Abstract
Nuclear factor of activated T cells (NFAT) is activated by calcineurin in response to calcium signals derived by metabolic and inflammatory stress to regulate genes in pancreatic islets. Here, we show that NFAT targets MAPKs, histone acetyltransferase p300, and histone deacetylases (HDACs) to gene promoters to differentially regulate insulin and TNF-α genes. NFAT and ERK associated with the insulin gene promoter in response to glucagon-like peptide 1, whereas NFAT formed complexes with p38 MAPK (p38) and Jun N-terminal kinase (JNK) upon promoters of the TNF-α gene in response to IL-1β. Translocation of NFAT and MAPKs to gene promoters was calcineurin/NFAT dependent, and complex stability required MAPK activity. Knocking down NFATc2 expression, eliminating NFAT DNA binding sites, or interfering with NFAT nuclear import prevented association of MAPKs with gene promoters. Inhibiting p38 and JNK activity increased NFAT-ERK association with promoters, which repressed TNF-α and enhanced insulin gene expression. Moreover, inhibiting p38 and JNK induced a switch from NFAT-p38/JNK-histone acetyltransferase p300 to NFAT-ERK-HDAC3 complex formation upon the TNF-α promoter, which resulted in gene repression. Histone acetyltransferase/HDAC exchange was reversed on the insulin gene by p38/JNK inhibition in the presence of glucagon-like peptide 1, which enhanced gene expression. Overall, these data indicate that NFAT directs signaling enzymes to gene promoters in islets, which contribute to protein-DNA complex stability and promoter regulation. Furthermore, the data suggest that TNF-α can be repressed and insulin production can be enhanced by selectively targeting signaling components of NFAT-MAPK transcriptional/signaling complex formation in pancreatic β-cells. These findings have therapeutic potential for suppressing islet inflammation while preserving islet function in diabetes and islet transplantation.
Nutrients and hormones are coupled to calcium signaling in pancreatic β-cells to regulate insulin production in response to metabolic demand (1–3). Increases in the intracellular ATP to ADP ratio by glucose and other nutrients result in cell depolarization and intracellular calcium fluxes in β-cells (4). These calcium transients induce insulin secretion and increase insulin gene expression in β-cells in response to metabolic fuels, which are amplified by gut-derived incretin hormones, such as glucose-dependent insulinotropic polypeptide and glucagon-like peptide 1 (GLP-1) (5–8).
In response to elevated intracellular calcium, the calcium/calmodulin-dependent protein phosphatase calcineurin (CN) dephosphorylates its downstream target nuclear factor of activated T cells (NFAT) to regulate genes required for β-cell proliferation and function (9–15). Islet-specific genes controlled by CN/NFAT signaling include insulin, glucose transporter isoform-2, glucokinase, and transcription factors pancreatic and duodenal homeobox 1 and neurogenic differentiation 1 (11, 12). Selective deletion of CN or NFAT genes from β-cells in transgenic mice results in diabetes characterized by decreased β-cell mass and function (12, 15). Conditional expression of constitutively nuclear NFAT in CN-deficient mice can rescue them from diabetes (12). Moreover, the CN inhibitor tacrolimus (FK506), widely used to prevent allograft rejection in clinical transplantation, is associated with reduced insulin secretory capacity and a high incidence of diabetes mellitus (16–18). Hence, CN/NFAT is a critical signaling component for β-cells to produce appropriate amounts of insulin to maintain glucose homeostasis.
CN/NFAT also induces expression of inflammatory and apoptotic genes in β-cells. β-Cells produce IL-1β when chronically exposed to high glucose in isolated human islets and type 2 diabetic patients (19, 20). We recently showed that IL-1β activates CN/NFAT to induce multiple inflammatory genes, including TNF-α, IL-1β, interferon-γ, and monocyte chemotactic protein-1, in β-cells (21). These cytokines are associated with islet inflammation and contribute to innate immune and alloimmune mediated islet graft destruction (22–29). IL-1β can also induce β-cell apoptosis by CN-dependent activation of inducible nitric oxide synthase expression (30). Thus, in addition to regulating genes that support β-cell function, CN/NFAT also potentially contributes to β-cell-mediated islet destruction during metabolic and inflammatory stress.
We previously showed that CN/NFAT signaling is integrated with 3 major MAPK pathways (ERK1/2, p38 MAPK [p38], and Jun N-terminal kinase [JNK]) in β-cells (11, 21). GLP-1 enhances glucose-induced activation of CN/NFAT and ERK1/2 in β-cells (31). Blockade of either signaling pathway inhibits up-regulation of the insulin gene by preventing NFAT and v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog A (MafA) to bind to the insulin promoter (11). In contrast, IL-1β induces TNF-α gene expression in β-cells via CN/NFAT and p38/JNK signaling pathways. Activation of both p38 and JNK is required for basic leucine zipper domain-containing transcription factors activating transcription factor (ATF)2 and c-Jun to cooccupy the TNF-α promoter with NFAT and induce expression (21). Moreover, ERK-dependent activation of NFAT-MafA competitively inhibits ATF2/c-Jun induction of the TNF-α promoter. Lastly, all 3 MAPK pathways can activate CCAAT/enhancer binding protein (C/EBP)-β, which represses both insulin and TNF-α promoters (11, 21). These studies show that ERK1/2, p38, and JNK signaling has both opposing and overlapping effects on downstream gene targets in β-cells.
In this study, we sought to determine mechanisms by which MAPKs differentially activate downstream transcription factors required to selectively regulate genes. We now show that NFAT directs MAPKs, histone acetyltransferase p300 (p300), and histone deacetylases (HDACs) to downstream gene targets to selectively regulate the insulin and TNF-α gene promoters. Although NFAT was required for recruitment of MAPKs to gene promoters in response to GLP-1 or IL-1β, MAPKs in turn stabilized transcriptional complexes containing p300 and HDACs to activate and repress gene promoter activity, respectively. These studies provide insight into how cytokine and hormonal signaling is transduced by CN/NFAT to differentially regulate expression of genes in β-cells in response to metabolic and inflammatory stress.
Materials and Methods
Cell and tissue culture
MIN6 β-cells were cultured in DMEM (Gibco) containing 25mM glucose, 10% heat-inactivated fetal bovine serum, 10mM HEPES (pH 7.4), 2mM L-glutamine, 1mM sodium pyruvate, 50μM β-mercaptoethanol, 100-U/mL penicillin, and 100-μg/mL streptomycin at 37°C in 5% CO2 humidified air. Isolated human islets from multiple donors (>90% purity) were obtained from the Islet Cell Resource Center Basic Science Islet Distribution Program and from the Islet Cell Processing Laboratory at Baylor University Medical Center and cultured in RPMI 1640 (Gibco) or Krebs-Ringer bicarbonate HEPES buffer media according to experimental design. Cells were transferred to medium containing 4.5mM glucose overnight and cultured with 2.8mM glucose 2 hours before experimental treatments unless specified otherwise. All experimentation using human islets have been performed a minimum of 3 times using independent donors.
Materials, recombinant DNA constructs, and transfections
Antibodies used were as follows: CN and p300 (Santa Cruz Biotechnology, Inc); NFATc2 (Thermo Scientific Pierce); ERK1/2 and Caspase-3 (Cell Signaling Technology); human TNF-α monoclonal antibody (R&D Systems); and ERK1/2, p38, and JNK as previously described (21). Vectors expressing green fluorescent protein (GFP) and mouse NFAT small hairpin RNA (shRNA) isoforms were obtained from OriGene. Plasmid expression vector dominant negative NFAT PxIxIT motif (dnNFAT) and promoter-reporter constructs harboring the mouse TNF-α and rat I insulin promoters pTNF(−1300)-Luc and rInsI(−410)-Luc, respectively, were previously described (11, 21). Mutation of NFAT DNA-binding sites was performed by site-directed mutagenesis as previously described (9). DNA transfections were performed by Lipofectamine 2000 (Invitrogen) 24 hours before cell treatments. Human islets (1500 IEQ) were pretreated with Accutase (Gibco) at 37°C for 2 minutes before transfection.
Immunofluorescent staining
Cells were plated onto 24-well dishes in Krebs-Ringer bicarbonate HEPES buffer and exposed to glucose under conditions specified for each experiment. Cells were fixed with ice-cold methanol, washed with 1 mL of PBS, and permeabilized with 0.5 mL cold PBS containing 0.2% Triton X-100 for 15 minutes. Before addition of antibodies, cells were washed with PBS containing 0.1% Triton X-100 and 4% BSA. Primary antibody (1:250) in the same solution was incubated with cells for 2 hours. After washing, cells were incubated with secondary antibody (1:5000) in cold PBS containing 0.1% Triton X-100 and 1% BSA for 1 hour.
Flow cytometry
MIN6 cells were transfected with shRNA constructs sh-NFAT-GFP and sh-scramble-GFP control, trypsinized after 24 hours, and subjected to flow cytometry. Fluorescent cells were purified on a MoFlo high-speed cell sorter. Cells were plated at 1 × 105 cells per sample and cultured in RPMI 1640 for an additional 24 hours before indicated treatments.
Luciferase assays
MIN6 cells transfected with promoter-reporter constructs and Renilla luciferase simian virus 40 promoter control plasmid (Promega) were treated under specified conditions and harvested with 1× Reporter Lysis buffer and assayed for luciferase and Renilla enzyme activity by the Dual-Luciferase reporter assay system (Promega) using a TD20/20 bioluminometer (Turner Designs).
Chromatin immunoprecipitation (ChIP) assays
ChIP assays were performed as previously described (21). MIN6 cells were fixed, and chromatin DNA-protein was cross-linked with 1% formaldehyde and sonicated with a Bioruptor 200 (Diagenode) to produce DNA fragments. DNA-protein complexes were immunoprecipitated with indicated antibodies and extensively washed, and the cross-links were reversed by heating to 65°C for 4 hours in Tris-HCl (pH 6.5), 5M NaCl, and 0.5M EDTA. DNA was extracted by phenol/CHCl3 and precipitated with ethanol. Precipitated DNA and 1% control inputs were analyzed by gel electrophoresis of radiolabeled deoxycytidine triphosphate α-32P-labeled end-point PCR products detected by autoradiography and by real-time quantitative PCR (qPCR).
