2-lacO-gfp). Septation was determined by calcofluor staining. Error bars are 95% confidence interval. Scale bar is 3 μm.
F. Images of cnp1Δ cells expressing GFP-tagged Cnp1 or Tailswap (left). The graph (right) plots the mean signal intensity of the GFP centromeric focus measured in large (lateG2) cells, normalized relative to wild-type GFP-Cnp1, for the indicated variants. Error bars are the standard deviation. Scale bar is 5 μm.
G. Images of Ndc80-GFP in cnp1Δ cells expressing untagged Cnp1 or Tailswap. The graph (right) plots the mean signal intensity of the Ndc80-GFP centromeric focus measured in large (late G2) cells of the indicated variants, normalized relative to wild-type Cnp1. Error bars are the standard deviation. Scale bar is 5 μm.
H. (left) Assay used to monitor spindle checkpoint-dependent arrest. Cold-sensitive nda3-KM311 (nda3cs) strains, containing indicated deletions and integrations, were grown at 30°C, shifted to non-permissive 18°C, and samples were fixed and processed for DAPI staining every 2 hours for 8 hours. (right) Graph plotting the percentage of cells with condensed chromatin, a marker for mitotic arrest, at different times following shift to the non-permissive temperature.
We next assessed chromosome segregation fidelity using drug sensitivity and minichromosome loss assays. N-tail variants exhibited increased sensitivity to the microtubule-destabilizing drug thiabendazole (TBZ; Fig. 1C), with growth retardation approaching that observed for clr4Δ, a mutant of the H3K9 methyltransferase in which pericentromeric heterochromatin formation and cohesin enrichment are disrupted [17,18]. TBZ sensitivity was not rescued by overexpression of Cnp1 N-tail variants (Fig. S1D). Consistent with the increased TBZ sensitivity, Cnp1 N-tail variants exhibited high rates of loss of a non-essential 27 kb minichromosome (Fig. 1D; the minichromosome carries an opal suppressor tRNA that complements a chromosomal ade6-704 mutation; loss of the minichromosome results in red or sectored colonies [19]); this phenotype was not observed when endogenous Cnp1 was present (Fig. 1D).
To monitor endogenous chromosome segregation, we imaged septated cells (i.e. mostly S-phase cells with two nuclei and calcofluor staining of the septum) harboring a GFP-marked LacO array insertion adjacent to the centromere of Chr II (referred to as pericen2-lacO-gfp). This analysis revealed elevated missegregation of endogenous Chr II in N-tail variants (Fig. 1E). Thus, while N-tail variants of Cnp1 support viability of a cell population, they exhibit significantly elevated chromosome loss rates.
Cnp1 N-Tail Variants are Normally Loaded at Centromeres and Support Outer Kinetochore Assembly
One explanation for the missegregation defect caused by alterations in the Cnp1 N-tail is a reduction in loading at centromeres. To test this possibility, we quantified the fluorescence at clustered centromeres of GFP-Cnp1, GFP-Halftail, and GFP-Tailswap in a cnp1Δ background. Both GFP-Halftail and GFP-Tailswap localized equivalently to GFP-Cnp1 at centromeres (Fig. 1F,S1E). In addition, equivalent localization was observed for GFP-Cnp1 and N-tail variants, with the exception of Deltatail, in the presence of endogenous Cnp1 (Fig. S1F). Consistent with their normal loading, overexpression of all Cnp1 N-tail variants rescued the temperature-sensitive growth defect of scm3-139 (Fig. S1G), which is caused by perturbation of the interaction between Cnp1 and its specialized chaperone Scm3 [20]. These results suggest that the elevated chromosome missegregation observed in the N-tail variants is not due to a defect in Cnp1 loading.
We next tested outer kinetochore assembly in the Cnp1 N-tail variants. First, we quantified centromere localization of the Ndc80 subunit of the conserved Ndc80 complex that directly mediates end-coupled microtubule attachments [21,22]. Ndc80 localization was unaffected in all tested N-tail Cnp1 variants (Fig. 1G). Second, we monitored spindle checkpoint activity, which requires an intact outer kinetochore to generate a signal that prevents cell cycle progression [23]. Analysis of the checkpoint-mediated arrest, performed using a cold-sensitive tubulin mutation to disrupt microtubules [24], revealed normal arrest in Cnp1 N-tail variants (Fig. 1H); in contrast, the checkpoint pathway mutant mad1Δ failed to arrest. Thus, Cnp1 N-tail variants cause significant chromosome segregation defects but these do not appear to arise from a problem in loading Cnp1 at centromeres or building an outer kinetochore with normal Ndc80 recruitment and checkpoint signaling activity.
