Abstract
Context:
Elevated LDL cholesterol (LDL-C) is an important risk factor for atherosclerosis and cardiovascular disease. Variants in the proprotein convertase subtilisin/kexin type 9 (PCSK9) gene have been associated not only with plasma LDL-C concentration, but also with ischemic heart disease. Little is known about the genetic architecture of PCSK9 and its influence on LDL-C in American Indians.
Objective:
We aimed to investigate the genetic architecture in the 1p32 region encompassing PCSK9 and its influence on LDL-C in American Indians.
Design:
The Strong Heart Family Study (SHFS) is a family-based genetic study.
Participants:
Two thousand four hundred fifty eight American Indians from Arizona, Oklahoma, North Dakota, and South Dakota, who were genotyped by Illumina MetaboChip.
Results:
We genotyped 486 SNPs in a 3.9 Mb region at chromosome 1p32 encompassing PCSK9 in 2458 American Indians. We examined the association between these SNPs and LDL-C. For common variants (MAF ≥ 1%), meta-analysis across the three geographic regions showed common variants in PCSK9 were significantly associated with higher LDL-C. The most significant SNP rs12067569 (MAF = 1.7 %, β = 16.9 ± 3.7, P = 5.9 × 10−6) was in complete LD (r2 = 1) with a nearby missense SNP, rs505151 (E670G) (β = 15.0 ± 3.6, P = 3.6 × 10−5). For rare variants (MAF < 1%), rs11591147 (R46L, MAF = 0.9%) was associated with lower LDL-C (β = − 31.1 ± 7.1, P = 1.4 × 10−5). The mean (SD) of LDL-C was 76.9 (7.8) and 107.4 (1.0) mg/dL for those with and without the R46L mutation, respectively. One person who was homozygous for R46L had LDL-C levels of 11 mg/dL. In one family, 6 out of 8 members carrying the R46L mutation had LDL-C levels below the lower 10% percentile of LDL-C among all study participants.
Conclusions:
Both rare and common variants in PCSK9 influence plasma LDL-C levels in American Indians. Follow-up studies may disclose the influence of these mutations on the risk of CVD and responses to cholesterol-lowering medications.
Elevated low-density lipoprotein cholesterol (LDL-C) is an important risk factor for atherosclerosis and cardiovascular disease (CVD). The current guidelines from the National Cholesterol Education Program (NCEP) Adult Treatment Panel III (ATP III) recommend LDL-C as the primary target of therapy for prevention of CVD (1). The estimated heritability for LDL-C is quite high, approximately 40–60% (2). Recent GWAS have identified many loci associated with LDL-C variation at the population level (3). However, common variants at these loci account for only 25% of LDL-C total heritability (3).
Since the first mutation in PCSK9 was identified in families of autosomal dominant hypercholesterolemia (4), PCSK9 has been the subject of many scientific queries. The PCSK9 gene encodes the proprotein convertase subtilisin/kexin 9 (PCSK9) protein. The major function of PCSK9 protein is to regulate cholesterol metabolism by binding to the LDL receptor (LDLR) and promoting its degradation (5, 6). Overexpression of PCSK9 in mice causes a reduction in LDLR level and LDL-C uptake from circulation, and thus leads to hypercholesterolemia and atherosclerosis (5–7).
In humans, variants in the PCSK9 gene have been associated with variability in LDL-C levels and ischemic heart disease (8–10). Several gain-of-function (GOF) mutations in PCSK9 have been found in cases of familial hypercholesterolemia while loss-of-function (LOF) mutations have been found in persons with hypocholesterolemia (4, 8). Anti-PCSK9 agents are being developed as cholesterol-lowering drugs. Several preclinical trials have shown encouraging results (11).
Previous studies have found lower LDL-C levels in American Indian populations in comparison to the general U.S. population (12). Genetic variation could contribute to these differences. Allelic heterogeneity as well as differences in allele frequencies at the PCSK9 gene have been noted across ancestrally diverse populations (13). Little is known about the genetic architecture of PCSK9 in American Indians. Accordingly, we aimed to investigate the genetic architecture in the 1p32 region encompassing PCSK9 and its influence on LDL-C in American Indians.
Materials and Methods
Study population
The Strong Heart Family Study (SHFS) is a family-based cohort study derived from the parent Strong Heart Study (SHS) aimed to investigate the genetics of CVD among the American Indian population (14). Briefly, over 3800 American Indians from more than 90 multigenerational families living in Arizona, North Dakota, South Dakota, and Oklahoma were included in the SHFS. Approximately 2650 participants were genotyped using the Illumina MetaboChip. After excluding participants missing LDL-C level information, the final sample size for this study was 2458.