Nuclear and cytosolic fractionation
MIN6 cells and isolated human islets were lysed in buffer solution containing 10mM Tris·HCl (pH 7.8), 10mM KCl, 0.2-mg/mL phenylmethylsulfonyl fluoride (PMSF), 0.1M NaF, 2mM Na3VO4, 10-μg/mL aprotinin, 5-μg/mL pepstatin A, 5-μg/mL leupeptin, and 0.05% Nonidet P-40. Nuclei were isolated by centrifugation on a 0.6M sucrose cushion at 600g in a table top centrifuge, washed, and resuspended in 10mM Tris·HCl (pH 7.8), 10mM KCl, 0.5mM MgCl2, 0.1mM EDTA, and 40% glycerol.
Immunoblot
Cells or nuclear fractions were harvested in lysis buffer containing 50mM HEPES (pH 7.5), 150mM NaCl, 1% Triton X-100, 0.2-mg/mL PMSF, 0.1M NaF, 2mM Na3VO4, 10-μg/mL aprotinin, 5-μg/mL pepstatin A, and 5-μg/mL leupeptin. Islets were sonicated for complete cell lysis. Equal protein lysates were subjected to SDS-PAGE, electrotransferred to nitrocellulose membranes, and blocked with 5% milk in Tris-buffered saline containing 0.1% Tween 20 (pH 7.4). Primary antibody was then added, and the membrane was incubated for 1 hour or overnight at 4°C and washed 3 times with Tris-buffered saline containing 0.1% Tween 20. Membranes were then incubated with secondary antibodies conjugated to DyLight 680 and 800 fluorescent dyes (Cell Signaling) or horseradish peroxidase (Abcam) to detect proteins by ODYSSEY Infrared Imaging System (LI-COR) or SuperSignal West Dura ECL (Thermo Scientific) on a G:BOX (Syngene), respectively.
Nuclear run-on assays
Intact fractionated nuclei were incubated at 30°C for 30 minutes in 10mM Tris·HCl (pH 7.8), 5mM MgCl2, 5mM KCl, 5mM dithiothreitol, 0.1mM EGTA, 0.2mM PMSF, 2-U RNasin recombinant ribonuclease inhibitor (Promega), and 30% glycerol in the presence of 200μM each of rATP, rGTP, rUTP, and rCTP. Reactions were stopped with TRI reagent. RNA was isolated and transcribed to cDNA. Relative changes in insulin, TNF-α, and 18S mRNA were quantified by qPCR.
cDNA synthesis
Total RNA was isolated from cells by using TRI reagent (Ambion). Islets were subjected to multiple passes through a syringe needle for complete cell lysis. RNA was reverse transcribed to cDNA by random primers using the high-capacity cDNA reverse transcription kit (Invitrogen).
Real-time qPCR
qPCR was performed using the Bio-Rad CFX-96 and Stratagene Mx3000P systems with RT2 SYBR Green qPCR Mastermix (SABiosciences). The Mx3000P thermal profile and cycling parameters were 95°C for 10 minutes for 1 cycle followed by 40 cycles of 95°C for 10 seconds and 60°C for 30 seconds. The next primers were used to detect immunoprecipitated 5′-flank DNA gene promoters: mouse insulin II, 5′-AACTGGTTCATCAGGCCATC and 5′-ACTGGGTCCCCACTACCTTT; rat insulin I, 5′-CTGGGAAATGAGGTGGAAAA and 5′-AGGAGGGGTAGGTAGGCAGA; human insulin, 5′-GTCCTGAGGAAGAGGTGCTG and 5′-CCATCTCCCCTACCTGTCAA; mouse TNF-α, 5′-CACACACACCCTCCTGATTG and 5′-CTCATTCAACCCTCGGAAAA; and human TNF-α, 5′-GCCCCTCCCAGTTCTAGTTC and 5′-CTCATTCAACCCTCGGAAAA. The next primers were used to detect cDNA from nuclear run-on assays: insulin, 5′-CTGTGGATGCGCCTCCTGCCCCTGCTGGC and 5′-TCTAGTTGCAGTAGTTCTCCAGCTGGTAGA; TNF-α, 5′-CGAGTGACAAGCCTGTAGCCCA and 5′-CGGCTGATGGTGTGGGTGAGGAGCAC; and 18S, 5′-CGCGGTTCTATTTTGTTGGT and 5′-AGTCGGCATCGTTTATGGTC.
Islet cell viability assay
Islets were stained with Hoechst 33342 and propidium iodide (PI) for 10 minutes before imaging by fluorescence microscopy. Micrographs were merged by ImageJ software (NIH) to identify necrotic cells.
Insulin secretion assay
Isolated human islets were treated for 48 hours in experimental conditions and then cultured in low (2.8mM) glucose for 60 minutes before stimulating in static incubation conditions with high (16.7mM) glucose for 60 minutes. Insulin protein concentration released in the medium was measured by insulin ELISA (ALPCO).
Results
Glucose and GLP-1 synergistically enhance association of NFAT with the insulin gene promoter
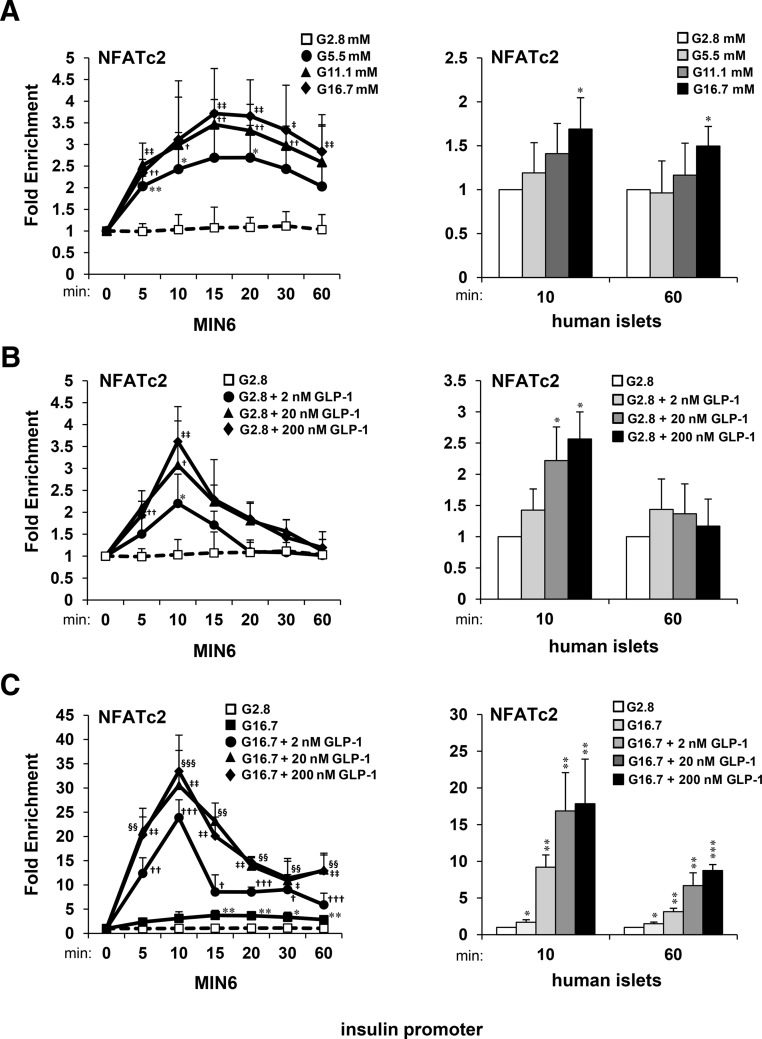
CN and NFAT were previously shown to bind to the insulin promoter in human islets in response to GLP-1 in the presence of glucose (32). However, NFAT was not detected on the insulin promoter by ChIP assay when islets were stimulated with GLP-1 or high glucose alone. In this study, we used ChIP-qPCR as an alternative to ChIP assay by end-point PCR to determine kinetics of effects of glucose and GLP-1 to stimulate association of NFAT with the insulin gene promoter in MIN6 β-cell lines and isolated human islets. ChIP-qPCR kinetic analysis indicated that both glucose and GLP-1 induced a rapid accumulation of NFATc2 upon the insulin promoter within 10–15 minutes (Figure 1, A and B). Although the effects of glucose were relatively sustained for up to 60 minutes, the acute effects of GLP-1 were largely diminished. When combined, glucose and GLP-1 enriched NFATc2 upon the insulin promoter up to 25-fold, which was sustained for up to 60 minutes (Figure 1C). Hence, both glucose and GLP-1 provide signaling components to synergistically enhance CN/NFAT signaling and sustain association of NFAT with the insulin promoter.
Figure 1.
Time- and concentration-dependent effects of glucose and GLP-1 on NFAT association with the insulin gene. ChIP-qPCR assays measuring relative fold enrichment of NFATc2 protein on insulin gene promoter DNA in MIN6 cells and human islets in response to increasing concentrations of glucose (A) or GLP-1 in the presence of (B) low (G 2.8mM) and (C) high (G 16.7mM) glucose. Graphed results are expressed as mean ± SD determined from at least 3 independent experiments. Asterisks above bars or next to plots indicate statistically significant differences (*, †, ‡, and §, P < .05; **, ††, ‡‡, and §§, P < .01; ***, †††, ‡‡‡, and §§§, P < .001) in mean values for treatments compared with 2.8mM glucose controls at corresponding time points based on a one-way ANOVA and Dunnett's multiple comparison post hoc test.
