Abstract
Ctp1 (aka CtIP or Sae2) collaborates with Mre11–Rad50–Nbs1 to initiate repair of DNA double strand breaks (DSBs), but its function(s) remain enigmatic. We report that tetrameric Schizosaccharomyces pombe Ctp1 harbors multivalent DNA-binding and bridging activities. Through structural and biophysical analyses of the Ctp1 tetramer we define the salient features of Ctp1 architecture: an N-terminal interlocking tetrameric helical dimer-of-dimers (THDD) domain and a central intrinsically disordered region (IDR) linked to C-terminal “RHR” DNA interaction motifs. The THDD, IDR and RHR are required for Ctp1 DNA bridging activity in vitro and both the THDD and RHR are required for efficient DSB repair in S. pombe. Our results establish non-nucleolytic roles for Ctp1 in binding and coordination of DSB repair intermediates and suggest that ablation of human CtIP DNA binding by truncating mutations underlie the CTIP-linked Seckel and Jawad syndromes.
INTRODUCTION
DNA double strand breaks (DSBs) arising from exposure to DNA damaging agents including ionizing radiation (IR) or DNA break-inducing chemotherapeutic drugs cause genomic instability, chromosome rearrangements and carcinogenesis1,2. Ctp1 (Ctp1 in fission yeast; CtIP in vertebrates and Sae2 in budding yeast) acts in conjunction with the Mre11–Rad50–Nbs1 (MRN) nuclease complex to coordinate the detection, signaling and repair of cytotoxic DSBs3–7. Together, Ctp1 and MRN initiate homologous recombination (HR)-mediated DSB repair to restore genome integrity. Ctp1(CtIP,Sae2)-dependant orchestration of DSB repair processing occurs through cell cycle regulated modulation of Ctp1 transcript levels in fission yeast5 or via phosphorylation of Sae2 (ref. 6) or CtIP in mammals8. Mutation of Ctp1(CtIP,Sae2) or MRN subunits in budding yeast (Saccharomyces cerevisiae) and fission yeast (Schizosaccharomyces pombe) causes genotoxin sensitivity and meiotic deficiencies resulting from impaired programmed DSB processing1,9. Critical genome maintenance functions of Ctp1(CtIP,Sae2) are underscored by the facts that CtIP is a tumor suppressor in mice10, and human CtIP mutations are linked to the microcephaly and dwarfism Seckel and Jawad syndromes11. However, the reasons that Ctp1(CtIP,Sae2) deficiencies alter biological responses remain unclear.
An early step in the onset of HR-mediated DSB repair is nucleolytic processing, which generates 3′ single strand DNA (ssDNA) tails to initiate strand invasion. Ctp1(CtIP,Sae2) is required for resection, although there may be differences between “clean” and “dirty” DSBs12–15. Also, Ctp1(CtIP,Sae2) is required for removal of proteins attached to the 5′-end of DSBs16–18, including Topoisomerase 2 DNA–protein conjugates induced by anticancer drugs such as Etoposide18. This is consistent with a reported Mg2+-dependent endonuclease activity of Sae2 (ref. 19) and its role in processing hairpin-capped DSBs in vivo20. DSB repair also requires structural DNA scaffolding for coordination of DSB processing intermediates. Intriguingly, Sae2 facilitates coincident “two-ended” DSB resection of IR-induced DSBs13. Thus, Ctp1(CtIP,Sae2) might coordinate DNA intermediates as well. However, Ctp1(CtIP,Sae2) bears no apparent nuclease or DNA interaction domains, and the molecular basis for Ctp1(CtIP,Sae2)-regulated DNA transactions are currently unknown.
We set out to better define Ctp1(CtIP,Sae2) molecular architecture and function as our understanding of Ctp1CtIP/Sae2 structure remains limited. Nbs1 recruits phosphorylated Ctp1 to DSBs via binding of the Nbs1 FHA domain to a Ctp1 SXT motifs (Ctp1 residues 74–94)4,21. The crystal structure of an Nbs1–Ctp1 phosphoprotein recognition element, chromatin immunoprecipitation (ChIP) and biophysical analyses suggested that Ctp1 tethering to a flexible Nbs1 arm restricts Ctp1(CtIP,Sae2) activities to the immediate vicinity of DSBs4. Outside of Nbs1-interacting SXT motifs, additional conserved Ctp1(CtIP,Sae2) regions map to its extreme N- and C-termini4,5. The N-terminal region has a predicted coiled-coil structure and mediates self-interaction in Ctp1 (ref. 5) and CtIP22. A third region of high homology is a C-terminal CxxC-RHR motif of poorly understood function. Here, we report an integrated structural, biophysical and genetic study of S. pombe Ctp1. Our collective results illuminates novel functional roles for Ctp1 as a flexible, DNA sensing and scaffolding factor in DSB repair.
RESULTS
Ctp1 structured domains and regions of intrinsic disorder
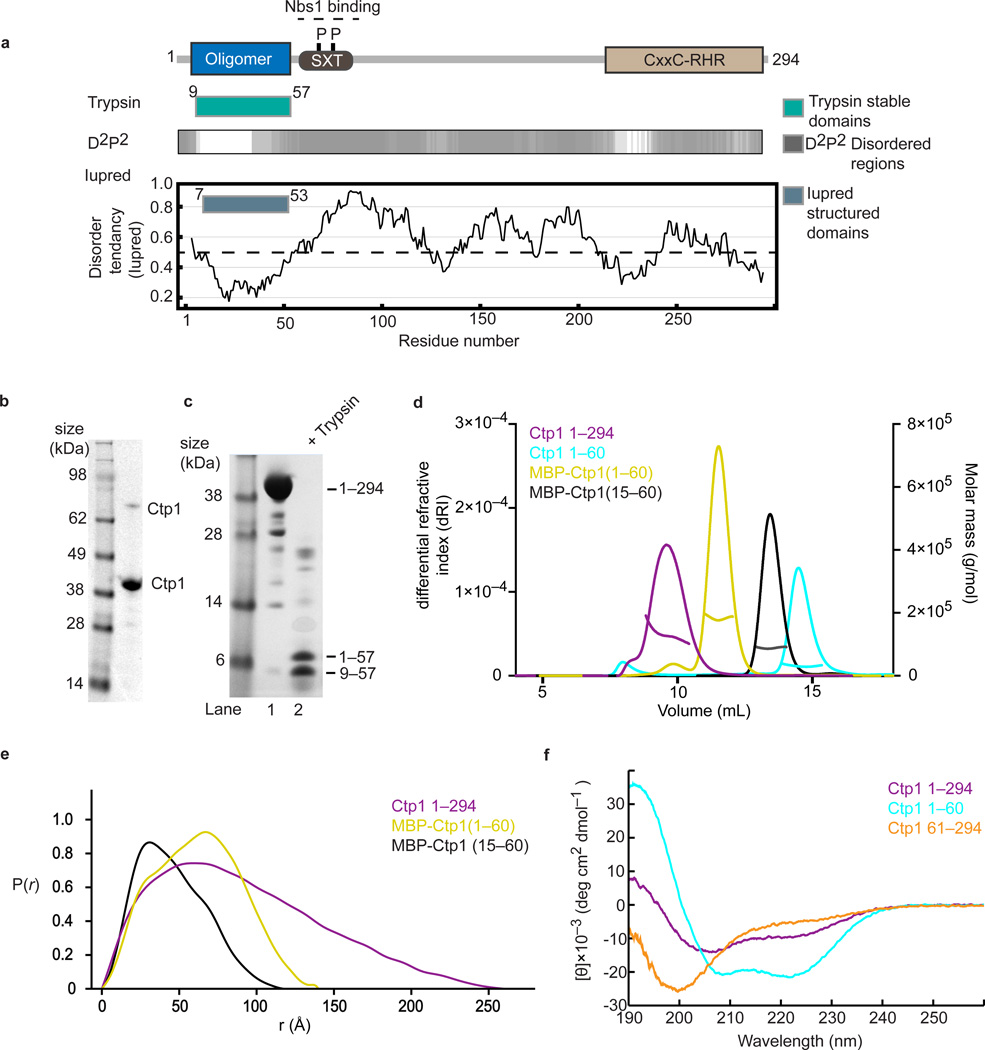
To understand Ctp1 protein architecture, we analyzed the Ctp1 protein sequence using predictors of protein order and disorder with Iupred23 and D2P2 (ref. 24), which derives a consensus disorder prediction from an array of protein order prediction algorithms (Fig. 1a). Both approaches suggest that Ctp1 harbors a structurally ordered region at its N-terminus (Fig. 1a), but is otherwise comprised of expanded stretches of low complexity, commonly referred to as intrinsically disordered regions (IDRs)25. To validate in silico protein order predictions for Ctp1, we purified recombinant full-length S. pombe Ctp1 (Ctp1) (Fig. 1b) and mapped structured domains with limited trypsin proteolysis (Fig. 1c). Purified recombinant Ctp1 migrated aberrantly in SDS-PAGE (> 38 kDa , predicted MW of 33.1 kDa) (Fig. 1b), a hallmark of intrinsically disordered proteins (IDPs)26. Consistent with protein disorder predictions, Ctp1 was highly susceptible to limited proteolytic cleavage, but contains a trypsin-resilient N-terminal core domain (residues 9–57) (Fig. 1c).
Figure 1.
Tetrameric Ctp1 is an intrinsically disordered protein (a) Domain structure of Ctp1 with proteolytic mapping and order or disorder predictions. (b) Purified full-length recombinant Ctp1. Monomeric and dimeric Ctp1 species are labeled. (c) Limited trypsin proteolysis of Ctp1. Trypsinolysis produced two stable fragments that mass spectrometry identified as Ctp11–57 and Ctp19–57 (d) SEC-MALS traces of differential refractive index and molar mass for Ctp1, Ctp11–60, N-terminal maltose-binding protein (MBP)-tagged Ctp11–60 and N-terminal MBP-tagged Ctp115–60 (e) Ctp1 SAXS pair distribution functions of Ctp1, MBP-tagged Ctp11–60 and Ctp115–60 (f) Circular dichroism spectra of Ctp1, Ctp1 1–60 and Ctp161–294 . θ, mean residue ellipticity. Original gels can be found in Supplementary Data Set 1.
Analysis of Ctp1 molecular mass using size-exclusion chromatography with multi-angle light scattering (SEC-MALS) showed Ctp1 is predominantly tetrameric (SEC-MALS molecular mass = 129.1 kDa, Ctp1 monomer mass = 33.1 kDa) in solution (Fig. 1d, Table 1). Small angle X-ray scattering (SAXS) analysis revealed the Ctp1 tetramer is a highly elongated molecule with a maximum particle dimension (Dmax) of 260 Å (Fig. 1e, Supplementary Fig.1). SAXS and SEC-MALS data are consistent with Ctp1 containing extended regions of low structural complexity. Similarly, circular dichroism (CD) measurements of Ctp1 exhibited low abundance of α or β secondary structure (Fig. 1f and Supplementary Table 1). A Ctp161–294 fragment encompassing the predicted disordered region had limited secondary structure as analyzed by CD. In contrast, an amino-terminal fragment Ctp1–60 has ~89 % alpha-helical content by CD (Fig. 1f, Supplementary Table 1), consistent with predictions that the Ctp1 N-terminus contains coiled-coil structure5,22.