Cnp1 Tail Variants Exhibit Synthetic Lethality In the Presence of an Altered Central Core Sequence
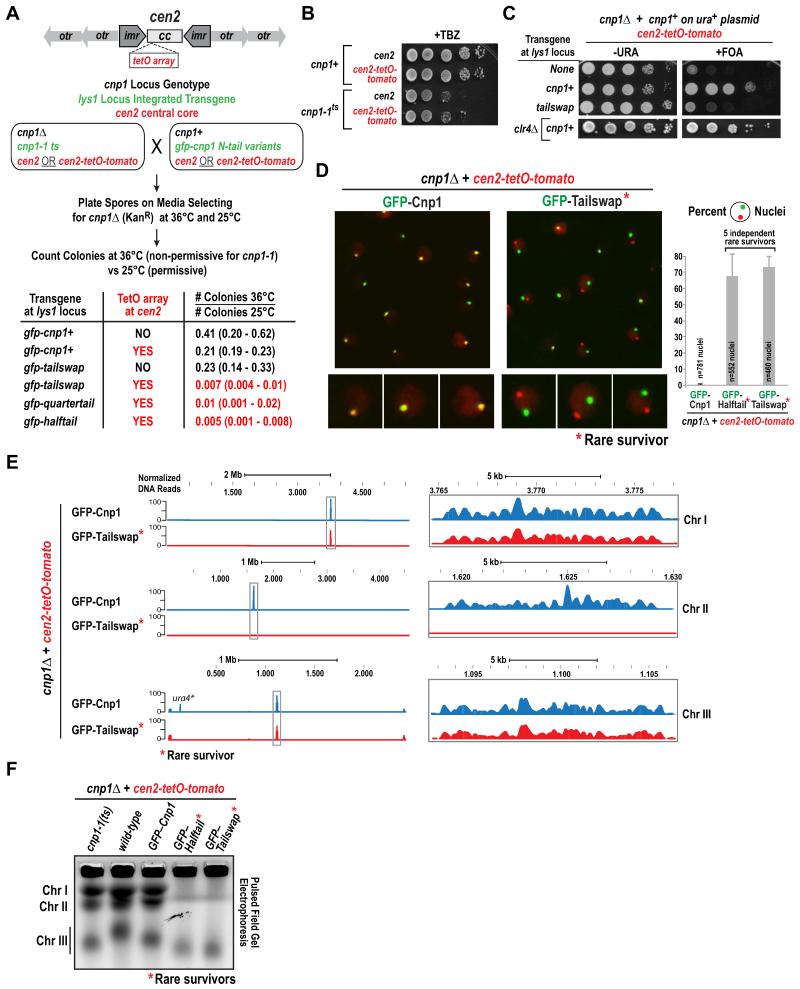
A clue as to the origin of the Cnp1 N-tail variant missegregation defect came from a serendipitous observation made while introducing N-tail variants into strains harboring operator array insertions. While we were able to construct strains expressing N-tail variants in a cnp1Δ background with operator array insertions outside the central core of the centromere (e.g. Fig. 1E; Cnp1 is concentrated in the central core [20, 25]), we were unable to obtain strains with a TetO array insertion in the central core (Fig. 2A; the strain also expresses a TetR-tomato fusion to label the array and is referred to as cen2-tetO-tomato [26]).
Figure 2. Cnp1 N-tail variants are unable to propagate a centromere harboring a repetitive TetO array insertion in the central core.
A. Schematic and results of mating and sporulation assay employed to assess synthetic lethality between GFP-tagged Cnp1 N-tail variants and the TetO array (cen2-tetO-tomato) insertion in the central core of centromere 2. Note that both partners in each mating harbored either unaltered cen2 or cen2-tetO-tomato.
B. TetO array insertion in the central core does not enhance TBZ sensitivity of cnp1-1ts. Serial dilutions (10-fold) of indicated strains were plated on rich YES medium containing 10 μg/ml TBZ and grown at 30°C.
C. Plasmid shuffle assay (10-fold serial dilutions) performed on the indicated strains. clr4Δ in combination with cen2-tetO-tomato and wildtype Cnp1 is shown below.
D. Images of cnp1Δ cen2-tetO-tomato strains expressing GFP-Cnp1 (left) or GFP-Tailswap (right); the GFP-Tailswap and other N-tail variant strains analyzed were rare survivors isolated from the experiment in Fig. 2A. Scale bar is 5 μm; images on the bottom are magnified a further 2-fold. Graph on right plots percentage of cells displaying delocalization of green (GFP) and red (Tomato) foci. 5 independent GFP-Halftail and GFP-Tailswap survivors were analyzed; error bars are the standard deviation.
E. GFP ChIP-Seq of a control (GFP-Cnp1) and a GFP-Tailswap survivor in the cnp1Δ cen2-tetO-tomato background. All three S. pombe chromosomes are displayed. The y-axis plots normalized read counts, with normalization relative to the number of mapped reads; the x-axis is chromosomal position. The centromere regions (boxed) are enlarged on the right. Conventional duplex ChIP-PCR was also performed (see Fig. S2C). The signal detected at the ura4 locus (ura4*) in GFP-Cnp1 is due to mapping of reads from the ura4 marker inserted adjacent to cen2-tetO-tomato; as this centromere is inactivated in the GFP-Tailswap survivor, no ura4 reads are mapped.
F. Undigested chromosomal DNA samples from the indicated strains analyzed by pulsed-field gel electrophoresis. See also Fig. S2D.