Genotyping methods and quality control (QC)
The study participants were genotyped using the MetaboChip array, an Illumina custom panel designed to replicate and fine-map known GWAS signals for metabolic and cardiovascular traits (15). Samples were genotyped at the Texas Biomedical Research Institute. For this study we interrogated 486 SNPs in a 3.9 Mb region from the MetaboChip that fine-mapped the BSND/PCSK9/USP24/PPAP2B region on the chromosome 1p32 region (chromosome 1, 52854173–56793533, NCBI build 36.3). We excluded SNPs with a marker call rate < 98% and individual samples with a call rate < 95%. Additional SNP quality controls included checks for concordance in duplicated samples, gender inconsistency and Mendelian errors. All reported SNPs were compatible with Hardy-Weinberg equilibrium at P > .0001.
Laboratory measurements
Total triglycerides, HDL-C, LDL-C, and total cholesterol were measured by standardized methods in blood samples obtained after a 12-h fast.
Statistical analysis methods
We conducted linear regression analysis for LDL-C with each SNP assuming an additive genetic model while adjusting for age, sex, and their interaction. The association analysis was performed using variance components models to account for familial correlation, as implemented in the SOLAR software package. We stratified the association analysis by geographical location (Arizona, Oklahoma, North Dakota, and South Dakota) to account for possible differences between the three locations. We conducted a fixed-effect meta-analysis across the center-specific association results and used I2 to assess heterogeneity across centers. For rare variants, we performed single variant association analysis and two variant collapsing tests (aka burden tests). We coded individuals as having at least one copy of a rare variant (or not) to perform the burden test. We also coded individuals by the number of rare variants carried in PCSK9 and tested this summary estimate for association with LDL-C values. For single SNP association tests, statistical significance was defined as P-value <10−4 for each SNP according to Bonferroni correction for multiple testing. For collapsing tests, a P-value <.05 was considered statistically significant.
Results
The mean (SD) age of study participants was 36.6 (15.7) years. The mean (SD) plasma level of LDL-C and total cholesterol were 103.3 (30.1) and 182.7(37.5) mg/dL, respectively (Supplemental Table 1). Only 7.6% of the participants had received lipid-lowering therapy.
The association between common variants in PCSK9 and LDL-C
Both common and rare variants were included among the 486 SNPs genotyped in a 3.9 MB region at chromosome 1p32 with SNPs densely tagging the genes BSND/PCSK9/USP24/PPAP2B. For variants with minor allele frequency (MAF) ≥ 1%, we conducted a meta-analysis of associations across the three geographic regions. The meta-analysis showed common variants in PCSK9 were significantly associated with higher LDL-C, and the most significant SNP was rs12067569 (MAF = 1.7%, β = 16.9 ± 3.7, P = 5.9 × 10−6) (I2 = 0%) (Supplemental Figure 1). SNP rs12067569 was in complete LD (r2 = 1) with a nearby missense SNP in PCSK9, rs505151(E670G), which was also significantly associated with higher LDL-C (β = 15.0 ± 3.6, P = 3.6 × 10−5). The mean (SD) of LDL-C was 102.9 (0.6) mg/dL for persons without the E670G mutation and 118.1(3.2) mg/dL for those with one G allele at the E670G variant. The estimated variation in LDL-C explained by the rs505151 (E670G) mutation was 1.2%. In a sensitivity analysis, we further adjusted our models for lipid-lowering medication and the association results were similar or slightly stronger (data not shown).
The association between rare variants in PCSK9 and LDL-C
Among the 23 coding SNPs in PCSK9 that were genotyped, we excluded16 SNPs from analysis (14 SNPs were monomorphic and 2 SNPs had low call rates) and we tested seven coding SNPs for association with LDL-C. Among the 14 monomorphic loci in American Indians (R104H, L112L, L118F, Y142X, L253F, P279T, Q344K, D432N, G466E, H553R, P576L, Q619P, D660H, C679X), some were previously reported as associated with hypocholesterolemia in other populations, such as Y142X (rs67608943) and C679X (rs28362286) (Table 1). Four of the seven coding SNPs tested were rare variants in American Indians (MAF < 1%) (Table 1). Only rs11591147 (R46L) (MAF = 0.9%) was associated with lower LDL-C (β = −31.1 ± 7.1, P = 1.4 × 10−5). The mean (SD) of LDL-C was 107.4 (1.0) mg/dL for persons without the R46L mutation and 76.9 (7.8) mg/dl for those carrying one R46L mutation. One person homozygous for the R46L mutation (TT genotype) had an extremely low LDL-C level of 11 mg/dL. The estimated variance in LDL-C explained by the R46L mutation was 2.6%. According to pedigree information, the R46L mutation segregated in seven families; in one family, 6 out of 8 members carrying the R46L mutation had a LDL-C level below the lower 10th percentile of LDL-C among all study participants (< 70 mg/dL) (Figure 1).