Table 1.
Oligomeric state of Ctp1 constructs
| Ctp1 Construct |
Predicted MW*(kDa) |
MALS MW (kDa) |
Oligomeric State |
|---|---|---|---|
| 1–294 | 33.1 | 129.1 | Tetramer |
| 1–60 | 7.2 | 30.8 | Tetramer |
| MBP(1–60) | 49.3 | 197.3 | Tetramer |
| MBP(15–60) | 46.5 | 88.2 | Dimer |
MW, Molecular weight
Both Ctp11–60 and its maltose binding protein (MBP) fusion equivalent (MBP-Ctp11–60) are tetrameric, indicating that Ctp11–60 harbors minimal determinants of Ctp1 oligomerization (Fig. 1d, Table 1). However, despite having ~50 % larger MALS molecular mass than Ctp1 (Table 1), MBP-Ctp11–60 migrated as an apparently smaller species in SEC (Fig. 1d) and had a decreased SAXS maximum particle dimension (Dmax= 140 Å) as compared to Ctp1 (Dmax= 260 Å). This data highlights the large hydrodynamic radius of the intrinsically disordered Ctp1 C-terminus. Thus, altogether the SAXS, SEC-MALS, CD, in silico order predictions and limited proteolytic analyses reveal Ctp1 is largely an IDP, but bears a folded tetramerization core domain at its N- terminus.
Structural basis for Ctp1 tetramerization
To establish the molecular basis for Ctp1 tetramerization we crystallized and determined the X-ray crystal structure of Ctp15–60 to 2.20 Å resolution (Table 2, Supplementary Figs. 2,3). The structure was phased with a brute force molecular replacement strategy using 16 coiled-coil models as search templates in 1276 molecular replacement trials. Search models included monomer, dimer, trimer and tetrameric coiled-coils (see Online Methods).
Table 2.
Data collection and refinement statistics (molecular replacement)
| Ctp1 native* | |
|---|---|
| Data collection | |
| Space group | P21 |
| Cell dimensions | |
| a, b, c (Å) | 49.07, 156.68, 48.93 |
| α,β,γ (°) | 90, 118.55, 90 |
| Resolution (Å) | 50–2.2(2.28–2.20)** |
| Rsym | 0.066(0.543) |
| I / σI | 20.7(2.3) |
| Completeness (%) | 99.9(100) |
| Redundancy | 3.8(3.7) |
| Refinement | |
| Resolution (Å) | 40.7–2.2 |
| No. reflections | 32678 |
| Rwork / Rfree | 19.8/24.6 |
| No. atoms | |
| Protein | 3214 |
| Ligand/ion | 52 |
| Water | 158 |
| B-factors | |
| Protein | 63.8 |
| Ligand/ion | 52 |
| Water | 158 |
| r.m.s. deviations | |
| Bond lengths (Å) | 0.009 |
| Bond angles (°) | 0.952 |
Data derived from single crystal
Values in parentheses are for highest-resolution shell.
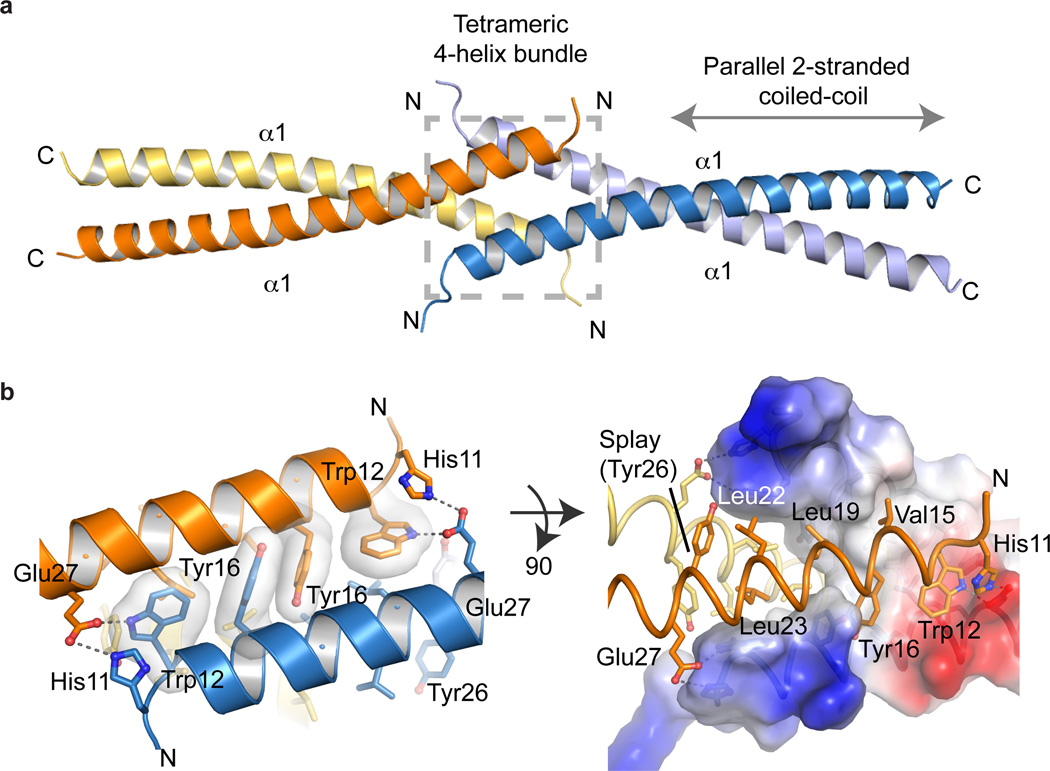
The Ctp1 N-terminal domain assembles as an interlocked tetrameric helical dimer of coiled-coil dimers, hereafter referred to as the THDD domain (Fig. 2a). Each Ctp1 monomer participates in a dimeric two-stranded parallel coiled-coil interaction bounded by residues 28–60. Two parallel coiled-coils engage one another via a 4-helix bundle at the Ctp1 N-terminus (residues 10–25). Within the tetramer, each coiled-coil dimer is oriented ~180° relative to the opposing dimer, creating an overall extended scaffold ~ 120 Å long.
Figure 2.
X-ray crystal structure of the Ctp1 N-terminal tetramerization domain (a) Cartoon representation of Ctp1 highlighting the tetrameric 4-helix bundle (gray dotted-line box) and the parallel 2-stranded coiled-coil (gray arrow). (b) The Ctp1 aromatic hooks. His11, Trp12 and Tyr16 interact with opposing helices to stabilize the tetrameric interface. The electrostatic surface representation of Ctp1 illustrates the transition from tetrameric 4-helix bundle to dimeric coiled-coil by the Tyr26 splay. Blue, electropositive surface; red, electronegative surface; white, hydrophobic.
The interlocked Ctp1 tetramer is unlike other coiled-coils previously characterized. Residues 27–60 of Ctp1 adopt a canonical coiled-coil heptad repeat with hydrophobic side chains occupying “a” and “d” positions of an “a” → “g” heptad repeat (Fig. 2b, Supplementary Fig. 2b). The exception is the 3rd–4th Ctp1 heptads, which participate in charged “a” → “d” position inter-chain salt bridging networks involving Glu40 and Arg44. At the dimer-to-tetramer transition (Tyr26), the coiled-coil repeat is interrupted by a bulky aromatic substitution at the “d”-position. Here, splaying of the dimeric coiled-coil facilitates tetramerization by an array of hydrophobic side chains at the N-terminus of helix α1. Seven residues (W12, V15, Y16, L19, L22, L23 and Y26) form the core of the N-terminal tetrameric interface, burying ~2900 Å2 of solvent accessible surface area. The four intertwined helices resemble two linked fishhooks with Trp12 and Tyr16 making up the “aromatic-hook”. Glu27 and His11 mediate additional inter-subunit salt-bridging contacts across the interface, thereby reinforcing the tetramer.
To test if the Ctp1 N-terminus is critical for Ctp1 tetramerization, we generated a deletion mutation (ΔN14, Ctp115–60), removing the first helical turn of α1 but retaining the Ctp1 dimeric coiled-coil region (residues 27–57). Consistent with predictions from the X-ray structure, MBP-Ctp115–60 is dimeric (Fig. 1d, Table 1). By comparison, deletion of the N-terminal 60 residues of Ctp1 encompassing the THDD (Ctp161–294) yielded a monomeric protein (Supplementary Fig. 4a). SAXS analysis of MBP-Ctp115–60 further affirmed observations from SEC-MALS, demonstrating the decreased size of MBP-Ctp115–60 (Dmax= 115 Å), compared to MBP-Ctp11–60 (Dmax= 140 Å). To validate roles for the aromatic-hook in mediating Ctp1 oligomerization, we generated a triple mutant (H11A W12A Y16A) that disrupts key inter-subunit tetramer contacts. Ctp1 (H11A W12A Y16A) also exhibited defective oligomerization (Supplementary Fig. 4b).
Thus, structural and mutational studies reveal the Ctp1 N-terminus assembles as a tetrameric helical dimer of dimers. We note two additional interfaces observed in the crystal lattice that may mediate alternative Ctp1 tetramerization interfaces. However, these interfaces do not employ the conserved N-terminal 16 residues of the protein that are required for stable oligomerization, which likely form during crystallization.
