Abstract
CDH13 encodes T-cadherin, a receptor for high molecular weight (HMW) adiponectin and low-density lipoprotein, promoting proliferation and migration of endothelial cells. Genome-wide association studies have mapped multiple variants in CDH13 associated with cardiometabolic traits (CMT) with variable effects across studies. We hypothesized that this heterogeneity might reflect interplay with DNA methylation within the region. Resequencing and EpiTYPER™ assay were applied for the HYPertension in ESTonia/Coronary Artery Disease in Czech (HYPEST/CADCZ; n = 358) samples to identify CDH13 promoter SNPs acting as methylation Quantitative Trait Loci (meQTLs) and to investigate their associations with CMT. In silico data were extracted from genome-wide DNA methylation and genotype datasets of the population-based sample Estonian Genome Center of the University of Tartu (EGCUT; n = 165). HYPEST–CADCZ meta-analysis identified a rare variant rs113460564 as highly significant meQTL for a 134-bp distant CpG site (P = 5.90 × 10−6; β = 3.19 %). Four common SNPs (rs12443878, rs12444338, rs62040565, rs8060301) exhibited effect on methylation level of up to 3 neighboring CpG sites in both datasets. The strongest association was detected in EGCUT between rs8060301 and cg09415485 (false discovery rate corrected P value = 1.89 × 10−30). Simultaneously, rs8060301 showed association with diastolic blood pressure, serum high-density lipoprotein and HMW adiponectin (P < 0.005). Novel strong associations were identified between rare CDH13 promoter meQTLs (minor allele frequency <5 %) and HMW adiponectin: rs2239857 (P = 5.50 × 10−5, β = −1,841.9 ng/mL) and rs77068073 (P = 2.67 × 10−4, β = −2,484.4 ng/mL). Our study shows conclusively that CDH13 promoter harbors meQTLs associated with CMTs. It paves the way to deeper understanding of the interplay between DNA variation and methylation in susceptibility to common diseases.
Electronic supplementary material
The online version of this article (doi:10.1007/s00439-014-1521-6) contains supplementary material, which is available to authorized users.
Introduction
The relevance of the Cadherin-13 gene (CDH13; 1.2 Mb) in a wide spectrum of biomedical fields—oncology, neurology, cardiovascular physiology—was recognized over a decade ago (Takeuchi and Ohtsuki 2001). CDH13 encodes T-cadherin, which belongs to the cadherin gene family of cell adhesion molecules (Ranscht and Dours-Zimmermann 1991). Its expression was first described in the developing chicken embryo and shown to be widely distributed throughout the entire avian and mammalian nervous systems (Rivero et al. 2013). Subsequent studies in vascular tissue identified high expression of T-cadherin in endothelial and smooth muscle cells, as well as specifically in cardiac myocytes (Philippova et al. 2009). Localized in membrane lipid rafts, T-cadherin functions in promoting survival, proliferation and migration of endothelial cells and in protecting cells from oxidative stress-induced apoptosis (Philippova et al. 2009; Joshi et al. 2005). In cardiovascular metabolism, it exhibits ligand-binding ability uncommon to classical cadherins, acting as the third receptor for high molecular weight (HMW) adiponectin and also binding low-density lipoprotein (LDL) (Tkachuk et al. 1998; Hug et al. 2004). Low circulating adiponectin levels (hypoadiponectinemia: <4 μg/mL) are associated with not only various cardiovascular and metabolic phenotypes [e.g., type 2 diabetes (T2D), hypertension, dyslipidemia, atherosclerosis, coronary artery disease and stroke], but also with gastrointestinal diseases, osteoporosis and cancers (Kishida et al. 2014). A considerable number of human cancer genomes are characterized by hypermethylated CDH13 promoter, and down-regulation of its transcription promotes tumor growth and invasiveness (Andreeva and Kutuzov 2010).
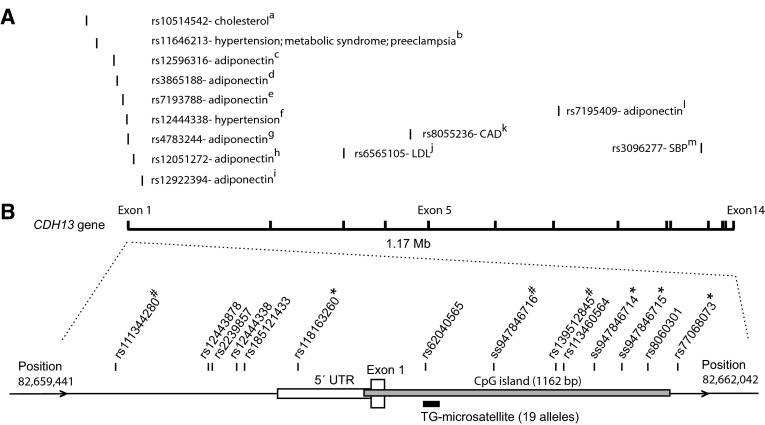
The era of genome-wide association studies (GWAS) has brought further evidence of pleiotropic effects attributed to CDH13. Genetic risk variants in CDH13 have been identified for cancer (Thomas et al. 2008) and neuropsychiatric disorders such as attention-deficit/hyperactivity disorder (ADHD), autism and dependence on psychotic substances (Rivero et al. 2013; Redies et al. 2012). However, the most notable genetic association signals in the CDH13 gene have been detected for a spectrum of cardiovascular and metabolic traits (Fig. 1). The strongest and the largest number of associations, mainly for a cluster of SNPs in the promoter region, have been reported for serum adiponectin levels and many of these findings have been replicated in diverse ethnic populations (Ling et al. 2009; Jee et al. 2010; Wu et al. 2010; Chung et al. 2011; Morisaki et al. 2012; Dastani et al. 2012; Gao et al. 2013). Decreased serum adiponectin levels have recently been showed for ADHD patients suggesting its possible involvement in the pathophysiology of ADHD (Mavroconstanti et al. 2014). SNPs in CDH13 have been associated with total cholesterol and LDL levels (Dong et al. 2011; Lee et al. 2013), coronary artery disease (CAD) (Wellcome Trust Case Control Consortium 2007), hypertension and blood pressure (Org et al. 2009; Levy et al. 2007; Lee et al. 2013), hyperlipidemia and myocardial infarction (Shia et al. 2011), metabolic syndrome (Fava et al. 2011) and preeclampsia (Wan et al. 2013).
Fig. 1.
Resequencing of the CDH13 promoter region in the HYPEST and CADCZ sample sets. a Illustrative map of previously identified genetic associations between cardiometabolic traits and SNPs in the CDH13 genomic region (1.17 Mb; GRCh37/hg19 Chr.16: 82,660,399–83,830,215; exons 1–14) is shown at the relative genomic scale. Numbers in superscript indicate the respective publications in the Reference List. b The CDH13 promoter region targeted for resequencing (2,602 bp; Chr.16: 82,659,441–82,662,042; bordered by arrows ‘>’) is zoomed, comprising the 5′ UTR (horizontal box), exon 1 (vertical box) and CpG island (grey box) of the gene. The location of SNPs and the polymorphic (TG)n microsatellite detected in the current study is shown; population-specific SNPs are marked with ‘*’ (HYPEST) or ‘#’ (CADCZ). CAD coronary artery disease, LDL low-density lipoprotein, SBP systolic blood pressure. aWallace et al. (2008); bOrg et al. (2009), Fava et al. (2011), Wan et al. (2013); cJee et al. (2010); dJee et al. (2010), Wu et al. (2010), Jo et al. (2012); eJee et al. (2010), Chung et al. (2011); fLee et al. (2013); gChung et al. (2011), Morisaki et al. (2012), Gao et al. (2013), Uetani et al. (2014); hMorisaki et al. (2012), Dastani et al. (2012); iDastani et al. (2012); jLee et al. (2013); kWellcome Trust Case Control Consortium (2007); lLing et al. (2009); mLevy et al. (2007)
Despite convincing evidence of the contribution of genetic variation in CDH13 to cardiometabolic traits, the primary causative SNP has not been identified and a multitude of contributing common variants with variable effects across studies has been reported (Fig. 1). DNA methylation has been suggested as a potential mediator of genetic risk for common diseases (Liu et al. 2013; Koestler et al. 2014). Low DNA methylation level in genomic regions associated with T2D in GWA studies was reported as an early marker of T2D suggesting early-onset, inter-individual methylation variation at isolated genomic sites that modify the predisposition to T2D (Toperoff et al. 2012). For the CDH13 gene, seminal reports have shown significant inter-individual variation in DNA methylation and described SNPs in the CDH13 gene that affect the methylation of nearby CpG sites (Flanagan et al. 2006, 2009; Zhi et al. 2013).