To assess if the N-tail variants and cen2-tetO-tomato were synthetic lethal, we used the mating-based random sporulation assay schematized in Fig. 2A. In this assay, synthetic lethality is measured by the ratio of the number of colonies at 36°C (which prevents growth of cnp1Δ spores that inherited the cnp1-1ts mutant transgene integrated at the lys1 locus) versus 25°C (where cnp1Δ spores that inherit either the cnp1-1ts mutant or a gfp-cnp1 N-tail transgene integrated in the lys1 locus form colonies). In the absence of a central core TetO array insertion and with a gfp-cnp1+ transgene the ratio was ~0.4, near the expected ratio of 0.5. The cen2-tetO-tomato insertion in combination with gfp-cnp1+ reduced this ratio to ~0.2 indicating a mild synthetic defect. The gfp-tailswap transgene in the absence of a TetO insertion exhibited a ratio of 0.23. Strikingly, for the combination of gfp-tailswap and the TetO insertion, the ratio was <0.01, indicating strong synthetic lethality. Similar magnitude synthetic lethality was observed with cen2-tetO-tomato and gfp-halftail and gfp-quartertail transgenes (Fig. 2A). Importantly, neither cnp1-1ts (Fig. 2B; Fig. S2A) nor clr4Δ (Fig. 2C), which exhibits similar magnitude chromosome missegregation as the N-tail variants (Fig. 1D), exhibited synthetic lethality with cen2-tetO-tomato. In addition, the observed synthetic lethality is not due to temperature sensitivity, as it was also observed in plasmid shuffle assays performed at 30°C (Fig. 2C). Finally, synthetic lethality was not observed when N-tail variants were combined with an operator array integrated outside the central core (Fig. S2B; Table S1). Thus, N-tail variants of Cnp1 exhibit strong synthetic lethality with a centromeric DNA sequence harboring an operator array insertion in the central core.
Rare Survivors Expressing Cnp1 N-Tail Variants and Harboring an Altered Central Core Sequence Exhibit Centromere Inactivation
While the majority of cells expressing GFP N-tail variants in the presence of cen2-tetO-tomato were inviable, a small number of survivors were recovered (0.5-1.0%; Fig. 2A). To determine how these cells maintained viability, we imaged >10 independent survivor colonies for two variants and found that the TetO-tomato focus was dissociated from the GFP focus and devoid of GFP signal (Fig. 2D), suggesting loss of the N-tail variant Cnp1 from cen2. In agreement with the imaging data, anti-GFP ChIP-PCR (Fig. S2C) and ChIP-Seq analysis of a rare gfp-tailswap;cen2-TetO-tomato survivor colony showed complete loss of Cnp1 at the central core of cen2 (Fig. 2E). In addition, evidence for a neocentromere on Chr II was not observed in the ChIP-Seq data, suggesting that these cells survive due to fusion of centromere-inactivated Chr II with one of the other two chromosomes, as previously observed following excision of a centromere [27]. To test this possibility, we performed pulsed-field gel electrophoresis, which indicated that Chr II had fused with Chr I in independent survivor colonies harboring different GFP-fused N-tail variants (Fig. 2F; S2D).
Thus, combination of a Cnp1 N-tail variant with an array insertion at the central core results in centromere inactivation that in the majority of cases is lethal but in rare cases is tolerated through chromosome fusion. Similar synthetic lethality is not observed with cnp1-1ts or clr4Δ, both of which compromise chromosome segregation. These observations suggest that Cnp1 N-tail variants increase the probability of centromere inactivation and that this effect is magnified by insertion of the TetO array in the central core.
Heterochromatin Occupies Inactivated Centromeres But is Not Required for Centromere Inactivation
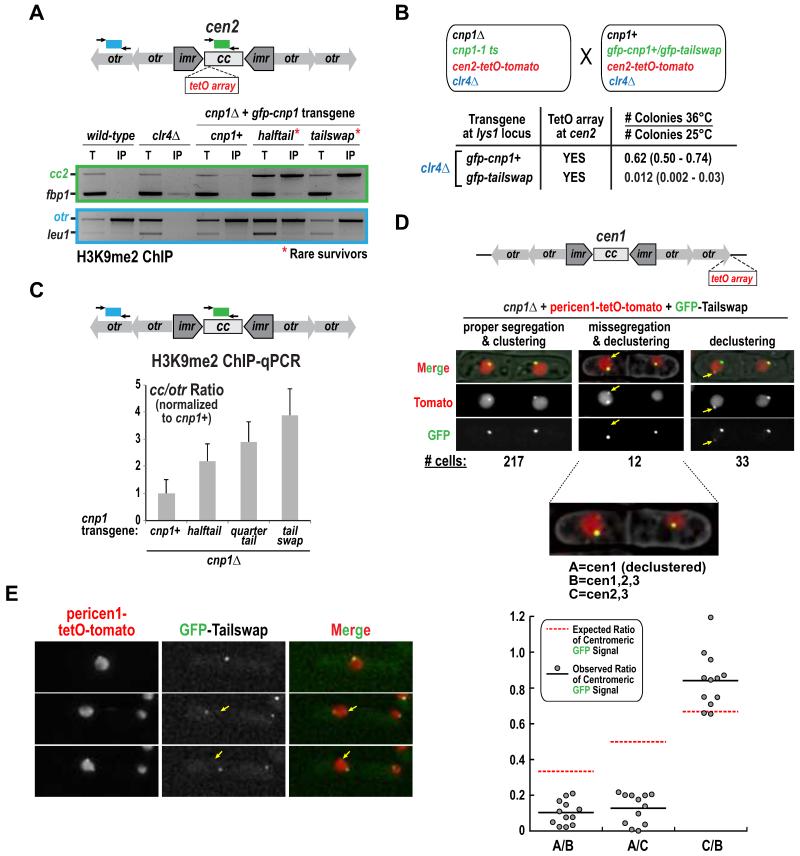
