Background: SorLA binds APP and decreases the production of Aβ; however, the molecular mechanisms controlling these processes are poorly understood.
Results: We identified CR(5–8) as an APP-binding site in SorLA.
Conclusion: The CR-cluster is essential for the SorLA-dependent decrease in APP proteolysis.
Significance: Details regarding the function of SorLA in APP metabolism might lead to an understanding of the genetic association of SorLA with Alzheimer disease.
Keywords: Alzheimer Disease, Amyloid Precursor Protein (APP), Golgi, Intracellular Trafficking, Membrane Protein, CR-domains, O-Glycosylation, Fingerprint Residues, SorLA
Abstract
SorLA is a neuronal sorting receptor that is genetically associated with Alzheimer disease. SorLA interacts directly with the amyloid precursor protein (APP) and affects the processing of the precursor, leading to a decreased generation of the amyloid-β peptide. The SorLA complement-type repeat (CR) domains associate in vitro with APP, but the precise molecular determinants of SorLA·APP complex formation and the mechanisms responsible for the effect of binding on APP processing have not yet been elucidated. Here, we have generated protein expression constructs for SorLA devoid of the 11 CR-domains and for two SorLA mutants harboring substitutions of the fingerprint residues in the central CR-domains. We generated SH-SY5Y cell lines that stably express these SorLA variants to study the binding and processing of APP using co-immunoprecipitation and Western blotting/ELISAs, respectively. We found that the SorLA CR-cluster is essential for interaction with APP and that deletion of the CR-cluster abolishes the protection against APP processing. Mutation of identified fingerprint residues in the SorLA CR-domains leads to changes in the O-linked glycosylation of APP when expressed in SH-SY5Y cells. Our results provide novel information on the mechanisms behind the influence of SorLA activity on APP metabolism by controlling post-translational glycosylation in the Golgi, suggesting new strategies against amyloidogenesis in Alzheimer disease.
Introduction
Alzheimer disease (AD)2 is the most prevalent form of dementia and affects more than 50% of the elderly population above the age of 85 in Western societies. However, no medical cure has yet been discovered. The disease causes neurodegeneration and is pathologically characterized by intraneuronal fibrillary tangles and extracellular amyloid plaques in the brain (1). The main constituent of amyloid plaques is a peptide named amyloid-β (Aβ), which is generated by the sequential cleavage of the amyloid precursor protein (APP) by β- and γ-secretases. Alternative processing in the anti-amyloidogenic pathway can also occur. Here, APP is cleaved in the middle of the Aβ region by α-secretase, generating a large soluble sAPPα fragment and preventing the formation of Aβ. Secretase activity is not evenly distributed throughout neurons, as each of the involved enzymes is a target of highly specific polarized sorting events. Accordingly, the cellular trafficking of APP influences whether it enters the amyloidogenic or the anti-amyloidogenic processing pathway. Therefore, the APP sorting mechanisms have recently become essential for understanding the balance of APP processing, providing renewed hope for novel therapeutic strategies (2–4). APP is subjected to several post-translational modifications, including both N- and O-glycosylation, depending on the sorting pathway (5, 6).
SorLA (also known as SORL1 and LR11) is an ∼250-kDa type-1 membrane protein that is highly expressed in the neurons of both the developing and the adult central nervous system (7, 8). Neuropathological studies have identified reduced SorLA expression in vulnerable neurons in the brains of AD patients (9–13). Recently, converging lines of evidence from basic biochemistry and cell biology to genetics have further implicated SorLA in AD pathology, and numerous variants of the SorLA gene (SORL1) have been shown to correlate with the risk of developing the late-occurring form of AD (14–16). Interestingly, pathogenic SorLA protein variants have also been reported in early-onset AD patients, providing further evidence that SorLA dysfunction is an important AD risk factor (17).
Our group and others have worked to identify the mechanisms linking SorLA activity with AD onset and/or progression. Using SorLA-deficient mice, we have found that in the absence of SorLA, AD pathology is strongly accelerated, suggesting a proximal role of SorLA in AD (18, 19) that has led to the current model in which SorLA protects against AD (20).
SorLA functions as a sorting receptor for APP. SorLA binds directly to APP and slows down its cellular transport out of the Golgi; thus, less of the precursor protein enters secretase-containing compartments, decreasing the processing of APP to both nonamyloidogenic (sAPPα) and amyloidogenic (Aβ) products (10, 11, 21–23). This function is dependent on receptor localization to the Golgi, establishing a direct link to the trafficking pathway from endosomes to the trans-Golgi network/Golgi, a pathway suspected to be altered in sporadic AD (2, 24, 25).
The extracellular part of SorLA is a complex mosaic structure comprising a VPS10p domain, a YWTD repeat, a six-bladed β-propeller, a cluster containing 11 complement-type repeat (CR) domains, and a cassette of six fibronectin type-3 domains (26–28). We previously identified an interaction between the isolated SorLA CR-cluster and APP (21). However, as the VPS10p domain of the SorLA-related proteins sortilin (29) and SorCS1 (30, 31) have also been reported to interact with APP, we wanted to determine whether the CR-cluster is the only site for APP binding within SorLA. Therefore, we generated a SorLA variant without the CR-domains to further study APP binding. We found that the CR-cluster is essential both for APP binding and for the protective activity against Aβ production, and we identified a novel function of SorLA in influencing the glycosylation of APP. This study thus provides novel insight into the mechanisms that determine APP trafficking and maturation, factors that strongly regulate the generation of Aβ and are thus risk factors for AD.
EXPERIMENTAL PROCEDURES
DNA Constructs
The SorLA-ΔCR construct (deletion of residues Cys-1050–Glu-1524 and substitution with an Ser-Asp-Gly insert) was generated by three-step PCR. Using the primer pair 1A, 5′-cggctagcaagacaaacgtgtacatctctagc-3′, and 1B, 5′-ggtgttctcttctttgacacaggtatg-3′, we amplified an upstream fragment from an internal NheI site to the N-terminal part of the CR-cluster. Using the primer pair 2A, 5′-gctcaagaacaatacctgtgtcaaagaagagaacaccagtgatgagttgactgtgtacaaagtac-3′, and 2B, 5′-cgactagtcaccgcagccactctgacg-3′, we amplified a downstream fragment from the C-terminal end of the CR-cluster to an internal SpeI site, where the first 37 bases of primer 2A define a sequence overlap with the fragment of PCR number 1. The third round of PCR was performed using a mixture of the two obtained fragments as the template together with primer pairs 1A and 2B to generate the SorLA-ΔCR-fragment between the internal NheI and SpeI sites. The wild type SorLA region in the full-length SorLA cDNA within a pBluescript cloning vector was replaced by the SorLA-ΔCR-fragment using the NheI and SpeI restriction enzymes, and the entire SorLA-ΔCR coding frame was finally transferred from the cloning vector with the NotI and XhoI restriction enzymes to the pcDNA3.1/zeo(+) expression vector. SorLA-DDD and SorLA-KKK were generated by site-directed mutagenesis of human cDNA in the pcDNA3.1/zeo(+) expression vector (10) using a PCR-based cloning strategy that allows multiple substitutions, according to the manufacturer's protocol (Stratagene). Primers were obtained from DNA Technology. The APP-FLAG vector (32) was kindly provided by D. Kaden (Freie Universität, Berlin).
Transfected Cell Lines
SH-SY5Y cells were cultured and transfected with FuGENE (Roche Diagnostics) or HiFect (Amaxa) as described previously (23). Stably transfected clones were maintained by adding 300 μg/ml Zeocin (Invitrogen).
Recombinant Proteins
The CR-fragments of SorLA corresponding to CR(1–11) (Val-1044–Thr-1526), CR(1–8) (Val-1044–Leu-1381), CR(1–4) (Val-1044–Lys-1210), CR(5–11) (Glu-1208–Thr-1526), CR(9–11) (Ser-1384–Thr-1526), and CR(5–8) (Glu-1208–Leu-1381) were cloned into the eukaryotic expression vector pCEP-Pu-sp-his-myc-fXa following similar procedures to those described previously (21). Constructs for CR(5–8)-DDD and CR(5–8)-KKK were generated by site-directed mutagenesis similar to the protocol for introduction of the mutations in the full-length SorLA plasmid. To produce recombinant CR-domain proteins, the expression vectors were introduced into EBNA 293 cells to generate cell clones that secrete large amounts of the receptor fragments. The conditioned cell medium was harvested after 48 h of cultivation, and the proteins were purified by Ni2+-nitrilotriacetic acid affinity chromatography as described previously (33). The APP-d6A protein was produced in Escherichia coli and purified as described previously (21). The His-tagged extracellular domain of APP (APP-His6; from the APP751 isoform) was produced as described previously (29) (gift from P. Madsen, Aarhus University).
Immunocytochemistry and Confocal Microscopy
Cells were fixed in 4% paraformaldehyde for 10 min and then washed three times with PBS to remove the fixative. The cells were then permeabilized by incubation for 30 min in PBS containing 0.1% Triton X-100 and blocked for 30 min in PBS containing 10% fetal calf serum. After blocking, the cells were incubated with primary antibodies against mannosidase II (Millipore Bioscience Research Reagents), CD8 (Santa Cruz Biotechnology), or SorLA (a gift from C. M. Petersen, Aarhus University) either for 2 h at room temperature or overnight at 4 °C. The cells were then washed three times, followed by incubation with fluorescently labeled secondary anti-rabbit or anti-goat antibodies (Calbiochem and Invitrogen). Nuclei were visualized with DAPI (Sigma). Images were acquired on a Carl Zeiss confocal LSM 510 META laser microscope with a ×40, NA 1.2 C-Apochromat objective (Carl Zeiss).
Internalization of sAPP
Soluble APP was labeled using the Alexa Fluor 488 microscale protein labeling kit (Invitrogen), according to the manufacturer's protocol. Cells were incubated with 20 nm sAPP for 40 min at 37 °C before fixation, and immunofluorescence staining for receptor expression was performed as described previously (29).
Surface Protein Determination
Biotinylation of cell surface proteins in SH-SY5Y cells stably expressing SorLA-WT/variants was performed as described previously (23).
Surface Plasmon Resonance (SPR) Analysis
Before immobilization of SorLA CR-fragments, the recombinant proteins were dialyzed against sodium acetate, pH 4.0, and then coupled to CM5 chips from BIAcore at a concentration of 10 mg/ml after activation of the chip surface using a 1:1 mixture of 0.2 m N-ethyl-N-[3(dimethylamino)propyl]carbodiimide and 0.05 m N-hydroxysuccinimide in water. The remaining binding sites were blocked by passage of 1 m ethanolamine, pH 8.5. The full-length SorLA receptor was purified as described previously (34) and attached to the biosensor surface after dilution in sodium acetate buffer, pH 4.0, by standard procedures as described above.
Determination of direct binding of APP-d6A to the CR-fragments was performed on a BIAcore2000 instrument (BIAcore) using CaHBS (10 mm HEPES, pH 7.4, 150 mm NaCl, 5 mm CaCl2, and 0.005% Tween 20) as both a sample and running buffer. Anti-histidine antibody (H-1029, Sigma) was applied as a control. After each cycle of the ligand binding experiment, the flow cell was regenerated using two 10-μl pulses of regeneration buffer (10 mm glycine, pH 4.0, 500 mm NaCl, 20 mm EDTA, and 0.005% Tween 20) and a single injection of 0.001% SDS.
The effect of pH on sAPP binding to SorLA was determined either by injecting 500 nm sAPP using CaHBS at pH 5.0 or 7.4 as the sample and running buffer or by injecting 40 μl of 500 nm sAPP in CaHBS, pH 7.4, as a running buffer followed by a second injection of CaHBS with pH 6.0, 7.0, or 7.4 to determine off-rates for the complex dissociation. Similarly, SPR studies for CR(5–8)-WT and CR(5–8)-DDD were conducted by injection of 500 nm sAPP in pH 7.4 running buffer, followed by two consecutive injections first with dissociation buffer with pH 5.0, 6.0, or 7.4 and then with pH 7.4 dissociation buffer. The BIAevaluation software was used to estimate the binding kinetics and affinity constants.