In the current study, we hypothesized that the heterogeneity of the identified genetic associations in CDH13 with a number of cardiometabolic traits might reflect interplay of inter-individual differences in DNA methylation variation. The study aimed (i) to identify SNPs in the CDH13 promoter region modulating DNA methylation of nearby CpG sites referred as methylation Quantitative Trait Loci (meQTLs); (ii) to investigate the genetic association of the identified meQTLs with serum adiponectin, lipids and blood pressure and (iii) to address the effect of DNA methylation level per se on the studied quantitative cardiometabolic parameters (Figure S1). We relied on the investigation of DNA extracted from whole blood, as the basic inter-individual variations in DNA methylation levels stemming from the difference in the genetic composition among study subjects are expected to be detectable across the majority of the cell types.
Materials and methods
HYPEST and CADCZ subjects for CDH13 DNA methylation analysis and resequencing
Participants of HYPertension in ESTonia (HYPEST; full sample: n = 1,966) have been recruited across Estonia during 2004–2007 with the aim to analyze genetic-epidemiological risk factors for cardiovascular disease (CVD) (Org et al. 2011). Subjects of Coronary Artery Disease in Czech study (CADCZ; full sample: n = 869) have been recruited across Czech Republic during 1998–2000 with the aim to study genetic factors related to homocysteine metabolism in coronary artery disease (CAD) (Janosíková et al. 2003). The recruitment has been carried out in compliance with the Helsinki Declaration and all participants have given written informed consent. The HYPEST study has been approved by the Ethics Committee on Human Research of the University of Tartu (permissions 122/13, 22.12.2003; 137/20, 25.04.2005). The CADCZ study has been approved by the Ethics Committee of Charles University—First Faculty of Medicine (December 1996).
The current (epi)genetic study used a subset of middle-aged patients (total, n = 358) from HYPEST (n = 192; aged 34–62, mean 50.5 ± 5.2 years) and CADCZ (n = 166; aged 33–61, 50.1 ± 4.6 years) sample sets (Table 1). Study inclusion criteria, measurement of blood pressure and serum lipids and correction of the measured values prior to the genetic data analysis for the subjects under medication are described in the Supplemental Methods. Serum samples of HYPEST subjects were stored at −86 °C immediately after blood draw. The concentration of high molecular weight (HMW) adiponectin in serum was measured for the study subjects with available stored serum samples (n = 184) using the Human HMW Adiponectin/Acrp30 ELISA assay (R&D Systems) according to the manufacturer’s protocol (Supplemental Methods).
Table 1.
Characteristics of study samples for (epi)genetic analysis
| Parameter (mean ± SD) | CVD study participants | Population cohort | |
|---|---|---|---|
| HYPEST | CADCZ | EGCUT | |
| No of individuals | 192 | 166 | 165 |
| Men/women | 93/99 | 114/52# | 72/93* |
| Age (years) | 50.5 ± 5.2 | 50.1 ± 4.6 | 41.6 ± 22.7* |
| BMI (kg/m2) | 30.2 ± 5.4 | 27.3 ± 4.1# | 25.0 ± 4.8* |
| SBP (mmHg) | 140.1 ± 18.7 | 127.1 ± 16.2# | 123.1 ± 17.4* |
| DBP (mmHg) | 88.6 ± 11.7 | 81.5 ± 9.5# | 76.5 ± 10.6* |
| Total cholesterol (mmol/L) | 5.6 ± 1.2 | 5.3 ± 0.9# | 5.3 ± 1.1 |
| LDL (mmol/L) | 3.7 ± 1.0 | 3.1 ± 0.8# | 3.2 ± 0.9* |
| HDL (mmol/L) | 1.5 ± 0.4 | 1.3 ± 0.4# | 1.6 ± 0.4* |
| Triglycerides (mmol/L) | 1.7 ± 1.2 | 1.9 ± 1.2 | 1.4 ± 0.9* |
| HMW adiponectin (ng/mL) | 4,564.3 ± 3,602.9a | NA | NA |
| Clinically diagnosed hypertension (%) | 86.4 | 25.5# | 11.5* |
| Coronary artery disease (%) | 22.2 | 50# | 6.1* |
| Early (<58 years) myocardial infarction (%) | 14.6 | 44.6# | 0.6* |
| Antihypertensive treatment (%) | 86.4 | 25.5 # | 24.2* |
| Antilipidemic treatment (%) | 15.6 | 32.5# | 5.5* |
HMW high molecular weight, CADCZ Coronary Artery Disease in Czech, CVD cardiovascular disease, EGCUT University of Tartu Estonian Genome Center, HYPEST HYPertension in ESTonia
#HYPEST versus CADCZ, * HYPEST + CADCZ versus EGCUT, P < 0.05; the Chi-square test for categorical and the Student’s t test or Mann–Whitney test for continuous variables that had a normal or skewed distribution, respectively
aMen 3,268.3 ± 2,811.5 ng/mL; women 5,701.6 ± 3,842.8 ng/mL
EGCUT sample subjected to genome-wide DNA methylation profiling and genotyping
The population-based biobank of the Estonian Genome Center of the University of Tartu (EGCUT) has been recruited across Estonia in 2003–2010 (http://www.geenivaramu.ee/en/). EGCUT includes epidemiological–clinical datasets and DNA samples extracted from blood for Estonian adults across all age groups (n = 51,515; Leitsalu et al. 2014). Measurement of blood pressure and serum lipids in the EGCUT samples are provided in the Supplemental Methods. In this study, in silico data for the CDH13 region were extracted and analyzed for the EGCUT samples with available datasets for both genome-wide DNA methylation and genome-wide imputed genotypes (n = 165; aged 18–84; 41.6 ± 22.7 years).
EpiTYPER™ analysis of DNA methylation in the CDH13 promoter in HYPEST and CADCZ samples
The suitability of DNA extracted from whole blood for the reliable CpG methylation profiling at the CDH13 locus was assessed using a published genome-wide DNA methylation dataset of purified human blood cells (Reinius et al. 2012) (Supplemental Methods). All analyzed blood cell types demonstrated similar CpG methylation profile across the CDH13 region (Figure S2).
The MassARRAY EpiTYPER™ assay (Sequenom, San Diego, CA, USA) was applied to measure DNA methylation in the CDH13 promoter for the HYPEST/CADCZ subjects. The targeted CpG sites were located within the CpG island (1,162 bp; GRCh37/hg19, Chr16: 82,660,652–82,661,813) and in the flanking 361 bp 5′ upstream region (Figure S3). Four EpiTYPER™ assays were designed to cover 110 CpG sites (13–40 CpG sites/assay) within a ~1.5 kb target region using the EpiDesigner software as instructed by the manufacturer (Table S1; Figure S3). Measurement of DNA methylation at targeted CpG sites followed the established experimental and analytical protocols (Ehrich et al. 2005) (http://bioscience.sequenom.com/sites/bioscience.sequenom.com/files/EpiTYPER%20Application%20Note.pdf) and was implemented using MassARRAY Analyzer 4 (Sequenom Inc.). The analyzed sequence fragments (1–57 bp) containing >1 CpG sites were named as CpG units and the methylation value for a CpG unit was calculated as average methylation across the CpG sites forming a unit. Singleton CpG sites per fragment were assessed individually. Experimental details of the EpiTYPER™ assay and quality control (QC) steps of CpG methylation measurements performed prior to the statistical association testing are provided in the Supplemental Methods. After stringent QC, DNA methylation at 66 CpG sites in the CDH13 promoter (33 CpG sites clustered in 13 CpG units and 33 individual CpG sites; Table S2) was subjected to the association testing with nearby SNPs and phenotypic traits.
Resequencing of CDH13 promoter region in HYPEST and CADCZ samples
The promoter region of CDH13 was resequenced in HYPEST (n = 192) and CADCZ (n = 166) samples. The resequenced region spanned 2,602 bp (GRCh37/hg19, Chr16: 82,659,441–82,662,042) and included the entire CpG island of the CDH13 gene (1,162 bp, Chr:16: 82,660,652–82,661,813) (Fig. 1, Figure S3). The region was amplified by long-range PCR (Supplemental Methods) and resequenced as described by Hallast et al. (2005) on both forward and reverse strands using 10 sequencing primers (Table S1). Sequences were assembled and SNPs were identified using CodonCode Aligner (http://www.codoncode.com/aligner/). Estimation of allele frequencies and conformance to Hardy–Weinberg Equilibrium (HWE; χ2, P > 0.05) were implemented in the PLINK v1.07 software (Purcell et al. 2007).