As the Cnp1-containing central core is flanked by pericentric heterochromatin, one possible mechanism for centromere inactivation is that heterochromatin encroaches into the N-tail variant-containing central core. ChIP analysis revealed high levels of H3K9 methylation in the inactive centromere central core of the rare survivors (Fig. 3A); in addition, a marker cassette inserted in this region was hyper-silenced in the survivors (Fig. S3A,B). To test if heterochromatin was required for centromere inactivation, we performed the mating-based assay in Fig. 2A, but with clr4Δ in both strains. No significant suppression of lethality was observed with clr4Δ (Fig. 3B). Thus, heterochromatin is not required for centromere inactivation but elevation of H3K9me2 in the central core provides an easy-to-measure readout for centromere inactivation events in a cell population.
Figure 3. Evidence for centromere inactivation in Cnp1 N-tail variants in the absence of any alterations in centromeric DNA sequence.
A. H3K9me2 ChIP-PCR analysis for the indicated strains. Centromere 2 (cc2) and heterochromatic outer repeats (otr) products were compared to non-centromeric controls fbp1 and leu1, respectively. A clr4Δ control, which lacks H3K9me2, is also shown.
B. Analysis of the effect of clr4Δ on synthetic lethality of GFP-Tailswap and cen2-tetO-tomato, conducted as in Fig. 2A.
C. H3K9me2 ChIP-qPCR for the indicated strains harboring unaltered centromeres. The normalized ratio between qPCR products from Cnp1 domains (central core 1&3; cc) and heterochromatic outer repeats (otr) is shown. Error bars represent the SD (n=3). See also Fig. S3C.
D. Images of septated cells expressing GFP-Tailswap in a cnp1Δ background with a pericentromeric TetO array insertion (pericen1-tetO-tomato) on chromosome 1. The three classes of cells observed are indicated with the numbers for each class shown below the images. The yellow arrow marks the TetO-tomato focus that is declustered and has low GFP signal. In cells with missegregation and declustering, GFP intensity was measured at each red (TetO-Tomato) focus and the indicated ratios are plotted below. The expected ratio, assuming equal amount of GFP-Cnp1 is present at each centromere, is shown with a red dashed line. Scale bars are 3 μm.
E. Representative time-lapse images providing evidence for centromere inactivation. Arrow indicates a TetO-tomato focus that has very low GFP signal. Scale bar is 3 μm.
Evidence for Centromere Inactivation in Cnp1 N-Tail Variant Cell Populations Without Alterations in the Central Core Sequence
To test if centromere inactivation occurs with Cnp1 N-tail variants in the absence of any insertions in the central core, albeit with lower penetrance, we first performed ChIP followed by quantitative PCR for presence of H3K9me2. This approach revealed a significant 2 to 4-fold increase in H3K9me2 in the central core in the presence of N-tail variants (Fig. 3C, Fig. S3C). Based on the analysis in Fig. 2, we suggest this elevation reflects inactivation of endogenous centromeres in a subset of the cell population that presumably leads to eventual lethality of the cells in which the inactivation event has occurred.
To assess centromere inactivation in single cells, we imaged GFP-Tailswap septated cells harboring a TetO array inserted adjacent to cen1 (referred to as pericen1-tetO-tomato), which is not synthetically lethal with N-tail variants (Fig. S2B; Table S1). We observed a significant frequency of two phenotypes: missegregation of cen1 (12/262) and declustering of cen1 from the other centromeres (33/262). Following missegregation, one of the two cen1 foci was always declustered (12/12; Fig. 3D). Notably, the missegregated and declustered centromeres exhibited highly reduced GFP signal, compared to the amount expected, indicating loss of Cnp1 from that centromere (Fig. 3D). In a complementary approach, we imaged fields of cells over time. No missegregation of the pericen1-tetO-tomato labeled chromosome was observed from imaging 175 GFP-Cnp1 divisions. From 457 GFP-Tailswap divisions, we could unambiguously score 6 events where both chromatids for Chr I segregated into one cell (Fig. 3E; the low signal-to-noise of the pericen1-tetO-tomato marker makes this number an underestimate). In all 6 cases, one of the TetO-marked centromeres was declustered and did not exhibit GFP signal (Fig. 3E, arrow), suggesting inactivation of the centromere on that chromatid.