Translocation of NFATc2 and ERK to the insulin gene promoter is blocked by CN inhibitor FK506
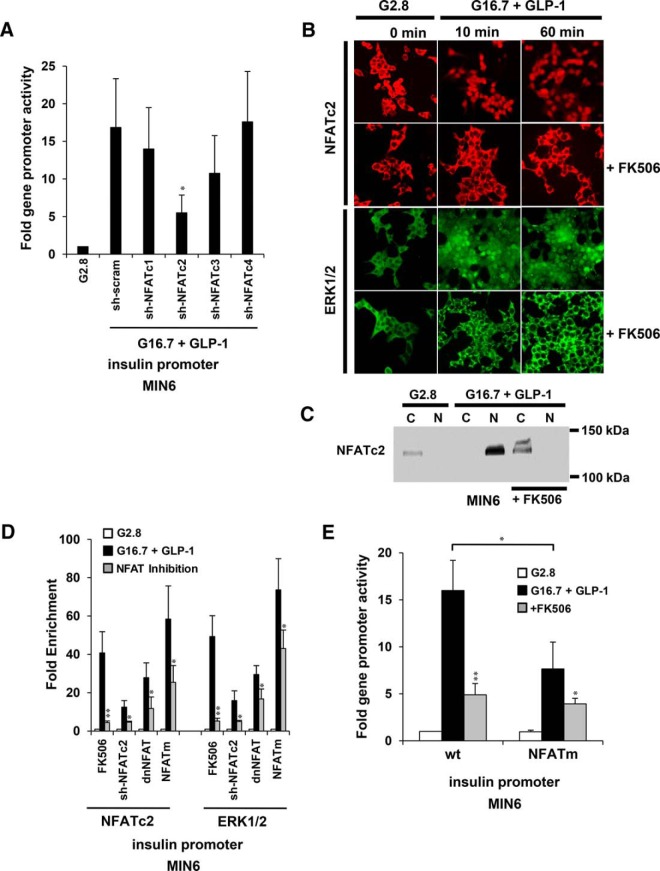
We previously showed that NFATc3 and ERK1/2 cooccupied the 5′ flanking insulin gene promoter in response to high glucose and GLP-1 (32). To identify requirements of NFAT isoforms for induction of insulin promoter activity, we cotransfected an insulin promoter-reporter construct with vectors expressing shRNA to silence gene expression of isoforms of the 4 CN-dependent NFAT transcription factor families in MIN6 cells. Reductions in insulin promoter activity were observed by knocking down NFAT isoform families c1–c3 (Figure 2A). Effects of NFATc2 knockdown, however, were most prominent, and no change was observed by knockdown of NFATc4. To determine the requirement of CN signaling for NFATc2 and ERK1/2 to be directed to the insulin gene promoter, we exposed MIN6 cells to 2.8mM glucose overnight and a pretreatment with 100nM CN inhibitor FK506 for 30 minutes and observed localization by immunofluorescence imaging and immunoblot (Figure 2, B and C). At low glucose, NFATc2 and ERK1/2 were primarily cytosolic. Subsequent treatment of MIN6 cells with 16.7mM glucose and 20nM GLP-1 resulted in a rapid translocation of both NFATc2 and ERK1/2 to the nucleus within 10 minutes and sustained for up to 60 minutes (Figure 2B). Both NFATc2 and ERK1/2 were restricted to the cytosol in the presence of CN inhibitor, FK506. These results correlated with the presence of an NFATc2 protein isoform of approximately 132 kDa detected primarily in cytosolic fractions from MIN6 cells under nonstimulatory conditions and exclusively in nuclear fractions upon stimulation with glucose and GLP-1 (Figure 2C). No distinct changes in molecular weight of NFATc2 from MIN6 extracts were observed by SDS-PAGE between stimulatory and nonstimulatory conditions as previously described in murine T cells (33). However, a shifted band of approximately 142 kDa was detected in cytosolic extracts from MIN6 cells pretreated with FK506, which is likely the result of the prevention of CN from dephosphorylating the protein. Lastly, nuclear translocation of NFATc2 and ERK1/2 corresponded with sustained association of NFAT, ERK1/2, and CN with the insulin gene promoter for up to 1 hour as previously shown by ChIP analysis (32). This association was effectively blocked by treatments with FK506. Collectively, these data indicate that CN activity is required for translocation and formation of a transcriptional complex containing NFATc2 and signaling enzymes ERK and CN upon the insulin gene.
Figure 2.
CN/NFAT signaling is required for ERK1/2 to translocate to the insulin gene promoter in response to glucose and GLP-1. A, Effect of shRNA targeting of NFAT family isoforms on insulin gene promoter activity in response to high (G 16.7mM) glucose and 20nM GLP-1 in MIN6 cells. B, Cellular localization by immunofluorescent staining of NFATc2 and ERK1/2 in MIN6 cells after 10 and 60 minutes of exposure to 16.7mM glucose and 20nM GLP-1 in the presence of CN inhibitor FK506. Cells were treated with 2.8mM glucose for 12 hours before stimulation. Images of nuclear DNA staining by 4′,6-diamidino-2-phenylindole are shown in Supplemental Figure 1. C, Cellular localization analysis of NFATc2 by immunoblot of cytosolic and nuclear fractions of MIN6 cells after 60 minutes of exposure to 16.7mM glucose and 20nM GLP-1 in the presence of CN inhibitor FK506. The immunoblot shown is representative of 3 independent experiments. D, Effects of NFAT inhibition by FK506, sh-NFATc2 RNAi, dnNFAT, and mutated NFAT binding site (NFATm) on fold enrichment of NFATc2 and ERK1/2 upon the insulin gene promoter with respect to 2.8mM glucose control treatments after 10 minutes of exposure of MIN6 cells to 16.7mM glucose and 20nM GLP-1. E, Effects of inhibition of CN/NFAT signaling on fold insulin gene promoter-reporter activity in response to a 6-hour treatment of 16.7mM glucose and 20nM GLP-1 relative to 2.8mM glucose controls in MIN6 cells. Graphed results are expressed as mean ± SD determined from at least 3 independent experiments. Asterisks above bars indicate statistically significant differences (*, P < .05; **, P < .01) in mean values for treatments compared with stimulatory groups (16.7mM glucose and 20nM GLP-1) of each experimental set based on a one-way ANOVA and Dunnett's multiple comparison post hoc test. The asterisk above the bracket (E) denotes statistical significance when comparing stimulatory groups between treatments.
NFAT signaling is required for ERK to associate with the insulin gene promoter
To further examine requirements of CN/NFAT signaling for translocation of ERK1/2 to the insulin gene, we analyzed effects of inhibiting NFAT expression, nuclear localization, and DNA binding on association of proteins with the insulin promoter in response to 16.7mM glucose and 20nM GLP-1 by ChIP-qPCR. Either inhibiting CN activity by FK506 or knocking down expression of NFATc2 by shRNA blocked association of NFAT and ERK1/2 with the endogenous insulin promoter within 10 minutes of stimulation (Figure 2D). Moreover, overexpression of the dnNFAT to inhibit NFAT nuclear translocation also reduced NFAT- and ERK-promoter association. Lastly, mutation of the NFAT binding site (5′-GGAAA to 5′-TCAAA) located at position −139 to −131 proximal to the start site within a rat I insulin gene promoter-reporter construct inhibited association of both NFATc2 and ERK1/2 on the transiently transfected exogenous insulin gene promoter. This correlated with blockade of induction of insulin promoter activity by glucose and GLP-1 (Figure 2E). These results indicate that NFAT activation, nuclear translocation, and DNA-binding are critical components of CN/NFAT signaling to target ERK1/2 to the insulin gene promoter.
CN/NFAT targets ERK to the insulin gene promoter to sustain DNA-binding activity
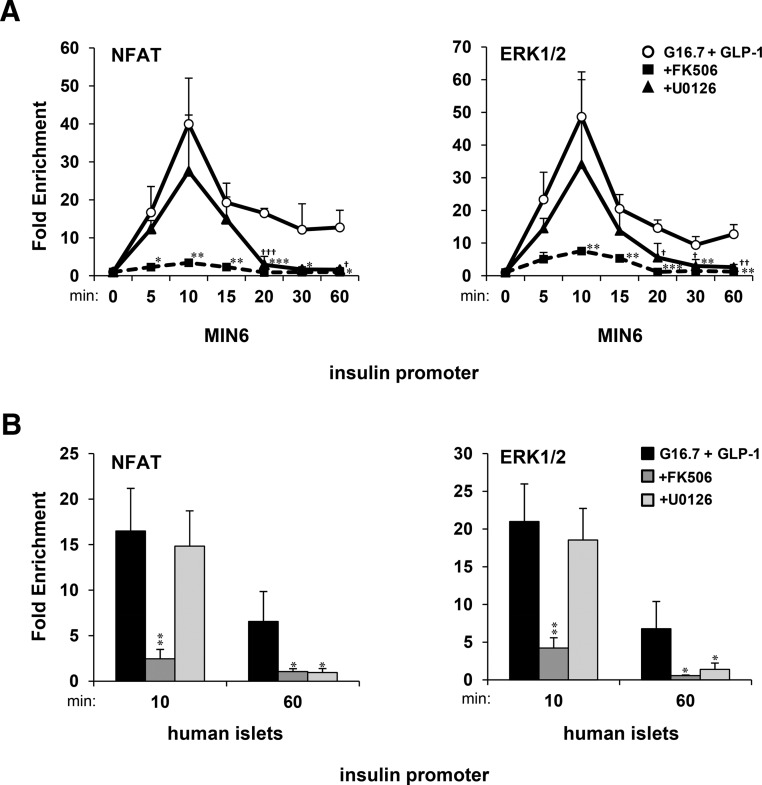
We sought to determine requirements of CN and MAPK activity for NFAT to recruit ERK1/2 to the insulin gene promoter in β-cells. ChIP-qPCR time courses showed that NFATc2 and ERK1/2 associated with the insulin gene promoter within 10-minute exposure of MIN6 cells and human islets to16.7mM glucose and 20nM GLP-1 and was sustained for up to 1 hour (Figure 3, A and B). Acute translocation of NFATc2 and ERK1/2 to the insulin promoter was blocked by FK506 but was relatively unaffected by 5μM MAPK kinase (MEK) inhibitor U0126. However, in the presence of U0126, the association of NFATc2 and ERK1/2 with the insulin promoter was lost within 20 minutes of glucose and GLP-1 exposure. These findings suggest that although CN activity is required for translocation of NFAT-ERK to the insulin promoter, ERK1/2 activity is required for sustained NFAT-ERK-promoter association.
Figure 3.