Extraction of CpG methylation data for the CDH13 promoter region from EGCUT genome-wide DNA methylation dataset
Genome-wide DNA methylation profiling (Infinium HumanMethylation450 BeadChips) of EGCUT samples (n = 165) was performed according to the manufacturer’s recommendations (Supplemental Methods). The original IDAT files were extracted from the HiScanSQ scanner. Data pre-processing and QC analyses were performed in R using the Bioconductor package minfi version 3.0.1 (Aryee et al. 2014) (Supplemental Methods). After QC steps, DNA methylation levels at the 57 CpG sites across the CDH13 genic region (GRCh37/hg19, Chr16: 82,474,489–83,829,911) were tested in association with allelic profile of the SNPs at the CDH13 promoter. Eight promoter CpG sites analyzed in EGCUT overlapped with the HYPEST–CADCZ dataset (EpiTYPER™ assays; Table S3).
Extraction of CDH13 genotyping data from EGCUT genome-wide dataset
Genomic DNA of EGCUT subjects (n = 165) was genotyped using HumanOmniExpress BeadChips (Illumina) according to the manufacturer’s instructions. The following QC filters were applied: sample call rate > 0.95, SNP call rate > 0.95, MAF > 0.01 and HWE P value > 0.00001. Genotype imputation is detailed in the Supplemental Methods.
In the current study, four SNPs in the CDH13 promoter region, which overlapped between HYPEST/CADCZ and EGCUT datasets were targeted in the genetic association testing of CpG methylation levels and cardiometabolic parameters. One of these SNPs had been genotyped (rs12443878) and the other three imputed (rs12444338, rs62040565, rs8060301). EGCUT genotype data was also exploited to calculate linkage disequilibrium (LD; r 2) between these SNPs and previously reported SNPs associated with cardiometabolic traits (Table S4).
Genetic association testing with CpG methylation levels: meQTL analysis
Seven CDH13 promoter SNPs shared by HYPEST and CADCZ (Fig. 1c) were tested for association with DNA methylation levels at the 46 studied cis-CpG sites/units (Figure S3). Prior to association analysis, the effect of age as a potential confounder of CpG methylation was assessed and no correlations remained significant after multiple testing correction (Table S5). Tests were performed using linear regression with an additive model (age, gender and experiment series as covariates) implemented in PLINK v1.07 (Purcell et al. 2007) and the combined meta-analysis was carried out using the inverse-variance method under a fixed-effects model implemented in R, ver. 3.0.2 (R Development Core Team 2014, http://www.r-project.org/). Meta-analysis of HYPEST and CADCZ results was used instead of joint analysis of the study subjects to eliminate confounding factors due to potential population stratification. Only those associations (nominal P value < 0.05) supported by enhanced statistical significance in the meta-analysis compared to separate tests for both HYPEST and CADCZ datasets were considered as potentially true results. Bonferroni threshold was calculated: α = 0.05/[2 (studies) × 7 (SNPs) × 46 (CpG sites/units)] = 7.76 × 10−5.
For meQTL confirmation analysis in the EGCUT sample set, the SNPs in the CDH13 promoter region overlapping with the HYPEST/CADCZ dataset (rs12443878, rs12444338, rs62040565, rs8060301) were assessed for the effect on the DNA methylation level at the CpG sites measured across the CDH13 gene (n = 57) in EGCUT. Linear regression coefficients were calculated to detect association between SNPs and the variation in the methylation levels. To correct for multiple testing, Benjamin–Hochberg false discovery rate (FDR) was estimated at 5 %, implemented using the p.adjust package in R.
Genetic association testing with cardiometabolic parameters
The effect of genotypes on cardiometabolic traits [total cholesterol, low- and high-density lipoproteins (LDL, HDL), triglycerides, systolic and diastolic blood pressure (SBP, DBP)] was tested in HYPEST, CADCZ and ECGUT meta-analysis using linear regression implemented in PLINK v1.07 (Purcell et al. 2007), and models were adjusted for age and gender. Multiple testing threshold was estimated: α = 0.05/[3 (studies) × 4 (SNPs) × 5 (SBP, DBP, HDL, LDL and triglycerides] = 8.33 × 10−4.
Genetic association testing with serum HMW adiponectin levels was applicable for 11 SNP detected in resequencing the HYPEST samples. In addition to gender and age, BMI [obesity affects adiponectin level (Arita et al. 1999)] and plate batch in the ELISA assay (to minimize the inter-assay effect) were incorporated into the model as covariates. Tests were implemented with natural logarithm-transformed adiponectin values.
Analysis of association between DNA methylation levels and cardiometabolic traits
The effect of methylation levels at the 46 CDH13 CpG sites/units on serum lipids and BP in HYPEST and CADCZ subjects was tested using linear regression in R incorporating age, gender and experiment series in the model. The results were combined in a meta-analysis as described above and the Bonferroni threshold was estimated: α = 0.05/[2 × (studies) × 5 (parameters) × 46 (CpG sites/units)] = 1.09 × 10−4.
Results
CDH13 promoter meQTLs
The methylation profiling of the CDH13 promoter in the HYPEST/CADCZ (HYPertension in ESTonia/Coronary Artery Disease in Czech) study sample (n = 358) resulted in the methylation levels of 46 CpG sites/units within 1,162 bp of its CpG island and flanking 361 bp 5′ upstream region (Fig. 1, Figure S3). Resequencing of the CDH13 promoter (2,602 bp) resulted in 14 SNPs, seven shared among the HYPEST and CADCZ samples (Fig. 1; Table S6). The rest of the SNPs were population-specific, including one and two novel variants for CADCZ and HYPEST, respectively. In addition, a highly polymorphic TG microsatellite (19 length variants) within the CpG island was detected with similar allelic distribution in the two sample sets (Supplemental Methods, Figure S4). One SNP, rs62040565 was located inside the TG microsatellite (Fig. 1).
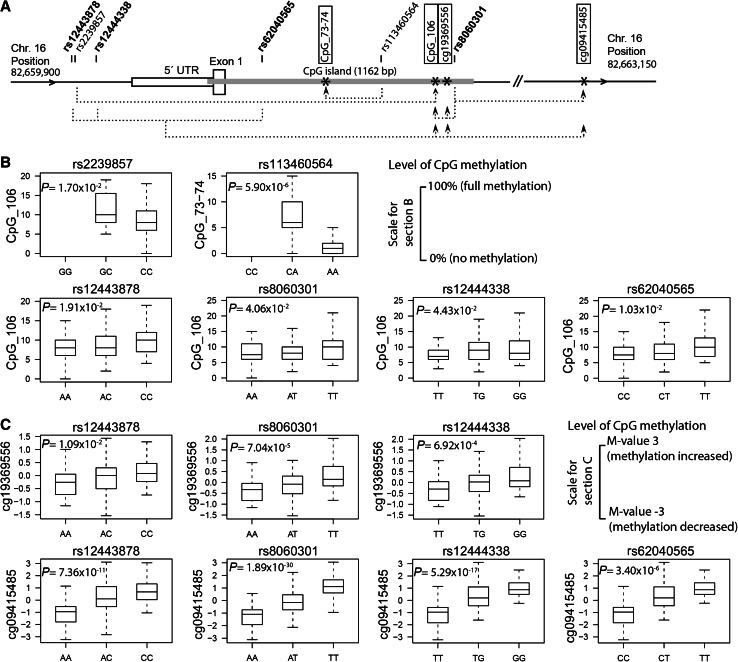
Association testing was performed between the seven shared SNPs and DNA methylation levels at the 46 CpG sites/units measured in HYPEST/CADCZ. A rare variant rs113460564 (MAF: HYPEST 0.8 %, CADCZ 2.1 %) within the CpG island was identified as a statistically significant meQTL located 134 bp from the major modulated CpG site (HYPEST: P = 4.25 × 10−4; CADCZ: P = 2.64 × 10−3; meta-analysis: P = 5.90 × 10−6, resistant to multiple testing correction; Table 2; Fig. 2a, b). Carrying one copy of the rs113460564 minor allele accounted for 2.8 or 5.5 % increased methylation at the CpG unit CpG_73–74 in HYPEST and CADCZ, respectively [meta-analysis: β (SE) = 3.19 (0.70)]. Five additional SNPs showed a non-significant trend for acting as meQTLs, potentially affecting DNA methylation levels of up to 3 CpG sites located 74–1,737 bp from the respective SNPs (Table 3). The length of the TG microsatellite did not show any evidence of association with DNA methylation levels at the tested CpG sites (data not presented).
Table 2.