Overall, both H3K9me2 ChIP-qPCR and imaging of single cells indicate that there is an elevated frequency of centromere inactivation events in the presence of Cnp1 N-tail variants even in the absence of any alterations in central core sequence.
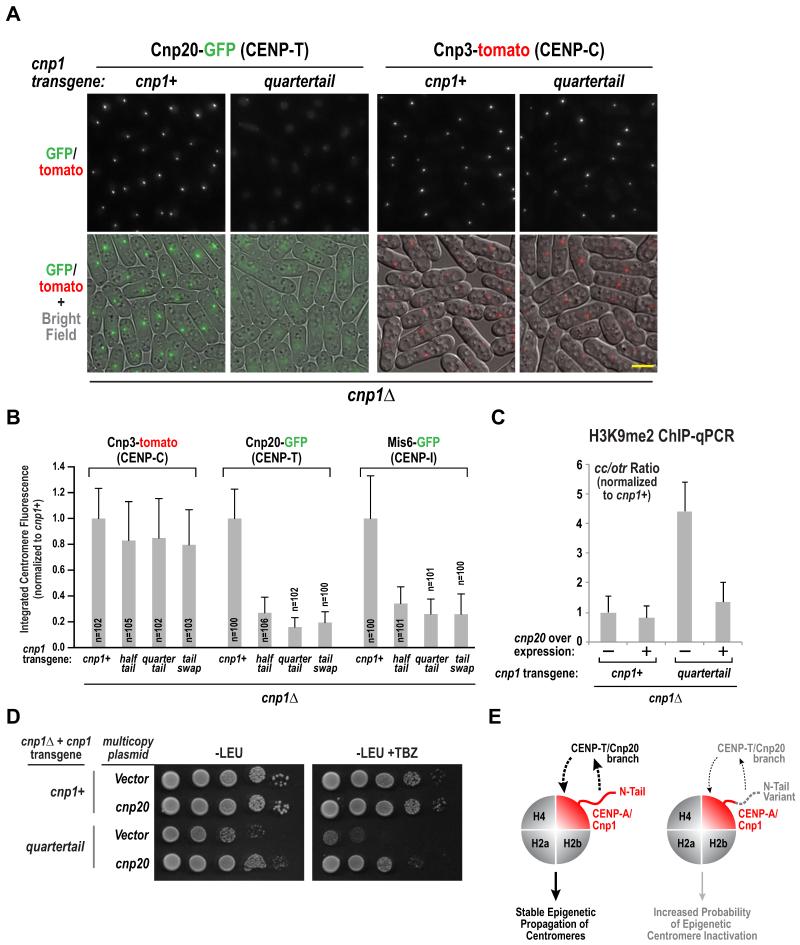
Cnp1 N-Tail Variants Selectively Reduce Centromeric Accumulation of the Cnp20/CENP-T Branch of the Inner Kinetochore
The increased probability of centromere inactivation in the absence of a loading defect led us to investigate the effect of Cnp1 N-tail variants on the chromatin-proximal region of the kinetochore. CENP-A nucleosomes primarily recruit Cnp3/CENP-C via their C-tail [28, 29] and, by an unknown mechanism, recruit the Cnp20/CENP-T branch of the CCAN [30]. Cnp3/CENP-C localization at centromeres was only mildly affected by the tested N-tail variants (Fig. 4A,B); in contrast, there was a striking and consistent reduction in Cnp20/CENP-T at centromeres (Fig. 4A,B). We next monitored localization of Mis6/CENP-I and found that its reduction was comparable to Cnp20 reduction (Fig. 4B, Fig. S4A). Thus, the CENP-T branch of the CCAN appears to be selectively diminished at centromeres in the Cnp1 N-tail variants, which likely underlies the increased probability of centromere inactivation and high rates of missegregation. As Ndc80 localization is unperturbed in the N-tail variants (Fig. 1G,H), and cnp20 mutants do not affect Ndc80 recruitment [31], this defect is potentially unrelated to the role of CENP-T family proteins in direct recruitment of the Ndc80 complex [32,33].
Figure 4. Cnp1 N-tail variants selectively reduce centromeric accumulation of the Cnp20/CENP-T branch of the CCAN.
A. Representative images of Cnp20-GFP and Cnp3-tomato in the indicated strains. Scale bar represents 5 μm. See also Fig. S4A.
B. Integrated fluorescence intensity of Cnp20-GFP, Mis6-GFP and Cnp3-tomato foci was measured and plotted for the indicate strains as in Fig. 1F&G. Error bars represent the standard deviation.
C. ChIP-qPCR with H3K9me2 antibody for the indicated conditions. The normalized ratio of products from Cnp1-enriched regions (cc) and heterochromatic outer repeats (otr) is displayed. See also Fig. S4B. Error bars represent the SD (n=3).
D. Serial dilutions of indicated strains harboring empty or Cnp20-overexpressing multicopy pREP1 plasmid were plated on minimal medium lacking leucine (PMG –LEU), with or without 15 μg/ml TBZ, and grown at 33°C. A ten-fold dilution series is shown for each strain.
