Background: COH1 is a peripheral membrane protein that is required for Golgi complex integrity and function.
Results: Association of COH1 with the Golgi complex is mediated by its interaction with RAB6 and is required for neurite outgrowth.
Conclusion: COH1 acts as downstream effector protein of RAB6.
Significance: Defective neuronal outgrowth due to loss of COH1 contributes to the neurological impairments found in Cohen syndrome patients.
Keywords: Genetic Disease, Golgi, GTPase, Neurite Outgrowth, Protein-Protein Interaction, COH1, Cohen Syndrome, RAB6
Abstract
Postnatal microcephaly, intellectual disability, and progressive retinal dystrophy are major features of autosomal recessive Cohen syndrome, which is caused by mutations in the gene COH1 (VPS13B). We have recently identified COH1 as a Golgi-enriched scaffold protein that contributes to the structural maintenance and function of the Golgi complex. Here, we show that association of COH1 with the Golgi complex depends on the small GTPase RAB6. RNAi-mediated knockdown of RAB6A/A′ prevents the localization of COH1 to the Golgi complex. Expression of the constitutively inactive RAB6_T27N mutant led to an increased solubilization of COH1 from lipid membrane preparations. Co-IP experiments confirmed the physical interaction of COH1 with RAB6 that preferentially occurred with the constitutively active RAB6_Q72L mutants. Depletion of COH1 in primary neurons negatively interfered with neurite outgrowth, indicating a causal link between the integrity of the Golgi complex and axonal outgrowth. We conclude that COH1 is a RAB6 effector protein and that reduced brain size in Cohen syndrome patients likely results from impaired COH1 function at the Golgi complex, causing decreased neuritogenesis.
Introduction
Autosomal recessive Cohen syndrome is characterized by postnatal microcephaly, intellectual disability (ID),2 typical facial features, progressive retinal dystrophy as well as intermittent neutropenia and is caused by loss of function mutations in the gene COH1 (also known as VPS13B) (1–5). COH1 is a protein of 3997 amino acids (aa), which harbors two short regions homologous to yeast vacuolar protein sorting-associated protein 13 (Vps13p), and was therefore classified as one of four mammalian VPS13 family members (2, 6). Vps13p, the presumable yeast homologue of COH1, is a peripheral membrane protein of 358 kDa that plays a role in the cycling of transmembrane proteins between the trans-Golgi network (TGN) and the prevacuolar compartment by interacting with Kex2p, Ste13p, and Vps10p (7, 8). Vps13p has been implicated in spindle pole organization by interacting with centrin Cdc31p (9). Moreover, after loss of Vps13p function an in increased cytotoxicity of an expanded polyQ domain in Rnq1p has been detected (10). Recently, Vps13p was shown to be required for prospore membrane morphogenesis through regulation of phosphatidylinositol phosphatases (11, 12). Our previous results have established the 450 kDa protein COH1 as an atypical Golgi matrix protein lacking coiled-coil domains that is crucial for Golgi complex organization. Here we aimed to further characterize the molecular mechanism by which COH1 associates with the Golgi complex and how this may relate to microcephaly and intellectual disability in Cohen syndrome.
We show that COH1 forms a physical and functional complex with RAB6. Our results point to a role of COH1 as a RAB6 effector protein. Depletion of COH1 leads to decreased neurite outgrowth in cultured primary hippocampal neurons. These results establish a critical role for RAB6-dependent function of COH1 in neuritogenesis by regulating Golgi complex organization. This provides a likely explanation for reduced postnatal brain size in Cohen syndrome patients.
EXPERIMENTAL PROCEDURES
Materials and Antibodies
All materials were purchased from Sigma-Aldrich unless otherwise stated. Polyclonal rabbit anti-COH1 peptide antibody to GEEDFVGNDPASTMHQ (COH1) was described previously (13). The following commercial antibodies were used in this study: mouse anti-clathrin (BD Bioscience), mouse anti-cortactin (Millipore), mouse anti-early endosome antigen 1 (EEA1, BD Bioscience), rabbit anti-FLAG (Invitrogen), mouse anti-FLAG M2 (Invitrogen), mouse anti-green fluorescent protein (GFP, Roche), rabbit anti-GFP (Abcam), rabbit anti-Giantin (Covance), mouse anti-GM130 (BD Bioscience), mouse anti-lysosome-associated membrane protein 2 (LAMP2, DSHB), mouse anti-myc (BD Bioscience), mouse anti-transferrin receptor (TfR, Zymed Laboratories Inc.), mouse anti-acetylated α-tubulin (Sigma). Monoclonal mouse anti-RAB6 antibody was a generous gift from Angelika Barnekow (University of Muenster). The following secondary antibodies were used for Western blot analysis: anti-mouse IgG-HRP or anti-rabbit IgG-HRP (both Cell Signaling) and for immunofluorescence: analysis anti-rabbit IgG Alexa Fluor 405, anti-rabbit IgG Alexa Fluor 488, anti-rabbit IgG-Alexa Fluor 555, anti-mouse IgG-Alexa Fluor 488, or anti-mouse IgG-Alexa Fluor 555 (all Invitrogen). 4′,6-Diamidino-2-phenylindole (DAPI) (Invitrogen) was used for nuclear staining. Restriction enzymes were purchased from NEB.
Human COH1 Constructs
For transient expression experiments of COH1, untagged full-length COH1 (according to NM_152564.3, NP_689777.3) was cloned into pcDNA3.1 V5/His as previously described. Moreover, by subcloning the following constructs were obtained: pEGFPC3_hCOH1 and pCMV-myc_hCOH1.
RAB6A/A′/B Constructs
RAB6A, A′, and B encoding cDNA was amplified from reverse transcribed (Fermentas cDNA kit) human tissue RNA (Takara Clontech) or from pGEM_RAB6A′ constructs (kindly provided by L. Johannes) and subcloned into KpnI and NotI restriction sites of N-terminally tagged pFLAG-CMV6 (Sigma). Using the resulting wild-type (wt) plasmids, T27N and Q72L mutants were obtained by overlap-PCR with primer combinations encoding the particular mutation and were subcloned into KpnI and NotI restriction sites of pFLAG-CMV6 (Sigma) or pFLAG-CMV5 (Sigma). For GFP-trap Co-IP, RAB6B wt and mutants were subcloned from pFLAG-CMV6 constructs into the restrictions sites HindIII and BamHI of pEGFPC1 (Clontech). RAB10 encoding cDNA was amplified from reverse transcribed (Fermentas cDNA kit) human tissue RNA (Takara Clontech) and subcloned into SacI and SalI restriction sites of N-terminally tagged pEGFPC2. RAB10 mutant contructs were subsequently generated by site-directed mutagenesis PCR using appropriate primers.