Results of HYPEST–CADCZ meta-analysis for genetic association tests between SNPs and DNA methylation at the CpG sites within the CDH13 promoter region
| SNP/tested allele | CpG site/unit: its position at chr. 16a | Distance between SNP and CpG (bp)b | HYPEST | CADCZ | Meta-analysisd | |||
|---|---|---|---|---|---|---|---|---|
| β (SE)c
95 % CI |
P value | β (SE) 95 % CI |
P value | β (SE) 95 % CI |
P value | |||
|
rs113460564 C |
CpG_73–74 82,661,285 |
134 |
2.77 (0.77) [1.27, 4.27] |
4.25 × 10−4 |
5.46 (1.78) [1.96, 8.95] |
2.64 × 10−3 |
3.19 (0.70) [1.81, 4.56] |
5.90 × 10−6 |
|
rs2239857 G |
CpG_13 82,660,554 |
611 |
−1.01 (0.38) [−1.76, −0.26] |
9.56 × 10−3 |
−0.97 (1.58) [−4.07, 2.14] |
5.42 × 10−1 |
−1.01 (0.37) [−1.74, −0.27] |
7.04 × 10−3 |
|
CpG_106 82,661,670 |
1,727 |
3.12 (2.04) [−0.88, 7.12] |
1.30 × 10−1 |
5.57 (2.84) [0.01, 11.13] |
5.17 × 10−2 |
3.96 (1.66) [0.71, 7.20] |
1.70 × 10−2 | |
|
CpG_3 82,660,376 |
433 |
−3.28 (1.61) [−6.43, −0.12] |
4.37 × 10−2 |
−3.68 (4.78) [−13.05, 5.69] |
4.43 × 10−1 |
−3.32 (1.53) [−6.31, −0.33] |
2.97 × 10−2 | |
|
rs12444338 T |
CpG_116 82,661,812 |
1,657 |
3.11 (1.50) [−6.04, −0.17] |
4.10 × 10−2 |
1.35 (0.70) [−0.02, 2.73] |
5.60 × 10−2 |
1.67 (0.64) [0.42, 2.91] |
8.61 × 10−3 |
|
CpG_106 82,661,670 |
1,515 |
−0.92 (0.75) [−0.55, 2.39] |
2.23 × 10−1 |
−0.74 (0.46) [−1.64, 0.16] |
1.11 × 10−1 |
−0.79 (0.39) [−1.56, −0.02] |
4.43 × 10−2 | |
|
rs62040565e C |
CpG_106 82,661,670 |
724 |
−1.18 (0.70) [−0.19, 2.54] |
9.59 × 10−2 |
−0.94 (0.48) [−1.88, 0.01] |
5.28 × 10−2 |
−1.02 (0.40) [−1.79, −0.24] |
1.03 × 10−2 |
|
CpG_3 82,660,376 |
570 |
0.82 (0.64) [−2.07, 0.44] |
2.04 × 10−1 |
1.50 (0.71) [0.10, 2.90] |
3.70 × 10−2 |
1.12 (0.48) [0.19, 2.05] |
1.85 × 10−2 | |
|
rs12443878 A |
CpG_106 82,661,670 |
1,737 |
−1.10 (0.69) [−0.25, 2.45] |
1.14 × 10−1 |
−0.83 (0.48) [−1.76, 0.10] |
8.35 × 10−2 |
−0.92 (0.39) [−1.68, −0.15] |
1.91 × 10−2 |
|
rs8060301 A |
CpG_106 82,661,670 |
74 |
−1.07 (0.73) [−0.35, 2.50] |
1.43 × 10−1 |
−0.73 (0.49) [−1.69, 0.24] |
1.44 × 10−1 |
−0.84 (0.41) [−1.64, −0.04] |
4.06 × 10−2 |
SE standard error, CI confidence interval
aNumbering and the precise localization of CpG sites/units within the CDH13 promoter region are provided in Figure S3. Promoter CpG sites 1–15 are located upstream of the CpG island, sites 16–116 are within the CpG island and sites 18–26 are in the first exon. Genomic position on chromosome 16 is according to GRCh37/hg19. In case of CpG units, the position of the first CpG site in the unit is given
bIn case of CpG units, distance between SNP and the closest CpG site in the unit is given
cTested allele effect on DNA methylation measured on a scale from 0 to 100 units; P values are calculated using linear regression, including age, gender and experiment series as covariates in the model
dOnly associations, which demonstrated enhanced statistical significance in meta-analysis compared to both sample-specific tests, are shown; results were combined using the inverse-variance method under a fixed-effects model; underlined P-value denotes statistically significant association after Bonferroni correction (α = 7.76 × 10−5)
eLocated within a polymorphic TG microsatellite and polymorphic substitution T to C creates an additional CpG site
Fig. 2.
Identified CDH13 promoter meQTLs modulating methylation levels in neighboring CpG sites. a Dotted lines connect meQTLs and CpG sites (boxed) showing significantly modulated methylation (*) in HYPEST–CADCZ (CpG_106, CpG_73–74) and EGCUT (cg19369556, cg09415485). Common SNPs are marked in bold. b Genotype effects of identified mQTLs on DNA methylation at selected CpG sites in HYPEST–CADCZ meta-analysis. Y-axis depicts measured DNA methylation level (%; 0 % no methylation, 100 % full methylation). P value was calculated using linear regression, including age, gender and experiment series as covariates in the model. c Genotype effects of tested mQTLs on DNA methylation variation in the EGCUT dataset. Y-axis represents relative DNA methylation level on transformed scale (M value). FDR-corrected P values were calculated using linear regression
Table 3.
Genetic association between four overlapping common SNPs identified in the HYPEST and CADCZ study groups and DNA methylation in the EGCUT sample set
| SNP | Tested allele | CpG sitea | CpG position at chr. 16b | Distance between SNP and CpG (bp) | β (SE)c | 95 % CI | P value | FDRc |
|---|---|---|---|---|---|---|---|---|
| rs8060301 | A | cg09415485 | 82,663,111 | 1,367 | −0.68 (0.06) | [−0.79, −0.57] | 1.53 × 10−32 | 1.89 × 10−30 |
| cg19369556 | 82,661,725 | 19 | −0.35 (0.07) | [−0.49, −0.20] | 2.84 × 10−6 | 7.04 × 10−5 | ||
| rs12444338 | T | cg09415485 | 82,663,111 | 2,956 | −0.57 (0.06) | [−0.70, −0.45] | 8.53 × 10−19 | 5.29 × 10−17 |
| cg19369556 | 82,661,725 | 1,570 | −0.31 (0.07) | [−0.46, −0.16] | 3.35 × 10−5 | 6.92 × 10−4 | ||
| rs12443878 | A | cg09415485 | 82,663,111 | 3,178 | −0.49 (0.07) | [−0.62, −0.35] | 1.78 × 10−12 | 7.36 × 10−11 |
| cg19369556 | 82,661,725 | 1,792 | −0.26 (0.08) | [−0.41, −0.11] | 6.15 × 10−4 | 1.09 × 10−2 | ||
| cg09044981 | 82,827,677 | 167,744 | 0.26 (0.08) | [0.11, 0.41] | 7.53 × 10−4 | 1.17 × 10−2 | ||
| rs62040565 | C | cg09415485 | 82,663,111 | 2,165 | −0.39 (0.07) | [−0.53, −0.24] | 1.10 × 10−7 | 3.40 × 10−6 |
SE standard error, CI confidence interval
aCpG site ID on Infinium HumanMethylation450 BeadChip
bGenomic position on chromosome 16 (GRCh37/hg19)
cFalse discovery rate (FDR) corrected P value; only associations, significant after FDR correction, are given. Effects (tested allele effect on DNA methylation on transformed scale, see “Materials and methods”) and P values are calculated using linear regression
Four SNPs (rs12443878, rs12444338, rs62040565 and rs8060301) showing evidence to act as meQTLs in HYPEST and CADCZ analysis overlapped with SNPs genotyped or imputed in the EGCUT (Estonian Genome Center of the University of Tartu) sample set. These SNPs were tested for association with methylation levels at 57 CpG sites measured across the CDH13 genic region in the EGCUT sample (Table S7). All four SNPs were confirmed as meQTLs and showed significant effects (FDR < 0.05) on the methylation levels of up to 3 CpG sites located 19–167,744 bp from the respective SNPs. The strongest association was detected between rs8060301 located within the CpG island and a CpG site cg09415485 at a distance of 1.3 kb (FDR 1.89 × 10−30; Table 3; Fig. 2a, c).