Sustained association of NFAT and ERK with the insulin gene promoter requires CN and ERK1/2 activity. ChIP-qPCR time-course analysis of fold enrichment of NFATc2 and ERK1/2 upon the insulin gene promoter in (A) MIN6 cells and (B) human islets in response to 16.7mM glucose and 20nM GLP-1 in the presence of FK506 and U0126 with respect to 2.8mM glucose (control). Graphed results are expressed as mean ± SD determined from at least 3 independent experiments. Asterisks above bars or next to plots indicate statistically significant differences (* and †, P < .05; ** and ††, P < .01; *** and †††, P < .001) in mean values for treatments compared with the stimulatory group (16.7mM glucose and 20nM GLP-1) at corresponding time points based on a one-way ANOVA and Dunnett's multiple comparison post hoc test.
NFAT signaling is required for MAPKs to associate with the TNF-α gene promoter
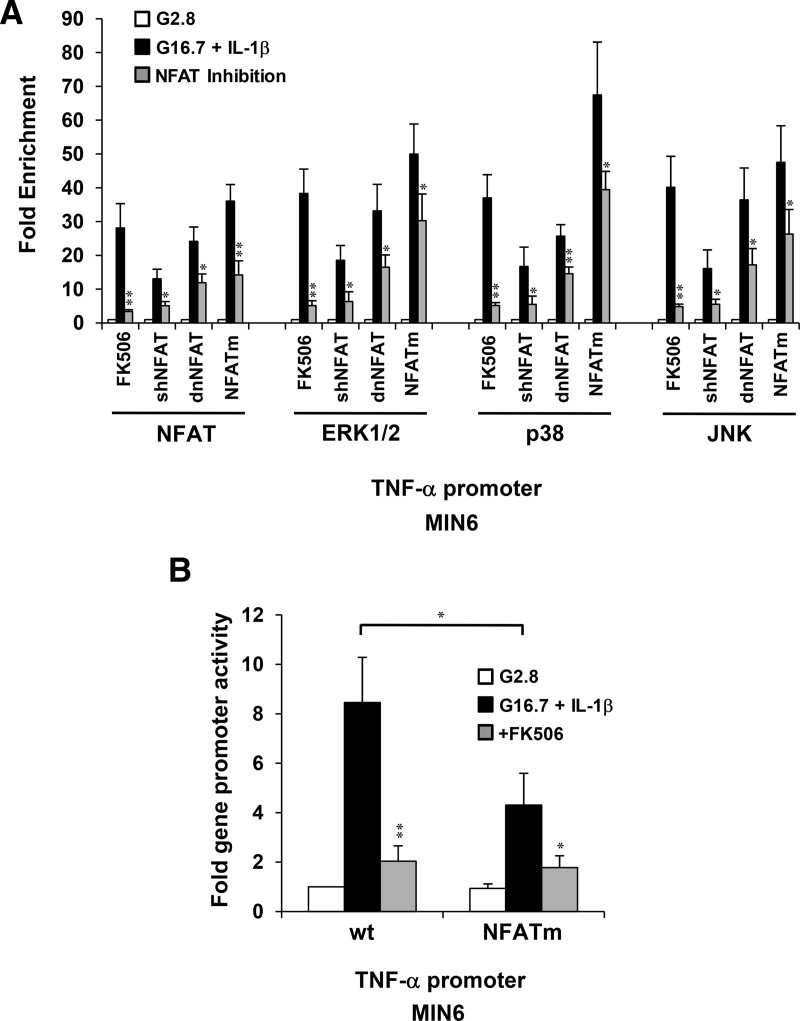
We previously showed that high glucose and IL-1β activate MAPKs ERK1/2, p38, and JNK to induce TNF-α gene expression in pancreatic β-cells (21). Therefore, we analyzed requirements of CN/NFAT signaling to direct MAPKs to the TNF-α gene promoter. Interfering with NFAT expression, nuclear localization, or DNA binding all inhibited association of MAPKs with the TNF-α gene in MIN6 cells in response to 16.7mM glucose and 20-ng/mL IL-1β (Figure 4A). Inhibiting CN or mutating the binding site of NFAT (5′-GGAAA to 5′-TCAAA) located at position −73 to −78 proximal to the start site within a mouse TNF-α gene promoter construct inhibited association of both NFATc2 and ERK1/2 on the transiently transfected exogenous insulin gene promoter, which correlated with reduced TNF-α promoter activity (Figure 4B). These data indicate that intact NFAT signaling is required for MAPKs to associate with the TNF-α gene promoter in response to glucose and IL-1β.
Figure 4.
NFAT mediates association of MAPKs with the TNF-α gene promoter and regulates promoter activity in response to IL-1β. A, Effects of NFAT inhibition by FK506, sh-NFAT RNAi, dnNFAT, and mutated NFAT binding site (NFATm) on fold enrichment of NFAT and MAPKs ERK1/2, p38, and JNK upon the TNF-α gene promoter with respect to 2.8mM glucose controls after 10 minutes of exposure of MIN6 cells to 16.7mM glucose and 20-ng/mL IL-1β. B, Effects of FK506 and NFATm on activation of the TNF-α gene promoter-reporter in response to 16.7mM glucose and 20-ng/mL IL-1β. Graphed results are expressed as mean ± SD determined from at least 3 independent experiments. Asterisks above bars indicate statistically significant differences (*, P < .05; **, P < .01) in mean values for treatments compared with stimulatory groups (16.7mM glucose and 20-ng/mL IL-1β) of each experimental set based on a one-way ANOVA and Dunnett's multiple comparison post hoc test. The asterisk above the bracket (B) denotes statistical significance when comparing stimulatory groups between treatments.
CN/NFAT differentially recruits ERK, p38, and JNK to the TNF-α gene promoter to stabilize complex formation
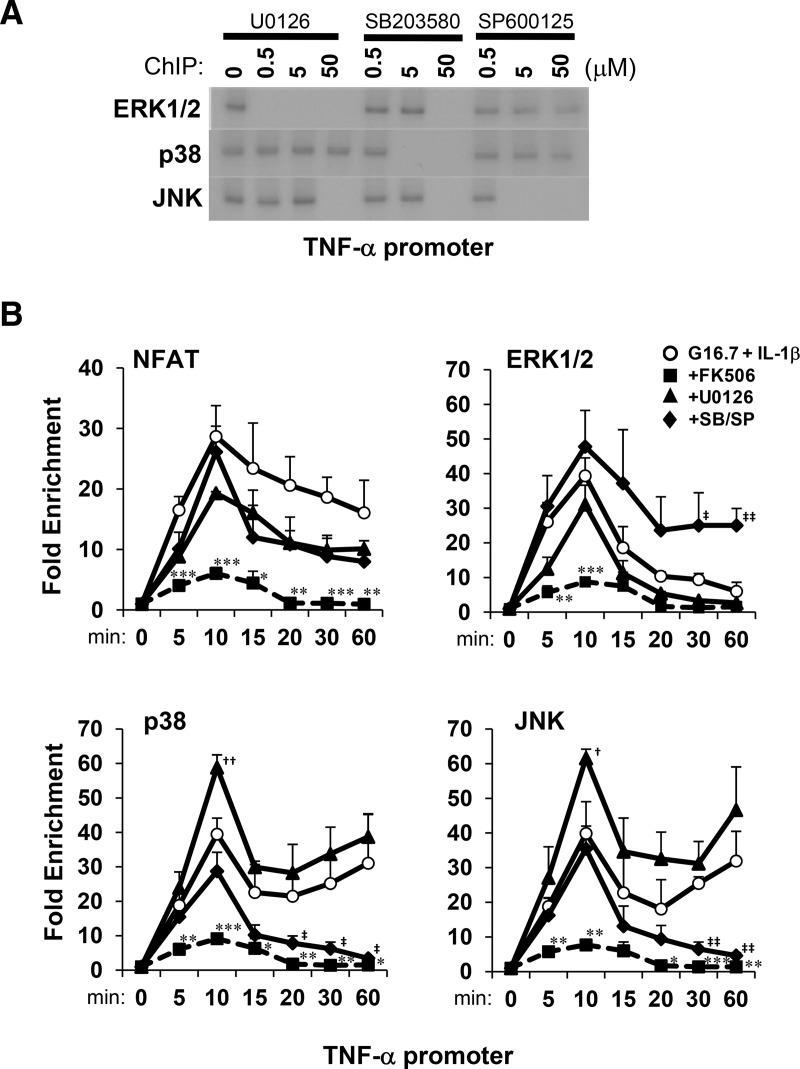
Requirements of CN and MAPK activity for NFAT to recruit MAPKs to the TNF-α gene promoter were also examined by ChIP time-course analysis. Glucose and IL-1β induced association of ERK1/2, p38, and JNK with the TNF-α gene promoter, which could be selectively inhibited within 30 minutes by 5μM U0126 (MEK inhibitor), 5μM SB203580 (p38 inhibitor), and 5μM SP600125 (JNK inhibitor) MAPK inhibitors, respectively (Figure 5A). This indicated a requirement of MAPK activity for MAPK-promoter association. Further examination of effects of inhibitors on MAPK targeting to the TNF-α gene by ChIP-qPCR showed that acute enrichment of NFATc2, ERK, p38, and JNK upon the TNF-α promoter within 10-minute stimulation required CN but was independent of MAPK activity (Figure 5B). In contrast, sustained association (up to 1 h) of MAPKs with the TNF-α promoter was blocked by MAPK inhibitors. Although ERK inhibition enhanced p38 and JNK association, p38 and JNK inhibition enhanced ERK association with the TNF-α promoter. This suggested that MAPK substrates selectively and competitively enhance sustained MAPK association with the TNF-α gene promoter. Collectively, the results indicate that although CN is required for NFAT to acutely direct MAPKs to the TNF-α promoter in response to IL-1β, MAPK activity selectively determines sustained stability of the NFAT-MAPK transcription/signaling factor-promoter complex.
Figure 5.
Selective and sustained association of MAPKs with the TNF-α gene promoter in response to glucose and IL-1β requires MAPK activity. A, ChIP analysis of selective effects of MAPK inhibitors U0126, SB203580, and SP600125 on MAPK association with the TNF-α gene promoter in MIN6 cells in response to 16.7mM glucose and 20-ng/mL IL-1β for 30 minutes. B, ChIP-qPCR time-course analysis of fold enrichment of NFATc2 and MAPKs upon the TNF-α promoter in MIN6 cells in response to IL-1β in the presence of FK506, U0126, or SB203580/SP600125 (SB/SP) relative to 2.8mM glucose controls. Graphed results are expressed as mean ± SD determined from at least 3 independent experiments. Asterisks next to plots indicate statistically significant differences (*, †, and ‡, P < .05; **, ††, and ‡‡, P < .01; ***, †††, and ‡‡‡, P < .001) in mean values for treatments compared with the stimulatory group (16.7mM glucose and 20-ng/mL IL-1β) at corresponding time points based on a one-way ANOVA and Dunnett's multiple comparison post hoc test.
