Abstract
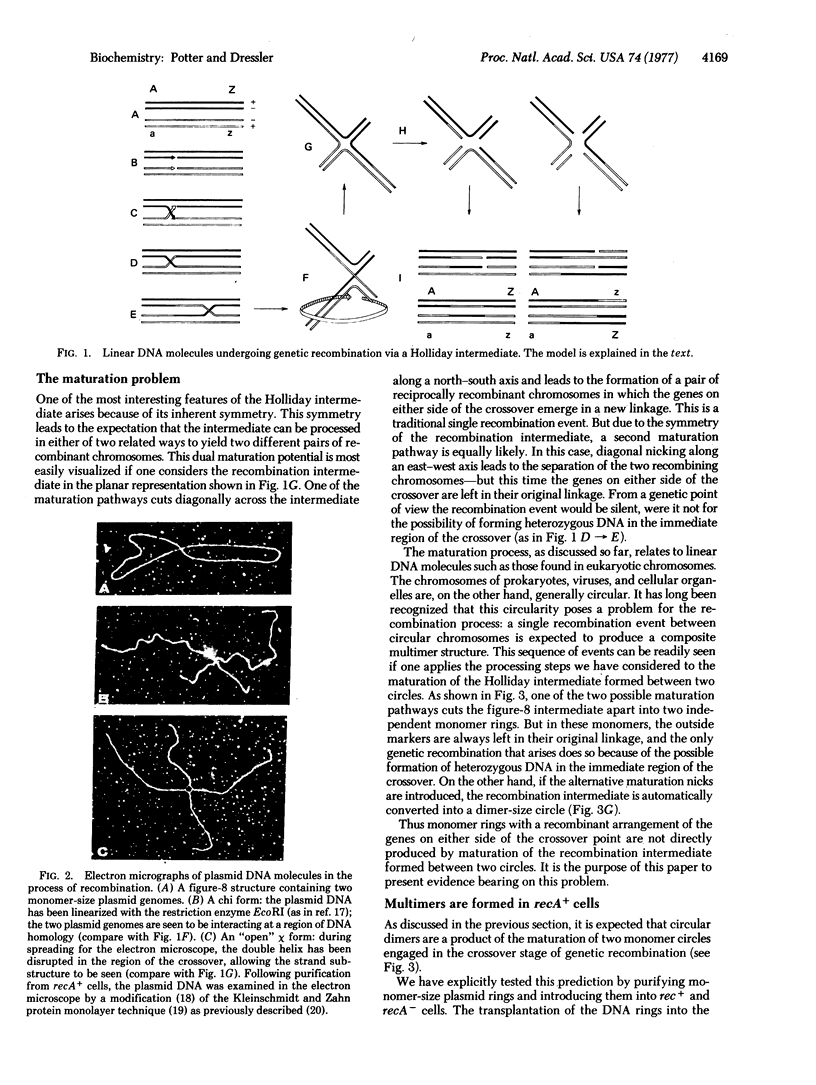
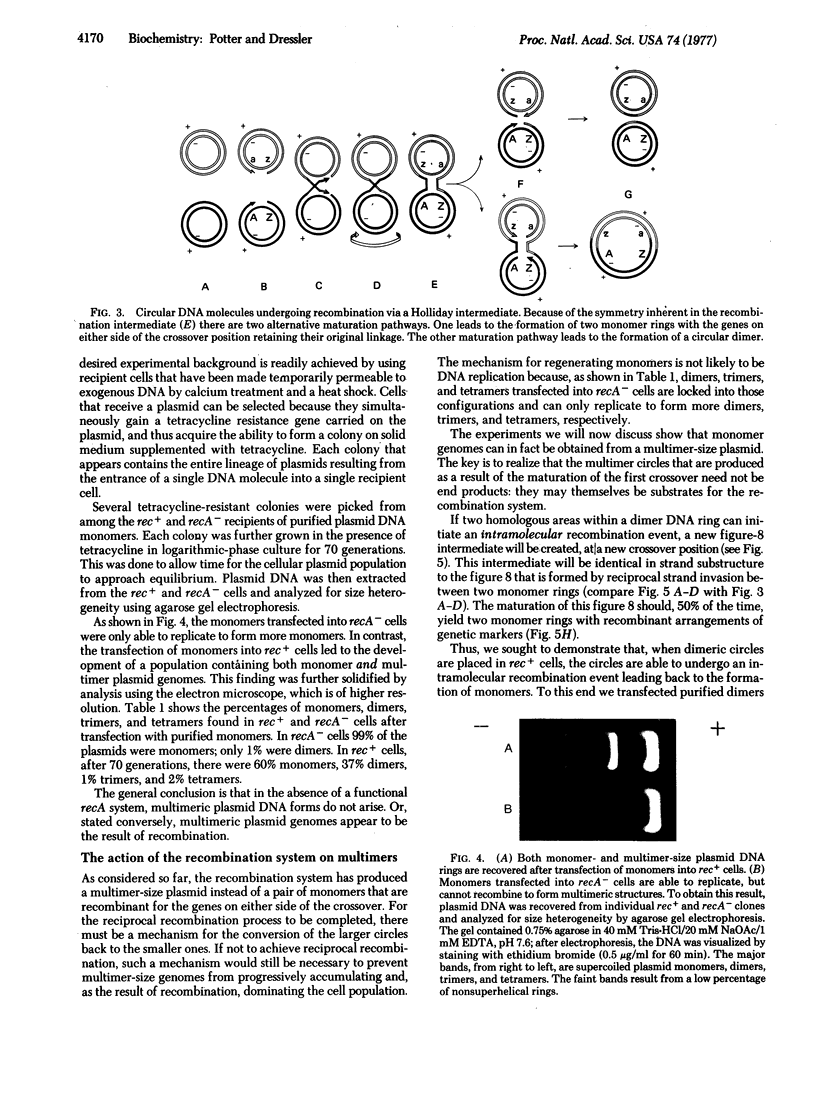
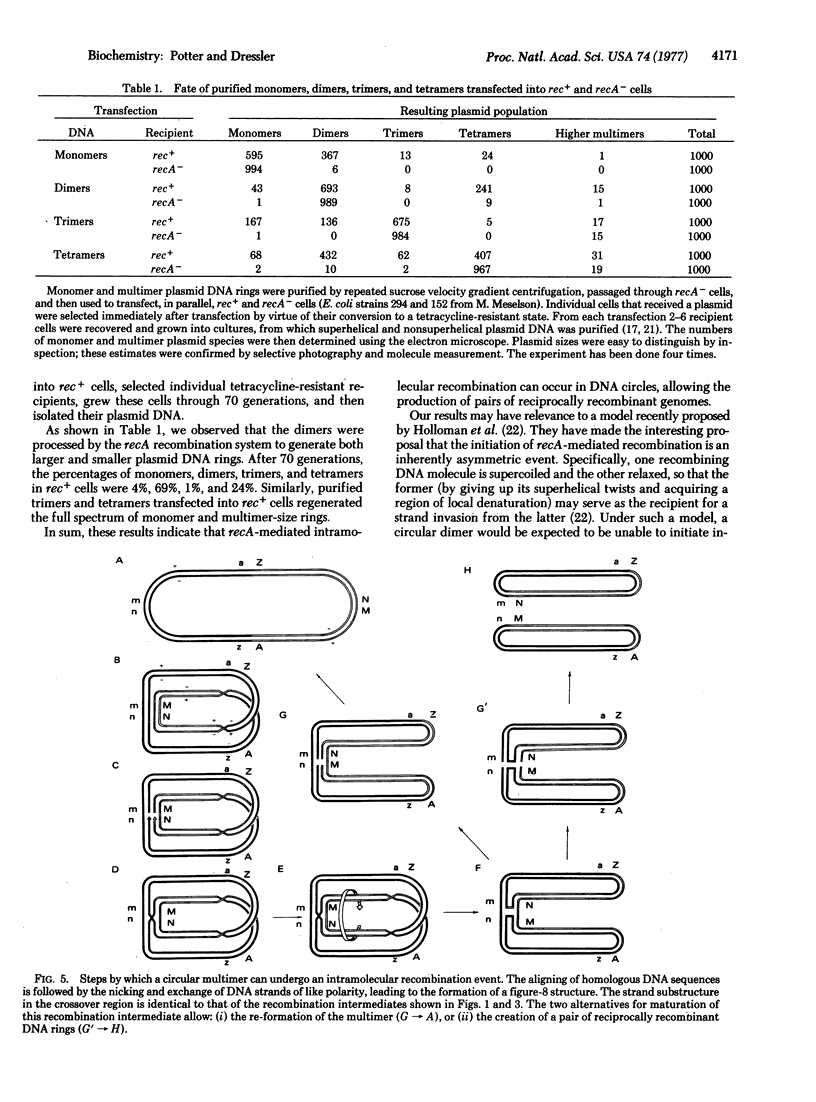
DNA molecules of the plasmid ColEl are normally recovered from wild-type cells as a set of monomer- and multimer-size rings. The data of this paper show that the multimer-size species are a product of genetic recombination. Multimer rings do not arise after transfection of purified monomers into bacterial host cells lacking a functional recA recombination system. Analogously, purified dimers, trimers, and tetramers, transfected into recA- cells, can replicate, but are constrained to remain in those conformations. Only upon transfection into rec+ cells can they regenerate the full spectrum of monomer- and multimer-size species. In this paper we trace the flow of genetic information from the monomer to the multimer state and back again under the guidance of the recA recombination system. The formation of multimer-size DNA rings is discussed as a natural consequence of the maturation of a Holliday recombination intermediate formed between two monomer plasmid genomes.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bedbrook J. R., Ausubel F. M. Recombination between bacterial plasmids leading to the formation of plasmid multimers. Cell. 1976 Dec;9(4 Pt 2):707–716. doi: 10.1016/0092-8674(76)90134-3. [DOI] [PubMed] [Google Scholar]
- Clewell D. B. Nature of Col E 1 plasmid replication in Escherichia coli in the presence of the chloramphenicol. J Bacteriol. 1972 May;110(2):667–676. doi: 10.1128/jb.110.2.667-676.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enea V., Zinder N. D. Heteroduplex DNA: a recombinational intermediate in bacteriophage f1. J Mol Biol. 1976 Feb 15;101(1):25–38. doi: 10.1016/0022-2836(76)90064-4. [DOI] [PubMed] [Google Scholar]