Cell Culture and Transient Transfection
HeLa cells were cultured at 37 °C and 5% CO2 in DMEM supplemented with 5% fetal calf serum (FCS) and 2 mm ultraglutamine. Hek293 cells were cultured at 37 °C and 5% CO2 in α-MEM supplemented with 5% FCS and 2 mm ultraglutamine. Transfection of plasmid DNA was performed using jetPEI (Polyplus transfection) according to the manufacturer's manual. Briefly, 0,003% (w/v) plasmid DNA in sterile 0.9% (w/v) NaCl was mixed with an equal volume of a 6% (v/v) jetPEI in sterile 0.9% (w/v) NaCl. After incubation for 20 min at room temperature the transfection mix was added dropwise into the cell culture dish and left for 24 h until subsequent analysis. All cell lines used in this study were from ATCC.
RNA Interference
All small interference RNAs (siRNA) specific for COH1/Coh1, Rab6A, Rab6A′, RAB6A/A′ (14), and negative controls were purchased from Ambion or Eurofins MWG Operon. siRNA target sequences are available on request. siRNAs were resuspended to 50 μm according to manufacturer's instructions and stored at −80 °C. For siRNA treatment, HeLa cells were grown on coverslips to ∼30% confluence in 6 well plates and transfected with 200 nm of each siRNA using INTERFERin (Polyplus transfection) and OptiMEM (Invitrogen) according to manufacturer's instructions. Briefly, cells were incubated in 1 ml cell culture medium, siRNAs were diluted to 200 nm in 100 μl final volume with OptiMEM and subsequently 2 μl INTERFERin was added. After 10 min incubation at room temperature the transfection mix was added dropwise to each well. Transfection was repeated after 12 h, medium was changed 12 h later, and cells were cultured for another 60 h. Finally, cells were prepared for subsequent analysis.
Quantitative PCR (qPCR)
Analysis was performed as decribed elsewhere (3). Briefly, total RNA was isolated from cell cultures using Trizol reagent (Invitrogen) according to the manufacturer's instructions. 1 μg isolated RNA was reverse transcribed using RevertAid H Minus First Strand cDNA Synthesis kit and random hexamer primers (Fermentas). Primer pairs for cDNA amplification COH1/Coh1, RAB6A/A′, and GAPDH/Gapdh were designed and cDNA primer sequences are available on request. mRNA expression levels were determined by quantitative PCR (qPCR) using cDNA from siRNA-treated cells. Each sample was analyzed as triplicate and amplified on an ABI PRISM7500 instrument (Applied Biosystems). Relative mRNA expression was quantified using the comparative Ct method (15). The different mRNA values were normalized against GAPDH/Gapdh mRNA.
FLAG M2 Co-immunoprecipitation (Co-IP)
Transiently transfected Hek293 cells were washed twice with cold PBS, harvested, and lysed in protein lysis buffer (20 mm HEPES, 50 mm NaCl, 5 mm MgCl2, 0.3% Triton X-100, and proteinase inhibitors). Cell lysates were centrifuged (10.000 × g, 15 min, 4 °C) and obtained supernatants were used for immunoprecipitation with FLAG M2 beads (Sigma), according to the manufacturer's manual. Briefly, supernatants were added to pre-washed FLAG M2 beads and incubated for 4 h under continuous shaking at 4 °C. Finally, beads were washed five times with 20 mm HEPES, 200 mm NaCl, 5 mm MgCl2, and proteinase inhibitors and for subsequent analysis prepared in 1x SDS loading buffer. pFLAG-CMV5_ANK (NM_054027.4, NP_473368.1) and pFLAG-CMV6_ACAP2 (NM_012287.5, NP_036419.3) were used as negative controls.
GFP-Trap®_A Co-IP
Transiently transfected Hek293 cells were washed twice with cold PBS, harvested, and lysed in protein lysis buffer (20 mm HEPES, 50 mm NaCl, 5 mm MgCl2, 0.5% Triton X-100, and proteinase inhibitors). Cell lysates were centrifuged (10.000 × g, 15 min, 4 °C) and obtained postnuclear supernatants (PNS) were used for immunoprecipitation with GFP-Trap®_A beads (Chromotek), according to the manufacturer's manual. Briefly, PNS was added to pre-washed GFP-Trap®_A beads and incubated for 2 h under continuous shaking at 4 °C. Finally, beads were washed five times with 20 mm HEPES, 200–1000 mm NaCl, 5 mm MgCl2, and proteinase inhibitors and prepared in 1× SDS loading buffer for subsequent analysis. pEGFPC1 (Clontech) was used as negative control. Membrane preparation was performed as previously described (13).
Western Blot Analysis
For Western blot analysis, all samples were diluted in 1× SDS loading buffer and resolved by gel electrophoresis in Bis-Tris SDS-polyacrylamide gels (self-made) or Tris-Acetate SDS 3–8% polyacrylamide gradient gels (Invitrogen). Protein concentrations were determined when required using the BCA protein assay kit (Pierce). After transfer on nitrocellulose membranes by tank blotting and blocking, blots were incubated with the appropriate primary and secondary antibodies in 5% block milk 0.2% Nonidet P-40 in 1× TBS. Final detection was performed using ECL PlusTM or ECL PrimeTM Western blotting Detection Reagents (Amersham Biosciences) and visualized on an x-ray film (Fujifilm Super RX).
Immunofluorescence and Image Analysis
For staining of overexpressed and endogenous proteins, cells were grown on glass coverslips (12 mm, Marienfeld). Cells were fixed with 4% (w/v) paraformaldehyde (PFA) in PBS at 4 °C, permeabilized in 0.5% (v/v) Triton X-100 in 3% (w/v) bovine serum albumin (BSA) in PBS, and blocked with 3% (w/v) BSA in PBS. Primary antibodies were applied in 3% BSA in PBS for 5 h at 4 °C, coverslips were washed in PBS, and secondary antibodies were applied in 3% BSA in PBS for 1 h at 4 °C. Coverslips were mounted on slides using Fluoromount-G (Southern Biotech). Images were taken with a confocal microscope LSM510 (Zeiss). Images for subsequent evaluation were acquired under identical exposure conditions.
Image analyses were performed using macros in ImageJ under identical threshold conditions. Statistical significance was calculated with Student's t test (two-sided, unpaired, homogenous variation).