E. Schematic summary of key findings. The Cnp1/CENP-A N-tail is required to recruit the Cnp20/CENP-T branch of the CCAN, which in turn is required for stable epigenetic propagation of centromeres (left). Cnp1 N-tail variants are normally loaded and support outer kinetochore assembly but the CENP-T branch of the CCAN is selectively reduced, increasing the probability of centromere inactivation and chromosome missegregation (right).
We next test if overexpression of Cnp20/CENP-T suppressed phenotypic defects of N-tail variants. We overexpressed Cnp20/CENP-T in the Quartertail and Tailswap variants (in a cnp1Δ background) and monitored H3K9me2 accumulation at the central core and TBZ sensitivity to assess suppression. Cnp20 overexpression suppressed elevation of H3K9me2 in the central core in the presence of Quartertail (Fig. 4C; Fig. S4B) and reduced TBZ sensitivity (Fig. 4D). In contrast, we did not observe suppression of Tailswap by Cnp20 overexpression (Fig. S4C,D). As Quartertail shares all of phenotypic features of the N-tail variants described here, these results suggest that the observed defects are primarily derived from reduced centromeric levels of the CENP-T branch of the CCAN. Tailswap may not be suppressed under the conditions tested either because it is more penetrant or because the presence of an H3 tail, a substrate for many modifications, has additional consequences.
Conclusions
The work described here implicates the N-tail of CENP-A in fission yeast in recruitment of the CENP-T branch of the CCAN and suggests that cooperation of Cnp1/CENP-A and Cnp20/CENP-T is important for stable centromere inheritance (Fig. 4E). While a prior study in human cells suggested that substitution of the CENP-A N-tail with the N-tail of H3 did not perturb CENP-T recruitment [11], this result may be due to a redundant contribution from the human centromeric alpha satellite sequence-specific DNA binding protein CENP-B [11,34-36].
As N-tail variants support viability, unlike a cnp20 mutant [31], and do not eliminate Cnp20/CENP-T centromere localization, there must be additional Cnp20/CENP-T localization mechanisms. Recent work suggests that monomethylation of lysine 20 on histone H4 (H4K20me1) of CENP-A nucleosomes may be a mark for CENP-T recruitment in vertebrates [37]. However, deletion of the only known H4K20 methyltransferase in fission yeast does not cause increased TBZ sensitivity [38] and H4K20me1 is not enriched at the CENP-A containing central core domain of the centromere (P. Svensson and K. Ekwall, personal communication). Future work is needed to assess if this modification of CENP-A nucleosomes plays a role in CENP-T recruitment outside of vertebrates. In addition, it will be important to elucidate the biochemical nature of the Cnp1/CENP-A N-tail – Cnp20/CENP-T connection, as well as determine the precise timing and mechanisms responsible for the inactivation events observed in the N-tail variants with reduced Cnp20/CENP-T recruitment.
Experimental Procedures
Details on strain and plasmid construction, genetic analysis, imaging, and chromatin immunoprecipitations are provided in the Supplemental Experimental Procedures.
Supplementary Material
Acknowledgments
We thank Shigeaki Saitoh and Kohta Takahashi (Kurume University), Alison Pidoux, Georgina Hamilton and Robin Allshire (University of Edinburgh), Takeshi Sakuno and Yoshinori Watanabe (University of Tokyo), Oliver Limbo and Paul Russell (Scripps Institute), and the National BioResource Project - Yeast (Japan) for their generosity in sharing strains, plasmid and expertise; Bing Ren for supporting acquisition of the ChIP-Seq datasets; Peter Svensson and Karl Ekwall (Karolinska Institute), and Ben Black and his colleagues (University of Pennsylvania) for communicating unpublished results. This work was supported by an NIH grant (GM074215) to A.D. and the Intramural Research Program of the National Institute of Health (S.I.S.G.). K.G.H. and K.M. were supported by a Wellcome Trust Programme Grant to K.G.H. [083610] and the Wellcome Trust Centre for Cell Biology core grant [092076]. C.A.E. was supported by a training grant NIH/NCI T32 CA009523-27S1. A.D. and K.O. receive salary and other support from the Ludwig Institute for Cancer Research.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions: H.D.F. designed and conducted the majority of the experiments with input from K.O. and A.D.; K.M.M. and K.G.H. performed the spindle checkpoint analysis; C.C. performed the timelapse imaging; C.A.E. performed the high-throughput sequencing and helped analyze the data; S.I.S.G. supported completion of the project. H.D.F. and A.D. wrote the manuscript, with input from all other authors.