Associations between CDH13 promoter meQTLs and cardiometabolic parameters
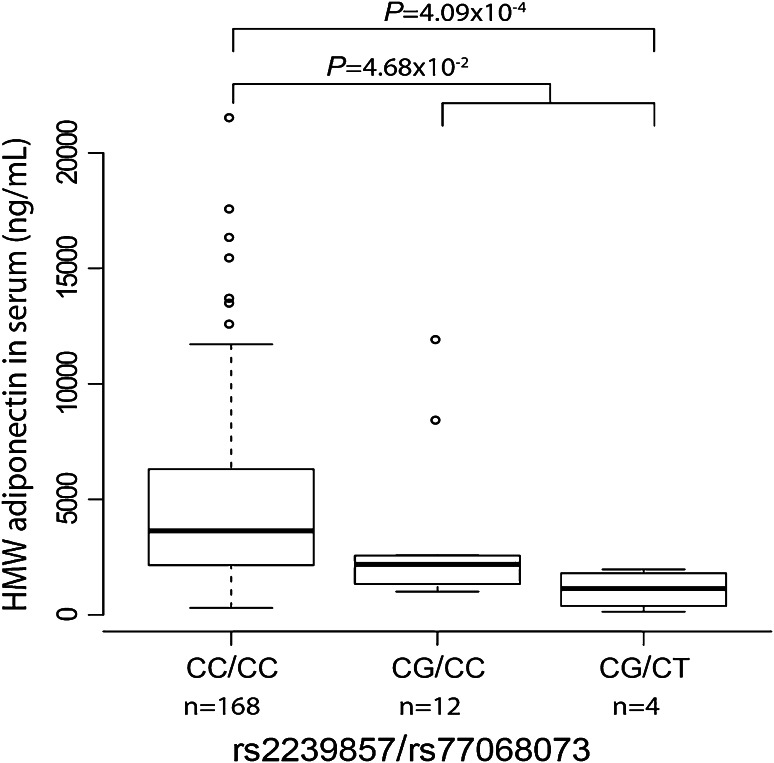
Next, genetic associations of identified meQTLs with cardiometabolic parameters were investigated. As the strongest associations of the CDH13 promoter region have been published for serum adiponectin, we firstly tested association with HMW adiponectin levels measured in HYPEST blood samples (unavailable for CADCZ, EGCUT). The strongest association was detected for two rare variants flanking the CpG island [rs2239857, C/G: MAF = 4.2 %, P = 5.50 × 10−5, β (SE) = −1,841.9 (711.9) ng/mL; rs77068073, C/T: MAF = 1 %, P = 2.67 × 10−4, β (SE) = −2,484.4 (1,463.1) ng/mL; Table 4; Fig. 1]. The lowest adiponectin levels (mean 1,095.1 ng/mL) were detected among the individuals who were heterozygous for both rs77068073 T-allele and rs2239857 G-allele (n = 4; Fig. 3). This represents approximately 4.5-fold lower adiponectin concentration compared to the study subjects carrying neither of these variants (mean 4,680.2 ng/mL). Subjects heterozygous for only rs2239857 (n = 12) also had reduced adiponectin levels (mean 3,356.8 ng/mL). In the SNP–CpG methylation association testing, rs2239857 had exhibited potential as a meQTL for 3 CpG sites (Table 2).
Table 4.
Association testing of CDH13 promoter SNPs with HMW adiponectin level in blood serum (ng/mL) in HYPEST
| SNP | Major/minor allelea | Tested allele | Tested allele frequency | β (SE) [ng/mL] |
95 % CI | P valueb |
|---|---|---|---|---|---|---|
| rs2239857 | C/G | G | 0.042 | −1,841.9 (711.7) | [−2,318.3, −1,147.9] | 5.50 × 10−5 |
| rs77068073 | C/T | T | 0.010 | −2,484.4 (1,463.1) | [−2,928.4, −1,582.3] | 2.67 × 10−4 |
| rs8060301 | T/A | A | 0.542 | −955.2 (273.2) | [−1,669.9, −342.2] | 1.63 × 10−3 |
| rs12444338 | G/T | T | 0.599 | −682.1 (278.3) | [−1,364.3, −98.4] | 2.13 × 10−2 |
| rs113460564 | A/C | C | 0.008 | 6,492.0 (3,684.4) | [−1,051.7, 38,677.6] | 1.48 × 10−1 |
| rs62040565 | T/C | C | 0.536 | −321.2 (264.9) | [−911.5, 187.6] | 2.31 × 10−1 |
| rs12443878 | C/A | A | 0.549 | −231.7 (267.2) | [−812.7, 268.5] | 3.85 × 10−1 |
| rs185121433 | G/T | T | 0.018 | 558.8 (1,107.1) | [−1,117.2, 3,488.5] | 5.90 × 10−1 |
| ss947846715 | C/G | G | 0.005 | −649.2 (2,362.3) | [−2,405.0, 4,336.8] | 6.87 × 10−1 |
| rs118163260 | A/T | T | 0.003 | −764.3 (3,777.2) | [−2,766.2, 8,006.3] | 7.32 × 10−1 |
| ss947846714 | A/G | G | 0.003 | −664.9 (3,715.4) | [−2,733.3, 8,235.9] | 7.68 × 10−1 |
HMW adiponectin in serum was measured for the study subjects with available stored HYPEST serum samples (n = 184) using Human HMW Adiponectin/Acrp30 ELISA assay (R&D Systems)
SE standard error, CI confidence interval
aMajor/minor alleles were defined according to UCSC Genome Browser (GRCh37/hg19)
bEffects and P values are calculated using linear regression, additive model including age, gender, BMI and ELISA plate as covariates in the model. Estimated significance threshold after correction for multiple testing P < 4.55 × 10−3
Fig. 3.
Novel genetic variants rs2239857 and rs77068073 associated with adiponectin level in blood serum. HMW adiponectin levels are shown for analyzed HYPEST subjects stratified based on their genotypes: heterozygotes for rs2239857 minor allele (n = 12, CG/CC), ‘double’ heterozygotes for both, rs2239857 and rs77068073 (n = 4, CG/CT) and homozygotes for major alleles of both SNPs (n = 168, CC/CC). The boxes represent the 25th and 75th percentiles. The median is denoted as the line that bisects the boxes. The whiskers are lines extending from each end of the box covering the extent of the data on 1.5× interquartile range. Circles represent the outlier values. P values from the t test comparing adiponectin levels of CC/CC group with CG/CT group and combined group of CG/CC and CG/CT are shown
Additionally, two common CDH13 SNPs (r 2 = 0.65) were associated with significantly lower adiponectin level: rs8060301 [T/A, A-allele = 54.2 %, P = 1.63 × 10−3, β (SE) = −955.2 (273.2) ng/mL] and rs12444338 [G/T, T-allele = 59.9 %, P = 2.13 × 10−2, β (SE) = −682.1 (278.3) ng/mL; Table 2]. These two SNPs are in strong LD (r 2 = 0.67–0.98; Table S4) with rs3865188 (9.7 kb upstream of CDH13) and rs4783244 SNPs (455 bp from CpG island) previously associated with serum adiponectin (Jee et al. 2010; Chung et al. 2011). Both rs8060301 and rs12444338 were shown to affect methylation of neighboring CpG sites in the HYPEST–CADCZ and EGCUT datasets (Tables 2, 3).
Four identified meQTLs in the CDH13 promoter represented common SNPs (rs8060301, rs1244438, rs62040565, rs12443878) with genotype and phenotype data available in HYPEST, EGCUT and CADCZ and enabled us to perform a meta-analysis (n = 523) of association tests for serum lipids (total cholesterol, LDL, HDL, triglycerides) and blood pressure (BP). The meQTL rs8060301 exhibited a suggestive pleiotropic effect on HDL and DBP (nominal P < 0.005; Table 5). It is noteworthy, that all SNPs showed a trend for association with serum HDL.
Table 5.