Co-immunoprecipitation
SH-SY5Y cells that were overexpressing SorLA-WT, SorLA-ΔCR, or SorLA-KKK were harvested and lysed for 1 h on ice in 100 μl of lysis buffer (10 mm Tris-HCl, pH 8.0, 5 mm EDTA, 1% Triton X-100, and 1% Nonidet P-40) supplemented with Complete Mini EDTA-free protease inhibitor tablets (Roche Diagnostics). Protein concentration was determined using the Bradford assay (Bio-Rad). Gamma Bind G-Sepharose beads (GE Healthcare) were washed in PBS and pre-coated with 2 μg of anti-His antibody (GenScript) for 2 h at room temperature. The beads were then washed with and resuspended in 300 μl of PBS. For each sample, 100 μg of total protein, 2 μg of APP-His6, and 500 μl of PBS were mixed, and 20 μl of this mixture was used for SDS-PAGE (input). The remaining preincubated Gamma Bind G-Sepharose beads were added to the rest of the lysate together with Complete Mini EDTA-free tablets (Roche Diagnostics). The reaction was incubated on a rotating wheel at 4 °C overnight. The beads were then washed five times with PBS + 0.05% Tween 20, and the immunoprecipitated proteins were eluted by adding 30 μl of reducing loading buffer and heating at 95 °C for 5 min. The co-immunoprecipitation was analyzed by Western blotting. Co-immunoprecipitation of SorLA with endogenous APP was performed essentially as above, except that a C-terminal APP antibody (1227) was coupled to the beads, and no APP-His6 was added to the lysates. The complex formed by SorLA-DDD and APP was studied in HEK293 cells transfected with APP-FLAG and either the SorLA-WT or SorLA-DDD expression vectors using the HiFect reagent. Cells were harvested and lysed 48 h post-transfection, and protein precipitations were performed using Gamma Bind beads precoated with sol-SorLA antibody.
APP Metabolism
SH-SY5Y cells were cultured in Dulbecco's modified Eagle's medium (DMEM) F-12 (Invitrogen) supplemented with 10% fetal bovine serum and 5% penicillin/streptomycin. Determination of APP metabolism was performed using the endogenous APP expression level in stably transfected SH-SY5Y cell lines. A total of 5 × 106 cells were cultured in T25 flasks (Nunc) and allowed to grow to >90% confluency. The medium was then changed to serum-free DMEM F-12. Both medium and cells were harvested after 48 h. Cells were lysed using 10 mm Tris-HCl, pH 8.0, 5 mm EDTA, 1% Triton X-100, and 1% Nonidet P-40. Cell-associated APP and secreted sAPPα were determined by Western blotting. Secreted Aβ40 from the conditioned medium was quantified by ELISA (KHB3481, Invitrogen). Significance was evaluated using two-tailed t tests. Surface levels of APP and SorLA were determined by biotinylation experiments as described previously using membrane-impermeable, EZ-linked sulfo-NHS-S-biotin (Pierce) and streptavidin beads (GE Healthcare) (23).
MS Identification of O-Glycosylated Peptides from APP
APP was immunoprecipitated from conditioned media according to a published procedure (35). Briefly, the 6E10 antibody (40 μg, Aβ epitope 6–9, Signet Laboratories) was immobilized on 250 μl of magnetic Dynabeads M-280 sheep anti-mouse IgG (Invitrogen). Conditioned media (50 ml) was added to the beads, and the samples were agitated for 12 h. A KingFisher magnetic particle separator (Thermo) was used for the washing steps and to release the bound fraction. Electrophoresis was performed via standard SDS-PAGE using gels that were cut into 15 pieces and subjected to in-gel trypsin digestion.
Nanoflow LC was performed on an Ettan MDLC (GE Healthcare) using a 150 × 0.075-mm C18 reverse-phase column (Zorbax; Agilent Technologies) and a 60-min elution time. The gradient ranged from 0 to 50% acetonitrile in 0.1% formic acid with a flow rate of 200–300 nl/min. The nano-ESI source was coupled to a hybrid linear quadrupole ion trap/FT ion cyclotron resonance mass spectrometer (LTQ-FT; Thermo). The mass spectrometer was operated in the data-dependent mode to automatically switch between MS1 and MS2 using collision-induced dissociation at a normalized collision energy of 30%. The LC-MS/MS files were converted to the Mascot general format (.mgf) using the Raw2 msm application, and Mascot searches were performed using the in-house Mascot server. LC-MS/MS files that contained peptide hits from APP were manually searched for the presence of glycosylated peptides, and the presence of diagnostic saccharide oxonium ions at m/z 366 (HexHexNAc+), m/z (292, Neu5Ac+), and m/z 274 (Neu5Ac-H2O) was specifically assessed.
Deglycosylation
To remove the terminal sialic acids, conditioned medium was incubated overnight with different neuraminidases and O-glycosidase in 60 mm sodium acetate, pH 5.5, at 16 °C before SDS-PAGE and Western blot analysis. The neuraminidases used were the Macrobdella decora α-2,3-neuraminidase (480706 Calbiochem), the Clostridium perfringens α-2,3/2,6-neuraminidase (480708 Calbiochem), the Vibrio cholerae α-2,3/2,6/2,8-neuraminidase (70364620 Roche Diagnostics), or the endoneuraminidase-N, which removes linear polymers of sialic acid with α-2,8-linkage with a minimum length of 7–9 residues (AbC0020 Eurobio Laboratories). The Diplococcus pneumoniae O-glycosidase was obtained from Roche Diagnostics (11347101001).
Western Blotting
Protein separation by SDS-PAGE analysis was performed using precast NuPAGE 4–12% BisTris gradient gels (Novex, Invitrogen) and MOPS gel electrophoresis buffer to optimize the separation of the two sAPPα glycosylation variants. For all other studies, we used standard 4–16% Tris-glycine gradient gels. Blotting of the proteins onto nitrocellulose membranes (Amersham Biosciences) was followed by incubation with the primary antibodies and the HRP-conjugated secondary antibodies. Bands were detected using the SuperSignal West Femto maximum sensitivity substrate (Thermo Scientific). Antibodies were used against the following factors: sAPPα (WO2, kindly provided by R. Cappai, University of Melbourne); APP (1227, O. M. Andersen (10)); actin (A5441, Sigma); His tag (A00186, GenScript); and FLAG tag (F3165, Sigma). SorLA-specific antibodies (sol-SorLA, tail-SorLA, VPS10p, and CR-cluster) were kindly provided by C. M. Petersen, Aarhus University.
Statistical Methods
Student's two-tailed t test was used (software PRISM 5.0) to compare differences and determine statistical significance between the control and experimental values. A p value above 0.05 was not considered significant (ns), whereas p < 0.05 (*), p < 0.01 (**), and p < 0.001 (***) were considered to be significantly different.
RESULTS
SorLA Mini Receptor with CR-cluster Deletion
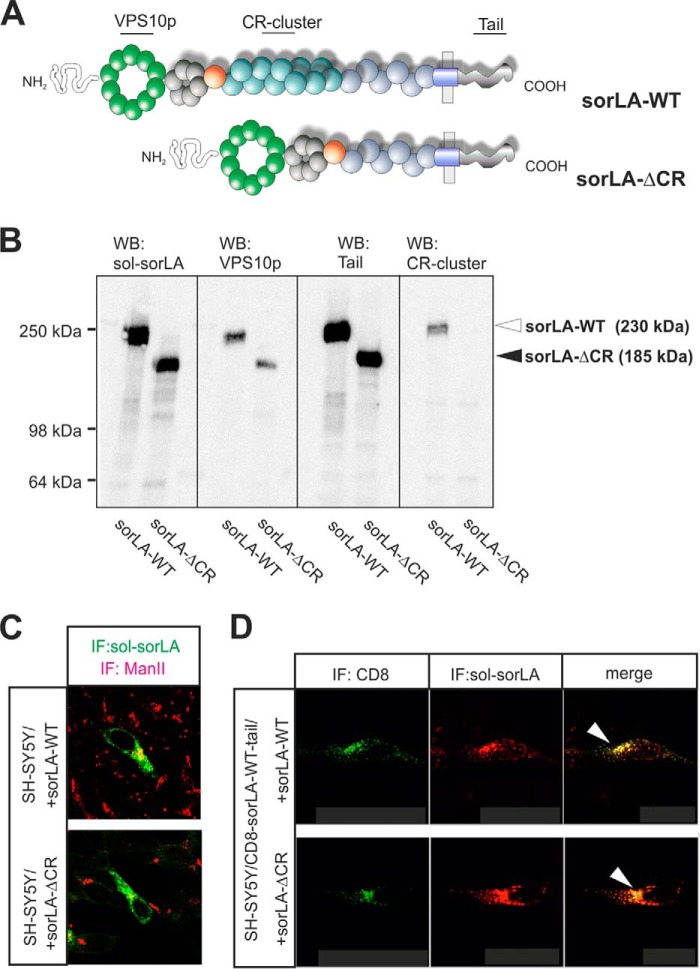
We produced an expression construct for a SorLA deletion mutant lacking all 11 CR-domains, termed SorLA-ΔCR (Fig. 1A). Using this construct, we produced an SH-SY5Y cell line that stably expresses the deletion mutant. Western blot analysis of cell extracts using an antiserum generated against the entire extracellular part of SorLA revealed similar expression levels of SorLA-ΔCR and SorLA-WT in a previously generated cell line (Fig. 1B) (10). The mutant receptor migrated in SDS-PAGE analysis with an aberrant molecular mass of 185 kDa, which is consistent with a 45-kDa reduction (∼11 × 4 kDa), compared with the full-length receptor (230 kDa). Verification of the N- and C-terminal domains of SorLA-ΔCR was demonstrated by Western blots using antibodies specific for the VPS10p domain and the cytoplasmic tail, respectively (Fig. 1B). However, an antibody against the CR-cluster region showed a signal for SorLA-WT but not for SorLA-ΔCR. These data verify the deletion of the 11 CR-domains in the mini-receptor but suggest an otherwise intact domain assembly.
FIGURE 1.
SorLA-ΔCR expression in SH-SY5Y cells. A, schematic presentation of full-length SorLA (SorLA-WT) and the CR-cluster deletion mutant (SorLA-ΔCR) showing the domains present in the composite receptors. B, Western blot (WB) analysis of SH-SY5Y cells transfected with SorLA-WT (∼230 kDa) or SorLA-ΔCR (∼185 kDa) using primary antibodies against either the entire extracellular domain (sol-sorLA) or specific for the VPS10p domain (VPS10p), the region with 11 CR-domains (CR-cluster), or the cytoplasmic domain (Tail). The positions of the antibody epitopes are indicated in A. C, immunostaining for SorLA (green) and the Golgi marker mannosidase II (red) in SH-SY5Y cells transfected with either SorLA-WT or SorLA-ΔCR. D, SH-SY5Y cells were co-transfected with constructs for CD8-SorLA-WT-tail and either SorLA-WT (top) or SorLA-ΔCR (bottom) and then stained for the extracellular domains of the CD8 reporter (green) or SorLA (red). Arrowheads indicate receptor co-localization for both tested combinations. IF, immunofluorescence.
To determine the cellular localization of SorLA-ΔCR in the transfected SH-SY5Y cells, we performed immunocytochemistry followed by confocal microscopy (Fig. 1C). The mini-receptor localized to perinuclear vesicles in a pattern indistinguishable from that of SorLA-WT; the expression of both receptor variants partly overlaps with the Golgi marker mannosidase II, previously found to co-localize with SorLA (23).
To confirm similar intracellular distributions of the mini-receptor and full-length receptors, we used a CD8-SorLA-WT-tail chimeric reporter construct encoding the extracellular part of CD8 and the transmembrane and cytoplasmic tails of SorLA-WT (23). SH-SY5Y cells were double-transfected with the CD8-SorLA-WT-tail and either SorLA-WT or SorLA-ΔCR (Fig. 1D). The cells were then stained with antibodies against the extracellular domains of SorLA and the CD8 reporter. Strong co-localization between the reporter and SorLA-WT to perinuclear Golgi-like compartments was observed, demonstrating that the tail of SorLA is sufficient to provide receptor localization to this region. Likewise, a similar strong overlap between the reporter and SorLA-ΔCR was found, suggesting that deletion of the CR-cluster does not affect SorLA's intracellular localization.