Selective targeting of p300/HDACs to gene promoters by CN/NFAT is dependent on MAPK activity
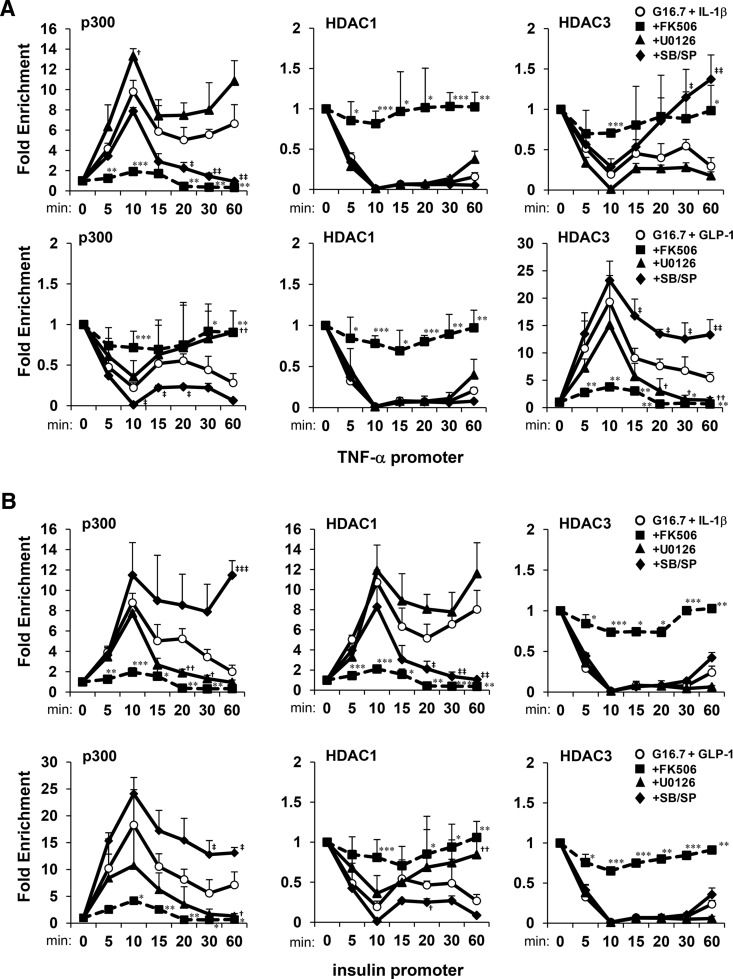
Histone acetyltransferase p300 has previously been shown to enhance expression of the insulin gene in pancreatic β-cells via interactions with transcription factors pancreatic and duodenal homeobox 1 and neurogenic differentiation 1 (34–36). We previously showed that p300 and acetylated histones H3 and H4 cooccupied the insulin gene promoter with NFAT and MAPKs in response to high glucose and IL-1β (37). We therefore tested requirements of CN/NFAT and MAPKs to recruit p300/HDACs to the TNF-α and insulin gene promoters. Time-course ChIP-qPCR analysis showed that p300 was rapidly enriched on the TNF-α gene promoter in response to 16.7mM glucose and 20-ng/mL IL-1β with similar kinetics observed for NFATc2 and MAPKs (Figure 6A). This contrasted with HDAC3, which was rapidly reduced in DNA-promoter complexes within 10 minutes. Similarly, GLP-1 induced rapid accumulation of p300 on the insulin gene promoter with concomitant exclusion of HDAC1 (Figure 6B). It was also observed that HDAC3 could be recruited to the TNF-α promoter as p300 was expelled after treatment with glucose and IL-1β. The effects of stimuli on p300/HDAC recruitment and expulsion required CN, but the opposing chromatin modifying enzymes were not mutually exclusive, because both p300 and HDAC1 were acutely enriched upon the insulin promoter in response to IL-1β. However, stable formation of DNA-protein complexes containing HDAC1 under these conditions significantly diminished promoter occupancy of p300 within 60 minutes.
Figure 6.
CN/NFAT and MAPK activity are required for stable formation of p300 and HDAC association with gene promoters in response to stimuli. ChIP-qPCR time-course analysis of fold enrichment of p300, HDAC1, and HDAC3 upon (A) TNF-α and (B) insulin gene promoters in MIN6 cells in response to 16.7mM glucose and 20nM GLP-1 or 20-ng/mL IL-1β in the presence of FK506, U0126, or SB203580/SP600125 (SB/SP) relative to 2.8mM glucose controls. Graphed results are expressed as mean ± SD determined from at least 3 independent experiments. Asterisks next to plots indicate statistically significant differences (*, †, and ‡, P < .05; **, ††, and ‡‡, P < .01; ***, †††, and ‡‡‡, P < .001) in mean values for treatments compared with the stimulatory group (16.7mM glucose and 20-ng/mL IL-1β or 20nM GLP-1) at corresponding time points based on a one-way ANOVA and Dunnett's multiple comparison post hoc test.
MAPKs also differentially influenced association of p300/HDACs with gene promoters. Although p38 and JNK activity was required for sustained accumulation of p300 and reduction of HDAC3 on the TNF-α gene promoter in response to IL-1β, ERK activity was required for the sustained presence of p300 and reduction of HDAC1 on the insulin promoter in response to GLP-1. Interestingly, p38/JNK and ERK inhibitors enhanced association of p300 with the insulin and TNF-α promoters, respectively. Although both GLP-1 and IL-1β could induce enrichment of p300 on the insulin promoter in the presence of glucose, the effect could not be sustained by IL-1β unless p38 and JNK were inhibited. Overall, these data suggest that CN/NFAT is required for recruitment of MAPKs, p300, and HDACs to the gene promoters, and MAPKs selectively influence stable formation and exclusion of promoter complexes containing p300, HDAC1, and HDAC3.
CN/NFAT-mediated recruitment of MAPKs, p300, and HDACs correlates with induction and repression of gene transcription
To examine requirements of CN/NFAT and MAPK signaling on gene transcription, we analyzed amounts of mRNA produced by insulin and TNF-α gene promoters.
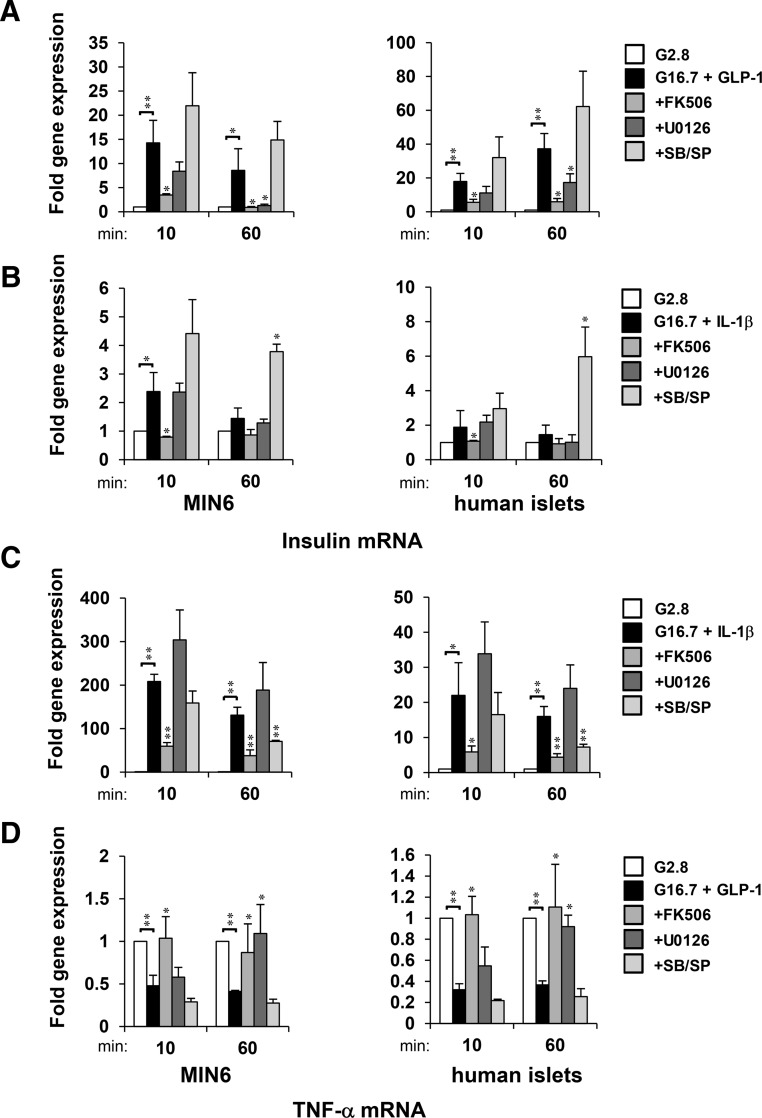
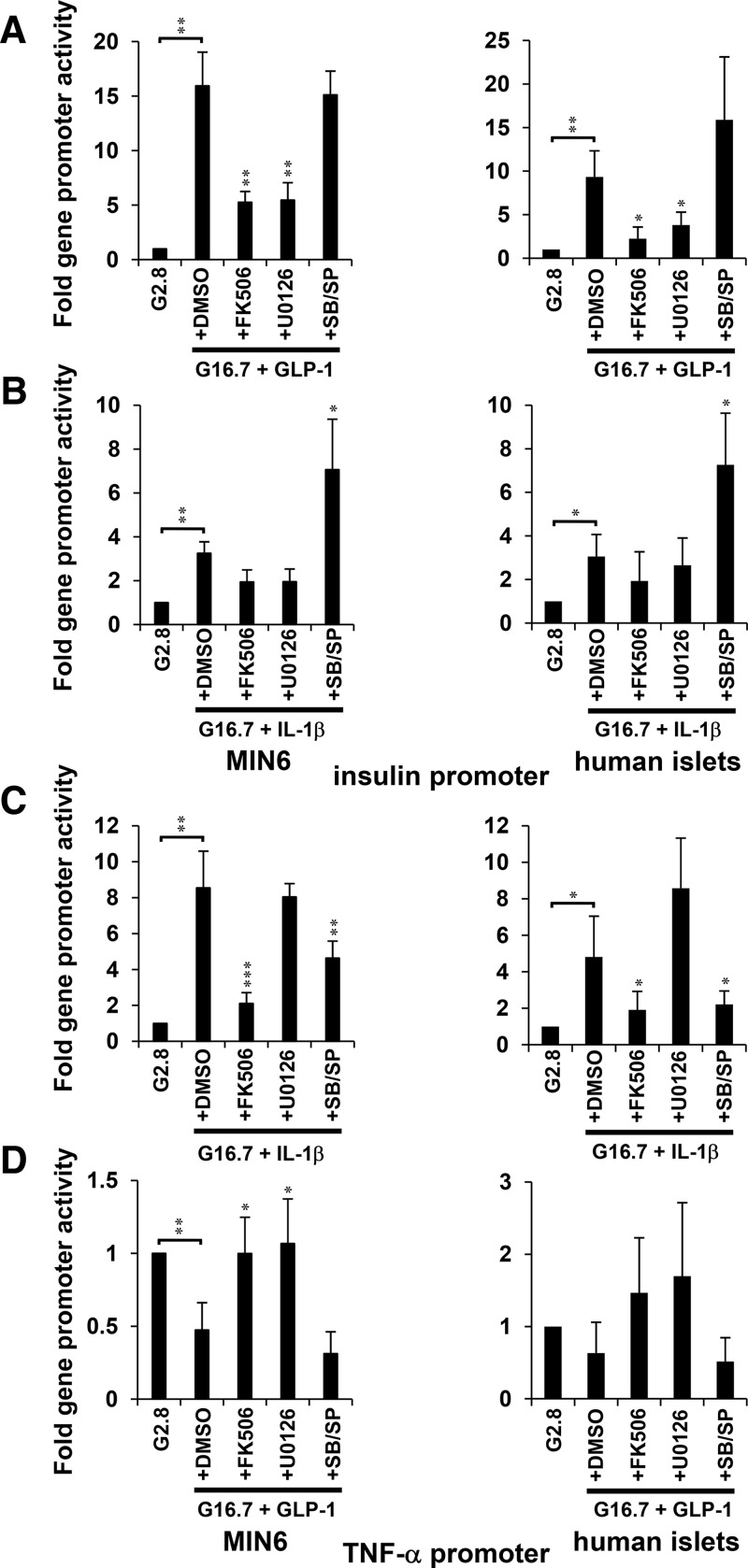
Nuclear run-on assays showed that glucose and GLP-1 induced insulin gene expression within 10 minutes, which was largely sustained for up to 60 minutes in both MIN6 and human islet cells (Figure 7A). Sustained induction of insulin mRNA by glucose and GLP-1 was repressed by addition of either CN or ERK1/2 inhibitor. The requirement of CN and ERK1/2 was also observed in the up-regulation of insulin promoter activity in MIN6 cells and human islets stimulated with glucose and GLP-1 (Figure 8A). IL-1β and glucose induced insulin mRNA to a much lesser extent, and the induction was diminished by 60 minutes (Figure 7B). However, the presence of p38 and JNK inhibitors enhanced and sustained the effects of glucose and IL-1β on insulin gene transcription (Figures 7B and 8B). Effects to activate the insulin gene correlated with the sustained recruitment of NFAT, ERK1/2, and p300 on the insulin promoter (Figures 3 and 6B). Although positive influences on insulin expression correlated with the sustained presence of NFAT-ERK-p300 on the insulin promoter, repression of insulin gene transcription was observed under conditions that promoted the sustained presence of HDAC1 (Figures 3A; 6B; 7, A and B; and 8, A and B). For example, ERK1/2 inhibition by U0126 prevented sustained occupancy of the insulin promoter by NFAT-ERK-p300 and stabilized occupancy by HDAC1 after 60 minutes of treatment with glucose and GLP-1 (Figure 6B). Inhibition of CN/NFAT signaling by FK506 prevented accumulation ERK1/2 or exchange of p300/HDAC signaling molecules upon the insulin gene promoter and prevented changes in insulin gene expression. The loss of association of HDAC3 with the insulin promoter was also dependent upon CN/NFAT signaling. However, changes in HDAC3 occupancy of the insulin promoter did not correlate with changes in insulin gene expression, suggesting that HDAC1 is a primary target of glucose and GLP-1-induced insulin gene regulation.