Neuronal Outgrowth
For neuronal outgrowth experiments rat hippocampal neurons were prepared as described elsewhere (16). Briefly, hippocampi of E18 rat embryos were dissected and digested for 15 min at 37 °C in trypsin solution. Afterward, the tissue was washed three times with warm HBSS and finally dissociated by pipetting with a Pasteur pipette. Directly after preparation aliquots of 500.000 neurons each were pelleted for 5 min at 800× rpm. Neurons were resuspended in 100 μl rat neuron nucleofector solution (amaxa). Into each aliquot, either 1.5 μg siRNA and 1.5 μg of the GFP expressing plasmid pmaxGFP (amaxa) or Rab6B expressing plasmids were added. The cells were then transferred into an amaxa cuvette and transfected using the nucleofector program O-003. After addition of 500 μl prewarmed medium, the cell suspension was plated on two 6-cm dishes containing 6 poly-l-lysine coated coverslips each. The next day coverslips were flipped onto dishes containing a 40% confluent astrocyte layer. The original dishes were further cultured and later processed for knock-down efficiency analysis by qPCR. 54 h after seeding the cells, coverslips were fixed with 4% PFA and subjected to immunofluorescence analysis. Neurons were imaged with a BX60 fluorescence microscope (Olympus). For neurite length measurements GFP expressing neurons were considered as silenced or as overexpressing the Rab6B constructs and their longest process was measured by AxioVision (Zeiss). Moreover, the number of neurites per cell and the Golgi orientation toward the longest process was determined. Statistical analysis was performed using Student's t test (two-sided, paired, n = 3 independent experiments, homogenous variation).
RESULTS
COH1 Co-localizes with RAB6 at the Golgi
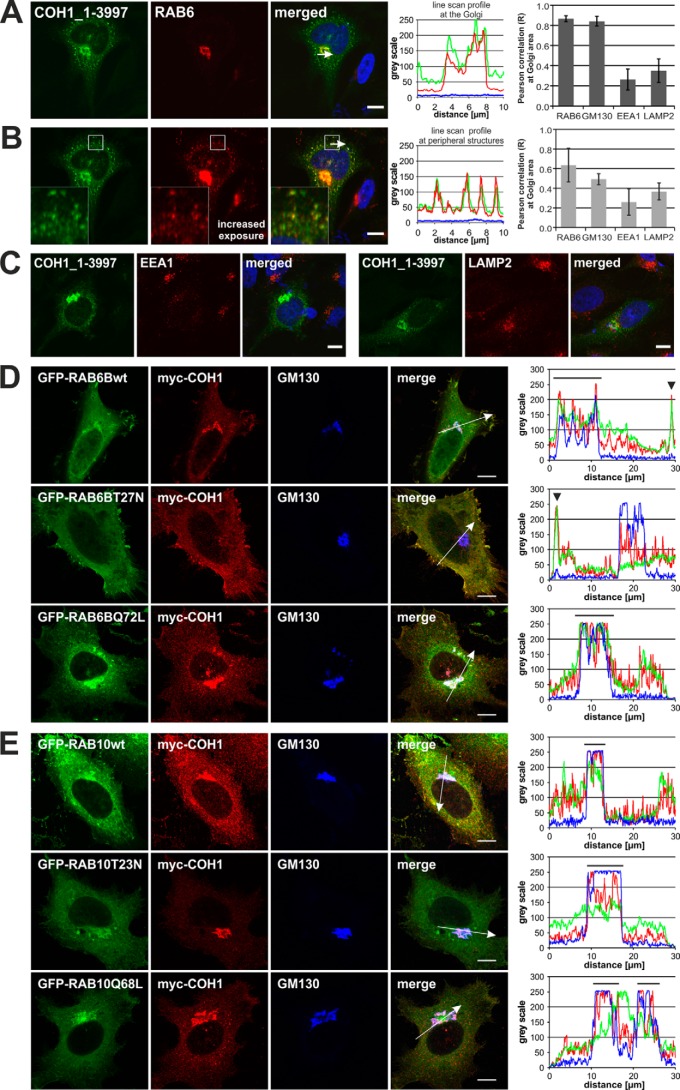
COH1 has been classified as mammalian homologue to yeast Vps13p (2, 6). Therefore, we screened the yeastgenome.org database for known interacting partners of yeast Vps13p. Vps13p has been shown to control membrane traffic at the TGN/endosomal interface (7, 8). Because of the comparable subcellular localization and predicted similar function of both, Vp13p and COH1, we focused on Golgi-associated proteins and filtered all Vps13p interactors for the term Golgi (35 out of 151). Among those were proteins of the cis oligomeric Golgi (COG)-complex, the Golgi-associated retrograde protein (GARP) complex, the coat protein (COP) complexes, soluble N-ethylmaleimide-sensitive-factor attachment receptor (SNARE) proteins and GTPases. Interestingly, Vps13p has been shown to associate with Ypt6p, the yeast homologue of the mammalian RAB6 GTPases (17, 18). Mammalian RAB6, which exists in three isoforms (A, A′, B), has been implicated in several Golgi-associated trafficking steps (19, 20) including the formation of exocytotic carriers (21). To analyze a potential association of COH1 with RAB6 in mammals, we investigated its subcellular distribution in transfected HeLa cells. Overexpressed COH1 indeed co-localized with endogenous RAB6 at the Golgi complex. At increased fluorescence excitation, RAB6 was further found to co-localize with COH1-positive punctate structures in the cell periphery, likely representing vesiculo-tubular transport carriers. Confirming previous results, Pearson correlation coefficient analysis proofed strong co-localization of COH1 at the Golgi complex, as shown here by the Golgi marker protein GM130 and with RAB6. COH1-positive punctate structures in the cell periphery were analyzed for co-localization with different subcellular markers. Pearson correlation coefficient analysis identified weak correlation between COH1 and the endosomal marker protein EEA1 as well as the lysosomal marker protein LAMP2, whereas strong correlation coefficients were found between COH1 and RAB6 (Fig. 1, A–C). These results were confirmed in HeLa cells co-expressing tagged COH1 and RAB6: myc-COH1 co-localized with EGFP-RAB6B_wt at the Golgi complex and was further detected in the cell periphery, with an intensified signal close to a subregion of the plasma membrane. Co-expression of COH1 together with constitutively active EGFP-RAB6B_Q72L led to recruitment of COH1 to the Golgi complex. By contrast, overexpression of GDP-locked inactive EGFP-RAB6B_T27N caused the redistribution of COH1 into the cell periphery including subplasma membrane regions (Fig. 1D, and supplemental Fig. S1A). In comparison, co-overexpression of another GTPase, RAB10, or their GDP- or GTP-locked variants did not alter the Golgi-enrichment of COH1 (Fig. 1E, and supplemental Fig. S1B). These data are consistent with a model whereby the localization of COH1 at the Golgi complex depends at least in part on active RAB6.
FIGURE 1.