CR-cluster Is Essential for SorLA Binding to APP and to Protect APP against Proteolysis
We previously identified a direct interaction between the extracellular domains of SorLA and APP and showed that a recombinant SorLA fragment containing the cluster of 11 CR-domains binds to APP in vitro (10, 21).
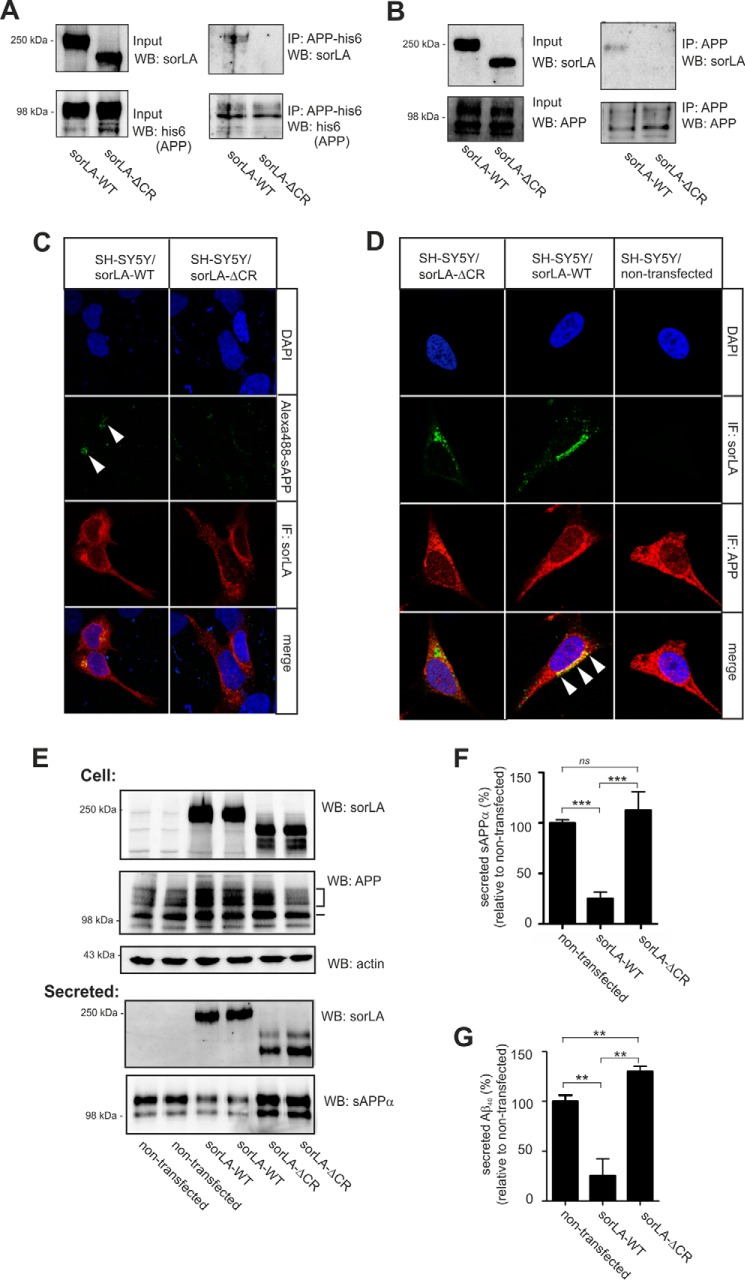
To determine whether the CR-cluster is the only region in the receptor ectodomain that interacts with APP, a ligand precipitation assay using SorLA-ΔCR with a recombinant variant of the sAPP extracellular domain was performed. Extracts of SH-SY5Y cells expressing either SorLA-WT or SorLA-ΔCR were mixed with His-tagged APP, and the ability of sAPP to bind SorLA-WT versus SorLA-ΔCR was compared by resolving the precipitates via SDS-PAGE and determining the amount of bound SorLA by Western blotting (Fig. 2A). SorLA was only precipitated from cells expressing the full-length receptor, but we were unable to detect any SorLA-ΔCR binding to APP. This result shows that deletion of the CR-cluster completely abolishes binding between the SorLA and APP extracellular regions.
FIGURE 2.
SorLA-ΔCR neither binds APP nor protects APP against processing. A, homogenates of SH-SY5Y cells expressing either SorLA-WT or SorLA-ΔCR were added to His-tagged sAPP (APP-his6), and protein complexes were immunoprecipitated using an antibody against polyhistidine. Lysates (Input) and precipitated proteins (IP) were subjected to SDS-PAGE and analyzed by Western blotting (WB) for either APP-His6 or SorLA as indicated. B, co-immunoprecipitation analyses of either SorLA-WT or SorLA-ΔCR with endogenous APP from SH-SY5Y cells using an antibody against the C-terminal part of APP. C, SH-SY5Y cells transfected with either SorLA-WT or SorLA-ΔCR were incubated with 20 nm Alexa Fluor 488-labeled sAPP for 40 min at 37 °C and stained with anti-SorLA (red). Internalized sAPP is indicated by the arrowheads and is only seen in cells expressing wild type SorLA. D, immunodetection of endogenous APP with SorLA in SH-SY5Y cells, indicating that SorLA-WT, but not SorLA-ΔCR, co-localizes with APP in the perinuclear region (arrowheads). E, SorLA and endogenous APP levels in extracts (Cell) and in the conditioned medium (Secreted) of SH-SY5Y cells stably transfected with either SorLA-WT or SorLA-ΔCR were analyzed by SDS-PAGE and Western blotting with the indicated antibodies. The actin level in cells was used as a control for loading equal amounts of protein in each lane. Migration of immature and mature APP is indicated by a dash and a bracket, respectively. F and G, secretion of sAPPα and Aβ40 to the medium was determined by either densitometric scanning of Western blots, as shown in E, or by ELISA and expressed as the percentage of processing products from nontransfected SH-SY5Y cells (set to 100%). The data are presented as the mean ± S.E. of four individual experiments. Statistical significance is as follows: **, p < 0.01; ***, p < 0.001; ns, not significant (p > 0.05).
Linkage between the SorLA and APP cytoplasmic domains by adaptor proteins has recently been suggested (36). Therefore, using SorLA-ΔCR, we tested whether association between the cytoplasmic tails is sufficient for complex formation. Accordingly, co-immunoprecipitations of endogenous full-length APP in SH-SY5Y cells expressing either SorLA-ΔCR or SorLA-WT were performed. Proteins from cell extracts were precipitated using an antibody against the APP C-terminal domain, and the precipitate was tested for the SorLA protein by Western blot (Fig. 2B). Similar levels of APP precipitation were verified using an APP mouse serum, but only SorLA-WT co-precipitated with APP.
As SorLA can mediate the endocytosis of bound sAPP at the cell surface, we also compared the internalization of sAPP in SH-SY5Y cells transfected with either SorLA-WT or SorLA-ΔCR as an independent assay to study the interaction between SorLA and APP. As reported previously (29), there was a specific uptake of fluorescently labeled sAPP in cells transfected with SorLA-WT (Fig. 2C). In contrast, no sAPP was internalized in cells transfected with SorLA-ΔCR (Fig. 2C), ruling out further APP-binding sites outside the CR-cluster. The observed differences in sAPP binding were not a result of differences in cell surface localization, as biotinylation experiments showed similar levels of SorLA-ΔCR (114.6 ± 24.1%, not significantly different) compared with SorLA-WT (set to 100%) at the plasma membrane (data not shown).
Co-localization between SorLA-WT and endogenous full-length APP in SH-SY5Y cells is observed in the perinuclear region. However, when we performed the same experiment using cells expressing SorLA-ΔCR, we could not detect a similarly strong co-localization, although both APP and SorLA mutants locate to the perinuclear region showing a partial overlap in their expression (Fig. 2D). The observed small degree of co-localization by the double fluorescent immunocytochemistry is not necessarily an indication of a direct protein-protein interaction between APP and SorLA-ΔCR, consistent with the lack of interaction between these two proteins shown by the two different co-immunoprecipitation analyses.
Accordingly, we conclude that the presence of a functional CR-cluster in SorLA is necessary for efficient binding to APP. None of our assays confirmed the suggested interaction between the cytoplasmic domains of APP and SorLA.
Numerous studies have indicated that SorLA activity decreases APP processing in several cell lines (10, 11, 18, 23). Although SorLA plays a direct role as a Golgi-localized retention factor that influences the amount of APP processed in subsequent cellular compartments (22), it remains unknown whether SorLA overexpression per se affects the processing machinery, i.e. whether SorLA also works as a competitive substrate of the secretases that cleave APP, thereby diminishing Aβ production. Therefore, we used cell lines expressing either SorLA-WT or the ΔCR mutant to study the processing of endogenous APP. Before analyzing the APP-processing products, we compared the expression levels of endogenous APP in our cell lines by Western blot analysis of total cell extracts using an antibody against the C terminus of APP, but we did not observe any significant changes in the APP levels (Fig. 2E). Next, we investigated the secretion of soluble sAPPα into medium collected after 48 h by quantifying Western blots. For these blots, we used the antibody WO2 that specifically recognizes the sAPPα but not sAPPβ. The signal for sAPPα displays as a doublet believed to represent the fragment from processing of the endogenous APP695 and APP751/770. However, although SorLA-WT significantly lowered the production of sAPPα by >50% compared with nontransfected cells (p < 0.0001), no decrease in sAPPα production was observed in the presence of SorLA-ΔCR (Fig. 2F). We also measured the secretion of Aβ using ELISA to determine the effects on the amyloidogenic processing pathway. Compared with the medium from nontransfected cells, a >50% reduction in the Aβ levels in the medium from cells overexpressing SorLA-WT was found (p < 0.005). In contrast, SorLA-ΔCR did not protect against amyloidogenic cleavages, as the level of secreted Aβ from cells expressing SorLA-ΔCR was not significantly lower than from nontransfected cells (Fig. 2G). Interestingly, an increase in Aβ was noticed, which could be a result of SorLA-ΔCR out-competing the low level of endogenous SorLA and abrogating its protective role in SH-SY5Y cells. In conclusion, these data show that the CR-cluster in SorLA is essential for APP binding and for the protective effects of SorLA activity against the proteolytic breakdown of APP.
Central CR-domains in SorLA Are Important for APP Binding
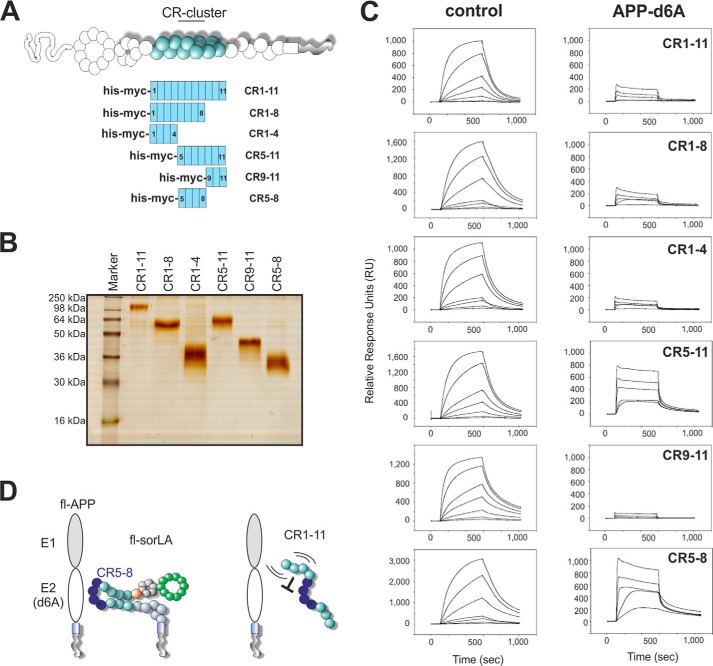
Next, we wanted to identify which of the 11 CR-domains bind to APP. Using expression in EBNA 293 cells, we produced recombinant CR-fragments spanning the eight N-terminal domains (CR(1–8)), the four N-terminal domains (CR(1–4)), the seven C-terminal domains (CR(5–11)), the three C-terminal domains (CR9–11), the central four domains (CR(5–8)), or the entire CR-cluster (CR(1–11)) (Fig. 3A). All of the protein fragments were effectively secreted from the cells in similar quantities and purified using a previously published method (21), generating clean protein preparations as indicated by SDS-PAGE followed by silver staining of the recombinant proteins (Fig. 3B). As expected, the CR-fragments showed reduced migration in SDS-PAGE analysis upon treatment with β-mercaptoethanol, indicating the presence of disulfide bridges (data not shown).