Table 1.
Association Analysis of Exonic SNPs in PCSK9 With LDL-C in SHFS
| SNP_ID | SNP | Ref. allele | Effect allele | P | β | se | MAF (%)c |
Ref.c | |||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Am. Indians | White | Black | Hisp. | ||||||||
| MAF<1% in American Indians | |||||||||||
| rs11591147 | R46L | G | T | 1.4 × 10−5 | −31.1 | 7.1 | 0.9a | 1.6–2.6a | 0.28 | 0.75 | 8, 9, 13 |
| rs28385701 | Synon | G | A | .6 | −7.8 | 16.8 | 0.6 | ||||
| rs28362261 | N425S | A | G | .7 | −4.7 | 11.5 | 0.2 | 0 | 1.9 | 0.2 | 13 |
| rs28362263 | A443T | G | A | .9 | −2.7 | 19.3 | 0.1 | 0.05 | 9.4a | 1.3 | 13 |
| MAF > 1% in American Indians | |||||||||||
| rs505151 | E670G | A | G | 3.6 × 10−5 | 15.0 | 3.6 | 1.8b | 3.6b | 26 | 4.2 | 13,16 ,18 |
| rs562556 | I474V | A | G | .04 | −2.9 | 1.4 | 10.7 | 18 | 22 | 9.7 | 13 |
| rs540796 | Synon | G | A | .04 | −2.9 | 1.4 | 12.4 | ||||
| No Mutation in American Indians | |||||||||||
| rs67608943 | Y142X | C | 0 | 0 | 0.8a | 0 | 8, 13 | ||||
| rs28362286 | C679X | C | 0 | 0 | 1.8a | 0.08 | 8, 13 | ||||
| rs72646508 | L253F | A | 0 | 0 | 0.25a | 0 | 13 | ||||
| rs28362270 | H553R | A | 0 | 0 | 1.3b | 0 | 13 | ||||
@14/23 coding SNPs in PCSK9 on Metabochip are monomorphic in American Indians (R104H, L112L, L118F, Y142X, L253F, P279T, Q344K, D432N, G466E, H553R, P576L, Q619P, D660H, C679X).
The mutation was associated with hypocholesterolemia.
The mutation was associated with hypercholesterolemia.
The allele frequencies of White, Black, and Hispanic are summarized from indicated references.
Abbreviations: Ref, reference; Am, American; Hisp, Hispanic.
Figure 1.
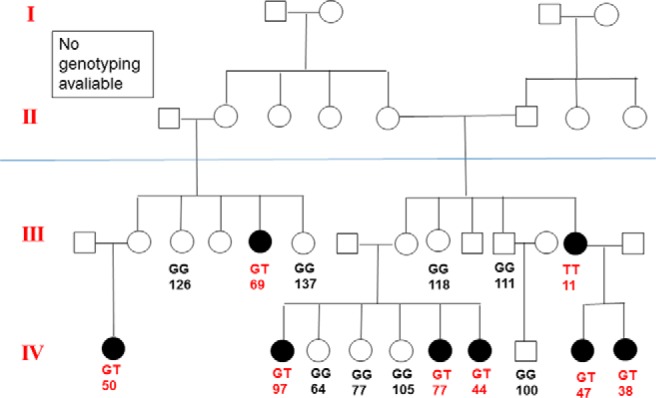
American Indian Family with R46L mutation in PCSK9. The notation indicates genotype and LDL-C levels.
Collapsing tests, performed by indicating individuals with at least one copy of a rare variant or not, also demonstrated evidence of association between rare variants in PCSK9 and lower LDL-C (β = −6.7 ± 3.2, P = .036). When recoding the four rare variants considering the number of mutations, 90 people had one mutation, 5 had two mutations, 3 had three mutations, and one person had all four mutations. The sum test showed persons with any mutation had significantly lower LDL-C than persons without such mutations (β = −6.4 ± 2.6, P = .013).