Ctp1 is a DNA binding factor
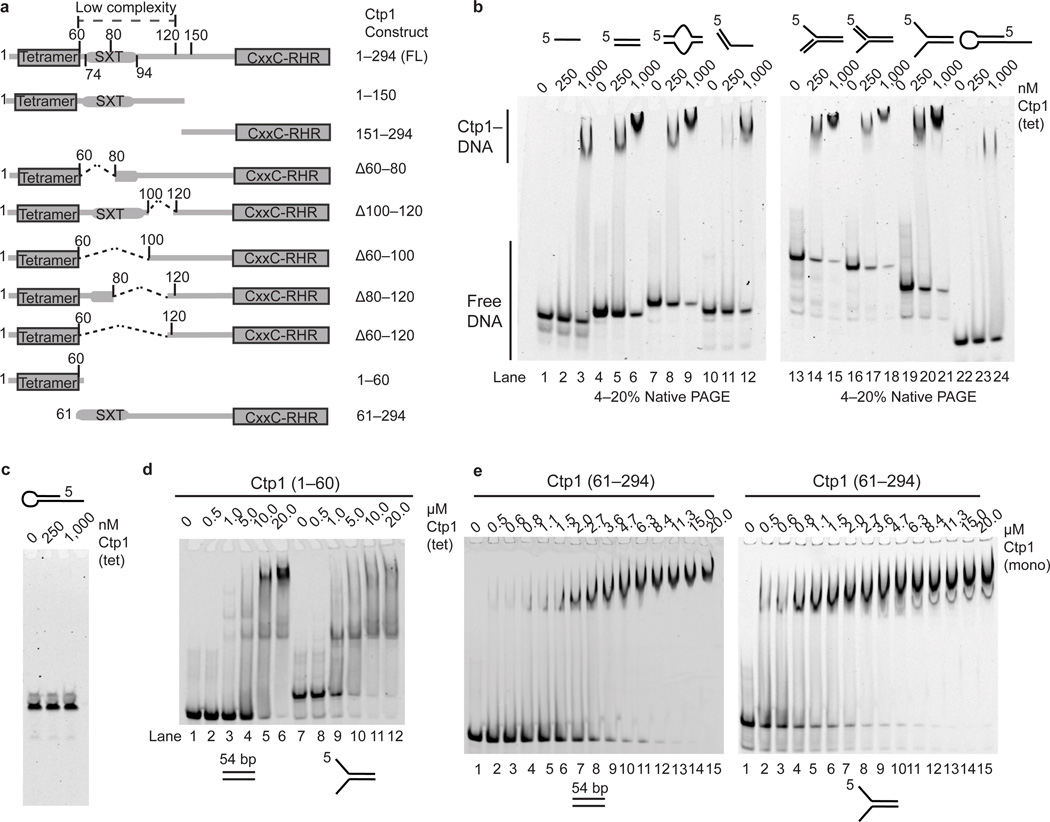
Proteins with extended regions of intrinsic disorder can act as flexible macromolecular interaction hubs25. Such is the case for Nbs1 that binds phosphorylated Ctp1, Mre11 and ATM(Tel1)4,21,27,28. Architecturally, the arrangement of the Ctp1 THDD suggests the tetrameric N-terminal bridge links binding events in the Ctp1 carboxyl-terminus. Given the central role of Ctp1 in HR DSB repair4,5,29, we reasoned that Ctp1 harbors intrinsic DNA binding activities. S. pombe Ctp1 is required for survival of agents causing dirty or random DSBs (e.g. IR) and replication fork stalling or collapse (e.g. Camptothecin, CPT)4,5. Thus, we evaluated the ability of full-length Ctp1 and Ctp1 truncation or deletion mutants to bind to variable DNA structures (Fig. 3a,b, Supplementary Fig. 5a, Supplementary Tables 2,3).
Figure 3.
Ctp1 binds DNA. (a) Domain organization and schematic of Ctp1 N-terminal, C-terminal and internal deletion constructs. (b) Ctp1 binds assorted DNA substrates. (c) Ctp1 displays no appreciable nuclease activity on a DNA hairpin substrate. (d) Ctp11–60 binding double-stranded and forked DNA. (e) Ctp161–294 binds double-stranded and forked DNA. Ctp1 concentrations expressed in terms of molarity of the Ctp1 tetramer (tet) or monomer (mono) as labeled. Experiments were repeated 3 times for (b), (d), and (e), and 2 times for (c). Original gels can be found in Supplementary Data Set 1.
Electrophoretic mobility shift assays (EMSAs) showed Ctp1 binds double-stranded DNA (dsDNA), ssDNA, and ssDNA-dsDNA junction substrates (Fig. 3b). Ctp1 favored binding to DNA forks and exhibited a preference for branched DNAs containing dsDNA-ssDNA junctions over dsDNA or ssDNA alone (Supplementary Fig. 5a). In addition to a preference for DNA secondary structures, Ctp1 displayed a preference for DNA length. Ctp1 titration on a 10 bp ladder revealed stable dsDNA binding for fragments greater than 40–50 bp (Supplementary Fig. 5b) but not for fragments < 20–30 bp. Thus, Ctp1 harbors structure-specific DNA binding. Notably, under conditions where Ctp1 is bound to DNA, in the presence of Mg2+ and ATP we did not observe nucleolytic activity, even at elevated enzyme concentrations (1.0 µM), suggesting that unlike Sae2 (ref. 19) purified recombinant S. pombe Ctp1 is not a nuclease, at least under the conditions examined here (Fig. 3c).
To map Ctp1–DNA interaction determinants, we expressed and purified Ctp1 truncations and internal deletions and evaluated their DNA-binding activities (Fig. 3a, Supplementary Fig. 5c). In isolation, the N- and C-terminal Ctp1 halves, (residues 1–150 and 151–294), displayed no appreciable binding to a 39 bp dsDNA substrate, even at 1.0 µM concentration where Ctp1 is > 90% bound (Supplementary Fig. 5d). To evaluate DNA-binding roles for the internal unstructured region of the protein, we engineered a series of internal deletions removing regions of predicted disorder (Fig. 3a, Supplementary Fig. 5e). In contrast to N- and C-terminal deletions, excision of large portions of the Ctp1 central IDR (e.g. Ctp1Δ60–120, Supplementary Fig. 5e, lanes 10–12) did not ablate dsDNA binding in vitro.
We hypothesized that DNA-binding elements localize to the most evolutionary conserved regions of the divergent Ctp1 orthologs, the N-terminal tetramerization (Fig. 2) and C-terminal CxxC-RHR homology regions. Consistent with this hypothesis, we observed weak sub-micromolar DNA binding by the isolated THDD (Ctp11–60), suggesting this region of Ctp1 contributes to Ctp1–DNA binding (Fig. 3d). A monomeric construct (Supplementary Fig. 4a) of the Ctp1 C-terminus encompassing the IDR and conserved CxxC-RHR (amino acids 61–294) region also bound DNA (Kd = 1.4µM dsDNA, 1.0 µM, forked substrate) (Fig. 3e). Given tetramerization of the N-terminal THDD domain scaffold, the N- and C-terminal Ctp1 DNA binding interactions likely contribute to high-affinity DNA interactions through avidity by linking multiple low-affinity DNA binding sites within the intact tetrameric Ctp1.
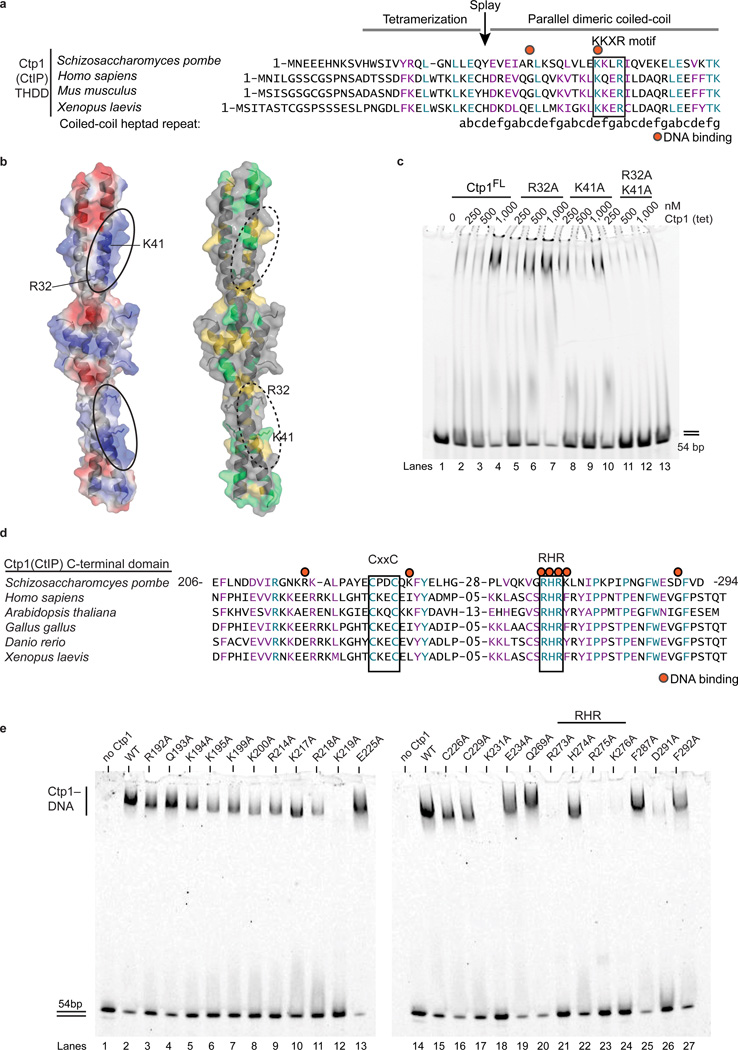
THDD and C-terminal RHR DNA binding motifs
To further interrogate Ctp1 DNA interactions, we mapped Ctp1 electrostatics and sequence conservation onto the THDD structure (Fig. 4a,b). A prominent positively charged surface marks a region of high sequence conservation (Fig. 4b). This includes a “KKxR” motif (residues 41–44 in S. pombe Ctp1) that is highly conserved between fission yeast Ctp1 and vertebrate CtIP. Arg32 in S. pombe Ctp1 abuts the KKxR and extends this putative electropositive DNA binding surface. Although Arg32 is not conserved in vertebrate CtIP, a conserved lysine residue in vertebrate CtIP (at the position of Gln36 in S. pombe Ctp1) would be located in a topologically related position on the coiled-coil surface. To test functional roles for the Ctp1 coiled-coil surface in DNA binding, we targeted the basic surface patches by mutagenesis either individually (R32A and K41A) or in combination (R32A K41A) and evaluated DNA-binding of purified recombinant Ctp1 mutants (Fig. 4c, Supplementary Figs. 6,7). Whilst the single mutants retained DNA-binding activity, an R32A K41A mutation ablated Ctp1–DNA binding in vitro (Fig. 4c). Importantly, the R32A K41A substitution did not disrupt Ctp1 tetramerization (Supplementary Fig. 4a). Thus, collectively our data support a model wherein direct DNA interactions mediated by the THDD are important for Ctp1 DNA-binding function.
Figure 4.
Identifying Ctp1–DNA interaction motifs. (a) Structure-based sequence alignment of the Ctp1 THDD domain. Conserved identical residues are highlighted blue, and similar residues are highlighted purple. Red dots indicate residues targeted for mutagenesis, and the black box outlines the conserved KKxR motif. The position of the Ctp1 coiled-coil “a”→“g” heptad repeat is indicated below the sequence. (b) Electrostatic surface potential (left) and conserved surface mapping (right) of the putative Ctp1 THDD DNA binding interface. Red, electropositive; blue, electronegative; green, conserved identical residues; yellow, conserved similar residues. (c) Ctp1 THDD mutants binding DNA. Ctp1 concentrations expressed in terms of molarity of the Ctp1 tetramer (tet). (d) Sequence alignment of the Ctp1 C-terminal domain. Same coloring as in (a). Red dots designate DNA-binding amino acids. Black boxes outline conserved CxxC and RHR motifs. (e) Alanine scanning mutagenesis to probe DNA binding amino acids in the Ctp1 C-terminal region. Experiments were repeated 3 times for (c), (e). Original gels can be found in Supplementary Data Set 1.