FIGURE 3.
SorLA CR(5–8) interacts with APP. A, schematic representation of the molecular dissection of the SorLA CR-cluster into six fragments, CR(1–11), CR(1–8), CR(1–4), CR(5–11), CR(9–11), or CR(5–8). B, silver staining and SDS-PAGE analysis of the CR-fragments expressed as recombinant secreted proteins from EBNA 293 cells. After secretion into the medium, the His-Myc-tagged proteins were purified by Ni2-nitrilotriacetic acid chromatography to homogeneous protein preparations and separated by SDS-PAGE under reducing conditions. The proteins migrate slower than expected according to their calculated molecular weights (values in parentheses below), suggesting the presence of post-translational glycosylation (CR(1–11) (61,189 Da); CR(1–8) (45,197 Da); CR(1–4) (25,937 Da); CR(5–11) (42,564Da); CR9–11 (22,675 Da); and CR(5–8) (26,572 Da)). C, surface plasmon resonance analysis of APP-d6A binding to an array of SorLA CR-fragments. Response units are relative to the signal from a flow cell with no immobilized proteins. Panels to the left represent sensorgrams from the application of a concentration series of anti-His antibody (control) to document the immobilization of similar receptor fragment densities. Representative sensorgrams from the SPR binding analysis of the APP-d6A fragment (0.1, 0.2, 0.5, 1.0, and 2.0 μm) are shown to the right. Increased response levels in each sensorgram series correspond directly to the ligand concentration. D, schematic model of the suggested binding between SorLA and APP.
To determine the APP-binding properties of the various CR-fragments, we immobilized similar femtomolar amounts of each fragment on biosensors. Comparable levels of bound receptor fragments were verified by applying a concentration series of an antibody against the hexahistidine polypeptide (His tag) in the N-terminal part of each protein (Fig. 3C, control). Every tested CR-fragment showed concentration-dependent binding via similar curvatures of the sensorgrams, suggesting that we had immobilized an approximately equal density of each receptor fragment. We then applied the d6A domain of APP at increasing concentrations (100, 200, 500, 1,000, and 2,000 nm) (Fig. 3C, APP-d6A). The CR(1–11) fragment bound to d6A with moderate affinity (KD ∼300 nm), as reported previously (21). Comparing the sensorgram series revealed that the proteins spanning either CR(5–8) (KD ∼27 nm) or CR(5–11) (KD ∼26 nm) bind most efficiently to APP-d6A (Fig. 3C). As the three C-terminal domains did not bind to APP-d6A, we conclude that the central four CR-domains (CR(5–8)) comprise an important APP binding region within CR(5–11).
Interestingly, more CR(5–8) associated with APP-d6A than either CR(1–8) or CR(1–11) in the described assay. As CR(1–4) and CR(1–8) showed similar binding to APP-d6A as CR(1–11), our hypothesis is that CR(1–4) binds to d6A in a way that dominates over the binding sites in CR(5–8) when they are present in the longer CR(1–8) and CR(1–11) constructs. Studies are ongoing to elucidate whether the overall conformation restricts access to CR(5–8), i.e. whether domains 1–4 provide steric hindrance that prevents access to the binding site in CR(5–8) in the isolated CR-fragments (Fig. 3D). However, no further attempt to address this question was pursued in this study. We decided to focus on the binding between APP and CR(5–8), as this fragment of SorLA seems ideal to elucidate the structure of a SorLA·APP complex.
APP Binding to SorLA under Acidic Conditions Is Linked to Unique Fingerprint Residues
The binding of extrinsic ligands to the CR-domains depends on a pair of fingerprint residues located at conserved positions in the primary structure of the receptor proteins. The acidic side chain of an aspartate residue (Asp) and the hydrophobic side chain of a tryptophan residue (Trp) point out from the molecular surface of many CR-domains, forming the primary ligand-binding site. The ligands bind to the receptor using side chains from a lysine and a hydrophobic residue (37–40). This simple motif has been identified in many complexes between ligands and CR-domains, including a minor group of human rhinovirus and three CR-domains from the very low density lipoprotein receptor (41), Reelin, in complex with two CR-domains from the apolipoprotein E receptor 2 (42, 43), β2-microglobulin in complex with the fourth CR-domain from LDLR (44, 45), and apolipoprotein E with CR17 from LRP1 (46, 47). These protein complexes represent classical extracellular ligands binding to endocytosis receptors; dissociation of the complex in the acidic endosome allows degradation of the ligands upon further transport to the lysosome and recycling of the receptors to the cell surface (48). This pH sensitivity seems to be dependent on the aspartic acid fingerprint residues (49).
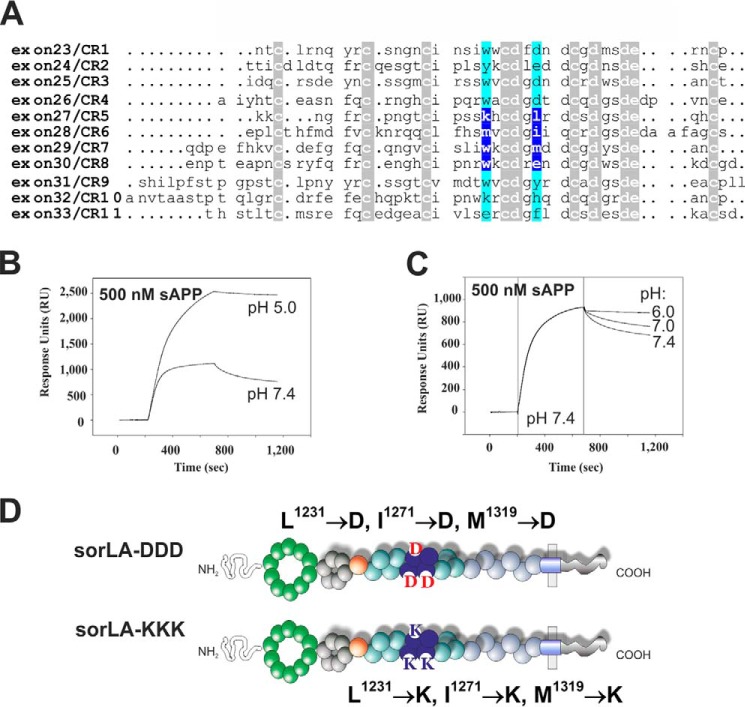
To characterize the role of the SorLA fingerprint residues in APP binding, we first prepared a sequence alignment of the 11 CR-domain sequences from the human SorLA protein to identify these amino acids in SorLA (Fig. 4A). In the four N-terminal CR-domains, the pair of fingerprint residues conformed to the described motif seen in endocytic receptors of an aromatic and an acidic amino acid as follows: Trp/AspCR1, Tyr/GluCR2, Trp/AspCR3, and Trp/AspCR4. Interestingly, further inspection of the alignment identified alterations to the classical fingerprint in the central CR-domains, as only CR8 contained a classical Trp/GluCR8 motif; the other three domains, CR5, CR6, and CR7, contained sets of lysine 1226 with leucine 1231 (Lys/LeuCR5), methionine 1266 with isoleucine 1271 (Met/IleCR6), and tryptophan 1314 with methionine 1319 (Trp/MetCR7) at the fingerprint positions (Fig. 4A). As APP apparently associates with CR(5–8), we speculated whether binding to a fragment containing an abnormal set of fingerprint residues would also provide an interaction that is sensitive to low pH.
FIGURE 4.
SorLA has a unique set of fingerprint residues and binds APP more avidly at acidic pH. A, alignment of the 11 CR-domain sequences from human SorLA. The ∼40 amino acids from each CR-domain are aligned according to 10 strictly conserved cysteines, aspartates, and glutamates (white letters with gray background). Residues written on a blue background correspond to amino acids that coordinate calcium by their backbone carbonyl oxygen and are termed “fingerprint” residues because their side chains present a primary recognition site for ligands. B, surface plasmon resonance binding analysis of 500 nm sAPP to immobilized SorLA CR(5–8) in HEPES-buffered saline with either pH 5.0 (kon 1.37 × 104 m−1 s−1; koff 4.89 × 10−5 s−1) or pH 7.4 (kon 2.84 × 104 m−1 s−1; koff 7.27 × 10−4 s−1). Kinetic parameters were obtained by fitting the sensorgrams using 1:1 Langmuir binding isotherms. C, SPR binding analysis of 500 nm sAPP to immobilized SorLA CR(5–8). The association of sAPP with CR(5–8) was conducted at pH 7.4 (200–700 s), and the dissociation of the complex was analyzed by injection of dissociation buffers with a pH of 6.0, 7.0 or 7.4 (700–1200 s). D, schematic representation of SorLA variants generated by site-directed mutagenesis to substitute the three hydrophobic fingerprint residues in CR5, CR6, and CR7 with either aspartic acid residues (L231→D, I1271→D, M1319→D) or lysine residues (L1231→K, I1271→K, M1319→K).
To test this hypothesis, we used surface plasmon resonance analysis to measure the binding of SorLA CR(5–8) to APP at various pH values. To our surprise, as evident from fitting of the sensorgrams representing 500 nm sAPP binding to SorLA CR(5–8), we found an ∼10-fold stronger interaction at pH 5.0 (KD ∼3.7 nm) than at pH 7.4 (KD ∼26 nm) (Fig. 4B). To provide further evidence for this unique pH binding profile, we measured the dissociation rates of a pre-formed SorLA-CR(5–8)·APP complex using our biosensor technique. Equal amounts of APP (500 nm) were injected onto a chip containing immobilized SorLA fragments. The pH was maintained at 7.4 during the association phase, but the pH of the running buffer was varied from 7.4 to 7.0 and 6.0 during the dissociation phase (Fig. 4C). Using this method, we confirmed a stronger binding interaction at low pH, as the dissociation rate is relatively fast at pH 7.4 (koff ∼3.78 × 10−4 s−1) compared with the 10-fold slower dissociation rate at pH 6.0 (koff ∼3.48 × 10−5 s−1).
In summary, the interaction between SorLA CR(5–8) and APP depends on the pH. However, the binding is favored by low pH, in contrast to what would be expected if APP bound to the CR-domains via the classical Trp/Asp fingerprint residues. As CR(5–8) contains a different set of fingerprint residues, these findings suggest that the unique fingerprint residues of CR(5–8) may be directly involved in APP binding.
APP Processing in the Presence of SorLA-DDD
To investigate the precise role of the leucine 1231 (CR5), isoleucine 1271 (CR6), and methionine 1319 (CR7) residues, we made two different expression constructs in which each of the three residues was mutated either to classical aspartate fingerprint amino acids (SorLA-DDD) or to lysines (SorLA-KKK) (Fig. 4D). For both constructs, we generated SH-SY5Y cell lines that stably express the SorLA variant.