Discussion
In this study, we identified SNPs in PCSK9 associated with LDL-C levels in American Indians. For common variants (MAF > 1%), the most significant SNP (rs12067569) and a nearby missense SNP, rs505151 (E670G), were associated with higher LDL-C. In contrast, for rare variants (MAF < 1%), we found a missense mutation rs11591147 (R46L) associated with lower LDL-C. This is the first study to comprehensively explore the genetic variants in PCSK9 and its relationship with LDL-C levels among American Indians.
Our results inferred that E670G (rs505151), which was in complete LD with the most significant SNP (rs12067569), is the causal variant in PCSK9 associated with higher LDL-C in American Indians. The allele frequency of E670G varies among different ethnic groups and the previously reported associations with LDL-C have been inconsistent across studies (13, 16–18). The frequency of the minor allele G is low in Caucasians (3.6%), Hispanics (4.2%), and Han Chinese (4.8%), but relatively high in African Americans (26%) (13, 18). In our study, only 3.7% of American Indians carried E670G mutation. This E670G mutation has been reported to be associated with hypercholesterolemia in two studies in populations of European ancestry (16). However, two other studies conducted in African Americans and Caucasians failed to replicate the association (13, 17). Interestingly, Ding et al suggested a positive selection on SNP rs505151 in African Americans although the underlying mechanism of selection was unclear (19).
Several loss-of-function mutations in PCSK9 have been associated with hypocholesterolemia in other populations, including R46L, G106R, Y142X, L253F, N157K, R237W, A443T, C679X (10, 13, 17). Only R46L was significantly associated with lower LDL-C in American Indians in this study. The association between R46L and hypocholesterolemia has been well established in Individuals of European ancestry (9, 10). Cohen et al showed R46L carriers had a 15% decrease in plasma LDL-C and a 50% reduction in coronary heart disease (CHD) risk in Caucasians (10). In meta-analyses among 66 698 Europeans, the R46L mutation was associated with a 12% reduction in LDL-C and a 28% reduction in risk of ischemic heart disease (9). The Y142X, C679X, and L253F mutations, which have been reported to be associated with low LDL-C in African Americans (10, 13), were not present in American Indians from the SHFS (Table 1).
In this study, we found both common and rare variants in PCSK9 influenced LDL-C level. The estimated variation in LDL-C explained by the common variant E670G was smaller (1.2%) than the rare variant mutation R46L (2.6%). Our results support the body of evidence that has shown that both common and rare variants can contribute to the genetic control of complex human traits. Using family data, we found significant association with modest numbers of families. We demonstrated the advantage of studying large families in that we can observe higher frequencies of rare variants in extended pedigrees compared to samples from the general population (20). As Figure 1 shows, 16 members in one extended family were genotyped and 50% (8 of 16) were carriers of R46L.
In conclusion, we expanded our knowledge of the genetic architecture of PCSK9 and identified both rare and common variants in PCSK9 influencing plasma LDL-C levels in American Indians with opposite direction of effect. We identified a relatively common variant rs505151 (E670G) and one rare variant rs11591147 (R46L) that were associated with LDL-C levels in American Indians. The large effect in reductions of LDL-C by the PCSK9 R46L mutation provides a target for development of PCSK9 inhibitors to treat hypercholesterolemia. Follow-up studies may disclose the influence of these mutations on the risk of cardiovascular disease and responses to cholesterol-lowering medications.
Acknowledgments
This study was supported by grants from the National Heart, Lung and Blood Institute (HL41642, HL41652, HL41654, and HL65521) and the National Institute of Environmental Health Sciences (R01ES021367).
Disclosure Summary: The authors have nothing to disclose.
Footnotes
- CVD
- cardiovascular disease
- LDL-C
- low-density lipoprotein cholesterol
- MAF
- minor allele frequency
- SHFS
- Strong Heart Family Study.