COH1 and RAB6 co-localize at the Golgi complex. A, pcDNA3.1_hCOH1-transfected (green) HeLa cells demonstrate co-localization of COH1 with endogenous RAB6 (red, anti-RAB6 Ab) at the Golgi complex and (B) in peripheral structures, most likely representing RAB6-postive transport carriers. Nuclei were stained using DAPI (blue). A and B, line scan graphs generated by ImageJ analysis show the immunofluorescence intensity along the arrow indicated in the corresponding image and demonstrate strong co-localization of COH1 and RAB6. Moreover Pearson correlation was performed on confocal images of HeLa cells overexpressing COH1 and co-staining with different subcellular marker proteins (RAB6, GM130, EEA1, and LAMP2) using Image correlation analysis plugin (ImageJ, WCIF). Two different ROIs (Golgi area and cell periphery) were analyzed. Error bars indicate S.D. C, HeLa cells were transiently transfected with the COH1_1–3997 plasmid construct (pcDNA3.1_hCOH1, green), and cells were processed for confocal imaging. Both, the early endosomal antigen (EEA1, red, left panel) and the lysosomal marker (LAMP1, red, right panel) do not co-localize with COH1. Nuclei were stained using DAPI (blue). D, confocal microscopy analysis of co-expressed pCMV-myc-COH1 (red, anti-myc Ab) and pEGFPC-RAB6B (green) constructs shows co-localization of COH1 and wt EGFP-RAB6B as well as constitutively active EGFP-RAB6B_Q72L at the Golgi complex (blue, anti-GM130 Ab). Moreover, COH1 (red, anti-myc AB) co-localizes with wt EGFP-RAB6B and constitutively inactive EGFP-RAB6B_T27N (green) at peripheral structures. E, confocal microscopy analysis showed that co-expression of pCMV-myc_COH1 (red, anti-myc Ab) and pEGFPC-RAB10 constructs (green) does not alter the localization of COH1 to the Golgi complex (blue, anti-GM130 Ab). C and D, line scan graphs generated by ImageJ analysis show the immunofluorescence intensity along the arrow indicated in the corresponding image. COH1 redistributes from Golgi localization (line above line scan) into peripheral structures (arrowhead) upon co-expression of EGFP-RAB6_T27N. All scale bars represent 10 μm.
COH1 Golgi Localization Is Mediated by Active RAB6
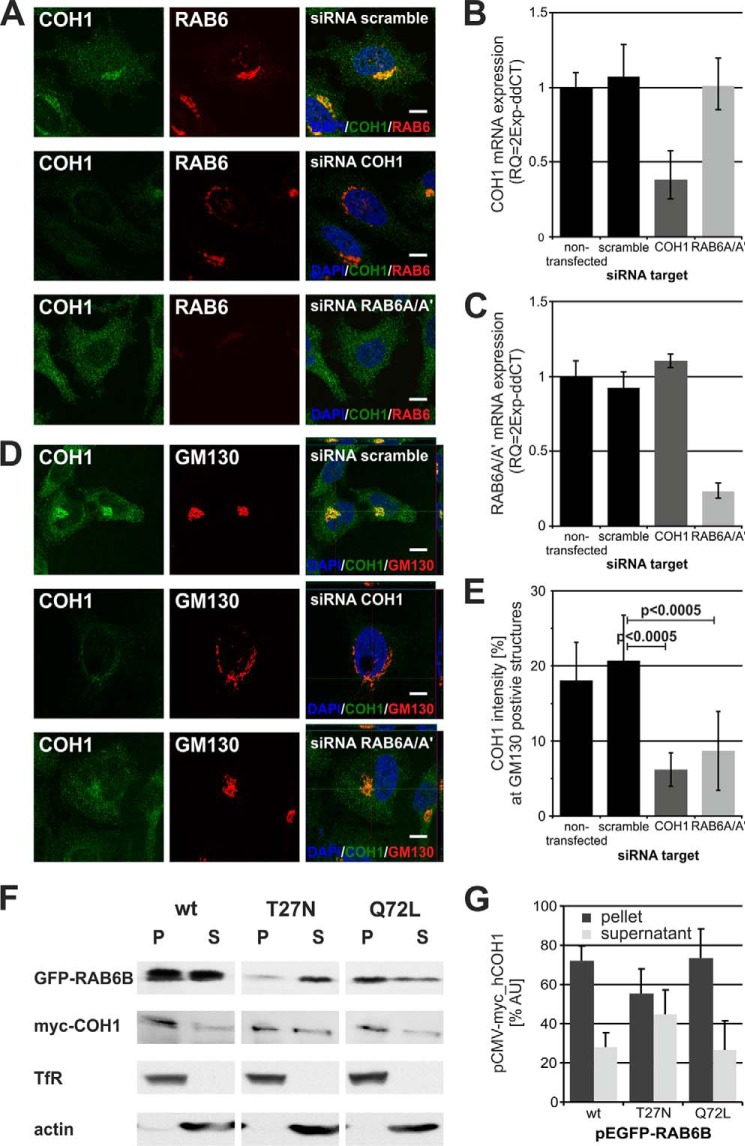
To test the hypothesis that COH1 association with the Golgi complex is mediated by active RAB6, we analyzed the localization of COH1 at the Golgi complex in HeLa cells depleted of endogenous RAB6 by specific siRNAs (Fig. 2, A–E). Efficient knockdown of COH1 and RAB6 was verified by qPCR (Fig. 2, B and C). Loss of COH1 induced fragmentation of the Golgi complex consistent with previous results (13), but did not alter RAB6 localization to the resulting Golgi fragments (Fig. 2A, middle panel), suggesting that RAB6 is recruited to the Golgi independent of COH1. In contrast, loss of RAB6A/A′ reduced the perinuclear enrichment of COH1 (Fig. 2A, lower panel), indicating that COH1 recruitment to the Golgi complex requires activation of RAB6. To validate these results, we quantified the COH1 enrichment by fluorescence intensity measurements. For this, all images were taken by confocal microscopy under identical acquisition, background correction, and threshold conditions. In scrambled siRNA transfected HeLa cells 21% of the total COH1 fluorescence intensity localizes to GM130-postive structures. As expected the fluorescence intensity at GM130-postive structures upon knockdown of COH1 was only 6%, most likely representing residual COH1 due to insufficient knockdown. Importantly, we found that depletion of RAB6A/A′ reduced the presence of COH1 at GM130-positive structures to 9%. Together with our previous characterization of COH1 as a soluble and peripheral membrane protein (13) these data suggest that COH1 recruitment to the Golgi membrane requires RAB6. This hypothesis was further substantiated by the analysis of membrane preparations from Hek293 cells transiently co-expressing myc-COH1 and active or inactive EGFP-RAB6B. While myc-COH1 was largely associated to the membrane in Hek293 cells expressing EGFP-RAB6B_wt or constitutively active EGFP-RAB6B_Q72L, its membrane association was reduced in cells expressing EGFP-RAB6B_T27N (Fig. 2, F and G). These collective data indicate that active RAB6 is required to recruit COH1 to the Golgi complex.
FIGURE 2.