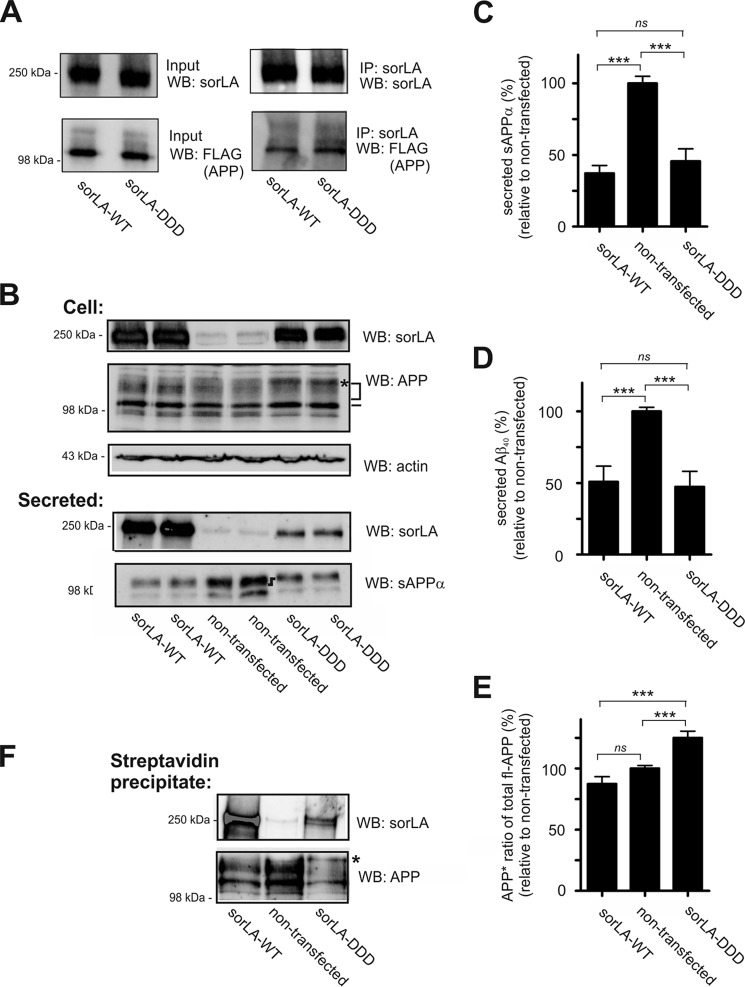
To determine whether mutating the Leu-1231–Ile-1271–Met-1319 residues to Asp-Asp-Asp influences the binding to APP, we performed co-immunoprecipitation studies using HEK293 cells expressing either SorLA-WT or SorLA-DDD. The cells were transfected with a FLAG-APP construct, and lysates were prepared 48 h later and incubated with anti-SorLA antibodies overnight. The bound protein complexes were isolated using Gamma Bind beads and separated by SDS-PAGE analysis. The amount of precipitated receptor was similar for SorLA-WT and SorLA-DDD, and surprisingly, the level of co-precipitated FLAG-APP was also similar (Fig. 5A). Notably, more APP tended to bind to the mutated SorLA than to the nonmutated receptor. Similar results were obtained when we precipitated proteins with an antibody against APP; equal amounts of SorLA-WT and SorLA-DDD were bound (data not shown). Our conclusion is that substitution of the three Leu-Ile-Met fingerprint residues to Asp-Asp-Asp does not impair APP binding per se. Nevertheless, we decided to investigate the processing of APP in the presence of SorLA-DDD using parallel studies to the ones already outlined for the detailed analysis of APP processing and SorLA-ΔCR.
FIGURE 5.
SorLA-DDD binds and protects APP as efficiently as SorLA-WT. A, HEK293 cells were co-transfected with FLAG-tagged APP695 together with constructs for either SorLA-WT or SorLA-DDD, and the protein complexes were immunoprecipitated using an antibody against SorLA. Lysates (Input) and precipitated proteins (IP) were subjected to SDS-PAGE and analyzed by Western blotting (WB) for either SorLA or APP (by anti-FLAG), as indicated. B, SorLA and endogenous APP levels in extracts (Cell) or in the conditioned medium (Secreted) of SH-SY5Y cells stably transfected with SorLA-WT or SorLA-DDD were analyzed by SDS-PAGE and Western blotting with the indicated antibodies. The actin level in cells was used as a control for loading equal amounts of protein in each lane. Migration of immature and mature full-length APP is indicated by a dash and a bracket, respectively. Differences in migration of sAPPα are indicated by a dark black symbol, and asterisks indicate an APP glycosylation variant specific for cells expressing SorLA-DDD. C and D, amount of sAPPα and Aβ40 products in the medium was determined by densitometric scanning of Western blots, as shown in B, or by ELISA, and expressed as the percentage of processing products from nontransfected SH-SY5Y cells (set to 100%). The data are presented as the mean ± S.E. of four individual experiments. E, ratio of APP* to total fl-APP was determined by Western blot quantification and expressed relative to the ratio observed for nontransfected cells. F, level of biotinylated SorLA and endogenous APP at the cell surface (Streptavidin precipitate), shown by representative Western blot. Statistical significance is as follows: ***, p < 0.001; ns, not significant (p > 0.05).
First, we verified that the expression level of SorLA-DDD was similar to SorLA-WT by Western blot analysis of cell extracts (Fig. 5B). Documentation by immunocytochemistry also showed an indistinguishable level of co-localization between mannosidase II and SorLA-DDD compared with SorLA-WT, suggesting that the mutation does not influence receptor localization in any major way (data not shown).
Further studies into APP processing showed an indistinguishable ability of SorLA-WT and SorLA-DDD to decrease the production of both sAPPα and Aβ (Fig. 5, C and D). Both the nonamyloidogenic and the amyloidogenic pathways were significantly decreased compared with the nontransfected SH-SY5Y cells, but quantification of the shed products revealed nearly identical levels of sAPPα and Aβ in the media of cells expressing either SorLA-WT or SorLA-DDD (Fig. 5C). As noted earlier, we observed an accumulation of full-length APP when SorLA-WT was expressed in SH-SY5Y cells (10). Surprisingly, although accumulation of mature APP was also seen in the presence of SorLA-DDD, the migration of the accumulated APP from cells expressing the mutant was altered, suggesting that the glycosylation pattern of APP varied from the nontransfected and SorLA-WT-expressing cells (APP*, Fig. 5B). Quantification (Fig. 5E) of this post-translationally modified APP band revealed a significant increase in the SorLA-DDD-expressing cells (125.1 ± 5.45%, p = 0.0009) compared with the nontransfected cells (level of APP* set as 100%). A similar increase in APP* was not observed in cells expressing SorLA-WT (87.34 ± 5.99%, not significant), suggesting that the introduction of the mutations had a specific effect on APP glycosylation. A change in the gel migration of sAPPα from SorLA-DDD-expressing cells was also observed (Fig. 5B).
We also looked for secretion of the SorLA extracellular domains into the conditioned medium and, unexpectedly, found a SorLA-DDD shedding reduction of 87.1 ± 2.4% (p < 0.0001) compared with SorLA-WT (Fig. 5B), suggesting altered SorLA-DDD trafficking despite the overlap with the Golgi-marker mannosidase II. We therefore tested whether the low level of mutant receptor shedding resulted from a loss of SorLA-DDD at the cell surface. To determine the surface expression of the SorLA variants, we incubated cell lines with a membrane-impermeable biotin reagent at 4 °C. A fraction of the total cell lysates and the biotinylated protein were resolved by SDS-PAGE analysis, and receptor expression was determined by Western blot (Fig. 5F). Consistent with the decreased shedding of SorLA-DDD, quantification of these blots revealed that SorLA-DDD expression at the plasma membrane was reduced by 50.0 ± 10.7% (p = 0.010) compared with the level of SorLA-WT. Interestingly, the level of APP at the plasma membrane did not differ between cells expressing SorLA-WT and SorLA-DDD, although both variants displayed lower surface levels of APP compared with nontransfected SH-SY5Y cells (as reported previously for SorLA-WT (10)). Notably, the migration change in mature APP was again clearly seen in cells expressing SorLA-DDD compared with SorLA-WT (Fig. 5F). From these experiments, we conclude that mutation of the Leu-Ile-Met residues to aspartates does not influence the role of SorLA in decreasing APP proteolysis but does affect APP maturation. Additionally, we conclude that the ability of SorLA to decrease APP processing is independent of the surface expression of SorLA, which is consistent with a model in which SorLA works predominantly as an APP retention factor at the Golgi.
APP Maturation Changes in the Presence of SorLA-KKK
We next analyzed the effect of mutating the three fingerprint residues, Leu-Ile-Met, to lysines. First, we confirmed similar expression levels of SorLA-KKK and SorLA-WT in the selected SH-SY5Y cell lines by Western blot analysis of cell lysates (Fig. 6, A and C).
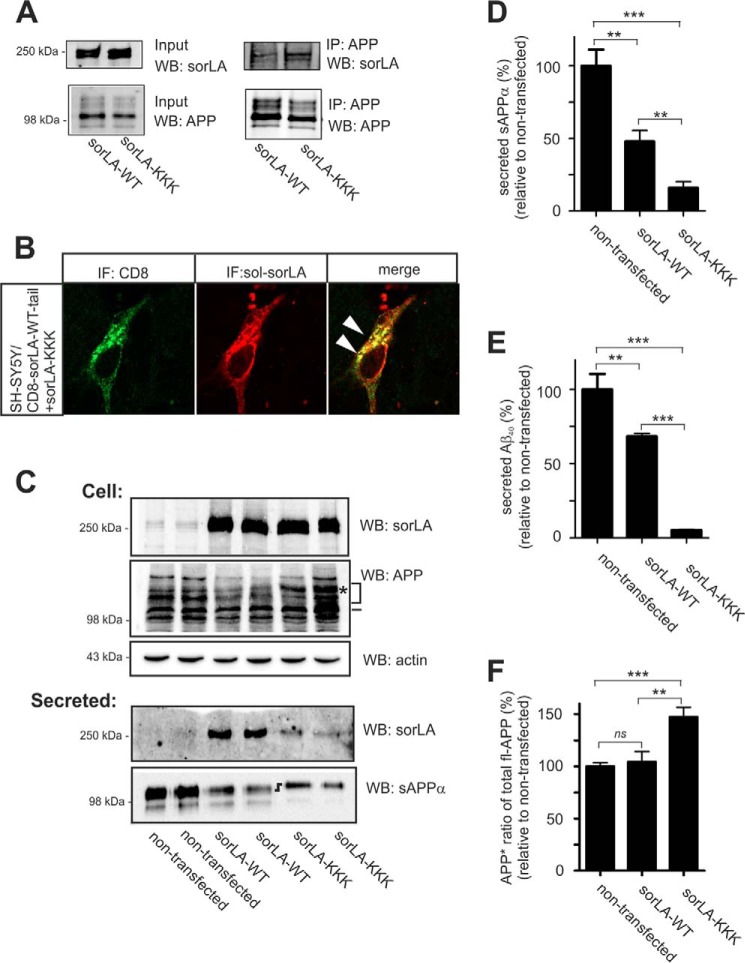
FIGURE 6.
SorLA-KKK associates with APP and influences its maturation and processing. A, extracts of HEK293 cells co-transfected with APP695 together with either the SorLA-WT or SorLA-KKK constructs were subjected to immunoprecipitation using an antibody against the C-terminal of APP. Lysates (Input) and precipitated proteins (IP) were subjected to SDS-PAGE and analyzed by Western blotting (WB) for APP or SorLA, as indicated. B, SH-SY5Y cells that were co-transfected with the CD8-SorLA-WT-tail and SorLA-KKK constructs were stained for the extracellular domains of the CD8 reporter (green) or for SorLA (red). Arrowheads indicate overlapping receptor distribution. IF, immunofluorescence. C, SorLA and endogenous APP levels in extracts (Cells) or in the conditioned medium (Secreted) of SH-SY5Y cells stably transfected with either SorLA-WT or SorLA-KKK were analyzed by SDS-PAGE and Western blotting with the indicated antibodies. The actin level in cells was used as a control for loading equal amounts of protein in each lane. Migration of immature and mature APP is indicated by a dash and a bracket, respectively. Asterisks indicate a unique APP glycosylation variant specific for the cells expressing the SorLA mutants. D and E, amount of sAPPα and Aβ40 products in the medium was determined by densitometric scanning of Western blots, as shown in C, or by ELISA, and was expressed as a percentage of processing products from nontransfected SH-SY5Y cells (set to 100%). The data are presented as the mean ± S.E. of four individual experiments. F, ratio of APP* to total fl-APP was determined by Western blot quantification and expressed relative to the ratio observed for nontransfected cells. Statistical significance is as follows: **, p < 0.01; ***, p < 0.001; ns, not significant (p > 0.05).
We then tested the cellular distribution of SorLA-KKK by immunocytochemistry. For this purpose, cells that had been transfected with the mutant construct and the CD8-SorLA-WT-tail reporter were studied. Cells that were co-stained for the luminal domains of SorLA and CD8 revealed a strong overlap, indicating that the main fraction of the two proteins localized to the same intracellular vesicles in the perinuclear region (Fig. 6B). Additionally, the clear co-localization of SorLA-KKK with mannosidase II suggested that the cellular distribution of the mutant was indistinguishable from SorLA-WT (data not shown). However, shedding of the mutant extracellular domain was reduced compared with SorLA-WT, which is similar to the situation observed for SorLA-DDD and indicates reduced trafficking of SorLA-KKK to the cell surface as well.