Results of HYPEST–CADCZ–EGCUT meta-analysis for genetic association tests between common SNPs within the CDH13 promoter region and cardiometabolic traits
| SNP | Test allele | Phenotype | Unit | HYPEST | CADCZ | EGCUT | Meta-analysisa | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| β (SE) [95 % CI] |
P value | β (SE) [95 % CI] |
P value | β (SE) [95 % CI] |
P value | β (SE) [95 % CI] |
P value | ||||
| rs8060301 | A | HDL | mmol/L |
0.06 (0.04) [−0.15, 0.03] |
1.77 × 10−1 |
0.09 (0.04) [0.01, 0.16] |
2.54 × 10−2 |
0.07 (0.04) [−0.01, 0.15] |
5.82 × 10−2 |
0.07 (0.02) [0.03, 0.12] |
1.25 × 10−3 |
| DBP | mmHg |
0.60 (1.65) [−3.84, 2.64] |
7.17 × 10−1 |
−3.45 (1.29) [−6.00, −0.92] |
8.34 × 10−3 |
−2.31 (1.10) [−4.47, −0.14] |
3.69 × 10−2 |
−2.10 (0.75) [−3.56, −0.63] |
4.97 × 10−3 | ||
| SBP | mmHg |
−1.18 (2.52) [−3.75, 6.12] |
6.39 × 10−1 |
−3.87 (2.17) [−8.12, 0.37] |
7.58 × 10−2 |
−1.71 (1.60) [−4.86, 1.45] |
2.87 × 10−1 |
−2.20 (1.15) [−4.45, 0.04] |
5.43 × 10−2 | ||
| rs12444338 | T | HDL | mmol/L |
0.04 (0.04) [−0.13, 0.05] |
3.67 × 10−1 |
0.09 (0.04) [0.02, 0.16] |
1.44 × 10−2 |
0.08 (0.04) [0.00, 0.15] |
4.99 × 10−2 |
0.07 (0.02) [0.03, 0.12] |
1.51 × 10−3 |
| rs62040565 | C | HDL | mmol/L |
0.06 (0.04) [−0.14, 0.02] |
1.41 × 10−1 |
0.07 (0.04) [−0.01, 0.14] |
8.88 × 10−2 |
0.03 (0.05) [−0.06, 0.12] |
5.10 × 10−1 |
0.06 (0.02) [0.01, 0.10] |
2.36 × 10−2 |
| rs12443878 | A | HDL | mmol/L |
0.06 (0.04) [−0.14, 0.03] |
1.94 × 10−1 |
0.05 (0.04) [−0.03, 0.12] |
2.24 × 10−1 |
0.03 (0.04) [−0.05, 0.11] |
4.13 × 10−1 |
0.04 (0.02) [−0.01, 0.09] |
5.48 × 10−2 |
SE standard error, CI confidence
aOnly associations, which demonstrated enhanced significance in meta-analysis compared to all sample-specific tests, are given. Effects and P values are calculated using linear regression, including age and gender as covariates in the model; results were combined using the inverse-variance method under a fixed-effects model. Estimated significance threshold after correction for multiple testing P < 8.33 × 10−4
No significant effect of CDH13 promoter CpG methylation level on cardiometabolic traits
The effect of methylation levels at the individual CpG sites within the CDH13 promoter (HYPEST, CADCZ: n = 46 CpG sites/units) was assessed on serum lipid and BP levels. Although 16 of 276 conducted tests reached nominal P < 0.05 (Table S8), no associations remained significant after multiple testing correction and none of the involved CpG sites were identified to be modulated by meQTLs.
Discussion
We hypothesized that the pleiotropy and heterogeneity of genetic associations in CDH13 with a number of cardiometabolic traits might reflect interplay of inter-individual differences in DNA methylation variation. It has recently been shown that genetic variability extensively impacts DNA methylation (Shi et al. 2014). Consistent with the study hypothesis, several SNPs in the CDH13 promoter region were significantly associated with the level of DNA methylation at nearby CpG sites (Tables 2, 3, 6; Fig. 3). These genetic variants showed simultaneous association signals with serum HMW adiponectin and lipid levels and were in LD with previously reported GWAS hits (Fig. 1; Tables 4, 5, 6).
Table 6.
Summary results of the study
| Analysis/trait | Study group | Significance level after correction for multiple testing | SNP showing statistically significant or suggestive for association |
|---|---|---|---|
| A . meQTL identification | |||
| CpG methylation | HYPEST–CADCZ meta-analysisa | P < 7.76 × 10−5 | rs113460564d, rs12443878, rs12444338, rs62040565, rs8060301, rs2239857 |
| EGCUTb | P < 7.53 × 10−4 | rs12443878d, rs12444338d, rs62040565d, rs8060301 d | |
| B . SNP and trait association testing | |||
| Adiponectinc | HYPEST | P < 4.55 × 10−3 | rs8060301 d, rs2239857d, rs77068073d, rs12444338 |
| HDL | HYPEST–CADCZ–EGCUT meta-analysis | P < 8.33 × 10−4 | rs12443878, rs12444338, rs62040565, rs8060301 |
| SBP, DBP | HYPEST–CADCZ–EGCUT meta-analysis | P < 8.33 × 10−4 | rs8060301 |
| C . DNA methylation-trait association | |||
| Methylation level of individual CDH13 promoter CpGs was not significantly modulating cardiometabolic traits | |||
SNP rs8060301 overlapping between the analysis results, is highlighted in bold
aGenotype data: targeted resequencing of the CDH13 promoter region; CpG methylation data: EpiTYPER™ assay covering majority of the CpG sites
bGenotype data: HumanOmniExpress BeadChips (Illumina); CpG methylation data: selected CpG sites at the Infinium HumanMethylation450 BeadChips
cHMW adiponectin measurements were only available for HYPEST
dSignificant after multiple testing correction
Among the identified meQTLs, rs8060301 located at the edge of the CDH13 CpG island within intron 1 modulated significantly DNA methylation in both study samples HYPEST/CADCZ and EGCUT (Table 6). Simultaneously, it exhibited the strongest genetic associations among the tested CDH13 SNPs with HDL (P = 1.25 × 10−3) and blood pressure levels (DBP = 4.97 × 10−3; SBP = 5.43 × 10−2) in the HYPEST–CADCZ–EGCUT meta-analysis. Association testing with serum HMW adiponectin in HYPEST highlighted rs8060301 as a common SNP (tested A-allele frequency 54.2 %) with the most significant and notable effect on this trait [P = 1.63 × 10−3, β (SE) = −955.2 (273.2) ng/mL]. Previously mapped serum adiponectin GWAS hit in Han Chinese (Chung et al. 2011) and Filipinos (Wu et al. 2010) rs4783244 is located only 524 bp from rs8060301 and these SNPs are in strong LD (Fig. 1; Table S4). However, only the rs8060301 (and not rs4783244) is located in the middle of the strongest binding site of RNA polymerase II (Pol2) within the CDH13 promoter (ENCODE Project Consortium 2012) and can thus exhibit direct effect on gene expression. The effect on the gene expression level of CDH13 may lead to altered protein expression and the affected sufficiency/abundance of T-cadherin molecules to bind adiponectin and lipids. Further functional studies need to be conducted to directly address its potential effect on CDH13 transcription level.
We used a complementary approach to identify meQTLs, which has both advantages and limitations. The HYPEST/CADCZ cardiovascular diseases samples were targeted to fine scale analysis of CpG sites at the CDH13 promoter using resequencing and the EpiTYPER™ assay. Using more dense assays provides additional value enabling to identify SNP–CpG associations that are not included in genome-wide assays. Data for a population-based cohort (EGCUT) were extracted from DNA methylation and genotyping arrays with highly standardized experimental and analytical methods. However, DNA methylation and genotyping chips do not cover dense runs of CpG sites within the CpG islands and rare SNPs, respectively. Thus, it was not possible to directly replicate the identified HYPEST/CADCZ top SNP–CpG site association pairs in EGCUT, although the same SNPs were confirmed as meQTLs in both study samples. An additional limitation in our study design was the not perfectly matched sample sets. Although both derived from Eastern/Central Europe, HYPEST (Estonia)/CADCZ (Czech) subjects have been recruited based on CVD (characterized by hypertension, CAD, MI), but EGCUT (Estonia) is a population-based cohort. This may introduce SNP-independent effects on the DNA methylation profiles and weaken the meQTL analysis.
GWAS studies have mapped the strongest genetic associations in the CDH13 gene with serum adiponectin levels. In the current study, we measured serum HMW adiponectin levels only for 184 HYPEST subjects and identified significant associations with rs8060301 and rs12444338 (Table 4), which are in strong LD with previously detected SNPs in GWASs [rs3865188 (Jee et al. 2010); rs4783244 (Chung et al. 2011)]. Additionally, our resequencing approach allowed identification of two novel rare (MAF < 5 %) variants (rs2239857 and rs77068073) with a highly significant, strong effect on serum HMW adiponectin (Fig. 3). Notably, rs2239857 also exhibited potential as a meQTL for 3 CpG sites (Table 2).
We did not identify any significant associations between DNA methylation levels in the CDH13 promoter and blood pressure or lipid levels. Alternative reasons (each one separately and also cumulatively), which may have affected achieving sufficient statistical power in testing these associations could be cross-sectional (whole blood cells) measurement of DNA methylation and its narrow range of variability within the promoter region, heterogenous study sample with regard to other variables modulating DNA methylation (e.g., age) and also possibly insufficient sample size failing to detect real correlations. In addition, investigation of direct effect of DNA methylation on cardiovascular phenotype trait would be more relevant using tissues with high and specific expression of CDH13, such as endothelium. In general, our study indicates that effects of SNPs on studied cardiometabolic traits could be primary, but due to strong meQTLs in the region, we suggest that these effects are dependent on DNA methylation levels, which further modulate the SNP trait effects.