Down-regulation of RAB6 activity disrupts COH1 recruitment to the Golgi complex. A, HeLa cells were depleted for COH1 as well as RAB6A/A′ and stained with a COH1 (green) and RAB6 (red) antibody. Loss of COH1 did not diminish RAB6 enrichment at the Golgi; however, depletion of RAB6A/A′ dispersed COH1 into the cytoplasm. Images were taken by confocal microscopy under identical exposure conditions. Nuclei were stained using DAPI. Scale bars represent 10 μm. B and C, qPCR confirmed efficient knockdown of either COH1 or RAB6A/A′ siRNA. D, Z-stack images of siRNA-treated HeLa cells stained with COH1 (green) and GM130 (red) specific antibodies were taken by confocal microscopy. Knockdown of COH1 but not RAB6A/A′ results in severe Golgi fragmentation. Moreover, data illustrate diminished COH1 recruitment to the Golgi complex upon RAB6A/A′ ablation. Nuclei were stained using DAPI. Scale bars represent 10 μm. E, images of siRNA-treated HeLa cells were processed by ImageJ to analyze COH1 enrichment at GM130 positive Golgi structures. Loss of RAB6A/A′ significantly diminishes COH1 recruitment to the Golgi complex. Image acquisition, background correction, and threshold were applied identically to all pictures. p values were determined using unpaired two-tailed t test. F and G, Hek293 cells were transiently co-transfected with constructs for myc-COH1 and EGFP-RAB6B wt, T27N or Q72L. Postnuclear cell lysates were processed for membrane preparations and equal portions of the obtained pellet (P) and supernatant (S) were analyzed by SDS-PAGE and Western blot (F). ImageJ analysis of three independent experiments show decreased COH1 membrane recruitment when constitutively inactive EGFP-RAB6B_T27N is co-expressed (G). Error bars indicate S.D.
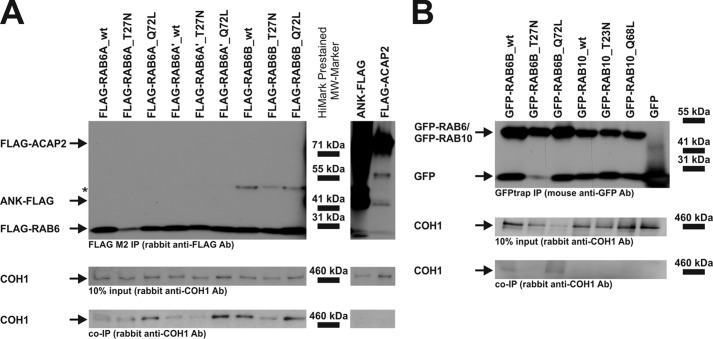
COH1 Interacts with All Three Mammalian RAB6 Homologues
To analyze whether the co-localization and functional interaction of COH1 with RAB6 reflects a physical association of both proteins, we performed co-immunoprecipitation (Co-IP) experiments using Hek293 cells co-expressing untagged COH1_1–3997aa together with FLAG-tagged wt or mutant RAB6A, RAB6A′, or RAB6B, respectively. COH1_1–3997aa was found to associate preferentially with the constitutively active mutants (Q72L) of all three RAB6 isoforms (Fig. 3A). In contrast, the transmembrane protein ANK and the soluble protein ACAP2 did not interact with COH1. Moreover, COH1 did not co-immunoprecipitate with the RAB6-related GTPase RAB10 (Fig. 3B). GFP-trap Co-IP confirmed the co-immunoprecipitation of COH1 with EGFP-RAB6B protein, and vice versa FLAG-RAB6B could be co-immunoprecipitated by GFP-COH1 (data not shown). This mirrors the findings from FLAG Co-IPs further highlighting the binding preference of COH1 to constitutively active RAB6_Q72L. We conclude that RAB6-GTP interacts with COH1 to recruit it to the Golgi complex.
FIGURE 3.
Complex formation between COH1 and the Golgi-associated small GTPase RAB6. A, in Hek293 cells different pFLAG-CMV-constructs, as indicated, were transiently co-transfected with untagged COH1 (pcDNA3.1_hCOH1). Cell lysates were processed for FLAG M2 (Sigma) Co-IP. Anti-FLAG co-immunoprecipitates (top) and input samples were analyzed by SDS-PAGE, and immunoblotting was performed with a COH1-specific antibody. COH1 was found to co-immunoprecipitate with both the wt and the constitutively active mutant (Q72L) RAB6 isoforms. Asterisk indicates unspecific signal. B, co-expression of untagged COH1 and GFP-RAB GTPase constructs in Hek293 and subsequent GFPtrap Co-IP showed preferential binding of COH1 to GFP-RAB6. No binding was detected between COH1 and GFP-RAB10.
Coh1 Depletion Inhibits Neurite Outgrowth in Vitro
Patients with Cohen syndrome demonstrate a broad spectrum of clinical features (2, 3, 5, 22). Most prominent is the neurological phenotype including progressive retinal dystrophy, nonprogressive intellectual disability, and postnatally developing microcephaly.
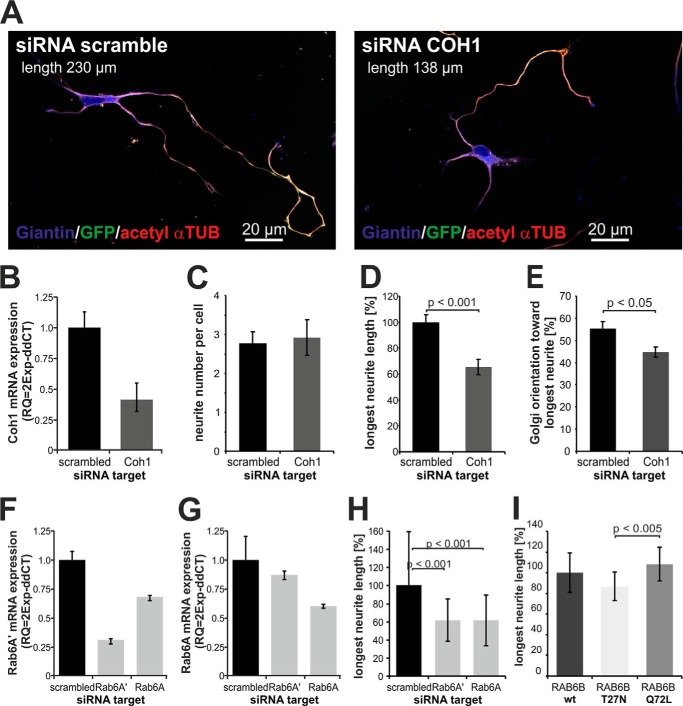
To analyze a potential function of COH1 in neuronal development, we depleted Coh1 from primary rat hippocampal neurons (E18) using RNAi (Fig. 4). Directly after dissociation, hippocampal neurons were co-transfected with rat Coh1-specific or scrambled (control) siRNAs together with a plasmid (pmaxGFP) encoding green fluorescent protein (GFP) as a marker to visualize neuronal morphology. Efficient depletion of Coh1 in rat hippocampus derived cultures was confirmed by qPCR (Fig. 4B). Down-regulation of the Coh1 mRNA level to ∼50% significantly reduced the length of the longest neurite (i.e. the axon) by ∼40% when compared with control-transfected neurons (siRNA scramble) (Fig. 4, A and D), while the number of forming neurites was unaffected (Fig. 4C). We also found a reduced alignment of the Golgi complex toward the longest neurite, indicating that loss of Coh1 not only disturbs Golgi complex maintenance but also influences orientation of the Golgi complex and thus polarization of cell (Fig. 4E). Given that COH1 function depends on active RAB6, depletion of RAB6 should phenocopy reduced neurite outgrowth observed in COH1 knockdown neurons. Consistent with this prediction neurite length was significantly reduced in rat hippocampal neurons depleted of endogenous Rab6A and Rab6A′ using specific siRNAs (Fig. 4, F–H). Consistent with a key role for active RAB6 in COH1-mediated neuritogenesis, we observed that expression of constitutively negative RAB6B_T27N akin to depletion of either RAB6A or A′ or COH1 decreased neurite outgrowth, whereas constitutively active RAB6B_Q72L led to an increased neurite outgrowth (Fig. 4I). These data further corroborate the functional interaction of COH1 with active RAB6 and establish a physiological function of both proteins in neuritogenesis.