- Holliday R. Molecular aspects of genetic exchange and gene conversion. Genetics. 1974 Sep;78(1):273–287. doi: 10.1093/genetics/78.1.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holloman W. K., Wiegand R., Hoessli C., Radding C. M. Uptake of homologous single-stranded fragments by superhelical DNA: a possible mechanism for initiation of genetic recombination. Proc Natl Acad Sci U S A. 1975 Jun;72(6):2394–2398. doi: 10.1073/pnas.72.6.2394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurst D. D., Fogel S., Mortimer R. K. Conversion-associated recombination in yeast (hybrids-meiosis-tetrads-marker loci-models). Proc Natl Acad Sci U S A. 1972 Jan;69(1):101–105. doi: 10.1073/pnas.69.1.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam S. T., Stahl M. M., McMilin K. D., Stahl F. W. Rec-mediated recombinational hot spot activity in bacteriophage lambda. II. A mutation which causes hot spot activity. Genetics. 1974 Jul;77(3):425–433. doi: 10.1093/genetics/77.3.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meselson M. S., Radding C. M. A general model for genetic recombination. Proc Natl Acad Sci U S A. 1975 Jan;72(1):358–361. doi: 10.1073/pnas.72.1.358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potter H., Dressler D. On the mechanism of genetic recombination: electron microscopic observation of recombination intermediates. Proc Natl Acad Sci U S A. 1976 Sep;73(9):3000–3004. doi: 10.1073/pnas.73.9.3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russo V. E. On the physical structure of lambda recombinant DNA. Mol Gen Genet. 1973 May 28;122(4):353–366. doi: 10.1007/BF00269436. [DOI] [PubMed] [Google Scholar]