Next, to pinpoint determinants of Ctp1 DNA-binding activity in the evolutionary conserved C-terminal CxxC-RHR region (Fig. 4d), we conducted comprehensive alanine scanning mutagenesis of the Ctp1 carboxyl terminus (Fig. 4e, Supplementary Fig. 6). Of 23 mutants evaluated, 5 mutants affecting positively charged residues (K219A, K231A, R273A, R275A, and K276A) ablated Ctp1-binding to a 54 bp dsDNA at a concentration of 200 nM mutant Ctp1. The mutations R273A and R275A directly targeted the “RHR” motif. Alanine substitution of the internal histidine (H274) of the RHR-motif also conferred a weak dsDNA-binding defect (Supplementary Fig. 7 compare lanes 1–7 and 15–21). Thus, we conclude the conserved C-terminal RHR motif is a critical Ctp1 DNA-binding determinant.
Ctp1 is a DNA bridging factor
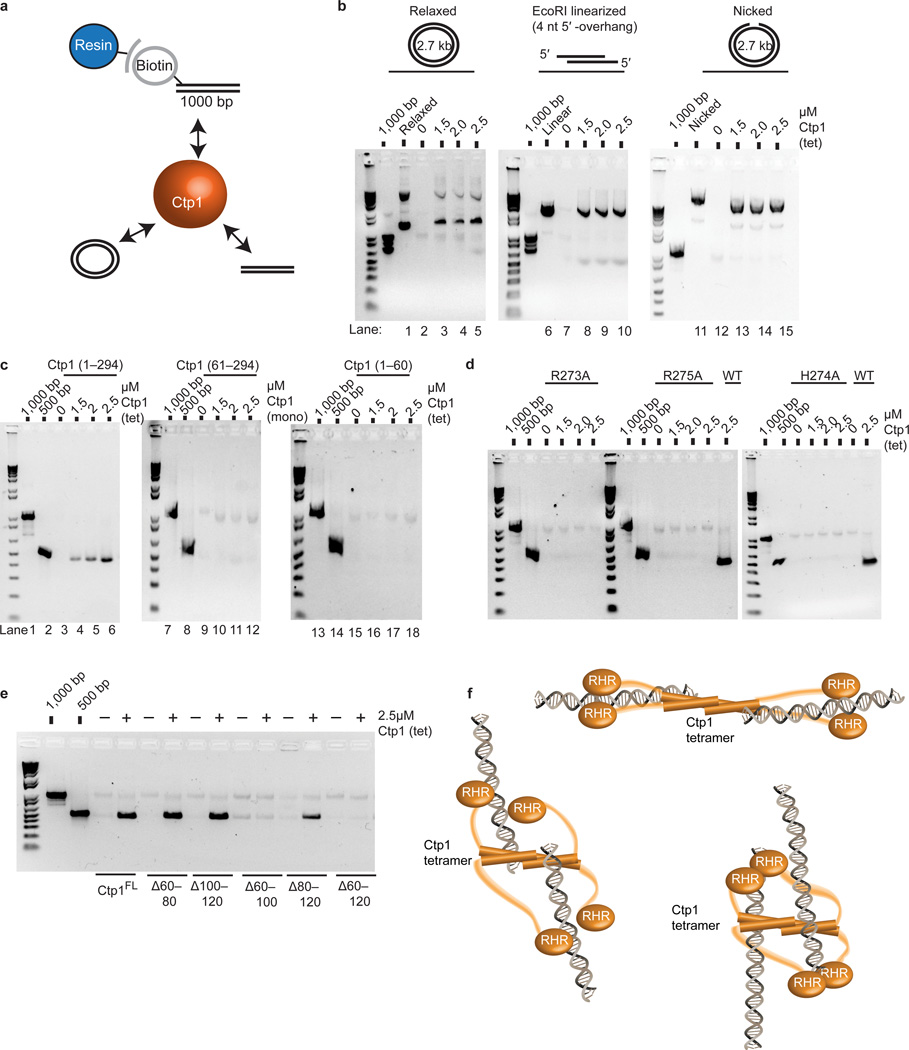
In the context of a Ctp1 tetramer, our structural and mutagenesis data indicates Ctp1 bears at least eight DNA interaction interfaces. What functions do multivalent Ctp1–DNA interactions serve? We observed that titration of Ctp1 on dsDNA super-shifts DNA-bound complexes in EMSAs (Fig. 3). These EMSA super-shifts may reflect the assembly of multiple Ctp1 tetramers onto DNA and/or Ctp1-dependant bridging of DNA termini, a property shared with the MRN core complex30–32. Given the roles for Ctp1 in early DSB repair processing, we reasoned that multivalent Ctp1 DNA-binding acts to bridge and coordinate strand break intermediates.
We adapted a magnetic bead DNA bridging assay33 for Ctp1, and evaluated the ability of recombinant Ctp1 variants to bridge different DNA structures to a resin-immobilized 1.0 kb dsDNA target (Fig. 5a, Online methods). Ctp1 bridged linearized 2.7 kb plasmid DNA (Fig. 5b) or 500 bp linear dsDNA (Fig. 5c) based on Ctp1-dependant recovery in a DNA-tethering pull down. Ctp1 also bridged both nicked and relaxed plasmid DNA substrates (Fig. 5b). Given that Ctp1 preferentially binds to abnormal structures at DNA ends (Supplementary Fig. 5a), these combined results indicate that Ctp1 can interact with both DNA ends and intact dsDNA. Neither the Ctp11–60 THDD domain nor the monomeric Ctp161–294 C-terminus alone tethered DNA (Fig. 5c). Similarly, Ctp1 point mutations that impaired DNA binding also lacked tethering abilities (Fig. 5d).
Figure 5.
Ctp1 bridges DNA (a) Bridging assay schematic. Ctp1 binds a biotinylated 1000 bp DNA substrate linked to magnetic beads and bridges to various substrates. (b) Bridging activity of Ctp1 to plasmid DNA substrates. “1000 bp” denotes biotinylated substrate prior to incubation with protein and DNA bridging targets. (c) DNA bridging activity of Ctp1, Ctp161–294 and Ctp11–60 to a 500bp dsDNA. (d) Bridging activity of Ctp1 C-terminal RHR mutants. WT, wildtype. (e) Bridging activity of Ctp1 internal deletions of the IDR region. (f) Model of Ctp1 binding and bridging a DNA DSB. Three possible Ctp1 DNA bridging architectures are diagrammed. Ctp1 concentrations expressed in terms of molarity of the Ctp1 tetramer (tet) or monomer (mono) as labeled. Experiments were repeated 3 times for (b), (c), (d), and (e), and twice for: EcoRI-linearized in (b), H274A in (d) and Ctp1Δ60–80 in (e). Original gels can be found in Supplementary Data Set 1.
Next, we tested roles for the IDR in facilitating DNA-bridging activity. Neither the 20-residue IDR deletion Ctp1Δ100–120 nor Ctp1Δ60–80, which encompasses the Nbs1-interacting region4, had an impact on bridging. However, the larger 40–60 residue deletion variants, Ctp1Δ60–100, Ctp1Δ80–120 and Ctp1Δ60–120, all impaired tethering of a 500 bp dsDNA to the 1.0 kb resin bound DNA target (Fig. 5e). As the Ctp1Δ60–100, Ctp1Δ80–120, and Ctp1Δ60–120 mutants had comparatively modest effects on dsDNA binding (Supplementary Fig. 5e), an intact IDR appears important for tethering, but not dsDNA binding. Thus, overall Ctp1 architecture appears suitable for orchestrating strand break coordination events (e.g. DNA tethering), and the Ctp1 THDD, IDR and RHR domains are all required for Ctp1 DNA bridging activity in vitro. Based on the requirement for the IDR and RHR for DNA bridging, we hypothesize that Ctp1 coordination of two DNA molecules requires a flexible reach imparted by the IDR, along with direct protein–DNA interactions of both the THDD and RHR. In principal, Ctp1 DNA-bridging interactions may involve any number of combinations of DNA-binding surfaces within a Ctp1 tetramer (Fig. 5f).
THDD and RHR mutations impair DSB Repair
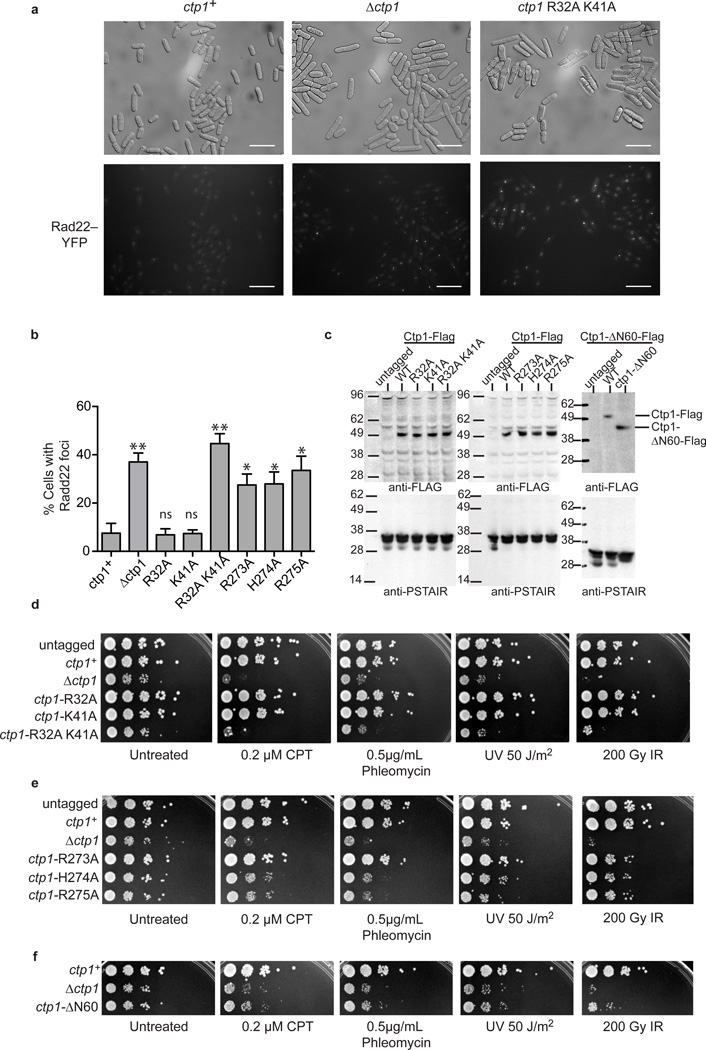
To assess the roles for Ctp1 DNA interaction motifs on DSB repair in vivo, we constructed Ctp1 mutants and monitored cellular growth in the presence or absence of genotoxins (Fig. 6). We replaced the genomic copy of ctp1 with ctp1+, which encodes a C-terminal 5×-Flag epitope-tagged Ctp1 protein (Supplementary Table 4). There was no apparent difference in survival between the ctp1+ and the untagged ctp1 strains. Based on immunoblotting, the mutant proteins were present at levels similar to wild type Ctp1 (Fig. 6c).