Figure 7.
Acute and sustained activation and repression of insulin and TNF-α mRNA expression is differentially regulated by MAPK activity. Nuclear run-on analysis of fold induction of insulin (A and B) and TNF-α (C and D) gene transcripts synthesized from nuclei isolated from MIN6 cells and human islets upon 10 and 60 minutes of treatment with IL-1β or GLP-1 as relative to 2.8mM glucose controls. Graphed results are expressed as mean ± SD determined from at least 3 independent experiments. Asterisks next to plots indicate statistically significant differences (*, P < .05; **, P < .01) in mean values for treatments compared with stimulatory groups (16.7mM glucose and 20nM GLP-1 or 20-ng/mL IL-1β) at t = 10- and 60-minute time points based on a one-way ANOVA and Dunnett's multiple comparison post hoc test.
Figure 8.
Regulation of insulin and TNF-α promoter activity is differentially regulated by MAPK activity. Luciferase reporter assay analysis of insulin (A and B) and TNF-α (C and D) promoter activity in response to 16.7mM glucose and 20nM GLP-1 or 20-ng/mL IL-1β in the presence of FK506, U0126, or SB203580/SP600125 (SB/SP) relative to 2.8mM glucose controls in MIN6 cells and human islets. Graphed results are expressed as mean ± SD determined from at least 3 independent experiments. Asterisks next to plots indicate statistically significant differences (*, P < .05; **, P < .01; ***, P < .001) in mean values for treatments compared with stimulatory groups (16.7mM glucose and 20nM GLP-1 or 20-ng/mL IL-1β) based on a one-way ANOVA and Dunnett's multiple comparison post hoc test.
Glucose and IL-1β induced TNF-α gene expression within 10 minutes, which was largely sustained for up to 60 minutes in both MIN6 and human islet cells (Figures 7C and 8C). Conversely, glucose and GLP-1 suppressed TNF-α gene transcription (Figures 7D and 8D). Although sustained induction of TNF-α gene expression by glucose and IL-1β was repressed by p38 and JNK inhibitors (Figures 7C and 8C), the suppressive effect of glucose and GLP-1 was prevented by ERK1/2 inhibitor (Figures 7D and 8D). These results indicate that p38 and JNK are required to activate the TNF-α gene and ERK1/2 contribute to its repression. Effects of glucose and IL-1β to activate the TNF-α gene correlated with recruitment of NFAT, p38, JNK, and p300 to the TNF-α gene promoter (Figures 5B and 6A). Suppression of TNF-α expression by glucose and GLP-1 correlated with ERK-dependent recruitment of HDAC3 (Figure 6A). Enrichment of HDAC3 upon the TNF-α gene promoter correlated with suppressed gene expression. In contrast, direct relationships between conditions that regulated TNF-α gene expression and HDAC1 promoter occupancy were not observed, indicating that HDAC3 is a primary regulator of TNF-α gene repression. Under each condition observed, CN inhibition prevented changes in promoter activity and mRNA from both TNF-α and insulin promoters in response to GLP-1 or IL-1β in the presence of high glucose (Figures 2E, 4B, 7, and 8). Overall, these data suggest that CN/NFAT can regulate induction and repression of gene transcription by targeting selectively activated MAPKs to promoters to produce stable transcriptional complexes containing p300 and HDACs, respectively.
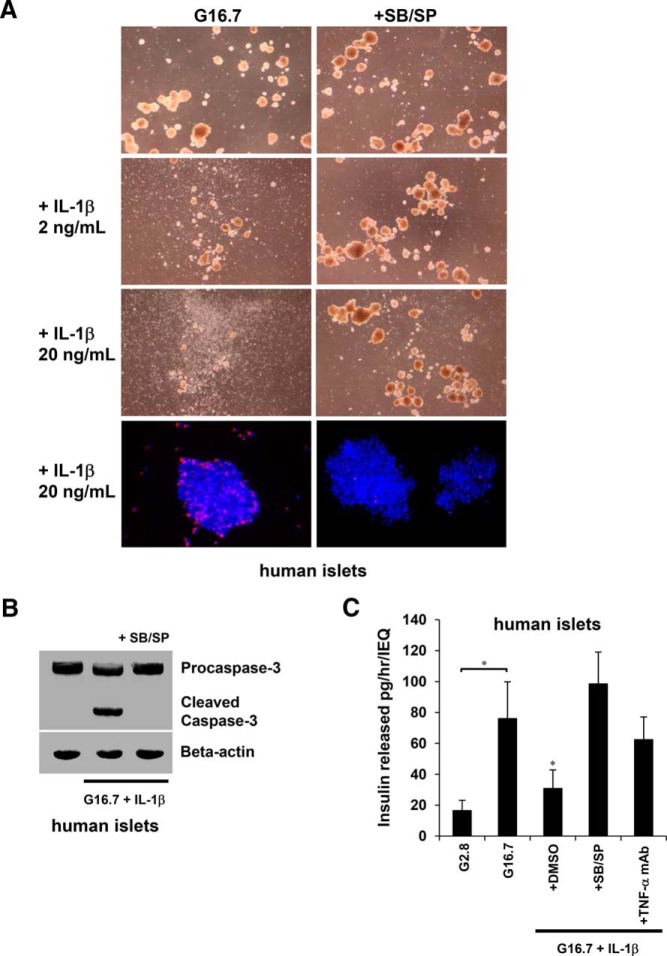
Blocking p38 and JNK activation enhances islet survival and function
The data indicate that expression of insulin and TNF-α genes in β-cells is differentially regulated by the selective targeting of MAPKs to gene promoters by NFAT. Most notably, selectively blocking p38 and JNK prevented expression of TNF-α and enhanced expression of insulin. These effects could have potential therapeutic implications in select cases of diabetes and islet transplantation, where IL-1β and TNF-α have been shown to contribute to loss of islets (20, 38–40). Therefore, we hypothesized that p38 and JNK would be prime signaling targets to improve islet survival under inflammatory conditions. To assess the effects on islet survival, we treated isolated human islets with glucose and IL-1β for 48 hours in the presence of p38 and JNK inhibitors. Although islets in high glucose alone remained mostly intact and maintained their structural integrity, increasing concentrations of IL-1β resulted in islet disintegration, which was prevented by p38 and JNK inhibitors (Figure 9A). Additionally, Hoechst 33342 and PI double staining showed islet cell death occurring within 12 hours of treatment with 16.7mM glucose and 20-ng/mL IL-1β, which was blocked by the p38/JNK inhibitors. Apoptosis was also observed by detection of Caspase-3 cleavage product within 8 hours of treatment of islets with glucose and IL-1β. To determine effects of the observed signaling mechanisms on islet function, we assessed the potential of blocking p38 and JNK on insulin secretion. Treating human islets for 24 hours with glucose and IL-1β resulted in impaired insulin release (Figure 9B). Islets pretreated with p38 and JNK inhibitors showed increased insulin secretion. Furthermore, islets pretreated with a TNF-α neutralizing monoclonal antibody partially restored the glucose-induced insulin response. These data suggest that p38 and JNK contribute to islet cell death and functional impairment, and impaired insulin secretion is contributed, at least in part, by IL-1β-induced expression of TNF-α. Furthermore, the findings indicate that these signaling mechanisms can be targeted to repress TNF-α expression in β-cells while maintaining islet integrity, viability, and function.