REFERENCES
- 1.Catania S, Allshire RC. Anarchic centromeres: deciphering order from apparent chaos. Current opinion in cell biology. 2014;26:41–50. doi: 10.1016/j.ceb.2013.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Muller S, Almouzni G. A network of players in H3 histone variant deposition and maintenance at centromeres. Biochimica et biophysica acta. 2014;1839:241–250. doi: 10.1016/j.bbagrm.2013.11.008. [DOI] [PubMed] [Google Scholar]
- 3.Fukagawa T, Earnshaw WC. The centromere: dhromatin foundation for the kinetochore machinery. Developmental cell. 2014;30:496–508. doi: 10.1016/j.devcel.2014.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Verdaasdonk JS, Bloom K. Centromeres: unique chromatin structures that drive chromosome segregation. Nature reviews. Molecular cell biology. 2011;12:320–332. doi: 10.1038/nrm3107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Perpelescu M, Fukagawa T. The ABCs of CENPs. Chromosoma. 2011;120:425–446. doi: 10.1007/s00412-011-0330-0. [DOI] [PubMed] [Google Scholar]
- 6.Cheeseman IM, Desai A. Molecular architecture of the kinetochore-microtubule interface. Nature reviews. Molecular cell biology. 2008;9:33–46. doi: 10.1038/nrm2310. [DOI] [PubMed] [Google Scholar]
- 7.Sullivan KF, Hechenberger M, Masri K. Human CENP-A contains a histone H3 related histone fold domain that is required for targeting to the centromere. The Journal of cell biology. 1994;127:581–592. doi: 10.1083/jcb.127.3.581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen Y, Baker RE, Keith KC, Harris K, Stoler S, Fitzgerald-Hayes M. The N terminus of the centromere H3-like protein Cse4p performs an essential function distinct from that of the histone fold domain. Molecular and cellular biology. 2000;20:7037–7048. doi: 10.1128/mcb.20.18.7037-7048.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Takayama Y, Sato H, Saitoh S, Ogiyama Y, Masuda F, Takahashi K. Biphasic incorporation of centromeric histone CENP-A in fission yeast. Molecular biology of the cell. 2008;19:682–690. doi: 10.1091/mbc.E07-05-0504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ravi M, Chan SW. Haploid plants produced by centromere-mediated genome elimination. Nature. 2010;464:615–618. doi: 10.1038/nature08842. [DOI] [PubMed] [Google Scholar]
- 11.Fachinetti D, Folco HD, Nechemia-Arbely Y, Valente LP, Nguyen K, Wong AJ, Zhu Q, Holland AJ, Desai A, Jansen LE, et al. A two-step mechanism for epigenetic specification of centromere identity and function. Nature cell biology. 2013;15:1056–1066. doi: 10.1038/ncb2805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Torras-Llort M, Medina-Giro S, Moreno-Moreno O, Azorin F. A conserved arginine-rich motif within the hypervariable N-domain of Drosophila centromeric histone H3 (CenH3) mediates BubR1 recruitment. PloS one. 2010;5:e13747. doi: 10.1371/journal.pone.0013747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pidoux AL, Allshire RC. Kinetochore and heterochromatin domains of the fission yeast centromere. Chromosome research: an international journal on the molecular, supramolecular and evolutionary aspects of chromosome biology. 2004;12:521–534. doi: 10.1023/B:CHRO.0000036586.81775.8b. [DOI] [PubMed] [Google Scholar]
- 14.Folco HD, Pidoux AL, Urano T, Allshire RC. Heterochromatin and RNAi are required to establish CENP-A chromatin at centromeres. Science. 2008;319:94–97. doi: 10.1126/science.1150944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ishii K. Conservation and divergence of centromere specification in yeast. Current opinion in microbiology. 2009;12:616–622. doi: 10.1016/j.mib.2009.09.007. [DOI] [PubMed] [Google Scholar]
- 16.Yamagishi Y, Sakuno T, Goto Y, Watanabe Y. Kinetochore composition and its function: lessons from yeasts. FEMS microbiology reviews. 2014;38:185–200. doi: 10.1111/1574-6976.12049. [DOI] [PubMed] [Google Scholar]
- 17.Bernard P, Maure JF, Partridge JF, Genier S, Javerzat JP, Allshire RC. Requirement of heterochromatin for cohesion at centromeres. Science. 2001;294:2539–2542. doi: 10.1126/science.1064027. [DOI] [PubMed] [Google Scholar]
- 18.Nonaka N, Kitajima T, Yokobayashi S, Xiao G, Yamamoto M, Grewal SI, Watanabe Y. Recruitment of cohesin to heterochromatic regions by Swi6/HP1 in fission yeast. Nature cell biology. 2002;4:89–93. doi: 10.1038/ncb739. [DOI] [PubMed] [Google Scholar]
- 19.Steiner NC, Clarke L. A novel epigenetic effect can alter centromere function in fission yeast. Cell. 1994;79:865–874. doi: 10.1016/0092-8674(94)90075-2. [DOI] [PubMed] [Google Scholar]