Figure 6.
Sensitivity of Ctp1 THDD and RHR mutations to DNA damaging agents (a) Representative images from n=3 cell culture replicates of Rad22-YFP foci in ctp1+, ctp1Ä, and ctp1-R32A K41A strains. Scale bars are 20 µm. (b) Levels of spontaneous Rad22-YFP foci in S. pombe harboring THDD and RHR mutations. Mean values shown. Error bars, s.d. (n=3 cell cultures) * P-value < 0.01; ** P-value < 0.001 compared to ctp1+ by two-tailed Student’s t test. ns, not significant. (c) Immunoblot (anti-Flag) of Ctp1-Flag tagged strains used in panels (a), (b), (d)–(f). Anti-PSTAIR is the loading control. (d) Sensitivity of Ctp1 N-terminal DNA binding mutants to DNA damaging agents. Representative images from n=3 technical replicates. (e) Sensitivity of Ctp1 C-terminal RHR mutants to DNA damaging agents. Images are representative of n=3 technical replicates . (f) Ctp1 N-terminal 60 amino acid deletion mutant sensitivity to DNA damaging agents . Representative images from n=3 technical replicates. Original blots can be found in Supplementary Data Set 1.
Strains with single amino acid substitutions in the Ctp1 THDD DNA-binding interface (R32A or K41A) were insensitive to all genotoxin treatments and exhibited normal growth. In contrast, a complete deletion of the THDD (ctp1-ΔN60) or the THDD double mutation R32A K41A strain mirrored ctp1Δ phenotypes. In the absence of exogenous DNA damage, these phenotypes included elongated cellular morphology, slow growth, and an increased frequency of spontaneous Rad22Rad52 foci (Fig. 6a,b). These phenotypes result from deficient repair of spontaneous DNA damage that leads to DNA damage checkpoint activation and cell death4,5,29. Furthermore, like the ctp1Δ knockout strain, R32A K41A and ctp1-ΔN60 strains were profoundly sensitive to all genotoxins tested (Fig. 6d,f) including 1) UV irradiation, which generates DNA photoproducts that may be converted to DSBs, 2) CPT, a Top1 inhibitor that causes replication fork breakage when the replisome collides with Top1–CPT covalent complexes34,35, 3) the radiomimetic drug phleomycin, which generates DSBs with damaged DNA termini36, and 4) IR. Strains with disruption of the RHR motif (R273A, H274A and R275A) all displayed intermediate phenotypes between ctp1+ and ctp1Δ (Fig. 6e), with R273A displaying the least sensitivity to clastogens. Variability in the RHR motif mutant phenotypes may reflect that additional factors (e.g. DNA binding by the Ctp1 binding partner MRN) differentially compensate for Ctp1 DNA binding defects in cells.
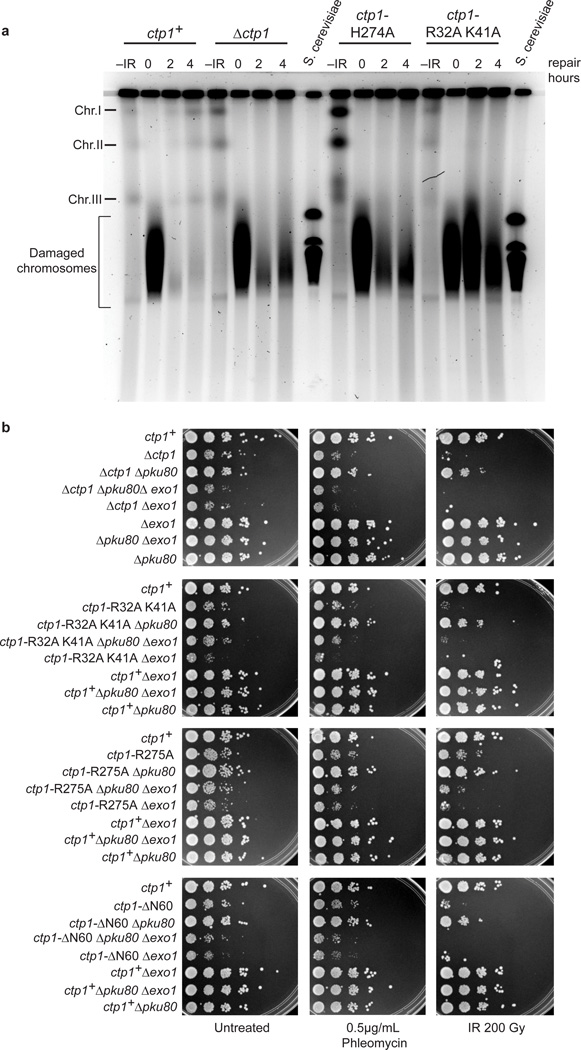
The genotoxin sensitivity suggests that the Ctp1 THDD and RHR motif variants are defective in DNA DSB repair. To directly assess impacts of Ctp1 mutations on chromosomal DSB repair, we exposed cells to 200 Gy IR and returned them to growth in rich medium to monitor repair of IR-induced DSBs by pulse field gel electrophoresis (PFGE) (Fig. 7a). In wildtype (ctp1+) cells, DSB repair resulted in reconstitution of full-length chromosomes within two hours. In stark contrast, no reconstituted chromosomes were detected in ctp1Δ, R32A K41A, or even H274A strains following a 4-hour recovery after IR exposure. Altogether, these results implicate critical roles for Ctp1 DNA–protein interaction interfaces in determining Ctp1 DSB repair functions in S. pombe.
Figure 7.
Ctp1 THDD and RHR mutations effects on S. pombe DSB repair. (a) Damage and repair of IR damaged S. pombe chromosomes. –IR, chromosomes prior to treatment with IR; S. cerevisiae chromosomes were used as size marker. Image is representative of n=2 cell culture replicates. (b) Suppression of Ctp1 RHR, THDD, and N-terminal deletion mutants by deleting Ku80, and rescue by Exo1. Representative images from n=3 technical replicates. Original full-size gel shown.
Published work has established that fission yeast Ctp1 is required for HR repair, but not for non-homologous end joining (NHEJ)5. To examine whether the Ctp1 THDD and RHR mutants are defective in 5′ to 3′ resection in HR repair, we genetically tested whether Exo1 can compensate for Ctp1 mutation. Exo1 is a 5′–3′ exonuclease that can partially substitute for the Mre11 nuclease and Ctp1 when relieved of inhibition by the Ku70–Ku80 complex5. The phenotypes of a ctp1Δ strain can be partially suppressed by deletion of the gene encoding Ku80 from the Ku70–Ku80 DNA end-binding complex that promotes NHEJ and protects DNA ends from the 5′ to 3′ resection activity of Exo1 (refs. 5,14). Similarly, a pku80Δ mutation suppresses the slow growth phenotypes, IR and phleomycin-sensitivities of all ctp1 mutant cells analyzed here including ctp1Δ, ctp1-R32A K41A, ctp1-R275A, and ctp1-ΔN60 (Fig. 7b). Furthermore, this suppression is dependent on Exo1 as the genotoxin-sensitivity of the triple mutants (ctp1-R32A K41A pku80Δ exo1Δ, ctp1-R275A pku80Δ exo1Δ, ctp1-R32A K41A pku80Δ exo1Δ, and ctp1-ΔN60 pku80Δ exo1Δ) was exacerbated compared to the exo1+ strains. Thus, elimination of Ku80 leads to substantial suppression of Ctp1 THDD and RHR mutants by a mechanism that requires the Exo1 5′–3′ nuclease. From these results, we conclude that the phenotypes of Ctp1 THDD and RHR mutants are largely attributable to an inability to properly resect DSBs. In this context, structure-specific Ctp1 DNA binding may act to promote HR by displacing Ku70–Ku80 from DNA, thereby blocking the Ku70–Ku80 complex from initiating NHEJ14. This idea is consistent with genetic data and ChIP experiments demonstrating that Ctp1 is required for Ku70–Ku80 eviction from DSB ends14.
DISCUSSION
Our combined structure-function studies of S. pombe Ctp1 establish novel roles for a flexible tetrameric Ctp1 DNA-tethering scaffold in the binding and coordination of DSB repair intermediates. The N-terminal interlocked Ctp1 THDD facilitates these activities and is unlike other coiled-coil interfaces previously identified. Oligomerization appears to be a conserved primordial feature of all characterized Ctp1 homologs22,37,38. Consistent with our structure-activity studies showing that the Ctp1 THDD is essential for DNA interactions and DSB repair, the S. cerevisiae Sae2 variant L25P also impacts the THDD, impairs Sae2 self-interaction, Sae2-dependant hairpin processing and spore viability37. Similarly, mutations within the predicted human CtIP THDD also impair CtIP DSB recruitment and microhomology-mediated DSB repair in U20S cells38. Sae2 has also been reported to cleave hairpin DNA substrates in vitro19, but this activity has recently been disputed39. We do not detect intrinsic nucleolytic activity for S. pombe Ctp1, even at elevated protein concentrations (Fig. 3c). Recombinant human CtIP also lacks nuclease activity7,40. Thus our data are consistent with non-nucleolytic DNA-binding and tethering functions for the Ctp1 tetramer.