Figure 9.
Blocking p38 and JNK activation enhances islet survival and function. Effects of high glucose and IL-1β on islet integrity and viability in the presence of SB203580/SP600125 (SB/SP). A, Islet integrity was observed after 48 hours of treatment with 16.7mM glucose and 2- to 20-ng/mL IL-1β by light microscopy, and cell death was observed within 12 hours by fluorescence microscopy of Hoechst/PI double-stained islets. B, Apoptosis in islets was detected within 8 hours by immunoblot of cleaved Caspase-3. C, Effects of high glucose and IL-1β on islet function in the presence of SB203580/SP600125 (SB/SP) or TNF-α neutralizing monoclonal Ab. After exposure of islets to 16.7mM glucose and 20-ng/mL IL-1β for 24 hours, insulin release was measured by static incubation from islets pretreated with 2.8mM glucose for 60 minutes and stimulated with 16.7mM glucose for 60 minutes. Graphed results are expressed as mean ± SD determined from at least 3 independent experiments. Asterisks above bars indicate statistically significant differences (*, P < .05) in mean values for treatments compared with the stimulatory group (16.7mM glucose) based on a one-way ANOVA and Dunnett's multiple comparison post hoc test.
Discussion
CN/NFAT signaling plays a critical role in β-cell proliferation, maturation, and function. It also induces inflammatory genes in β-cells that potentially contribute to islet inflammation and apoptosis. Our results showed that GLP-1 stimulated a rapid accumulation of NFATc2 and ERK1/2 upon the insulin gene promoter, and IL-1β induced enrichment of transcriptional complexes containing NFATc2 and p38/JNK upon the TNF-α promoter. We also observed that NFAT-ERK recruited a sustained enrichment of p300 upon the insulin promoter and HDAC3 upon the TNF-α gene. This correlated with increased insulin and decreased TNF-α gene expression, respectively. In contrast, NFAT-p38/JNK promoted sustained enrichment of p300 upon the TNF-α promoter and HDAC1 upon the insulin gene, correlating with increased TNF-α and reduced insulin gene expression, respectively. These data support a role of NFAT as a mediator that selectively recruits signaling enzymes to DNA promoter complexes in response to GLP-1 and IL-1β to differentially regulate gene transcription (Figure 10).
Figure 10.
Schematic model for NFAT-mediated activation and repression of insulin and TNF-α genes in β-cells in response to GLP-1 and IL-1β. GLP-1 in the presence of glucose activates CN/NFAT and ERK1/2 to induce enrichment of ERK-p300 and ERK-HDAC3 upon the insulin and TNF-α gene promoters, respectively. Activation of CN/NFAT and p38/JNK by IL-1β in the presence of glucose favors formation of p38/JNK-p300 and p38-HDAC1 upon the TNF-α and insulin gene promoters, respectively. Although NFAT-ERK enhances insulin gene expression, p38/JNK favors induction of TNF-α gene expression and insulin gene repression. Dashed arrows indicate activation. Green and red colors indicate activation and repression, respectively.
Most notably, MAPK activity largely contributed to differential effects on enrichment of NFAT-MAPK-p300/HDAC complexes upon gene promoters. MAPK inhibition had marginal effects on NFAT-mediated assembly of kinases and chromatin-modifying enzymes upon gene promoters, which was globally inhibited by the CN inhibitor FK506. However, MAPKs selectively contributed to the overall stability of the NFAT-MAPK-p300/HDAC DNA promoter complex for up to 60 minutes. The stable complexes sustained by MAPKs contributed to changes in gene transcription.
Although stabilizing transcription factor-signaling enzyme complexes enriched with p300 enhanced gene promoter activity, complexes favoring HDAC stabilization had a repressive effect on gene expression. Consistent with this concept, FK506 inhibited association of NFATc2 and HDAC3 with the TNF-α promoter, which prevented repression of the TNF-α transcription in response to glucose and GLP-1. These data indicate that NFAT signaling affects both gene activation and repression and that inhibiting NFAT prevents an overall net change in gene transcription in response to stimuli.
Formation of stable NFAT-MAPK transcriptional/signaling complexes likely occurs via MAPK-dependent phosphorylation of other transcription factors that bind to and regulate the gene promoter under these conditions. Previous studies in cardiomycytes support this concept, in which MEK1 and ERK1/2 enhance NFAT-dependent gene expression by forming a complex with CN/NFATc3 to induce AP-1 transcriptional activity (41). Indeed, we previously showed that p38 and JNK inhibitors prevented c-Jun and ATF2 from associating with and activating the TNF-α promoter in response to IL-1β in MIN6 cells (21). MafA/C/EBP-β could also competitively displace ATF2/c-Jun and repress TNF-α gene transcription in an ERK-dependent manner.
These observations correspond to results of the current study, where p38/JNK inhibition resulted in a switch from NFAT-p38/JNK-p300 to NFAT-ERK-HDAC3 promoter complex formation and repression of TNF-α gene promoter activity and mRNA expression. Interestingly, inhibition of p38 and JNK increased occupancy of p300 and reduced HDAC1 on the insulin promoter in response to both GLP-1 and IL-1β. We propose that p38 and JNK accomplish this by activating substrates that promote HDAC1 assembly upon the insulin promoter and concomitantly compete with ERK-dependent substrates that are required to form a stable p300 transcriptional complex. Kinase-dependent competitive cooccupancy of p38/JNK with ATF2/c-Jun (activator) and ERK1/2 with MafA/C/EBP-β (repressor) on the TNF-α promoter, for example, suggests that these transcription factors contribute to stabilization of NFAT-MAPK-p300/HDAC complex formation (21).
Scaffolding protein complexes may also contribute to formation of NFAT-MAPK transcription/signaling factor complexes in pancreatic β-cells. CN has been shown to modulate MAPK activity via controlling localization of the kinase suppressor of Ras 2 scaffold proteins to the membrane in response to Ca2+ signaling in rat insulinoma INS-1 β-cell lines (42). Thus, it possible that exchange and formation of signaling molecules between these complexes are in dynamic equilibrium with scaffolding that occurs both at membrane receptor systems and on chromatin located near gene promoters to control acute and sustained responses to metabolic and inflammatory stress. Although intact NFAT binding sites were required for association of NFAT and MAPKs with gene promoters, it should be noted that these experiments were performed with transiently transfected DNA promoter vectors. Hence, it remains unclear what contribution these binding sites may have for complex formation upon endogenous gene promoters. Moreover, direct interactions between NFAT, MAPKs, and other signaling factors upon gene promoters have not been fully described. Thus, further study is required to support these hypotheses.
Overall, the results elucidate CN/NFAT-mediated signaling events that control formation and stabilization of transcription/signaling factor complexes upon DNA promoters to distinctly regulate insulin and TNF-α genes. The data describe mechanisms whereby NFAT is required for 1) changes to occur within transcriptional/signaling complexes upon gene promoters in response to stimuli and 2) recruitment of signaling enzymes to gene promoters to contribute to stability of newly assembled complexes. The study also provides proof of concept that blocking p38 and JNK signaling pathways can protect islets and preserve β-cell function in the presence of inflammatory cytokines. Targeting these mechanisms may have potential for reducing local islet inflammation and improving islet function in therapeutic applications for diabetes and islet transplantation.
Additional material
Supplementary data supplied by authors.
Acknowledgments
We thank Isaac Klein for technical assistance and Cindy Orticio for critical reading of the manuscript.
This work was supported by the National Institutes of Health Grant R01 DK55310 (to M.H. Cobb for K.M.) and the Baylor Health Care System.
Disclosure Summary: The authors have nothing to disclose.
Footnotes
- ATF
- activating transcription factor
- C/EBP
- CCAAT/enhancer binding protein
- ChIP
- chromatin immunoprecipitation
- CN
- calcineurin
- dnNFAT
- dominant negative NFAT PxIxIT motif
- GFP
- green fluorescent protein
- GLP-1
- glucagon-like peptide 1
- HDAC
- histone deacetylase
- JNK
- Jun N-terminal kinase
- MafA
- v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog A
- MEK
- MAPK kinase
- NFAT
- nuclear factor of activated T cells
- p300
- histone acetyltransferase p300
- p38
- p38 MAPK
- PI
- propidium iodide
- PMSF
- phenylmethylsulfonyl fluoride
- qPCR
- quantitative PCR
- shRNA
- small hairpin RNA.