- Sarthy P. V., Meselson M. Single burst study of rec- and red-mediated recombination in bacteriophage lambda. Proc Natl Acad Sci U S A. 1976 Dec;73(12):4613–4617. doi: 10.1073/pnas.73.12.4613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson B. J., Escarmis C., Parker B., Slater W. C., Doniger J., Tessman I., Warner R. C. Figure-8 configuration of dimers of S13 and phiX174 replicative form DNA. J Mol Biol. 1975 Feb 5;91(4):409–419. doi: 10.1016/0022-2836(75)90269-7. [DOI] [PubMed] [Google Scholar]
- Valenzuela M. S., Inman R. B. Visualization of a novel junction in bacteriophage lambda DNA. Proc Natl Acad Sci U S A. 1975 Aug;72(8):3024–3028. doi: 10.1073/pnas.72.8.3024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White R. L., Fox M. S. On the molecular basis of high negative interference. Proc Natl Acad Sci U S A. 1974 Apr;71(4):1544–1548. doi: 10.1073/pnas.71.4.1544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfson J., Dressler D., Magazin M. Bacteriophage T7 DNA replication: a linear replicating intermediate (gradient centrifugation-electron microscopy-E. coli-DNA partial denaturation). Proc Natl Acad Sci U S A. 1972 Feb;69(2):499–504. doi: 10.1073/pnas.69.2.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worcel A., Burgi E. On the structure of the folded chromosome of Escherichia coli. J Mol Biol. 1972 Nov 14;71(2):127–147. doi: 10.1016/0022-2836(72)90342-7. [DOI] [PubMed] [Google Scholar]