FIGURE 4.
COH1 and RAB6 regulate neurite outgrowth in primary neurons. A, primary rat hippocampal neurons were co-transfected with a scrambled (control) or Coh1-specific siRNA and pmaxGFP. GFP-expressing cells were considered as Coh1-depleted neurons. Neurons were stained for giantin (blue) and acetylated α-tubulin (red). Images were taken by confocal microscopy. B, efficient siRNA-mediated mRNA depletion was confirmed by quantitative PCR (qPCR). Coh1 mRNA level was reduced by ∼50%. Delta-CT was normalized to Gapdh. C–E, neurite development was analyzed 54h post siRNA transfection (stage 3 of neuron development). Number of neurites per cell, length of the longest neurite and Golgi orientation were measured in images (n = 3, at least 50 cells were counted per condition and experiment), which were taken by fluorescence microscopy (Olympus BX60) and processed for data analysis by AxioVision (Zeiss). C, Coh1 depletion does not affect initiation of neurites as shown by normal average of ∼3 neurites per neuron. D, loss of Coh1 reduces outgrowth of the longest forming neurite to ∼60%. E, Golgi orientation toward the longest neurite is significantly changed in Coh1 depleted neurons. F–H, primary rat hippocampal neurons were co-transfected with a scrambled (control), Rab6A- or Rab6A′-specific siRNA and pmaxGFP. GFP-expressing cells were considered as transfected neurons. Efficient knockdown was confirmed by quantitative PCR (qPCR) (F and G). Similar to overexpression of RAB6B mutants, siRNA-mediated knockdown of either RAB6A or RAB6A′ led to significantly reduced neurite outgrowth (n = 1, at least 50 cells were counted per condition). I, rat hippocampal neurons were co-transfected with RAB6B (pCMV5_RAB6B wt, T27N, or Q72L) and pmaxGFP. After 54 h, neurons were fixed and further processed. All GFP expressing cells were considered as RAB6B-overexpressing neurons. Measurements of the longest neurite (n = 3, at least 40 cells were counted per condition and experiment) were performed using AxioVision software (Zeiss). Constitutively inactive RAB6B_T27N mutant overexpression decreases neurite outgrowth significantly compared with its constitutively active RAB6B_Q72L mutant. C–E, I: p value was determined using paired two-tailed t test among the three independent experiments. H: p value was determined using unpaired two-tailed t test among the 50 measured cells. Error bars indicate S.D.
DISCUSSION
Brain development is a complex process comprising neuron proliferation, migration, polarization, and functional synaptic integration (23–25). Diverse genetic defects affecting neuronal proliferation (26, 27), neuronal progenitor migration (28) or neuritogenesis are important determinants of the postmitotic brain size (29). MRI studies in Cohen syndrome patients revealed a reduced postnatal brain size but show an otherwise normal brain structure with appropriate cerebral as well as cortical patterning (30). In situ hybridization experiments have demonstrated expression of murine Coh1 in neurons and highest expression has been found in Purkinje cells and the internal granular layer of the cerebellum as well as in cortical layers II-VI (4). Together, these observations suggest that loss of COH1 function has no significant impact on mitosis of progenitor cells and most likely not on radial migration of neuroblasts. Thus, we hypothesized that COH1 regulates the differentiation and integration of neurons into a functional network.
Here, we show that overexpressed COH1 and endogenous RAB6 co-localized at the Golgi complex and in peripheral punctate structures. This suggests interplay of COH1 and the Golgi-associated small GTPase RAB6, which is in line with previous results for Golgi localization of both proteins (13, 20). COH1 association with the Golgi complex critically depends on active RAB6, since the overexpression of constitutively inactive RAB6_T27N or siRNA-mediated depletion of RAB6 negatively interfered with COH1 recruitment to lipid membrane preparations and at the Golgi complex, respectively. In accordance to the functional interaction, different Co-IPs established a physical interaction of COH1 with RAB6 that preferentially occurred with the constitutively active RAB6_Q72L mutant. As RAB6 is involved in different Golgi-associated trafficking steps our findings support the previously described impact of COH1 for Golgi-associated membrane transport processes (2, 6, 13). Previous studies have indicated a potential neuroprotective effect of RAB6. In particular, one report suggested a positive role for RAB6 in neurite outgrowth (31). We therefore analyzed the role of COH1 and RAB6 to neuritogenesis. Both proteins were shown to promote neurite outgrowth. Moreover, COH1 is critical for the orientation of the Golgi toward the longest neurite. Positioning of the centrosome and the Golgi complex in postmitotic neurons determines the accelerated outgrowth of one neurite by which the prospective axon is specified (32). Our finding suggests that reduced COH1 attachment to the Golgi membrane upon loss of RAB6 subsequently disturbs Golgi maintenance and orientation. This finally inhibits efficient polarization and targeted membrane transport toward the developing axon. However, the characteristics by which COH1 controls Golgi maintenance and orientation as well as membrane transport awaits further characterization.
The impact of directed microtubule-dependent transport and small GTPase activity for neurite outgrowth was recently highlighted by the identification of the RAB6 effector Bicaudal-d-related protein 1 (BICDR-1) (31). Depletion of RAB6A, RAB6A′, and RAB6B in rat hippocampal neurons has been shown to reduce neurite outgrowth (31). These data propose a critical role of RAB6-associated membrane transport for neuron development. In line with this, our results showed that inactive RAB6 negatively interfered with neurite outgrowth, while active RAB6 facilitated neurite outgrowth. Moreover, depletion of either COH1 or RAB6 led to significantly decreased neurite outgrowth. Together, COH1 likely functions as downstream effector protein of RAB6 and we hypothesize that this is partially important for proper neurite outgrowth.