We assessed the APP binding of SorLA-KKK by co-immunoprecipitation analysis. Extracts from HEK293 cells that were transfected with APP in combination with either SorLA-WT or SorLA-KKK were precipitated using anti-APP antibodies in combination with GammaBind beads. The precipitates were resolved by SDS-PAGE analysis, and bound SorLA was subsequently detected by Western blotting (Fig. 6A). We observed a strong SorLA signal from cells expressing SorLA-KKK, suggesting that substitution of residues Leu-Ile-Met to lysine residues leaves the ability to form complexes between APP and SorLA intact.
Next, we examined how the KKK mutant influences APP processing. Investigating the full-length APP by Western blotting of cell extracts revealed an accumulation of a mature and glycosylated variant in the presence of SorLA-KKK, similar to cells expressing SorLA-DDD, which was again distinctly different from the mature APP forms seen in SorLA-WT and nontransfected cells (Fig. 6C). Quantification of the Western blots (Fig. 6F) revealed a significant increase in APP* in the SorLA-KKK-expressing cells (147.4 ± 9.23%, p = 0.0007), whereas expression of SorLA-WT did not result in APP* glycosylation (104.5 ± 9.84%, not significant) compared with the nontransfected cells (level of APP* set to 100%). Again, these data suggest an effect on the APP metabolites. We therefore analyzed the processing products in conditioned medium from our cells. As expected, we observed a significant decrease in the amount of soluble sAPPα (Fig. 6D) secreted to the medium from SorLA-KKK-expressing cells compared with nontransfected cells.
The production of Aβ was strongly decreased in cells expressing SorLA-KKK compared with nontransfected and SorLA-WT-expressing cells (Fig. 6E). In fact, the level of Aβ was below the detection limit of our ELISA when we used identical volumes of medium. To reliably quantify the level Aβ in SorLA-KKK-expressing cells, we had to measure larger volumes of media, suggesting a very pronounced effect of the KKK mutation on the amyloidogenic cleavages, which had not been observed for any of our other cell lines expressing SorLA variants. Interestingly, similar to what we had observed for SorLA-DDD cells, we observed a shift by SDS-PAGE in the migration of sAPPα produced from SorLA-KKK-expressing cells compared with sAPPα from either nontransfected or SorLA-WT-expressing cells (Fig. 6C).
To investigate whether the observed changes in sAPPα were caused by different glycosylation patterns, we performed deglycosylation experiments using sAPPα-containing medium from our SH-SY5Y cell lines. O-Glycosidase was used in combination with different neuraminidases to release glycans attached to serine or threonine residues of sAPPα. Neuraminidase treatment is necessary because O-glycosidase activity is prevented by substituents on the GalNAc disaccharide directly attached to the amino acid. Accordingly, conditioned media from SH-SY5Y cells expressing either SorLA-WT or SorLA-KKK were treated with O-glycosidase and neuraminidases specific for different α-linkages of the second carbon of sialic acid to the sugar chains in the glycan, followed by separation by SDS-PAGE and Western blotting (Fig. 7A). Neither treatment with neuraminidases specific for α-2,3- or α-2,8-linkages nor treatment with an enzyme that cleaves both α-2,3- and α-2,6-linkages was able to remove the sialic acids, inhibiting further cleavage by O-glycosidase (Fig. 7A). However, O-glycosidase with a neuraminidase with a broad specificity capable of removing α-2,3-, α-2,6-, and α-2,8-sialic acids, produced sAPPα with identical migration from both SorLA-WT- and SorLA-KKK-expressing SH-SY5Y cells. Interestingly, others have also shown that the saccharides of sAPP require a neuraminidase that cleaves the sialic acids in α-2,3-, α-2,6-, and α-2,8-linkages (50). Therefore, the sAPPα products do not seem to differ in their amino acid composition. Instead, SorLA-KKK and SorLA-DDD change the terminal sialic acid modifications of the residues within the APP extracellular domain. As a control experiment, we also compared the glycosylation of sAPPα in media from SorLA-ΔCR-expressing and nontransfected cells. As seen in Fig. 7A, there is no difference between the migration of sAPPα from these two cell lines, which is consistent with our finding that SorLA-ΔCR neither binds to nor affects the processing of APP.
FIGURE 7.
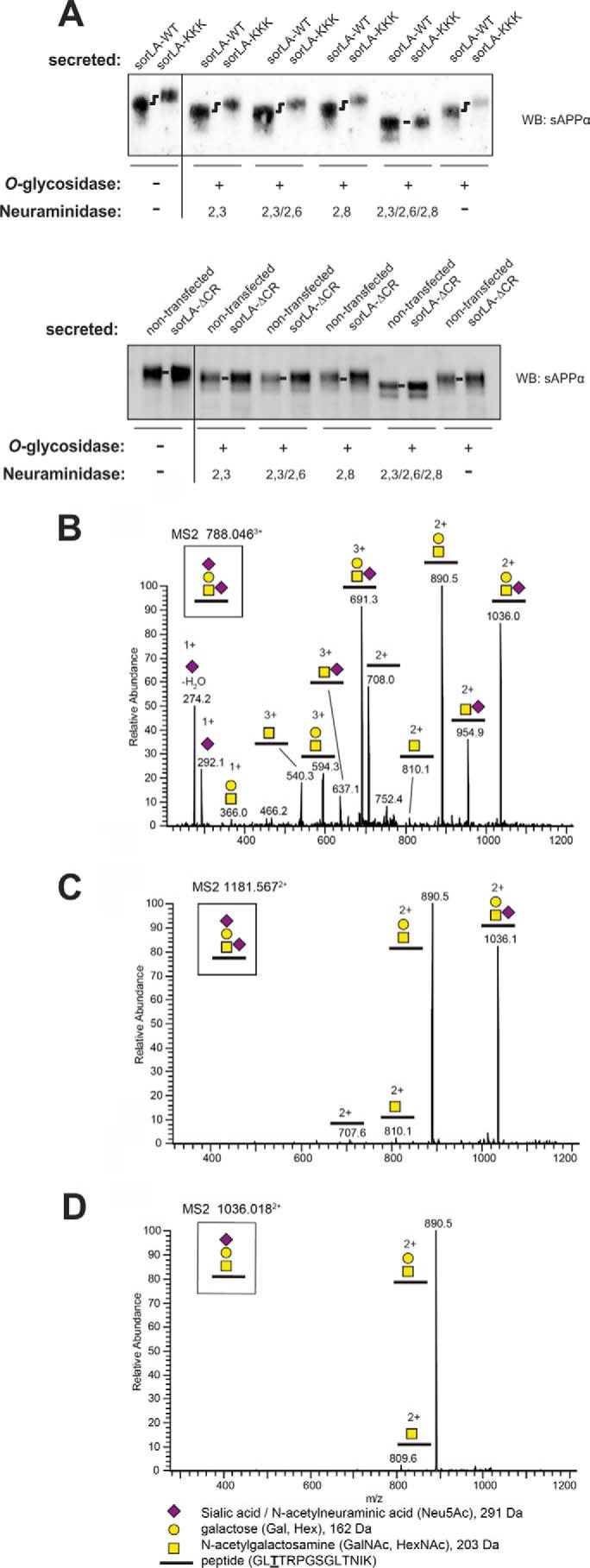
O-Linked glycosylation of sAPPα. A, deglycosylation analysis of the conditioned medium from nontransfected SH-SY5Y cells or from SH-SY5Y cells expressing SorLA-ΔCR (lower part), SorLA-WT, or SorLA-KKK (upper part). Samples were analyzed by BisTris SDS-PAGE and Western blotting (WB) for sAPPα, showing that treatment with O-glycosidase in combination with a 2,3/2,6/2,8-specific neuraminidase leads to identical migration patterns of sAPPα from SorLA-WT- and SorLA-KKK-expressing cell lines. B–D, annotated collision-induced dissociation MS2 spectra of O-glycosylated peptides from sAPP of the disialylated [M + 3H]3+ precursor at m/z 788.046 (B), the disialylated [M + 2H]2+ precursor at m/z 1181.567 (C), and monosialylated [M + 2H]2+ precursor at m/z 1036.018 (D). The precursor structures are shown in B.
To determine the composition and attachment site of the APP glycans, we immunoprecipitated APP from conditioned media using the 6E10 antibody, separated the precipitated fraction by SDS-PAGE, and performed in-gel trypsin digestion and liquid chromatography-tandem mass spectrometry (LC-MS/MS) to identify tryptic peptides and glycosylated peptides from APP. The major glycan on sAPPα from nontransfected SH-SY5Y cells contained a Neu5AcHex(Neu5Ac)HexNAc structure, which supports the disialylated core-1 structure (Neu5Acα3Galβ3(Neu5Acα6)GalNAcβ1-O-) (Fig. 7, B and C). A monosialylated core-1 glycoform was also observed (Fig. 7D). Only the O-glycosylated peptide GLTTRPGSGLTNIK, which probably corresponds to glycosylation of Thr-651 (underlined), was observed (the numbering is according to the APP770 isoform). GalNAc modification of Thr-651 has also been reported in APP produced from CHO cells, suggesting that this site is generally glycosylated (51). No other glycoforms were observed. Further glycosylation of Tyr-681 and the tryptic peptide Thr-652, Thr-659, and Thr-663 had previously been identified on sAPP in cerebrospinal fluid (52). Similar studies showed an identical glycan structure of Thr-651 using media from SorLA-DDD-transfected cells, but we were unable to detect other glycosylated peptides (data not shown). Accordingly, the combined data from the neuraminidase assay and the mass spectrometry analysis confirm differences in the O-linked glycosylation of APP but suggest the existence of a novel site of APP glycosylation exposed in cells transfected with SorLA-DDD and SorLA-KKK. Unfortunately, this site is located somewhere within the sAPPα sequence, which was not recoverable after the tryptic digests.
We were surprised by our findings that both SorLA-DDD and SorLA-KKK were able to change the glycosylation pattern of APP, but only SorLA-KKK significantly changed the processing of APP. Furthermore, both receptor mutants indicated a stronger binding to APP when tested in the immunoprecipitation studies performed at neutral pH. To understand this difference in mutant behavior, we produced CR(5–8) proteins carrying either the DDD or KKK mutation and tested the APP binding properties of these mutant proteins using SPR analysis. In a first set of experiments, we immobilized equal amounts of CR(5–8)-WT and CR(5–8)-DDD to biosensor chips and tested binding of 500 nm sAPP. In line with the stronger binding of SorLA-DDD to fl-APP at pH 7.4, we also found a slower dissociation of the complex between sAPP with CR(5–8)-DDD compared with CR(5–8)-WT at pH 7.4. However, when we repeated the assay using pH 6.0 and pH 5.0 for the dissociation, we observed a much more comparable interaction (Fig. 8A) where the dissociation becomes indistinguishable at pH 5.0. Running concentration series of sAPP to the biosensor chip at pH 5.0 confirmed that both SorLA fragments bind to sAPP with an identical affinity of 4 nm at pH 5.0 (data not shown).
FIGURE 8.