DNA methylation changes in the CDH13 promoter region are well characterized in the development of various cancers. In perspective, the meQTLs identified in this study in the context of CVD could be further subjected to studies of cancer patients. We speculate that meQTLs in the CDH13 promoter may serve as potential prognostic markers for an increased risk to trigger more extensive changes in the DNA methylation pattern. Recently, a comprehensive meQTL catalog was published containing DNA methylation associations for 21 % of interrogated cancer risk polymorphisms (Heyn et al. 2014).
In summary, our study shows conclusively that the CDH13 promoter harbors meQTLs associated with cardiometabolic traits. It paves the way to deeper understanding of the interplay between DNA variation and methylation in susceptibility to common diseases.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgments
We acknowledge all participants of the HYPEST, CADCZ and EGCUT studies. Wellcome Trust International Senior Research Fellowship (070191/Z/03/A) in Biomedical Science in Central Europe; Estonian Ministry of Education and Research Core Grants (SF0180022s12); European Union through the European Regional Development Fund (project HAPPY PREGNANCY, 3.2.0701.12-0047); Estonian Science Foundation Grants (ETF9030, ETF9353, ETF9293 and ETF7491); Center of Excellence in Genomics (EXCEGEN); University of Tartu (SP1GVARENG), the Estonian Research Council Grant (IUT20-60), and the Estonian Research Roadmap through the Estonian Ministry of Education and Research (3.2.0304.11-0312). V.K. was receiving institutional support by research programs of the Charles University in Prague PRVOUKP24/LF1/3 and UNCE 20401.
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical standards
Experiments comply with the current laws of the countries in which they were performed.
References
- Andreeva AV, Kutuzov MA. Cadherin 13 in cancer. Genes Chromosomes Cancer. 2010;49(9):775–790. doi: 10.1002/gcc.20787. [DOI] [PubMed] [Google Scholar]
- Arita Y, Kihara S, Ouchi N, Takahashi M, Maeda K, Miyagawa J, Hotta K, Shimomura I, Nakamura T, Miyaoka K, et al. Paradoxical decrease of an adipose-specific protein, adiponectin, in obesity. Biochem Biophys Res Commun. 1999;257(1):79–83. doi: 10.1006/bbrc.1999.0255. [DOI] [PubMed] [Google Scholar]
- Aryee MJ, Jaffe AE, Corrada-Bravo H, Ladd-Acosta C, Feinberg AP, Hansen KD, Irizarry RA. Minfi: a flexible and comprehensive Bioconductor package for the analysis of Infinium DNA methylation microarrays. Bioinformatics. 2014;30(10):1363–1369. doi: 10.1093/bioinformatics/btu049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung CM, Lin TH, Chen JW, Leu HB, Yang HC, Ho HY, Ting CT, Sheu SH, Tsai WC, Chen JH, et al. A genome-wide association study reveals a quantitative trait locus of adiponectin on CDH13 that predicts cardiometabolic outcomes. Diabetes. 2011;60(9):2417–2423. doi: 10.2337/db10-1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dastani Z, Hivert MF, Timpson N, Perry JR, Yuan X, Scott RA, Henneman P, Heid IM, Kizer JR, Lyytikäinen LP, et al. Novel loci foradiponectin levels and their influence on type 2 diabetes and metabolic traits: a multi-ethnic meta-analysis of 45,891 individuals. PLoS Genet. 2012;8(3):e1002607. doi: 10.1371/journal.pgen.1002607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong C, Beecham A, Wang L, Slifer S, Wright CB, Blanton SH, Rundek T, Sacco RL. Genetic loci for blood lipid levels identified by linkage and association analyses in Caribbean Hispanics. J Lipid Res. 2011;52(7):1411–1419. doi: 10.1194/jlr.P013672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrich M, Nelson MR, Stanssens P, Zabeau M, Liloglou T, Xinarianos G, Cantor CR, Field JK, van den Boom D. Quantitative high-throughput analysis of DNA methylation patterns by base-specific cleavage and mass spectrometry. Proc Natl Acad Sci. 2005;102(44):15785–15790. doi: 10.1073/pnas.0507816102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ENCODE Project Consortium An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489(7414):57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fava C, Danese E, Montagnana M, Sjögren M, Almgren P, Guidi GC, Hedblad B, Engström G, Lechi A, Minuz P, et al. A variant upstream of the CDH13 adiponectin receptor gene and metabolic syndrome in Swedes. Am J Cardiol. 2011;108(10):1432–1437. doi: 10.1016/j.amjcard.2011.06.068. [DOI] [PubMed] [Google Scholar]
- Flanagan JM, Popendikyte V, Pozdniakovaite N, Sobolev M, Assadzadeh A, Schumacher A, Zangeneh M, Lau L, Virtanen C, Wang SC, et al. Intra- and interindividual epigenetic variation in human germ cells. Am J Hum Genet. 2006;79(1):67–84. doi: 10.1086/504729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flanagan JM, Munoz-Alegre M, Henderson S, Tang T, Sun P, Johnson N, Fletcher O, Dos Santos Silva I, Peto J, Boshoff C, et al. Gene-body hypermethylation of ATM in peripheral blood DNA of bilateral breast cancer patients. Hum Mol Genet. 2009;18(7):1332–1342. doi: 10.1093/hmg/ddp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao H, Kim YM, Chen P, Igase M, Kawamoto R, Kim MK, Kohara K, Lee J, Miki T, Ong RT, et al. Genetic variation in CDH13 is associated with lower plasma adiponectin levels but greater adiponectin sensitivity in East Asian populations. Diabetes. 2013;62(12):4277–4283. doi: 10.2337/db13-0129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallast P, Nagirnaja L, Margus T, Laan M. Segmental duplications and gene conversion: human luteinizing hormone/chorionic gonadotropin beta gene cluster. Genome Res. 2005;15(11):1535–1546. doi: 10.1101/gr.4270505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyn H, Sayols S, Moutinho C, Vidal E, Sanchez-Mut JV, Stefansson OA, Nadal E, Moran S, Eyfjord JE, Gonzalez-Suarez E, et al. Linkage of DNA methylation quantitative trait loci to human cancer risk. Cell Rep. 2014;7(2):331–338. doi: 10.1016/j.celrep.2014.03.016. [DOI] [PubMed] [Google Scholar]
- Hug C, Wang J, Ahmad NS, Bogan JS, Tsao TS, Lodish HF. T-cadherin is a receptor for hexameric and high-molecular-weight forms of Acrp30/adiponectin. Proc Natl Acad Sci. 2004;101(28):10308–10313. doi: 10.1073/pnas.0403382101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janosíková B, Pavlíková M, Kocmanová D, Vítová A, Veselá K, Krupková L, Kahleová R, Krijt J, Kraml P, Hyánek J, et al. Genetic variants of homocysteine metabolizing enzymes and the risk of coronary artery disease. Mol Genet Metab. 2003;79(3):167–175. doi: 10.1016/S1096-7192(03)00079-9. [DOI] [PubMed] [Google Scholar]
- Jee SH, Sull JW, Lee JE, Shin C, Park J, Kimm H, Cho EY, Shin ES, Yun JE, Park JW, et al. Adiponectin concentrations: a genome-wide association study. Am J Hum Genet. 2010;87(4):545–552. doi: 10.1016/j.ajhg.2010.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jo J, Sull JW, Park EJ, Jee SH. Effects of smoking and obesity on the association between CDH13 (rs3865188) and adiponectin among Korean men: the KARE study. Obesity (Silver Spring). 2012;20(8):1683–1687. doi: 10.1038/oby.2011.128. [DOI] [PubMed] [Google Scholar]
- Joshi MB, Philippova M, Ivanov D, Allenspach R, Erne P, Resink TJ. T-cadherin protects endothelial cells from oxidative stress-induced apoptosis. FASEB J. 2005;19(12):1737–1739. doi: 10.1096/fj.05-3834fje. [DOI] [PubMed] [Google Scholar]
- Kishida K, Funahashi T, Shimomura I. Adiponectin as a routine clinical biomarker. Best Pract Res Clin Endocrinol Metab. 2014;28(1):119–130. doi: 10.1016/j.beem.2013.08.006. [DOI] [PubMed] [Google Scholar]