References
- 1. Expert Panel on Detection, Evaluation, and Treatment of High Blood Cholesterol in Adults. Executive summary of the third report of the national cholesterol education program (ncep) expert panel on detection, evaluation, and treatment of high blood cholesterol in adults (adult treatment panel iii). JAMA. 2001;285:2486–2497. [DOI] [PubMed] [Google Scholar]
- 2. Pilia G, Chen WM, Scuteri A, et al. Heritability of cardiovascular and personality traits in 6,148 sardinians. PLoS genetics. 2006;2:e132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Teslovich TM, Musunuru K, Smith AV, et al. Biological, clinical and population relevance of 95 loci for blood lipids. Nature. 2010;466:707–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Abifadel M, Varret M, Rabès JP, et al. Mutations in pcsk9 cause autosomal dominant hypercholesterolemia. Nat Genet. 2003;34:154–156. [DOI] [PubMed] [Google Scholar]
- 5. Seidah NG. Proprotein convertase subtilisin kexin 9 (pcsk9) inhibitors in the treatment of hypercholesterolemia and other pathologies. Current Pharm Des. 2013;19:3161–3172. [DOI] [PubMed] [Google Scholar]
- 6. Maxwell KN, Breslow JL. Adenoviral-mediated expression of pcsk9 in mice results in a low-density lipoprotein receptor knockout phenotype. Proc Natl Acad Sci U S A. 2004;101:7100–7105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Denis M, Marcinkiewicz J, Zaid A, et al. Gene inactivation of proprotein convertase subtilisin/kexin type 9 reduces atherosclerosis in mice. Circulation. 2012;125:894–901. [DOI] [PubMed] [Google Scholar]
- 8. Cohen J, Pertsemlidis A, Kotowski IK, Graham R, Garcia CK, Hobbs HH. Low ldl cholesterol in individuals of african descent resulting from frequent nonsense mutations in pcsk9. Nat Genet. 2005;37:161–165. [DOI] [PubMed] [Google Scholar]
- 9. Benn M, Nordestgaard BG, Grande P, Schnohr P, Tybjaerg-Hansen A. Pcsk9 r46l, low-density lipoprotein cholesterol levels, and risk of ischemic heart disease: 3 independent studies and meta-analyses. J Am College Cardiol. 2010;55:2833–2842. [DOI] [PubMed] [Google Scholar]
- 10. Cohen JC, Boerwinkle E, Mosley TH, Jr., Hobbs HH. Sequence variations in pcsk9, low ldl, and protection against coronary heart disease. New Eng J Med. 2006;354:1264–1272. [DOI] [PubMed] [Google Scholar]
- 11. Farnier M. Pcsk9: From discovery to therapeutic applications. Arch Cardiovasc Dis. 2014;107:58–66. [DOI] [PubMed] [Google Scholar]
- 12. Robbins DC, Welty TK, Wang WY, Lee ET, Howard BV. Plasma lipids and lipoprotein concentrations among american indians: Comparison with the us population. Current Opinion in Lipidology. 1996;7:188–195. [DOI] [PubMed] [Google Scholar]
- 13. Kotowski IK, Pertsemlidis A, Luke A, et al. A spectrum of pcsk9 alleles contributes to plasma levels of low-density lipoprotein cholesterol. Am J Human Genet. 2006;78:410–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. North KE, Howard BV, Welty TK, et al. Genetic and environmental contributions to cardiovascular disease risk in american indians: The strong heart family study. Am J Epidemiology. 2003;157:303–314. [DOI] [PubMed] [Google Scholar]
- 15. Voight BF, Kang HM, Ding J, et al. The metabochip, a custom genotyping array for genetic studies of metabolic, cardiovascular, and anthropometric traits. PLoS Genetics. 2012;8:e1002793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Chen SN, Ballantyne CM, Gotto AM, Jr., Tan Y, Willerson JT, Marian AJ. A common pcsk9 haplotype, encompassing the e670g coding single nucleotide polymorphism, is a novel genetic marker for plasma low-density lipoprotein cholesterol levels and severity of coronary atherosclerosis. J Am College Cardiol. 2005;45:1611–1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Huang CC, Fornage M, Lloyd-Jones DM, Wei GS, Boerwinkle E, Liu K. Longitudinal association of PCSK9 sequence variations with low-density lipoprotein cholesterol levels: The Coronary Artery Risk Development in Young Adults Study. Circ Cardiovasc Genet. 2009;2:354–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Aung LH, Yin RX, Miao L, et al. The proprotein convertase subtilisin/kexin type 9 gene E670G polymorphism and serum lipid levels in the Guangxi Bai Ku Yao and Han populations. Lipids Health Dis. 2011;10:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ding K, Kullo IJ. Molecular population genetics of pcsk9: A signature of recent positive selection. Pharmacogenetics and Genomics. 2008;18:169–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Dering C, Hemmelmann C, Pugh E, Ziegler A. Statistical analysis of rare sequence variants: An overview of collapsing methods. Genet Epidemiol. 2011;35 Suppl 1:S12–S17. [DOI] [PMC free article] [PubMed] [Google Scholar]