A more recent study has shown that Cohen syndrome patients display a tissue-specific glycosylation defect (18). This observation is consistent with our finding that COH1 is important for Golgi homeostasis and membrane transport (13). Most congenital glycosylation disorders are caused by mutations in Golgi-associated protein complexes regulating membrane partitioning and localization of glycosyltransferases (33). These Golgi-associated protein complexes comprise molecules required for docking and/or fusion of transport carriers, protein sorting, and luminal pH homeostasis (33). One such example is the COG complex (34), which is critically important for retrograde vesicle transport within the Golgi and for Golgi homeostasis downstream of RAB6 (35, 36). RAB6 preferentially interacts with the COG6 subunit and mutations in COG6 leads to a glycosylation disorder due to altered retrograde membrane transport (37, 38). However, for RAB6 depletion a glycosylation defect remains to be identified. In conclusion, we have established COH1 as a potential Rab6 effector protein and as positive regulator protein of neuritogenesis.
Acknowledgments
We thank Drs. Ludger Johannes for RAB6 plasmids and Angelika Barnekow for the RAB6 antibody. We thank Inge Walther for preparing hippocampal neurons.
This work was supported by grants from the Deutsche Forschungsgemeinschaft (to T. M., V. H., and W. S.).

This article contains supplemental Fig. S1.
- ID
- intellectual disability
- ROI
- region of interest
- DAPI
- 4′,6-diamidino-2-phenylindole
- SNARE
- N-ethylmaleimide-sensitive-factor attachment receptor
- TGN
- trans-Golgi network
- COG
- cis oligomeric Golgi
- GARP
- Golgi-associated retrograde protein
- COP
- coat protein.
REFERENCES
- 1. Cohen M. M., Jr., Hall B. D., Smith D. W., Graham C. B., Lampert K. J. (1973) A new syndrome with hypotonia, obesity, mental deficiency, and facial, oral, ocular, and limb anomalies. J. Pediatr 83, 280–284 [DOI] [PubMed] [Google Scholar]
- 2. Kolehmainen J., Black G. C., Saarinen A., Chandler K., Clayton-Smith J., Träskelin A. L., Perveen R., Kivitie-Kallio S., Norio R., Warburg M., Fryns J. P., de la Chapelle A., Lehesjoki A. E. (2003) Cohen syndrome is caused by mutations in a novel gene, COH1, encoding a transmembrane protein with a presumed role in vesicle-mediated sorting and intracellular protein transport. Am. J. Hum. Genet. 72, 1359–1369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Seifert W., Holder-Espinasse M., Kühnisch J., Kahrizi K., Tzschach A., Garshasbi M., Najmabadi H., Walter Kuss A., Kress W., Laureys G., Loeys B., Brilstra E., Mancini G. M., Dollfus H., Dahan K., Apse K., Hennies H. C., Horn D. (2009) Expanded mutational spectrum in Cohen syndrome, tissue expression, and transcript variants of COH1. Hum. Mutat. 30, E404–E420 [DOI] [PubMed] [Google Scholar]
- 4. Mochida G. H., Rajab A., Eyaid W., Lu A., Al-Nouri D., Kosaki K., Noruzinia M., Sarda P., Ishihara J., Bodell A., Apse K., Walsh C. A. (2004) Broader geographical spectrum of Cohen syndrome due to COH1 mutations. J. Med. Genet. 41, e87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hennies H. C., Rauch A., Seifert W., Schumi C., Moser E., Al-Taji E., Tariverdian G., Chrzanowska K. H., Krajewska-Walasek M., Rajab A., Giugliani R., Neumann T. E., Eckl K. M., Karbasiyan M., Reis A., Horn D. (2004) Allelic heterogeneity in the COH1 gene explains clinical variability in Cohen syndrome. Am. J. Hum. Genet. 75, 138–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Velayos-Baeza A., Vettori A., Copley R. R., Dobson-Stone C., Monaco A. P. (2004) Analysis of the human VPS13 gene family. Genomics 84, 536–549 [DOI] [PubMed] [Google Scholar]
- 7. Redding K., Brickner J. H., Marschall L. G., Nichols J. W., Fuller R. S. (1996) Allele-specific suppression of a defective trans-Golgi network (TGN) localization signal in Kex2p identifies three genes involved in localization of TGN transmembrane proteins. Mol. Cell. Biol. 16, 6208–6217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Brickner J. H., Fuller R. S. (1997) SOI1 encodes a novel, conserved protein that promotes TGN-endosomal cycling of Kex2p and other membrane proteins by modulating the function of two TGN localization signals. J. Cell Biol. 139, 23–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kilmartin J. V. (2003) Sfi1p has conserved centrin-binding sites and an essential function in budding yeast spindle pole body duplication. J. Cell Biol. 162, 1211–1221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Meriin A. B., Zhang X., Miliaras N. B., Kazantsev A., Chernoff Y. O., McCaffery J. M., Wendland B., Sherman M. Y. (2003) Aggregation of expanded polyglutamine domain in yeast leads to defects in endocytosis. Mol. Cell. Biol. 23, 7554–7565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Park J. S., Okumura Y., Tachikawa H., Neiman A. M. SPO71 encodes a developmental stage-specific partner for Vps13 in Saccharomyces cerevisiae. Eukaryot Cell 12, 1530–1537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Park J. S., Neiman A. M. VPS13 regulates membrane morphogenesis during sporulation in Saccharomyces cerevisiae. J. Cell Sci. 125, 3004–3011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Seifert W., Kühnisch J., Maritzen T., Horn D., Haucke V., Hennies H. C. (2011) Cohen syndrome-associated protein, COH1, is a novel, giant Golgi matrix protein required for Golgi integrity. J. Biol. Chem. 286, 37665–37675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Del Nery E., Miserey-Lenkei S., Falguières T., Nizak C., Johannes L., Perez F., Goud B. (2006) Rab6A and Rab6A′ GTPases play non-overlapping roles in membrane trafficking. Traffic 7, 394–407 [DOI] [PubMed] [Google Scholar]
- 15. Livak K. J., Schmittgen T. D. (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-ΔΔC(T)) Method. Methods 25, 402–408 [DOI] [PubMed] [Google Scholar]
- 16. Kaech S., Banker G. (2006) Culturing hippocampal neurons. Nat. Protoc. 1, 2406–2415 [DOI] [PubMed] [Google Scholar]