- Koestler DC, Chalise P, Cicek MS, Cunningham JM, Armasu S, Larson MC, Chien J, Block M, Kalli KR, Sellers TA, et al. Integrative genomic analysis identifies epigenetic marks that mediate genetic risk for epithelial ovarian cancer. BMC Med Genomics. 2014;7:8. doi: 10.1186/1755-8794-7-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Shin DJ, Park S, Kang SM, Jang Y, Lee SH. Association between CDH13 variants and cardiometabolic and vascular phenotypes in a Korean population. Yonsei Med J. 2013;54(6):1305–1312. doi: 10.3349/ymj.2013.54.6.1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leitsalu L, Haller T, Esko T, Tammesoo ML, Alavere H, Snieder H, Perola M, Ng PC, Mägi R, Milani L et al (2014) Cohort profile: Estonian Biobank of the Estonian Genome Center, University of Tartu. Int J Epidemiol. [Epub ahead of print] [DOI] [PubMed]
- Levy D, Larson MG, Benjamin EJ, Newton-Cheh C, Wang TJ, Hwang SJ, Vasan RS, Mitchell GF. Framingham Heart Study 100K Project: genome-wide associations for blood pressure and arterial stiffness. BMC Med Genet. 2007;8(Suppl 1):S3. doi: 10.1186/1471-2350-8-S1-S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling H, Waterworth DM, Stirnadel HA, Pollin TI, Barter PJ, Kesäniemi YA, Mahley RW, McPherson R, Waeber G, Bersot TP, et al. Genome-wide linkage and association analyses to identify genes influencing adiponectin levels: the GEMS Study. Obesity (Silver Spring). 2009;17(4):737–744. doi: 10.1038/oby.2008.625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Aryee MJ, Padyukov L, Fallin MD, Hesselberg E, Runarsson A, Reinius L, Acevedo N, Taub M, Ronninger M, et al. Epigenome-wide association data implicate DNA methylation as an intermediary of genetic risk in rheumatoid arthritis. Nat Biotechnol. 2013;31(2):142–147. doi: 10.1038/nbt.2487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mavroconstanti T, Halmøy A, Haavik J. Decreased serum levels of adiponectin in adult attention deficit hyperactivity disorder. Psychiatry Res. 2014;216(1):123–130. doi: 10.1016/j.psychres.2014.01.025. [DOI] [PubMed] [Google Scholar]
- Morisaki H, Yamanaka I, Iwai N, Miyamoto Y, Kokubo Y, Okamura T, Okayama A, Morisaki T. CDH13 gene coding T-cadherin influences variations in plasma adiponectin levels in the Japanese population. Hum Mutat. 2012;33(2):402–410. doi: 10.1002/humu.21652. [DOI] [PubMed] [Google Scholar]
- Org E, Eyheramendy S, Juhanson P, Gieger C, Lichtner P, Klopp N, Veldre G, Döring A, Viigimaa M, Sõber S, et al. Genome-wide scan identifies CDH13 as a novel susceptibility locus contributing to blood pressure determination in two European populations. Hum Mol Genet. 2009;18(12):2288–2296. doi: 10.1093/hmg/ddp135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Org E, Veldre G, Viigimaa M, Juhanson P, Putku M, Rosenberg M, Tomberg K, Uuetoa T, Laan M. HYPEST study: profile of hypertensive patients in Estonia. BMC Cardiovasc Disord. 2011;11:55. doi: 10.1186/1471-2261-11-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippova M, Joshi MB, Kyriakakis E, Pfaff D, Erne P, Resink TJ. A guide and guard: the many faces of T-cadherin. Cell Signal. 2009;21(7):1035–1044. doi: 10.1016/j.cellsig.2009.01.035. [DOI] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81(3):559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranscht B, Dours-Zimmermann MT. T-cadherin, a novel cadherin cell adhesion molecule in the nervous system lacks the conserved cytoplasmic region. Neuron. 1991;7(3):391–402. doi: 10.1016/0896-6273(91)90291-7. [DOI] [PubMed] [Google Scholar]
- Redies C, Hertel N, Hübner CA. Cadherins and neuropsychiatric disorders. Brain Res. 2012;1470:130–144. doi: 10.1016/j.brainres.2012.06.020. [DOI] [PubMed] [Google Scholar]
- Reinius LE, Acevedo N, Joerink M, Pershagen G, Dahlén SE, Greco D, Söderhäll C, Scheynius A, Kere J. Differential DNA methylation in purified human blood cells: implications for cell lineage and studies on disease susceptibility. PLoS One. 2012;7(7):e41361. doi: 10.1371/journal.pone.0041361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivero O, Sich S, Popp S, Schmitt A, Franke B, Lesch KP. Impact of the ADHD-susceptibility gene CDH13 on development and function of brain networks. Eur Neuropsychopharmacol. 2013;23(6):492–507. doi: 10.1016/j.euroneuro.2012.06.009. [DOI] [PubMed] [Google Scholar]
- Shi J, Marconett CN, Duan J, Hyland PL, Li P, Wang Z, Wheeler W, Zhou B, Campan M, Lee DS, et al. Characterizing the genetic basis of methylome diversity in histologically normal human lung tissue. Nat Commun. 2014;5:3365. doi: 10.1038/ncomms4365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shia WC, Ku TH, Tsao YM, Hsia CH, Chang YM, Huang CH, Chung YC, Hsu SL, Liang KW, Hsu FR. Genetic copy number variants in myocardial infarction patients with hyperlipidemia. BMC Genom. 2011;12(Suppl 3):S23. doi: 10.1186/1471-2164-12-S3-S23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi T, Ohtsuki Y. Recent progress in T-cadherin (CDH13, H-cadherin) research. Histol Histopathol. 2001;16(4):1287–1293. doi: 10.14670/HH-16.1287. [DOI] [PubMed] [Google Scholar]
- Thomas G, Jacobs KB, Yeager M, Kraft P, Wacholder S, Orr N, Yu K, Chatterjee N, Welch R, Hutchinson A, et al. Multiple loci identified in a genome-wide association study of prostate cancer. Nat Genet. 2008;40(3):310–315. doi: 10.1038/ng.91. [DOI] [PubMed] [Google Scholar]
- Tkachuk VA, Bochkov VN, Philippova MP, Stambolsky DV, Kuzmenko ES, Sidorova MV, Molokoedov AS, Spirov VG, Resink TJ. Identification of an atypical lipoprotein-binding protein from human aortic smooth muscle as T-cadherin. FEBS Lett. 1998;421(3):208–212. doi: 10.1016/S0014-5793(97)01562-7. [DOI] [PubMed] [Google Scholar]
- Toperoff G, Aran D, Kark JD, Rosenberg M, Dubnikov T, Nissan B, Wainstein J, Friedlander Y, Levy-Lahad E, Glaser B, et al. Genome-wide survey reveals predisposing diabetes type 2-related DNA methylation variations in human peripheral blood. Hum Mol Genet. 2012;21(2):371–383. doi: 10.1093/hmg/ddr472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uetani E, Tabara Y, Kawamoto R, Onuma H, Kohara K, Osawa H, Miki T. CDH13 genotype-dependent association of high-molecular weight adiponectin with all-cause mortality: the J-SHIPP study. Diabetes Care. 2014;37(2):396–401. doi: 10.2337/dc13-1658. [DOI] [PubMed] [Google Scholar]
- Wallace C, Newhouse SJ, Braund P, Zhang F, Tobin M, Falchi M, Ahmadi K, Dobson RJ, Marçano AC, Hajat C, et al. Genome-wide association study identifies genes for biomarkers of cardiovascular disease: serum urate and dyslipidemia. Am J Hum Genet. 2008;82(1):139–149. doi: 10.1016/j.ajhg.2007.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan JP, Zhao H, Li T, Li CZ, Wang XT, Chen ZJ. The common variant rs11646213 is associated with preeclampsia in Han Chinese women. PLoS One. 2013;8(8):e71202. doi: 10.1371/journal.pone.0071202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellcome Trust Case Control Consortium Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447(7145):661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Li Y, Lange EM, Croteau-Chonka DC, Kuzawa CW, McDade TW, Qin L, Curocichin G, Borja JB, Lange LA, et al. Genome-wide association study for adiponectin levels in Filipino women identifies CDH13 and a novel uncommon haplotype at KNG1-ADIPOQ. Hum Mol Genet. 2010;19(24):4955–4964. doi: 10.1093/hmg/ddq423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhi D, Aslibekyan S, Irvin MR, Claas SA, Borecki IB, Ordovas JM, Absher DM, Arnett DK. SNPs located at CpG sites modulate genome-epigenome interaction. Epigenetics. 2013;8(8):802–806. doi: 10.4161/epi.25501. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.