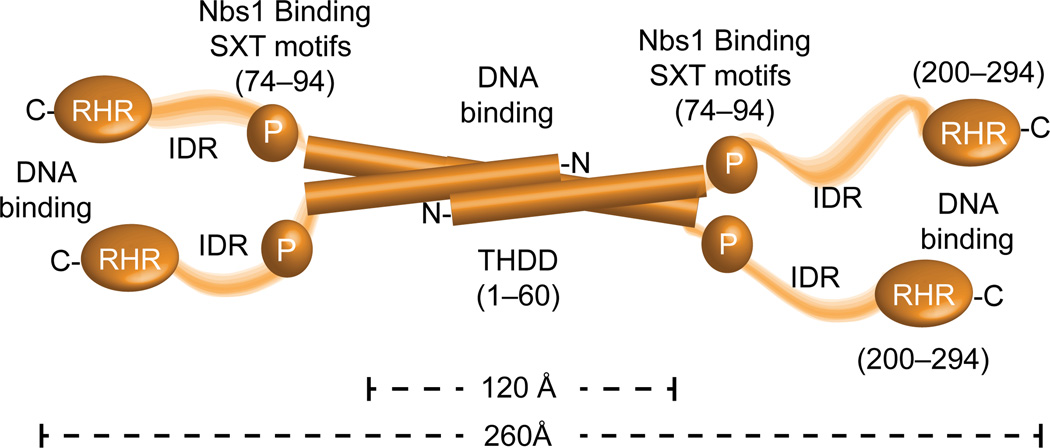
The physical attributes of the flexible Ctp1 architecture exhibit hallmarks of a macromolecular interaction hub25, consistent with roles for Ctp1 homologs as integrators of cell cycle signaling to dictate DSB repair pathway choice5,6,8,41. Current data and published work4,21 indicates four features that define Ctp1 molecular structure: 1) an N-terminal DNA-binding THDD tetrameric core (residues 1–60), 2) the Nbs1-binding phosphorylated Ctp1 SXT motifs4,21 (residues 74–94), 3) a central region of low sequence complexity and intrinsic disorder, and 4) the conserved DNA binding carboxyl-terminal CxxC-RHR region (Fig. 8). Establishing that Ctp1 binds and tethers DNA further reconciles roles for Ctp1 in DNA end processing pathways. Rather than acting as a nuclease, Ctp1 DNA binding and bridging may alternatively modulate the intrinsic nucleolytic activity of a flexibly linked MRN–DNA processing complex. In support of this notion, recent data have demonstrated that S. cerevisiae Sae2 promotes dsDNA endonucleolytic cleavage by MRN proximal to DSB ends39, and Sae2 is required for the coordinated DSB resection of IR-generated “dirty” DSBs13. Flexible associations of Ctp1 with both DNA and Nbs1 may coordinate the two DNA substrates undergoing nuclease processing. Notably, it is also well established that Mre11–Rad50 (MR) core complex can also bridge DNA 30–32,42–44. Although the precise roles for alternative DNA tethering activities within the MRN–Ctp1 complex needs to be established in future work , we hypothesize that alternate means for coordinating DNA substrates might be required initially during a DSB sensing event (e.g. MR mediated bridging) and then subsequently to facilitate DSB processing (e.g. Ctp1-mediated tethering).
Figure 8.
Overall model for Ctp1 molecular architecture from X-ray structures, SAXS and biophysical analysis.
Ctp1–DNA interactions are multivalent. Altogether our observations indicate that the THDD and RHR DNA-binding regions are both critical for Ctp1 function in DSB repair. Weak individual DNA interaction regions with apparent affinities in the micromolar range dictate the high overall affinity of Ctp1–DNA interactions. Such interactions are well suited for driving rapid structural transitions within the context of the MRN core DNA processing machinery. Indeed, blocking any single one of the Ctp1–DNA interfaces impairs Ctp1–DNA interactions in vitro, and has severe consequences for DSB repair at the organismal level in S. pombe (Figs. 6, 7). Intriguingly, two truncating mutations have been mapped in human CtIP that correspond to deletion of the RHR region11. Thus, loss of human CtIP DNA binding activity by nonsense mutation may underlie the CTIP-linked pathology in Seckel and Jawad syndromes11.
Online Methods
Ctp1 expression and purification
All Ctp1 proteins were expressed at 15°C in Escherichia coli Rosetta2 (DE3) (Novagen). Ctp1 point mutations were introduced with Quikchange Mutagenesis (Agilent Technologies) into pGEX-6P-1-Ctp11–294. Ctp11–294, Ctp11–150, and Ctp11–60 were expressed as GST-tagged proteins from pGEX-6P-1 (GE Healthcare) in Terrific Broth. Following cell lysis by sonication in lysis buffer (20 mM Tris pH 7.5, 0.1% β-mercaptoethanol, 1 M NaCl, 0.1 mg ml−1 lysozyme and EDTA-free Complete protease inhibitor cocktail tablet, Roche), GST-fusion proteins were affinity purified on Glutathione Sepharose 4 Fast Flow resin (GE Healthcare). For GST-Ctp11–150 and GST-Ctp11–294, the GST-tags were removed by on-resin PreScission Protease digestion, whereas Ctp11–60 was eluted with lysis buffer plus 20 mM reduced glutathione, and subsequently cleaved.
Ctp115–294, Ctp161–294, Ctp1151–294 were expressed and purified as 6×His-tagged fusions from pMCSG7 (ref. 45). Ctp1 IDR internal deletions were generated as codon optimized synthetic genes (GenScript) and expressed and purified as 6×His-tagged fusions from pMCSG7 (ref. 45). Ctp1206–294 was expressed as 6×His-tagged protein from pET-15b (Novagen). All 6×His-tagged proteins were affinity purified on HisPur Ni-NTA resin (Thermo Scientific) and eluted in lysis buffer plus 250 mM imidazole, followed by His-tag removal by TEV protease (pMCSG7 expressed proteins) or Thrombin protease (Sigma-Aldrich) (pET-15b expressed protein) digestion.
MBP-Ctp115–60 and MBP-Ctp11–60 were expressed as MBP-fusions in the pMALX vector. MBP-tagged proteins were affinity purified on amylose resin (New England Biolabs) and eluted in lysis buffer plus 40 mM maltose. For crystallization, a Ctp15–60- L51M mutant was cloned and expressed as a Histidine-MBP-tagged fusion from pMCSG9 (ref. 45). This mutation was introduced to enable Se-Met phasing and improved crystal quality.
Following affinity purification (above), Ctp1 proteins were additionally purified by combinations of size-exclusion chromatography on a 16/60 S-200 column (GE Healthcare) in size-exclusion column buffer (50 mM Tris pH 7.5, 0.1% β-mercaptoethanol, and 500 mM NaCl), anion exchange chromatography on a HiTrap Q HP column (GE Healthcare), and fractionation on a HiTrap Heparin HP column (GE Healthcare). All proteins were exchanged into storage buffer (20 mM Tris pH 7.5, 0.1 mM Tris(2-carboxyethyl)phosphine (TCEP), 50 mM NaCl and 10% glycerol). Uncropped gels are available in Supplementary Data Set 1.
Ctp1 Trypsinolysis
Purified Ctp1 was digested with 10 µg ml−1 trypsin for 30 minutes at 4°C, prior to quenching with 1 mM phenylmethylsulfonyl fluoride and analysis by SDS-PAGE. The Protein Microcharacterization Core Facility at NIEHS performed protein identification and mass analysis. Uncropped gels are available in Supplementary Data Set 1.
Circular Dichroism (CD) analysis
For CD analysis 0.1 mg ml−1 Ctp1 variants were buffer exchanged into 20 mM sodium phosphate pH 7.5 and 0.1 mM TCEP. CD data were collected on a Jasco J-810 Spectropolarimeter (Jasco Inc.) at 25°C and analyzed using the Selcon3 algorithm46,47 on the Dichroweb Server48.
Small angle X-ray scattering (SAXS)
SAXS data were collected at SIBYLS beamline 12.3.1 at the Advanced Light Source, Lawrence Berkeley National Laboratory. For SAXS studies, Ctp1 variants were buffer exchanged into SAXS buffer (20 mM Tris pH 7.5, 50 mM NaCl, 1 mM TCEP, 5% glycerol). SAXS data collection was performed at 0.5, 1, and 1.5 mg ml−1 for Ctp1 FL, and 2.5, 5.0 and 10.0 mg ml−1 for MBP-Ctp11–60 and MBP-Ctp115–60. SAXS data were analyzed as described previously4 using the ATSAS suite of SAXS data analysis tools (http://www.embl-hamburg.de/biosaxs/software.html).
Size-exclusion chromatography with multi-angle light scattering (SEC-MALS)
For SEC-MALS analysis, MBP-Ctp11–60 or MBP-Ctp115–60 proteins in buffer A (25 mM Tris pH 7.5, 200 mM NaCl, 5 mM maltose) or Ctp1 and Ctp11–60 proteins in buffer B (50 mM Tris pH 7.5, 500 mM NaCl, 0.1% β-mercaptoethanol) or Ctp161–294 and Ctp1 R32A K41A in buffer C (25 mM Tris pH 7.5, 200 mM NaCl, 0.1% β-mercaptoethanol) at 2–20 mg ml−1, were run on a Superdex 200 10/300 GL column (GE Healthcare) fit inline with a miniDAWN TREOS light scattering diode array and an Optilab rex differential refractive index detector (Wyatt Technology). Data were analyzed with ASTRA 5.3.4.14 (Wyatt Technology).
Crystallization of Ctp1
For crystallization, a Ctp15–60- L51M mutant was cloned and expressed with a Histidine-MBP fusion in pMCSG9. This mutation was introduced to enable Se-Met phasing and improve crystal quality, however Se-Met phasing was not required due to a successful molecular replacement solution. Ctp15–60-L51M crystals were grown at 20°C by hanging-drop vapor diffusion by mixing 2 µL protein solution (1 mg ml−1 Ctp15–60 in storage buffer), 2 µL precipitant A (0.2 M sodium acetate, 0.1 M Tris pH 8.5, 16% PEG 4000) and 0.4 µL of 0.1 M barium chloride dihydrate. To alter drop equilibration rates, and increase crystal size, 53 µL of 3 M NaCl was added to 800 µL of precipitant A in the well. Cells were cryo-protected by slow soaking in precipitant A supplemented with 25% ethylene glycol.
X-ray diffraction data collection, phasing and refinement
X-ray data (Table 2) was collected at 105 K on beamline 22-ID of the Advanced Photon Source at a wavelength of 1.000 Å. X-ray diffraction data were processed and scaled using the HKL2000 suite49. An array of 2-helix, 3-helix, and 4-helix coiled-coil bundle containing structures were mined from the RCSB Protein Data Bank and used in molecular replacement trials in PHASER50. A successful molecular replacement hit (PHASER TFZ=8.8, LLG=249) was obtained using 3 copies of a 37-residue parallel dimeric coiled-coil of Geminin (PDB entry 1T6F)51. Following refinement of a poly-alanine model of this MR solution, model building was guided by clear sigma-A weighted Fo-Fc and 2Fo-Fc difference electron density (Supplementary Fig. 3) showing positions of missing coil regions and side chains. Iterative model building and refinement in Refmac52,53 and Coot54 yielded difference electron density that allowed for unambiguous fitting of two Ctp1 tetramers (8 chains) in the monoclinic P21 crystallographic asymmetric unit. Phases and experimental electron density maps were improved by density modification in RESOLVE55,56. Subsequent iterative rounds of model building and refinement were carried out with PHENIX55 and Coot54. The final model displays excellent geometry. Ramachandran statistics: 99.0 % favored, 0.0 % outliers.
Preparation of oligonucleotide substrates
Oligonucleotide substrates and construction are summarized in Supplementary Tables 2–3. DNA substrates were purchased from Integrated DNA Technologies, and annealed by heating to 70°C followed by slow cooling to room temperature. Oligo 39bpf was purified by polyacrylamide gel electrophoresis prior to annealing. Oligonucleotides single-stranded, double-stranded, DNA bubble, 3′-overhang, 3′-flap, 5′-flap and fork were purified by analytical size-exclusion chromatography.