- 20.Pidoux AL, Choi ES, Abbott JK, Liu X, Kagansky A, Castillo AG, Hamilton GL, Richardson W, Rappsilber J, He X, et al. Fission yeast Scm3: A CENP-A receptor required for integrity of subkinetochore chromatin. Molecular cell. 2009;33:299–311. doi: 10.1016/j.molcel.2009.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cheerambathur DK, Desai A. Linked in: formation and regulation of microtubule attachments during chromosome segregation. Current opinion in cell biology. 2014;26:113–122. doi: 10.1016/j.ceb.2013.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tooley J, Stukenberg PT. The Ndc80 complex: integrating the kinetochore’s many movements. Chromosome research: an international journal on the molecular, supramolecular and evolutionary aspects of chromosome biology. 2011;19:377–391. doi: 10.1007/s10577-010-9180-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Musacchio A, Salmon ED. The spindle-assembly checkpoint in space and time. Nature reviews. Molecular cell biology. 2007;8:379–393. doi: 10.1038/nrm2163. [DOI] [PubMed] [Google Scholar]
- 24.Hiraoka Y, Toda T, Yanagida M. The NDA3 gene of fission yeast encodes beta-tubulin: a cold-sensitive nda3 mutation reversibly blocks spindle formation and chromosome movement in mitosis. Cell. 1984;39:349–358. doi: 10.1016/0092-8674(84)90013-8. [DOI] [PubMed] [Google Scholar]
- 25.Takahashi K, Chen ES, Yanagida M. Requirement of Mis6 centromere connector for localizing a CENP-A-like protein in fission yeast. Science. 2000;288:2215–2219. doi: 10.1126/science.288.5474.2215. [DOI] [PubMed] [Google Scholar]
- 26.Sakuno T, Tada K, Watanabe Y. Kinetochore geometry defined by cohesion within the centromere. Nature. 2009;458:852–858. doi: 10.1038/nature07876. [DOI] [PubMed] [Google Scholar]
- 27.Ishii K, Ogiyama Y, Chikashige Y, Soejima S, Masuda F, Kakuma T, Hiraoka Y, Takahashi K. Heterochromatin integrity affects chromosome reorganization after centromere dysfunction. Science. 2008;321:1088–1091. doi: 10.1126/science.1158699. [DOI] [PubMed] [Google Scholar]
- 28.Carroll CW, Milks KJ, Straight AF. Dual recognition of CENP-A nucleosomes is required for centromere assembly. The Journal of cell biology. 2010;189:1143–1155. doi: 10.1083/jcb.201001013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kato H, Jiang J, Zhou BR, Rozendaal M, Feng H, Ghirlando R, Xiao TS, Straight AF, Bai Y. A conserved mechanism for centromeric nucleosome recognition by centromere protein CENP-C. Science. 2013;340:1110–1113. doi: 10.1126/science.1235532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hori T, Amano M, Suzuki A, Backer CB, Welburn JP, Dong Y, McEwen BF, Shang WH, Suzuki E, Okawa K, et al. CCAN makes multiple contacts with centromeric DNA to provide distinct pathways to the outer kinetochore. Cell. 2008;135:1039–1052. doi: 10.1016/j.cell.2008.10.019. [DOI] [PubMed] [Google Scholar]
- 31.Tanaka K, Chang HL, Kagami A, Watanabe Y. CENP-C functions as a scaffold for effectors with essential kinetochore functions in mitosis and meiosis. Developmental cell. 2009;17:334–343. doi: 10.1016/j.devcel.2009.08.004. [DOI] [PubMed] [Google Scholar]
- 32.Malvezzi F, Litos G, Schleiffer A, Heuck A, Mechtler K, Clausen T, Westermann S. A structural basis for kinetochore recruitment of the Ndc80 complex via two distinct centromere receptors. The EMBO journal. 2013;32:409–423. doi: 10.1038/emboj.2012.356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nishino T, Rago F, Hori T, Tomii K, Cheeseman IM, Fukagawa T. CENP-T provides a structural platform for outer kinetochore assembly. The EMBO journal. 2013;32:424–436. doi: 10.1038/emboj.2012.348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Earnshaw WC, Sullivan KF, Machlin PS, Cooke CA, Kaiser DA, Pollard TD, Rothfield NF, Cleveland DW. Molecular cloning of cDNA for CENP-B, the major human centromere autoantigen. The Journal of cell biology. 1987;104:817–829. doi: 10.1083/jcb.104.4.817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Okada T, Ohzeki J, Nakano M, Yoda K, Brinkley WR, Larionov V, Masumoto H. CENP-B controls centromere formation depending on the chromatin context. Cell. 2007;131:1287–1300. doi: 10.1016/j.cell.2007.10.045. [DOI] [PubMed] [Google Scholar]
- 36.Hellwig D, Munch S, Orthaus S, Hoischen C, Hemmerich P, Diekmann S. Live-cell imaging reveals sustained centromere binding of CENP-T via CENP-A and CENP-B. Journal of biophotonics. 2008;1:245–254. doi: 10.1002/jbio.200810014. [DOI] [PubMed] [Google Scholar]
- 37.Hori T, Shang WH, Toyoda A, Misu S, Monma N, Ikeo K, Molina O, Vargiu G, Fujiyama A, Kimura H, et al. Histone H4 Lys 20 monomethylation of the CENP-A nucleosome is essential for kinetochore assembly. Developmental cell. 2014;29:740–749. doi: 10.1016/j.devcel.2014.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sanders SL, Portoso M, Mata J, Bahler J, Allshire RC, Kouzarides T. Methylation of histone H4 lysine 20 controls recruitment of Crb2 to sites of DNA damage. Cell. 2004;119:603–614. doi: 10.1016/j.cell.2004.11.009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.