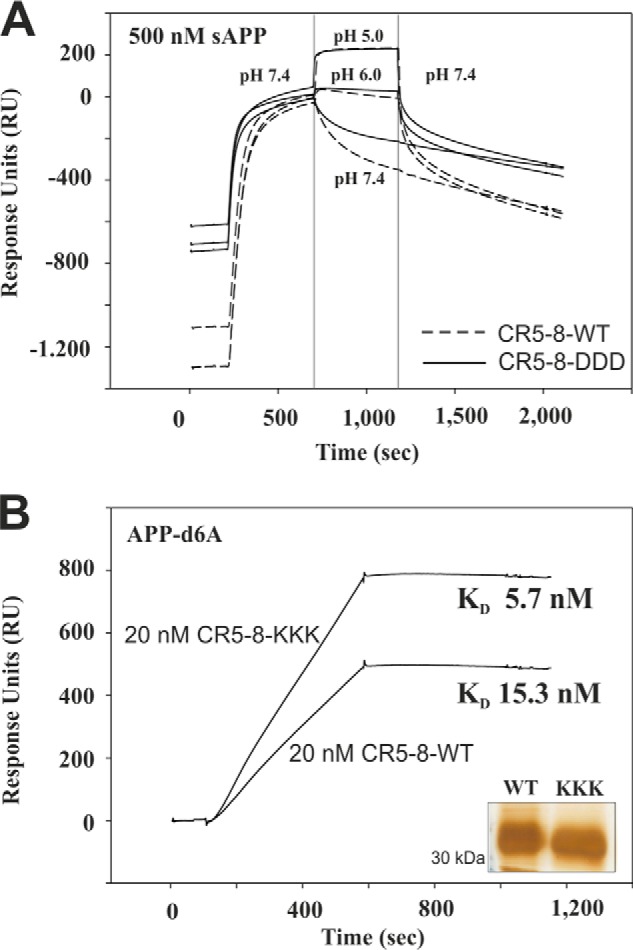
SPR analysis of CR(5–8) mutants binding to sAPP/APP-d6A at acidic pH. A, 500 nm sAPP was associated (200–700 s) to immobilized CR(5–8)-WT (broken lines) or CR(5–8)-DDD (solid lines) at pH 7.4. Dissociation was then followed at different pH values of 5.0, 6.0, and 7.4 (700–1200 s), before pH was adjusted to pH 7.4 for the remaining dissociation phase (1,200–2,000 s). All curves are zeroed at 700 s to more clearly highlight the strong difference in complex dissociation at pH 7.4, whereas there is no difference at pH 5.0. B, representative sensorgrams showing binding of 20 nm CR(5–8)-WT and 20 nm CR(5–8)-KKK to immobilized APP-d6A at pH 5.0. Concentration series of both proteins at 10, 20, 30, 40, 50, 100, and 200 nm (data not shown) were fitted using a 1:1 Langmuir binding model, showing a significant difference in their affinity at acidic pH with KD values 15.3 and 5.7 nm for CR(5–8)-WT and CR(5–8)-KKK, respectively. The inset shows silver-stained SDS-PAGE analysis of equal amounts of the purified CR(5–8)-WT and CR(5–8)-KKK proteins.
In contrast, CR(5–8)-KKK showed a stronger interaction with APP-d6A compared with CR(5–8)-WT at acidic pH (Fig. 8B). For this experiment, we used immobilized APP-d6A and tested concentration series of CR(5–8)-WT and CR(5–8)-KKK at 10, 20, 30, 40, 50, 100, and 200 nm binding at pH 5.0. Fitting of these sensorgrams using the 1:1 Langmuir binding model and BIAevaluation software showed that CR(5–8)-KKK binds 2.7-fold stronger to APP-d6A than CR(5–8)-WT, KD 5.7 nm, and KD 15.3 nm, respectively. These data collectively suggest that SorLA binding to APP at pH 5.0 determines the effect on APP processing, whereas the binding profiles at pH 7.4 could be more relevant for the observed change in APP glycosylation.
DISCUSSION
Binding Site Characterization
In this study, we investigated the binding between APP and SorLA, focusing on the role of the CR-domains. We found no other binding sites for APP outside the CR-cluster in SorLA either within the cytoplasmic tail, previously suggested as a possible linker between the two proteins (36), or within other regions of the composite SorLA extracellular domain. This result was surprising, as the VPS10p domains from the homologous receptors sortilin and SorCS1 have recently been suggested as novel APP interaction domains (29, 31, 53). For SorCS1, it is possible that the leucine-rich domain also contributes to the binding of APP. However, the limited sequence similarity between the VPS10p receptors might explain why APP only binds to some of the members via the VPS10p domains (54). The unique binding of APP to the SorLA CR-domains might explain the different roles of SorLA, sortilin, and SorCS1 in APP sorting (29, 53).
Spoelgen et al. (36) previously identified a binding site in the cytoplasmic tail. We speculate that the use of different cell lines (SH-SY5Y cells in our study versus N2a cells by Spoelgen et al. (36)) may explain why we see no interaction between the cytoplasmic tails. As mentioned by Spoelgen et al. (36), the cytosolic tails of SorLA and APP most likely associate with adaptor proteins that mediate the interaction, and the array of available adaptors may vary between different cell lines, suggesting a possible explanation for the observed difference.
LRP1 and LRP1B are examples of receptor proteins that also bind to APP via their CR-domains (55–58). The physiological function of these interactions is not fully understood, but the current model suggests that these receptors bind APP at the cell surface and release APP in the endosome to regulate its internalization (57–61). Based on our previous work, we have proposed a model in which SorLA does not contribute to APP endocytosis but instead associates with APP in intracellular vesicles (10, 22, 23). This suggestion raised the interesting question of how SorLA remains associated with APP in a more acidic environment, whereas APP dissociates from other CR-domains (e.g. LRP1) upon entry to the endosome, despite a strong structural conservation of all known CR-domain structures. Accordingly, we attempted to determine the mechanism by which SorLA acts as an intracellular sorting determinant for APP in the more acidic Golgi and endosome compartments, which have a pH range of 5.5–6.7 (62). We found that the binding of APP to the CR-domains of SorLA was indeed 10-fold stronger at pH 6.0 than at pH 7.4. Additionally, the affinity we determined at low pH indicates a more physiologically relevant interaction than the moderate affinity (KD ∼200 nm) we previously reported between SorLA and APP at neutral pH (10, 21).
We identified the central CR(5–8) region of SorLA as a binding site for APP. The CR(5–8) fragment is clearly distinct from the other CR-domains known to be involved in direct ligand binding, as the solved structures of such CR·ligand complexes all reveal negatively charged fingerprint residues (i.e. aspartates) binding to ligands with exposed basic residues (39, 40). The APP·CR(5–8) complex does not follow this minimal motif, as CR5, CR6, and CR7 contain hydrophobic side chains at the position where acidic fingerprint residues are normally found. However, substitution of these three fingerprint residues to either DDD or KKK did not abolish binding to APP, suggesting that other residues are also involved in complex formation between APP and SorLA. Therefore, it is highly interesting that a recent report identified a SorLA variant (N1358S) in patients with early-onset AD (17), as the Asn-1358 residue is located in CR7. We speculate that this residue might be part of the APP-binding site in SorLA and that the identified mutation might directly impair APP binding to SorLA, resulting in elevated Aβ production compared with neurons containing nonmutated SorLA. However, no attempts were made to investigate this hypothesis in this study.
Surprisingly, we observed that the presence of the first four CR-domains reduced binding to APP-d6A when we tested interactions with the isolated CR-clusters, suggesting that CR(1–4) might present steric hindrance to the CR(5–8)-binding site (Fig. 3). This possibility raises the question how APP binds to the full-length receptor. We hypothesize that the overall folding of full-length SorLA leads to exposure of CR(5–8) and that the other CR-domains may be engaged in intramolecular interactions to ensure the correct overall conformation. However, when expressed in the absence of flanking structural restrictions, CR(1–4) is not involved in maintaining the overall folded structure and may present steric hindrance for proper ligand binding to CR(5–8) (Fig. 3D). By SPR analysis, we found that an antibody directed against residues Asp-1193–Ile-1310, which encompasses CR5 and CR6, decreased the binding of APP to SorLA by ∼25%, suggesting that this region indeed plays an important role in the association of full-length SorLA with APP (data not shown).
Model of SorLA Activity in APP Metabolism
Using SorLA-ΔCR, we provide evidence here for a direct role of SorLA in APP metabolism and challenge the hypothesis that SorLA decreases Aβ production by competing as an alternative secretase substrate. The ectodomain of SorLA can be released by a metalloprotease with α-secretase activity (63, 64), and SorLA is processed by γ-secretase activity (65, 66) as well as proposed to be a substrate for BACE1 (67). Therefore, SorLA could potentially be a competitive substrate for the enzymes responsible for APP cleavage, suggesting one mechanism for how SorLA expression might inhibit APP proteolysis. However, SorLA-ΔCR is shed as efficiently from SH-SY5Y cells as SorLA-WT and shows no evidence of intracellular mislocalization. Nevertheless, we found that SorLA-ΔCR did not decrease the processing of APP, providing strong evidence of a specific role for SorLA in determining the intracellular trafficking and fate of APP. However, SorLA has also been shown to bind BACE (36), and it is therefore possible that APP and BACE compete for binding to the same region of SorLA. Thus, our mutants may also affect the ability to bind and sort BACE, although we believe that APP is the preferred binding partner of SorLA. This situation was not addressed in this study but would be very interesting to determine in future studies.
The exact mechanism of how SorLA activity influences APP metabolism is not completely understood. Thus far, two models have been proposed that account equally well for the observed protective function of SorLA in APP processing (20). One model suggests that SorLA regulates APP control and transport out of the Golgi, and the other model describes SorLA as a molecule that assists APP retrograde transport away from amyloidogenic processing in the endocytic pathway (68). As these models are not mutually exclusive, it is possible that they are both correct.
Our current data mainly support a model in which SorLA is best described as an APP retention factor at the Golgi, influencing release of the precursor into the regular processing pathway. Two observations from our study indicate that SorLA activity is related to APP localization in the secretory pathway. First, SorLA-DDD is as efficient as SorLA-WT in decreasing the processing of APP even though there is less mutant receptor at the plasma membrane, suggesting a function independent of the level available for retrograde trafficking. Second, both SorLA-DDD and SorLA-KKK influence APP glycosylation, a process that is highly regulated by passage through various Golgi stacks containing the enzymes responsible for glycosylation of the complex (69). However, although our data suggest a dominant function of SorLA in the Golgi apparatus where maturation of APP occurs, we cannot preclude that SorLA also plays an important role in the endosome to trans-Golgi network trafficking of APP.
Most interestingly, our data using CR(5–8)-DDD and CR(5–8)-KKK suggest that the strength of the interaction between SorLA and APP at acidic pH is a prime determinant for the protective effect on APP processing. However, the change in APP glycosylation is observed upon expression of both SorLA-DDD and SorLA-KKK and can thus be speculated to be an effect of disrupting a function normally held by the hydrophobic fingerprint residues.
In many cases, post-translation modifications such as glycosylation are important for regulating the intracellular sorting processes and stabilizing against proteolysis (70). Glycosylation, particularly sialylation, of APP has been proposed to influence the processing pathway of APP by determining its passage from the Golgi to the cell membrane (50, 71, 72). Additionally, a study by Tomita et al. (73) concluded that APP is only cleaved after maturation by O-glycosylation, which occurs during the transport of APP through the Golgi complex. These findings are all consistent with SorLA (and mutants) decreasing both the amyloidogenic and nonamyloidogenic processing of APP and functioning in localizing APP to the early steps of the secretory pathway. However, we find that SorLA binds to APP the most strongly at low pH, which is found in the vesicles of the Golgi network and within the endocytic pathway. Accordingly, further studies are necessary to resolve the role of SorLA in the retrograde sorting of APP.
Two recent studies have also reported that APP glycosylation is an important determinant of its processing. First, expression of the Golgi-localized protein TMEM59 induced APP retention in the Golgi, influencing complex glycosylation and decreasing Aβ generation (74). Second, a shift in APP glycosylation was observed for APP harboring a double mutation (K476A/K477A), leading to reduced processing (75). Interestingly, these findings are very similar to our data, which demonstrate both a change in APP glycosylation in the presence of SorLA mutants and a pronounced decrease of Aβ production. However, it is not yet known whether the two lysines in APP are important for binding to SorLA or whether SorLA associates with TMEM59.
The mechanism how SorLA mutants alter APP glycosylation is still not known, but the altered migration of both cellular APP and secreted sAPPα on SDS-PAGE, lost only after treatment with a combination of O-glycosidase and a mixture of 2,3/2,6/2,8-neuraminidase, suggests a difference in O-glycosylation. However, mass spectrometry identified a common sialylated glycan structure attached to Thr-651 of APP. We speculate that substitution of the three hydrophobic fingerprint residues leads to a change in the conformation of the complex between SorLA and APP, whereby a novel site for attachment of O-glycans is exposed. Alternatively, a novel structural isomer of the O-glycan, not revealed from our MS analysis, may be synthesized in the cells expressing mutated SorLA. Only further experiments will settle these issues.
Acknowledgments
We are indebted to Marit Nyholm Nielsen, Anne Marie Bundsgaard, and Sandra Bonnesen for expert technical assistance. We thank Anders Nykjær for critical reading of the manuscript and helpful discussions.