DNA binding assays
For assays involving 6-FAM labeled DNA substrates, purified Ctp1 proteins were incubated with 10 nM DNA substrate in 1X reaction buffer (20 mM Tris pH 7.5, 25 mM NaCl, 0.1 mM DTT, 10 µg ml−1 bovine serum albumin, 0.5% glycerol), for 20 minutes at 20°C. Assays examining mutants of the C-terminus (Fig. 4e) used 200 nM protein in each reaction. Reactions were resolved on Native PAGE 4–20% gradient TBE gels (Invitrogen) on ice and imaged using a Typhoon 9000 imager (GE Healthcare). For Ctp1 binding a 10 bp DNA ladder (Supplementary Fig.5b), purified Ctp1 was incubated with 1 µg µl−1 of a 10bp DNA ladder (Invitrogen), in 1X reaction buffer for 20 minutes at room temperature. Reactions were resolved on a 3% TBE agarose gel. Uncropped gels are available in Supplementary Data Set 1.
For quantitative DNA binding studies (Supplementary Fig. 5a), DNA binding and gel electrophoresis were carried out at 4°C. DNA binding experiments (n=3 for each DNA substrate) were analyzed using ImageJ (http://imagej.nih.gov/ij/). Dissociation constants were calculated using one-site specific binding with hill slope in GraphPad Prism 6.0a (GraphPad Software, Inc.).
DNA nuclease assay
Ctp1 nuclease activity on a hairpin substrate was carried out under the same reaction conditions as the DNA binding assays, with the addition of 0.5 mM ATP and 5 mM MgCl2 to the 1X reaction buffer. Reactions were resolved on a 15% TBE-Urea gel (Invitrogen). Uncropped gels are available in Supplementary Data Set 1.
DNA bridging assays
Biotinylated 1000 bp oligonucleotide substrates for the bridging assays were prepared as previously described33. Nicked plasmid bridging substrate was a modified pUC-19 plasmid (courtesy of Dr. M. Junop), containing an Nb.BbvCI restriction site to generate nicked plasmid. Relaxed circular pHOT-1 plasmid DNA is commercially available (TopoGEN). Supercoiled pHOT-1 was linearized using restriction enzymes indicated in Fig. 5b and gel-purified for bridging assays. For DNA bridging assays, 10 µL of Dynabeads M-280 Streptavidin (Invitrogen) were washed three times in bind buffer (20 mM Tris pH 7.5, 50 mM KCl, 5 mM EDTA, 1 mM DL-Dithiothreitol, 5% glycerol, 40 µg ml−1 bovine serum albumin) prior to incubation with 200 ng of the biotinylated 1000 bp DNA substrate for 15 minutes at room temperature. Beads were collected by magnet for 3 minutes, washed three times with bind buffer and resuspended in 10 µL bind buffer. 200 ng of the 500 bp DNA substrate, the indicated amount of Ctp1 protein variants, and bind buffer was added to total 40 µL and incubated for 30 minutes at 20°C. Beads were then collected by magnet for 3 minutes, washed twice with bind buffer and once with bind buffer lacking bovine serum albumin, before resuspension in 20 µL bind buffer minus bovine serum albumin. Ctp1 was removed from the reaction by adding 10 µg µl−1 Proteinase K and 0.5% SDS for 30 minutes at 65°C. Beads were then collected by magnet for 3 minutes and the supernatant removed and resolved by 1% TBE agarose gel electrophoresis. Uncropped gels are available in Supplementary Data Set 1.
Strain construction
Methods and growth media for S. pombe genetics were carried out using standard methods57. All strains used in this study are listed in Supplementary Table 4. Site-directed mutagenesis was performed using Quikchange Mutagenesis (Agilent). Strains were constructed as previously described4,30, except ctp1+ and mutant derivatives were engineered by PCR to have a C-terminal 5×Flag-tag. Gene replacements were confirmed by PCR, and sequencing was used to verify the presence of the correct point mutation(s).
Immunoblotting
Whole-cell extracts were prepared from 10 mL of mid-log phase cells lysed with 6.5% β-mercaptoethanol and 1.9 M sodium hydroxide. After a 10 minute incubation, 175 µL of 100% TCA was added, followed by a second 10 minute cold incubation. Lysate was pelleted, washed twice with acetone and resolved on a NuPAGE 4–12% Bis-Tris gel (Invitrogen). Proteins were transferred to a nitrocellulose membrane, blocked in 1× phosphate-buffered saline, 0.1% Tween-20 and 5% milk powder. The membrane was first probed with monoclonal anti-Flag M2 (mouse) antibody at 1:1000 dilution (Sigma F1804; for validation see Williams et al., 2008)30, followed by probing with peroxidase-affinipure rabbit anti-mouse IgG antibody (Jackson ImmunoResearch Laboratories 315-035-00) at 1:1000 dilution and developed with ECL Plus Western Blotting Kit (GE Life Sciences). The membrane was then stripped before re-probing with monoclonal anti-PSTAIR (mouse) antibody at 1:1000 dilution (Sigma P7962; for validation see https://www.sigmaaldrich.com/content/dam/sigma-aldrich/docs/Sigma/Datasheet/5/p7962dat.pdf and Tournier et al., 1997)58. Uncropped blots are available in Supplementary Data Set 1.
Clastogen sensitivity assays
Mid-log phase cell cultures grown in YES were serially diluted 10-fold and spotted onto YES agar plates containing clastogens as labeled in Figs. 6 and 7 or spotted onto YES agar plates that were then irradiated with IR at 200 Gy or UV at 50 J m−2. Images were taken after 72 hours of growth with an AlphaImager HP imaging system (Alpha Innotech).
Genomic instability measured by Rad22-YFP foci
The experiment was carried out as previously described, with some modifications59. Briefly, Rad22-YFP-expressing strains were grown in YES to mid-log phase. Live cells were photographed using a Leitz Diaplan microscope combined with a Zeiss AxioCam MRm Rev.3 camera and an 89 North PhotoFluor II light source (Chroma Technology Corp.). For each sample, foci were scored in at least 500 cells for n=3 cell culture replicates. Statistical analyses (student t-test, two-tailed) were performed by GraphPad Prism version 6.0a (GraphPad Software, Inc.).
Schizosaccharomyces pombe whole chromosome repair assay
Irradiation (200 Gy) of S. pombe cells and post-irradiation incubation was as described previously13, except that the irradiated cells were resuspended in YES media instead of YPDA. Plugs for PFGE were prepared as previously reported60, except that standard LE agarose (Lonza) was used rather than low melting point agarose. Samples for PFGE were run using a CHEF Mapper XA system (Bio-Rad) and 0.6% SeaKem Gold agarose (Lonza) in 0.5× TAE (20 mM Tris, 10 mM acetic acid, 0.5 mM EDTA), which was also used for the running buffer. Pulses were ramped linearly from 20 to 30 sec for 81 hours at 1.5 V/cm at 14 °C.
Supplementary Material
Acknowledgements
Our studies are supported by the US National Institute of Health Intramural Program: US National Institute of Environmental Health Sciences (NIEHS), 1Z01ES102765 (R.S.W.), and 1Z01ES021016 (M.A.R.). We thank L. Pedersen of the NIEHS Collaborative crystallography group and the Advanced Photon Source (APS) Southeast Regional Collaborative Access Team (SER-CAT) for beamline access. Use of the APS was supported by the U. S. Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract No. W-31–109-Eng-38. We thank M. Junop (University of Western Ontario) for nicked plasmid substrate, J. Williams of the NIEHS protein microcharacterization core for mass spectrometry analysis, R. Dutcher (NIEHS) for help with MALS analysis, and G. Mueller (NIEHS) and B. Wallace (NIEHS) for comments on the manuscript.
Footnotes
Accession codes
Coordinates and structure factors have been deposited in the RCSB Protein Data Bank under accession code 4×01.
Author Contributions
S.N.A., R.S.W., M.A.R., J.W., and J.S.W. designed the experiments. S.N.A. and P.D.R. performed crystallization experiments. S.N.A. and R.S.W solved and refined the Ctp1 X-ray structure. S.N.A and R.S.W. carried out SAXS experiments. S.N.A, Y.N. and C.D.A performed biochemical experiments. S.N.A, J.W., C.D.A. and J.S.W performed S. pombe experiments. R.S.W. and S.N.A wrote the manuscript with input from all authors. R.S.W. managed the project.