References
- 1. Frankel BJ, Atwater I, Grodsky GM. Calcium affects insulin release and membrane potential in islet β-cells. Am J Physiol. 1981;240(1):C64–C72. [DOI] [PubMed] [Google Scholar]
- 2. Weir GC, Mojsov S, Hendrick GK, Habener JF. Glucagonlike peptide I (7–37) actions on endocrine pancreas. Diabetes. 1989;38(3):338–342. [DOI] [PubMed] [Google Scholar]
- 3. Henquin JC, Dufrane D, Nenquin M. Nutrient control of insulin secretion in isolated normal human islets. Diabetes. 2006;55(12):3470–3477. [DOI] [PubMed] [Google Scholar]
- 4. Henquin JC. Triggering and amplifying pathways of regulation of insulin secretion by glucose. Diabetes. 2000;49(11):1751–1760. [DOI] [PubMed] [Google Scholar]
- 5. Lu M, Wheeler MB, Leng XH, Boyd AE., 3rd The role of the free cytosolic calcium level in β-cell signal transduction by gastric inhibitory polypeptide and glucagon-like peptide I(7–37). Endocrinology. 1993;132(1):94–100. [DOI] [PubMed] [Google Scholar]
- 6. Holz GG, 4th, Leech CA, Habener JF. Activation of a cAMP-regulated Ca(2+)-signaling pathway in pancreatic β-cells by the insulinotropic hormone glucagon-like peptide-1. J Biol Chem. 1995;270(30):17749–17757. [PMC free article] [PubMed] [Google Scholar]
- 7. Gromada J, Dissing S, Bokvist K, et al. Glucagon-like peptide I increases cytoplasmic calcium in insulin-secreting β TC3-cells by enhancement of intracellular calcium mobilization. Diabetes. 1995;44(7):767–774. [DOI] [PubMed] [Google Scholar]
- 8. Drucker DJ. The role of gut hormones in glucose homeostasis. J Clin Invest. 2007;117(1):24–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lawrence MC, Bhatt HS, Watterson JM, Easom RA. Regulation of insulin gene transcription by a Ca(2+)-responsive pathway involving calcineurin and nuclear factor of activated T cells. Mol Endocrinol. 2001;15(10):1758–1767. [DOI] [PubMed] [Google Scholar]
- 10. Lawrence MC, Bhatt HS, Easom RA. NFAT regulates insulin gene promoter activity in response to synergistic pathways induced by glucose and glucagon-like peptide-1. Diabetes. 2002;51(3):691–698. [DOI] [PubMed] [Google Scholar]
- 11. Lawrence MC, McGlynn K, Park BH, Cobb MH. ERK1/2-dependent activation of transcription factors required for acute and chronic effects of glucose on the insulin gene promoter. J Biol Chem. 2005;280(29):26751–26759. [DOI] [PubMed] [Google Scholar]
- 12. Heit JJ, Apelqvist AA, Gu X, et al. Calcineurin/NFAT signalling regulates pancreatic β-cell growth and function. Nature. 2006;443(7109):345–349. [DOI] [PubMed] [Google Scholar]
- 13. Soleimanpour SA, Crutchlow MF, Ferrari AM, et al. Calcineurin signaling regulates human islet β-cell survival. J Biol Chem. 2010;285(51):40050–40059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Demozay D, Tsunekawa S, Briaud I, Shah R, Rhodes CJ. Specific glucose-induced control of insulin receptor substrate-2 expression is mediated via Ca2+-dependent calcineurin/NFAT signaling in primary pancreatic islet β-cells. Diabetes. 2011;60(11):2892–2902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Goodyer WR, Gu X, Liu Y, Bottino R, Crabtree GR, Kim SK. Neonatal β cell development in mice and humans is regulated by calcineurin/NFAT. Dev Cell. 2012;23(1):21–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Al-Uzri A, Stablein DM, A Cohn R. Posttransplant diabetes mellitus in pediatric renal transplant recipients: a report of the North American Pediatric Renal Transplant Cooperative Study (NAPRTCS). Transplantation. 2001;72(6):1020–1024. [DOI] [PubMed] [Google Scholar]
- 17. Maes BD, Kuypers D, Messiaen T, et al. Posttransplantation diabetes mellitus in FK-506-treated renal transplant recipients: analysis of incidence and risk factors. Transplantation. 2001;72(10):1655–1661. [DOI] [PubMed] [Google Scholar]
- 18. Choi D, Kim BH, Lee MK, Cho H. Effects of water-soluble tacrolimus-PEG conjugate on insulin-dependent diabetes mellitus and systemic lupus erythematosus. Arch Pharm Res. 2011;34(8):1301–1310. [DOI] [PubMed] [Google Scholar]
- 19. Maedler K, Sergeev P, Ris F, et al. Glucose-induced β cell production of IL-1β contributes to glucotoxicity in human pancreatic islets. J Clin Invest. 2002;110(6):851–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Böni-Schnetzler M, Thorne J, Parnaud G, et al. Increased interleukin (IL)-1β messenger ribonucleic acid expression in β-cells of individuals with type 2 diabetes and regulation of IL-1β in human islets by glucose and autostimulation. J Clin Endocrinol Metab. 2008;93(10):4065–4074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lawrence MC, Naziruddin B, Levy MF, Jackson A, McGlynn K. Calcineurin/nuclear factor of activated T cells and MAPK signaling induce TNF-α gene expression in pancreatic islet endocrine cells. J Biol Chem. 2011;286(2):1025–1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Rabinovitch A. An update on cytokines in the pathogenesis of insulin-dependent diabetes mellitus. Diabetes Metab Rev. 1998;14(2):129–151. [DOI] [PubMed] [Google Scholar]
- 23. Piemonti L, Leone BE, Nano R, et al. Human pancreatic islets produce and secrete MCP-1/CCL2: relevance in human islet transplantation. Diabetes. 2002;51(1):55–65. [DOI] [PubMed] [Google Scholar]
- 24. Wachlin G, Augstein P, Schröder D, et al. IL-1β, IFN-γ and TNF-α increase vulnerability of pancreatic β cells to autoimmune destruction. J Autoimmun. 2003;20(4):303–312. [DOI] [PubMed] [Google Scholar]
- 25. Alexandraki K, Piperi C, Kalofoutis C, Singh J, Alaveras A, Kalofoutis A. Inflammatory process in type 2 diabetes: the role of cytokines. Ann NY Acad Sci. 2006;1084:89–117. [DOI] [PubMed] [Google Scholar]
- 26. Schwarznau A, Hanson MS, Sperger JM, et al. IL-1β receptor blockade protects islets against pro-inflammatory cytokine induced necrosis and apoptosis. J Cell Physiol. 2009;220(2):341–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zineh I, Beitelshees AL, Silverstein JH, Haller MJ. Serum monocyte chemoattractant protein-1 concentrations associate with diabetes status but not arterial stiffness in children with type 1 diabetes. Diabetes Care. 2009;32(3):465–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. McCall M, Pawlick R, Kin T, Shapiro AM. Anakinra potentiates the protective effects of etanercept in transplantation of marginal mass human islets in immunodeficient mice. Am J Transplant. 2012;12(2):322–329. [DOI] [PubMed] [Google Scholar]
- 29. Panee J. Monocyte chemoattractant protein 1 (MCP-1) in obesity and diabetes. Cytokine. 2012;60(1):1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zaitsev SV, Appelskog IB, Kapelioukh IL, et al. Imidazoline compounds protect against interleukin 1β-induced β-cell apoptosis. Diabetes. 2001;50(suppl 1):S70–S76. [DOI] [PubMed] [Google Scholar]
- 31. Arnette D, Gibson TB, Lawrence MC, et al. Regulation of ERK1 and ERK2 by glucose and peptide hormones in pancreatic β cells. J Biol Chem. 2003;278(35):32517–32525. [DOI] [PubMed] [Google Scholar]
- 32. Lawrence MC, McGlynn K, Shao C, et al. Chromatin-bound mitogen-activated protein kinases transmit dynamic signals in transcription complexes in β-cells. Proc Natl Acad Sci USA. 2008;105(36):13315–13320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ruff VA, Leach KL. Direct demonstration of NFATp dephosphorylation and nuclear localization in activated HT-2 cells using a specific NFATp polyclonal antibody. J Biol Chem. 1995;270(38):22602–22607. [DOI] [PubMed] [Google Scholar]
- 34. Qiu Y, Sharma A, Stein R. p300 mediates transcriptional stimulation by the basic helix-loop-helix activators of the insulin gene. Mol Cell Biol. 1998;18(5):2957–2964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Qiu Y, Guo M, Huang S, Stein R. Insulin gene transcription is mediated by interactions between the p300 coactivator and PDX-1, BETA2, and E47. Mol Cell Biol. 2002;22(2):412–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Mosley AL, Corbett JA, Ozcan S. Glucose regulation of insulin gene expression requires the recruitment of p300 by the β-cell-specific transcription factor Pdx-1. Mol Endocrinol. 2004;18(9):2279–2290. [DOI] [PubMed] [Google Scholar]
- 37. Lawrence MC, Shao C, McGlynn K, Naziruddin B, Levy MF, Cobb MH. Multiple chromatin-bound protein kinases assemble factors that regulate insulin gene transcription. Proc Natl Acad Sci USA. 2009;106(52):22181–22186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ehses JA, Lacraz G, Giroix MH, et al. IL-1 antagonism reduces hyperglycemia and tissue inflammation in the type 2 diabetic GK rat. Proc Natl Acad Sci USA. 2009;106(33):13998–14003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Butcher MJ, Hallinger D, Garcia E, et al. Association of proinflammatory cytokines and islet resident leucocytes with islet dysfunction in type 2 diabetes. Diabetologia. 2014;57(3):491–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Potter KJ, Westwell-Roper CY, Klimek-Abercrombie AM, Warnock GL, Verchere CB. Death and dysfunction of transplanted β-cells: lessons learned from type 2 diabetes? Diabetes. 2014;63(1):12–19. [DOI] [PubMed] [Google Scholar]
- 41. Sanna B, Bueno OF, Dai YS, Wilkins BJ, Molkentin JD. Direct and indirect interactions between calcineurin-NFAT and MEK1-extracellular signal-regulated kinase 1/2 signaling pathways regulate cardiac gene expression and cellular growth. Mol Cell Biol. 2005;25(3):865–878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Dougherty MK, Ritt DA, Zhou M, et al. KSR2 is a calcineurin substrate that promotes ERK cascade activation in response to calcium signals. Mol Cell. 2009;34(6):652–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.