- 17. Costanzo M., Baryshnikova A., Bellay J., Kim Y., Spear E. D., Sevier C. S., Ding H., Koh J. L., Toufighi K., Mostafavi S., Prinz J., St Onge R. P., VanderSluis B., Makhnevych T., Vizeacoumar F. J., Alizadeh S., Bahr S., Brost R. L., Chen Y., Cokol M., Deshpande R., Li Z., Lin Z. Y., Liang W., Marback M., Paw J., San Luis B. J., Shuteriqi E., Tong A. H., van Dyk N., Wallace I. M., Whitney J. A., Weirauch M. T., Zhong G., Zhu H., Houry W. A., Brudno M., Ragibizadeh S., Papp B., Pál C., Roth F. P., Giaever G., Nislow C., Troyanskaya O. G., Bussey H., Bader G. D., Gingras A. C., Morris Q. D., Kim P. M., Kaiser C. A., Myers C. L., Andrews B. J., Boone C. (2010) The genetic landscape of a cell. Science 327, 425–431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hoppins S., Collins S. R., Cassidy-Stone A., Hummel E., Devay R. M., Lackner L. L., Westermann B., Schuldiner M., Weissman J. S., Nunnari J. (2011) A mitochondrial-focused genetic interaction map reveals a scaffold-like complex required for inner membrane organization in mitochondria. J. Cell Biol. 195, 323–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Martinez O., Antony C., Pehau-Arnaudet G., Berger E. G., Salamero J., Goud B. (1997) GTP-bound forms of rab6 induce the redistribution of Golgi proteins into the endoplasmic reticulum. Proc. Natl. Acad. Sci. U.S.A. 94, 1828–1833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Martinez O., Schmidt A., Salaméro J., Hoflack B., Roa M., Goud B. (1994) The small GTP-binding protein rab6 functions in intra-Golgi transport. J. Cell Biol. 127, 1575–1588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Grigoriev I., Splinter D., Keijzer N., Wulf P. S., Demmers J., Ohtsuka T., Modesti M., Maly I. V., Grosveld F., Hoogenraad C. C., Akhmanova A. (2007) Rab6 regulates transport and targeting of exocytotic carriers. Dev Cell 13, 305–314 [DOI] [PubMed] [Google Scholar]
- 22. Seifert W., Holder-Espinasse M., Spranger S., Hoeltzenbein M., Rossier E., Dollfus H., Lacombe D., Verloes A., Chrzanowska K. H., Maegawa G. H., Chitayat D., Kotzot D., Huhle D., Meinecke P., Albrecht B., Mathijssen I., Leheup B., Raile K., Hennies H. C., Horn D. (2006) Mutational spectrum of COH1 and clinical heterogeneity in Cohen syndrome. J Med. Genet. 43, e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Barkovich A. J., Kuzniecky R. I., Jackson G. D., Guerrini R., Dobyns W. B. (2005) A developmental and genetic classification for malformations of cortical development. Neurology 65, 1873–1887 [DOI] [PubMed] [Google Scholar]
- 24. Bielas S., Higginbotham H., Koizumi H., Tanaka T., Gleeson J. G. (2004) Cortical neuronal migration mutants suggest separate but intersecting pathways. Annu. Rev. Cell Dev. Biol. 20, 593–618 [DOI] [PubMed] [Google Scholar]
- 25. Guerrini R., Dobyns W. B., Barkovich A. J. (2008) Abnormal development of the human cerebral cortex: genetics, functional consequences and treatment options. Trends Neurosci. 31, 154–162 [DOI] [PubMed] [Google Scholar]
- 26. Jackson A. P., Eastwood H., Bell S. M., Adu J., Toomes C., Carr I. M., Roberts E., Hampshire D. J., Crow Y. J., Mighell A. J., Karbani G., Jafri H., Rashid Y., Mueller R. F., Markham A. F., Woods C. G. (2002) Identification of microcephalin, a protein implicated in determining the size of the human brain. Am. J. Hum. Genet. 71, 136–142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Rauch A., Thiel C. T., Schindler D., Wick U., Crow Y. J., Ekici A. B., van Essen A. J., Goecke T. O., Al-Gazali L., Chrzanowska K. H., Zweier C., Brunner H. G., Becker K., Curry C. J., Dallapiccola B., Devriendt K., Dörfler A., Kinning E., Megarbane A., Meinecke P., Semple R. K., Spranger S., Toutain A., Trembath R. C., Voss E., Wilson L., Hennekam R., de Zegher F., Dörr H. G., Reis A. (2008) Mutations in the pericentrin (PCNT) gene cause primordial dwarfism. Science 319, 816–819 [DOI] [PubMed] [Google Scholar]
- 28. Sheen V. L., Ganesh V. S., Topcu M., Sebire G., Bodell A., Hill R. S., Grant P. E., Shugart Y. Y., Imitola J., Khoury S. J., Guerrini R., Walsh C. A. (2004) Mutations in ARFGEF2 implicate vesicle trafficking in neural progenitor proliferation and migration in the human cerebral cortex. Nat. Genet. 36, 69–76 [DOI] [PubMed] [Google Scholar]
- 29. Pfenninger K. H. (2009) Plasma membrane expansion: a neuron's Herculean task. Nat Rev. Neurosci. 10, 251–261 [DOI] [PubMed] [Google Scholar]
- 30. Kivitie-Kallio S., Autti T., Salonen O., Norio R. (1998) MRI of the brain in the Cohen syndrome: a relatively large corpus callosum in patients with mental retardation and microcephaly. Neuropediatrics 29, 298–301 [DOI] [PubMed] [Google Scholar]
- 31. Schlager M. A., Kapitein L. C., Grigoriev I., Burzynski G. M., Wulf P. S., Keijzer N., de Graaff E., Fukuda M., Shepherd I. T., Akhmanova A., Hoogenraad C. C. (2010) Pericentrosomal targeting of Rab6 secretory vesicles by Bicaudal-D-related protein 1 (BICDR-1) regulates neuritogenesis. EMBO J. 29, 1637–1651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zmuda J. F., Rivas R. J. (1998) The Golgi apparatus and the centrosome are localized to the sites of newly emerging axons in cerebellar granule neurons in vitro. Cell Motil. Cytoskeleton 41, 18–38 [DOI] [PubMed] [Google Scholar]
- 33. Rosnoblet C., Peanne R., Legrand D., Foulquier F. (2013) Glycosylation disorders of membrane trafficking. Glycoconj. J. 30, 23–31 [DOI] [PubMed] [Google Scholar]
- 34. Miller V. J., Ungar D. (2012) Re'COG'nition at the Golgi. Traffic 13, 891–897 [DOI] [PubMed] [Google Scholar]
- 35. Majeed W., Liu S., Storrie B. (2014) Distinct sets of Rab6 effectors contribute to ZW10- and COG-dependent Golgi homeostasis. Traffic 15, 630–647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sun Y., Shestakova A., Hunt L., Sehgal S., Lupashin V., Storrie B. (2007) Rab6 regulates both ZW10/RINT-1 and conserved oligomeric Golgi complex-dependent Golgi trafficking and homeostasis. Mol. Biol. Cell 18, 4129–4142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Fukuda M., Kanno E., Ishibashi K., Itoh T. (2008) Large scale screening for novel rab effectors reveals unexpected broad Rab binding specificity. Mol. Cell Proteomics 7, 1031–1042 [DOI] [PubMed] [Google Scholar]
- 38. Lubbehusen J., Thiel C., Rind N., Ungar D., Prinsen B. H., de Koning T. J., van Hasselt P. M., Korner C. (2010) Fatal outcome due to deficiency of subunit 6 of the conserved oligomeric Golgi complex leading to a new type of congenital disorders of glycosylation. Hum. Mol. Genet. 19, 3623–3633 [DOI] [PubMed] [Google Scholar]