References
- 1.Williams RS, Williams JS, Tainer JA. Mre11-Rad50-Nbs1 is a keystone complex connecting DNA repair machinery, double-strand break signaling, and the chromatin template. Biochem. Cell Biol. 2007;85:509–520. doi: 10.1139/O07-069. [DOI] [PubMed] [Google Scholar]
- 2.Pommier Y. Drugging topoisomerases: lessons and challenges. ACS Chem. Biol. 2013;8:82–95. doi: 10.1021/cb300648v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.You Z, et al. CtIP links DNA double-strand break sensing to resection. Mol. Cell. 2009;36:954–969. doi: 10.1016/j.molcel.2009.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Williams RS, et al. Nbs1 flexibly tethers Ctp1 and Mre11-Rad50 to coordinate DNA double-strand break processing and repair. Cell. 2009;139:87–99. doi: 10.1016/j.cell.2009.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Limbo O, et al. Ctp1 is a cell-cycle-regulated protein that functions with Mre11 complex to control double-strand break repair by homologous recombination. Mol. Cell. 2007;28:134–146. doi: 10.1016/j.molcel.2007.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huertas P, Cortes-Ledesma F, Sartori AA, Aguilera A, Jackson SP. CDK targets Sae2 to control DNA-end resection and homologous recombination. Nature. 2008;455:689–692. doi: 10.1038/nature07215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sartori AA, et al. Human CtIP promotes DNA end resection. Nature. 2007;450:509–514. doi: 10.1038/nature06337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huertas P, Jackson SP. Human CtIP mediates cell cycle control of DNA end resection and double strand break repair. J. Biol. Chem. 2009;284:9558–9565. doi: 10.1074/jbc.M808906200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stracker TH, Petrini JH. The MRE11 complex: starting from the ends. Nat. Rev. Mol. Cell Biol. 2011;12:90–103. doi: 10.1038/nrm3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen PL, et al. Inactivation of CtIP leads to early embryonic lethality mediated by G1 restraint and to tumorigenesis by haploid insufficiency. Mol. Cell. Biol. 2005;25:3535–3542. doi: 10.1128/MCB.25.9.3535-3542.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Qvist P, et al. CtIP Mutations Cause Seckel and Jawad Syndromes. PLoS Genet. 2011;7:e1002310. doi: 10.1371/journal.pgen.1002310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhou Y, Caron P, Legube G, Paull TT. Quantitation of DNA double-strand break resection intermediates in human cells. Nucleic Acids Res. 2014;42:e19. doi: 10.1093/nar/gkt1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Westmoreland JW, Resnick MA. Coincident resection at both ends of random, gamma-induced double-strand breaks requires MRX (MRN), Sae2 (Ctp1), and Mre11-nuclease. PLoS Genet. 2013;9:e1003420. doi: 10.1371/journal.pgen.1003420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Langerak P, Mejia-Ramirez E, Limbo O, Russell P. Release of Ku and MRN from DNA ends by Mre11 nuclease activity and Ctp1 is required for homologous recombination repair of double-strand breaks. PLoS Genet. 2011;7:e1002271. doi: 10.1371/journal.pgen.1002271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mimitou EP, Symington LS. Sae2, Exo1 and Sgs1 collaborate in DNA double-strand break processing. Nature. 2008;455:770–774. doi: 10.1038/nature07312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Neale MJ, Pan J, Keeney S. Endonucleolytic processing of covalent protein-linked DNA double-strand breaks. Nature. 2005;436:1053–1057. doi: 10.1038/nature03872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Farah JA, Cromie GA, Smith GR. Ctp1 and Exonuclease 1, alternative nucleases regulated by the MRN complex, are required for efficient meiotic recombination. Proc. Natl. Acad. Sci. USA. 2009;106:9356–9361. doi: 10.1073/pnas.0902793106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hartsuiker E, Neale MJ, Carr AM. Distinct requirements for the Rad32(Mre11) nuclease and Ctp1(CtIP) in the removal of covalently bound topoisomerase I and II from DNA. Mol. Cell. 2009;33:117–123. doi: 10.1016/j.molcel.2008.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lengsfeld BM, Rattray AJ, Bhaskara V, Ghirlando R, Paull TT. Sae2 is an endonuclease that processes hairpin DNA cooperatively with the Mre11/Rad50/Xrs2 complex. Mol. Cell. 2007;28:638–651. doi: 10.1016/j.molcel.2007.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lobachev KS, Gordenin DA, Resnick MA. The Mre11 complex is required for repair of hairpin-capped double-strand breaks and prevention of chromosome rearrangements. Cell. 2002;108:183–193. doi: 10.1016/s0092-8674(02)00614-1. [DOI] [PubMed] [Google Scholar]
- 21.Lloyd J, et al. A supramodular FHA/BRCT-repeat architecture mediates Nbs1 adaptor function in response to DNA damage. Cell. 2009;139:100–111. doi: 10.1016/j.cell.2009.07.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dubin MJ, et al. Dimerization of CtIP, a BRCA1- and CtBP-interacting protein, is mediated by an N-terminal coiled-coil motif. J. Biol. Chem. 2004;279:26932–26938. doi: 10.1074/jbc.M313974200. [DOI] [PubMed] [Google Scholar]
- 23.Dosztanyi Z, Csizmok V, Tompa P, Simon I. IUPred: web server for the prediction of intrinsically unstructured regions of proteins based on estimated energy content. Bioinformatics. 2005;21:3433–3434. doi: 10.1093/bioinformatics/bti541. [DOI] [PubMed] [Google Scholar]
- 24.Oates ME, et al. D(2)P(2): database of disordered protein predictions. Nucleic Acids Res. 2013;41:D508–D516. doi: 10.1093/nar/gks1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Babu MM, Kriwacki RW, Pappu RV. Structural biology Versatility from protein disorder. Science. 2012;337:1460–1461. doi: 10.1126/science.1228775. [DOI] [PubMed] [Google Scholar]
- 26.Tompa P. Intrinsically unstructured proteins. Trends Biochem. Sci. 2002;27:527–533. doi: 10.1016/s0968-0004(02)02169-2. [DOI] [PubMed] [Google Scholar]
- 27.Schiller CB, et al. Structure of Mre11-Nbs1 complex yields insights into ataxia-telangiectasia-like disease mutations and DNA damage signaling. Nat. Struct. Mol. Biol. 2012;19:693–700. doi: 10.1038/nsmb.2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.You Z, Chahwan C, Bailis J, Hunter T, Russell P. ATM activation and its recruitment to damaged DNA require binding to the C terminus of Nbs1. Mol. Cell. Biol. 2005;25:5363–5379. doi: 10.1128/MCB.25.13.5363-5379.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Akamatsu Y, et al. Molecular characterization of the role of the Schizosaccharomyces pombe nip1+/ctp1+ gene in DNA double-strand break repair in association with the Mre11-Rad50-Nbs1 complex. Mol. Cell. Biol. 2008;28:3639–3651. doi: 10.1128/MCB.01828-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Williams RS, et al. Mre11 dimers coordinate DNA end bridging and nuclease processing in double-strand-break repair. Cell. 2008;135:97–109. doi: 10.1016/j.cell.2008.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hopfner KP, et al. The Rad50 zinc-hook is a structure joining Mre11 complexes in DNA recombination and repair. Nature. 2002;418:562–566. doi: 10.1038/nature00922. [DOI] [PubMed] [Google Scholar]
- 32.Deshpande RA, et al. ATP-driven Rad50 conformations regulate DNA tethering, end resection, and ATM checkpoint signaling. EMBO J. 2014;33:482–500. doi: 10.1002/embj.201386100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Andres SN, et al. A human XRCC4-XLF complex bridges DNA. Nucleic Acids Res. 2012;40:1868–1878. doi: 10.1093/nar/gks022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hsiang YH, Lihou MG, Liu LF. Arrest of replication forks by drug-stabilized topoisomerase I-DNA cleavable complexes as a mechanism of cell killing by camptothecin. Cancer Res. 1989;49:5077–5082. [PubMed] [Google Scholar]
- 35.Pommier Y, Cherfils J. Interfacial inhibition of macromolecular interactions: nature's paradigm for drug discovery. Trends Pharmacol. Sci. 2005;26:138–145. doi: 10.1016/j.tips.2005.01.008. [DOI] [PubMed] [Google Scholar]
- 36.Povirk LF. Processing of damaged DNA ends for double-strand break repair in mammalian cells. ISRN Mol. Biol. 2012;2012 doi: 10.5402/2012/345805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim HS, et al. Functional interactions between Sae2 and the Mre11 complex. Genetics. 2008;178:711–723. doi: 10.1534/genetics.107.081331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang H, et al. CtIP protein dimerization is critical for its recruitment to chromosomal DNA double-stranded breaks. J. Biol. Chem. 2012;287:21471–21480. doi: 10.1074/jbc.M112.355354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cannavo E, Cejka P. Sae2 promotes dsDNA endonuclease activity within Mre11-Rad50-Xrs2 to resect DNA breaks. Nature. 2014;514:122–125. doi: 10.1038/nature13771. [DOI] [PubMed] [Google Scholar]
- 40.Eid W, et al. DNA end resection by CtIP and exonuclease 1 prevents genomic instability. EMBO Rep. 2010;11:962–968. doi: 10.1038/embor.2010.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang Y, Jasin M. An essential role for CtIP in chromosomal translocation formation through an alternative end-joining pathway. Nat. Struct. Mol. Biol. 2011;18:80–84. doi: 10.1038/nsmb.1940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Moreno-Herrero F, et al. Mesoscale conformational changes in the DNA-repair complex Rad50/Mre11/Nbs1 upon binding DNA. Nature. 2005;437:440–443. doi: 10.1038/nature03927. [DOI] [PubMed] [Google Scholar]
- 43.Lobachev K, Vitriol E, Stemple J, Resnick MA, Bloom K. Chromosome fragmentation after induction of a double-strand break is an active process prevented by the RMX repair complex. Curr. Biol. 2004;14:2107–2112. doi: 10.1016/j.cub.2004.11.051. [DOI] [PubMed] [Google Scholar]
- 44.Chen L, Trujillo K, Ramos W, Sung P, Tomkinson AE. Promotion of Dnl4-catalyzed DNA end-joining by the Rad50/Mre11/Xrs2 and Hdf1/Hdf2 complexes. Mol. Cell. 2001;8:1105–1115. doi: 10.1016/s1097-2765(01)00388-4. [DOI] [PubMed] [Google Scholar]
Online Methods References
- 45.Stols L, et al. A new vector for high-throughput, ligation-independent cloning encoding a tobacco etch virus protease cleavage site. Protein Expr. Purif. 2002;25:8–15. doi: 10.1006/prep.2001.1603. [DOI] [PubMed] [Google Scholar]
- 46.Sreerama N, Woody RW. A self-consistent method for the analysis of protein secondary structure from circular dichroism. Anal. Biochem. 1993;209:32–44. doi: 10.1006/abio.1993.1079. [DOI] [PubMed] [Google Scholar]
- 47.Sreerama N, Venyaminov SY, Woody RW. Estimation of the number of alpha-helical and beta-strand segments in proteins using circular dichroism spectroscopy. Protein Sci. 1999;8:370–380. doi: 10.1110/ps.8.2.370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Whitmore L, Wallace BA. DICHROWEB, an online server for protein secondary structure analyses from circular dichroism spectroscopic data. Nucleic Acids Res. 2004;32:W668–W673. doi: 10.1093/nar/gkh371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in osciallation mode. In: Carter CW Jr, Sweet RM, editors. Methods in Enzymology. Vol. 276. Academic Press; 1997. pp. 307–326. [DOI] [PubMed] [Google Scholar]
- 50.McCoy AJ, et al. Phaser crystallographic software. J. Appl. Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Thepaut M, et al. Crystal structure of the coiled-coil dimerization motif of geminin: structural and functional insights on DNA replication regulation. J. Mol. Biol. 2004;342:275–287. doi: 10.1016/j.jmb.2004.06.065. [DOI] [PubMed] [Google Scholar]
- 52.Winn MD, et al. Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 2011;67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D. Biol. Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 54.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D. Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 55.Adams PD, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D. Biol. Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Terwilliger TC. Maximum-likelihood density modification. Acta Crystallogr. D. Biol. Crystallogr. 2000;56:965–972. doi: 10.1107/S0907444900005072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Moreno S, Klar A, Nurse P. Methods in Enzymology. Vol. 194. Academic Press; 1991. Molecular Genetic Analysis of Fission Yeast Schizosaccharomyces pombe; pp. 795–823. [DOI] [PubMed] [Google Scholar]
- 58.Tournier S, Gachet Y, Hyams JS. Identification and preliminary characterization of p31, a new PSTAIRE-related protein in fission yeast. Yeast. 1997;13:727–734. doi: 10.1002/(SICI)1097-0061(19970630)13:8<727::AID-YEA134>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 59.Williams JS, et al. gammaH2A binds Brc1 to maintain genome integrity during S-phase. EMBO J. 29:1136–1148. doi: 10.1038/emboj.2009.413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Birren B, Lai E. Pulsed Field Gel Electrophoresis: A Practical Guide. San Diego: Academic Press; 1993. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.