This work was supported by grants from the Lundbeck Foundation, the Novo Nordisk Foundation, the Augustinus Foundation, the Danish Alzheimer Research Foundation, the Aase and Einar Danielsen Foundation, the Hartman Foundation, and the Hørslev Foundation.
- AD
- Alzheimer disease
- APP
- amyloid precursor protein
- CR
- complement-type repeat
- Aβ
- amyloid-β
- sAPP
- soluble APP
- SPR
- surface plasmon resonance
- BisTris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- BACE
- β-site APP-cleaving enzyme.
REFERENCES
- 1. Blennow K., de Leon M. J., Zetterberg H. (2006) Alzheimer's disease. Lancet 368, 387–403 [DOI] [PubMed] [Google Scholar]
- 2. Small S. A., Gandy S. (2006) Sorting through the cell biology of Alzheimer's disease: intracellular pathways to pathogenesis. Neuron 52, 15–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Sannerud R., Annaert W. (2009) Trafficking, a key player in regulated intramembrane proteolysis. Semin. Cell Dev. Biol. 20, 183–190 [DOI] [PubMed] [Google Scholar]
- 4. Haass C., Kaether C., Thinakaran G., Sisodia S. (2012) Trafficking and proteolytic processing of APP. Cold Spring Harb. Perspect Med. 2, a006270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Weidemann A., König G., Bunke D., Fischer P., Salbaum J. M., Masters C. L., Beyreuther K. (1989) Identification, biogenesis, and localization of precursors of Alzheimer's disease A4 amyloid protein. Cell 57, 115–126 [DOI] [PubMed] [Google Scholar]
- 6. Oltersdorf T., Ward P. J., Henriksson T., Beattie E. C., Neve R., Lieberburg I., Fritz L. C. (1990) The Alzheimer amyloid precursor protein. Identification of a stable intermediate in the biosynthetic/degradative pathway. J. Biol. Chem. 265, 4492–4497 [PubMed] [Google Scholar]
- 7. Motoi Y., Aizawa T., Haga S., Nakamura S., Namba Y., Ikeda K. (1999) Neuronal localization of a novel mosaic apolipoprotein E receptor, LR11, in rat and human brain. Brain Res. 833, 209–215 [DOI] [PubMed] [Google Scholar]
- 8. Hermans-Borgmeyer I., Hampe W., Schinke B., Methner A., Nykjaer A., Süsens U., Fenger U., Herbarth B., Schaller H. C. (1998) Unique expression pattern of a novel mosaic receptor in the developing cerebral cortex. Mech. Dev. 70, 65–76 [DOI] [PubMed] [Google Scholar]
- 9. Scherzer C. R., Offe K., Gearing M., Rees H. D., Fang G., Heilman C. J., Schaller C., Bujo H., Levey A. I., Lah J. J. (2004) ApoE receptor LR11 in Alzheimer's disease: gene profiling of lymphoblasts mirrors changes in the brain. Arch. Neurol. 61, 1200–1205 [DOI] [PubMed] [Google Scholar]
- 10. Andersen O. M., Reiche J., Schmidt V., Gotthardt M., Spoelgen R., Behlke J., von Arnim C. A., Breiderhoff T., Jansen P., Wu X., Bales K. R., Cappai R., Masters C. L., Gliemann J., Mufson E. J., Hyman B. T., Paul S. M., Nykjaer A., Willnow T. E. (2005) SorLA/LR11, a neuronal sorting receptor that regulates processing of the amyloid precursor protein. Proc. Natl. Acad. Sci. U.S.A. 102, 13461–13466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Offe K., Dodson S. E., Shoemaker J. T., Fritz J. J., Gearing M., Levey A. I., Lah J. J. (2006) The lipoprotein receptor LR11 regulates amyloid b production and amyloid precursor protein traffic in endosomal compartments. J. Neurosci. 26, 1596–1603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Dodson S. E., Gearing M., Lippa C. F., Montine T. J., Levey A. I., Lah J. J. (2006) LR11/SorLA expression is reduced in sporadic Alzheimer disease but not in familial Alzheimer disease. J. Neuropathol. Exp. Neurol. 65, 866–872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Sager K. L., Wuu J., Leurgans S. E., Rees H. D., Gearing M., Mufson E. J., Levey A. I., Lah J. J. (2007) Neuronal LR11/SorLA expression is reduced in mild cognitive impairment. Ann. Neurol. 62, 640–647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Rogaeva E., Meng Y., Lee J. H., Gu Y., Kawarai T., Zou F., Katayama T., Baldwin C. T., Cheng R., Hasegawa H., Chen F., Shibata N., Lunetta K. L., Pardossi-Piquard R., Bohm C., Wakutani Y., Cupples L. A., Cuenco K. T., Green R. C., Pinessi L., Rainero I., Sorbi S., Bruni A., Duara R., Friedland R. P., Inzelberg R., Hampe W., Bujo H., Song Y. Q., Andersen O. M., Willnow T. E., Graff-Radford N., Petersen R. C., Dickson D., Der S. D., Fraser P. E., Schmitt-Ulms G., Younkin S., Mayeux R., Farrer L. A., St George-Hyslop P. (2007) The neuronal sortilin-related receptor SORL1 is genetically associated with Alzheimer disease. Nat. Genet. 39, 168–177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Reitz C., Cheng R., Rogaeva E., Lee J. H., Tokuhiro S., Zou F., Bettens K., Sleegers K., Tan E. K., Kimura R., Shibata N., Arai H., Kamboh M. I., Prince J. A., Maier W., Riemenschneider M., Owen M., Harold D., Hollingworth P., Cellini E., Sorbi S., Nacmias B., Takeda M., Pericak-Vance M. A., Haines J. L., Younkin S., Williams J., van Broeckhoven C., Farrer L. A., St George-Hyslop P. H., Mayeux R. (2011) Meta-analysis of the association between variants in SORL1 and Alzheimer disease. Arch. Neurol. 68, 99–106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Reitz C., Tosto G., Vardarajan B., Rogaeva E., Ghani M., Rogers R. S., Conrad C., Haines J. L., Pericak-Vance M. A., Fallin M. D., Foroud T., Farrer L. A., Schellenberg G. D., George-Hyslop P. S., Mayeux R. (2013) Independent and epistatic effects of variants in VPS10-d receptors on Alzheimer disease risk and processing of the amyloid precursor protein (APP). Transl. Psychiatry 3, e256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Pottier C., Hannequin D., Coutant S., Rovelet-Lecrux A., Wallon D., Rousseau S., Legallic S., Paquet C., Bombois S., Pariente J., Thomas-Anterion C., Michon A., Croisile B., Etcharry-Bouyx F., Berr C., Dartigues J. F., Amouyel P., Dauchel H., Boutoleau-Bretonnière C., Thauvin C., Frebourg T., Lambert J. C., Campion D., and PHRC GMAJ Collaborators. (2012) High frequency of potentially pathogenic SORL1 mutations in autosomal dominant early-onset Alzheimer disease. Mol. Psychiatry 17, 875–879 [DOI] [PubMed] [Google Scholar]
- 18. Rohe M., Carlo A. S., Breyhan H., Sporbert A., Militz D., Schmidt V., Wozny C., Harmeier A., Erdmann B., Bales K. R., Wolf S., Kempermann G., Paul S. M., Schmitz D., Bayer T. A., Willnow T. E., Andersen O. M. (2008) Sortilin-related receptor with A-type repeats (SORLA) affects the amyloid precursor protein-dependent stimulation of ERK signaling and adult neurogenesis. J. Biol. Chem. 283, 14826–14834 [DOI] [PubMed] [Google Scholar]
- 19. Dodson S. E., Andersen O. M., Karmali V., Fritz J. J., Cheng D., Peng J., Levey A. I., Willnow T. E., Lah J. J. (2008) Loss of LR11/SORLA enhances early pathology in a mouse model of amyloidosis: evidence for a proximal role in Alzheimer's disease. J. Neurosci. 28, 12877–12886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Willnow T. E., Andersen O. M. (2013) Sorting receptor SORLA–a trafficking path to avoid Alzheimer disease. J. Cell Sci. 126, 2751–2760 [DOI] [PubMed] [Google Scholar]
- 21. Andersen O. M., Schmidt V., Spoelgen R., Gliemann J., Behlke J., Galatis D., McKinstry W. J., Parker M. W., Masters C. L., Hyman B. T., Cappai R., Willnow T. E. (2006) Molecular dissection of the interaction between APP and its neuronal trafficking receptor SorLA/LR11. Biochemistry 45, 2618–2628 [DOI] [PubMed] [Google Scholar]
- 22. Schmidt V., Sporbert A., Rohe M., Reimer T., Rehm A., Andersen O. M., Willnow T. E. (2007) SorLA/LR11 regulates processing of amyloid precursor protein via interaction with adaptors GGA and PACS-1. J. Biol. Chem. 282, 32956–32964 [DOI] [PubMed] [Google Scholar]
- 23. Fjorback A. W., Seaman M., Gustafsen C., Mehmedbasic A., Gokool S., Wu C., Militz D., Schmidt V., Madsen P., Nyengaard J. R., Willnow T. E., Christensen E. I., Mobley W. B., Nykjær A., Andersen O. M. (2012) Retromer binds the FANSHY sorting motif in SorLA to regulate amyloid precursor protein sorting and processing. J. Neurosci. 32, 1467–1480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Small S. A., Kent K., Pierce A., Leung C., Kang M. S., Okada H., Honig L., Vonsattel J. P., Kim T. W. (2005) Model-guided microarray implicates the retromer complex in Alzheimer's disease. Ann. Neurol. 58, 909–919 [DOI] [PubMed] [Google Scholar]
- 25. Bhalla A., Vetanovetz C. P., Morel E., Chamoun Z., Di Paolo G., Small S. A. (2012) The location and trafficking routes of the neuronal retromer and its role in amyloid precursor protein transport. Neurobiol. Dis. 47, 126–134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Jacobsen L., Madsen P., Moestrup S. K., Lund A. H., Tommerup N., Nykjaer A., Sottrup-Jensen L., Gliemann J., Petersen C. M. (1996) Molecular characterization of a novel human hybrid-type receptor that binds the α2-macroglobulin receptor-associated protein. J. Biol. Chem. 271, 31379–31383 [DOI] [PubMed] [Google Scholar]
- 27. Yamazaki H., Bujo H., Kusunoki J., Seimiya K., Kanaki T., Morisaki N., Schneider W. J., Saito Y. (1996) Elements of neural adhesion molecules and a yeast vacuolar protein sorting receptor are present in a novel mammalian low density lipoprotein receptor family member. J. Biol. Chem. 271, 24761–24768 [DOI] [PubMed] [Google Scholar]
- 28. Mörwald S., Yamazaki H., Bujo H., Kusunoki J., Kanaki T., Seimiya K., Morisaki N., Nimpf J., Schneider W. J., Saito Y. (1997) A novel mosaic protein containing LDL receptor elements is highly conserved in humans and chickens. Arterioscler. Thromb. Vasc. Biol. 17, 996–1002 [DOI] [PubMed] [Google Scholar]
- 29. Gustafsen C., Glerup S., Pallesen L. T., Olsen D., Andersen O. M., Nykjær A., Madsen P., Petersen C. M. (2013) Sortilin and SorLA display distinct roles in processing and trafficking of amyloid precursor protein. J. Neurosci. 33, 64–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lane R. F., Raines S. M., Steele J. W., Ehrlich M. E., Lah J. A., Small S. A., Tanzi R. E., Attie A. D., Gandy S. (2010) Diabetes-associated SorCS1 regulates Alzheimer's amyloid-β metabolism: evidence for involvement of SORL1 and the retromer complex. J. Neurosci. 30, 13110–13115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Reitz C., Tokuhiro S., Clark L. N., Conrad C., Vonsattel J. P., Hazrati L. N., Palotás A., Lantigua R., Medrano M., Z Jiménez-Velázquez I., Vardarajan B., Simkin I., Haines J. L., Pericak-Vance M. A., Farrer L. A., Lee J. H., Rogaeva E., George-Hyslop P. S., Mayeux R. (2011) SORCS1 alters amyloid precursor protein processing and variants may increase Alzheimer's disease risk. Ann. Neurol. 69, 47–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kaden D., Voigt P., Munter L. M., Bobowski K. D., Schaefer M., Multhaup G. (2009) Subcellular localization and dimerization of APLP1 are strikingly different from APP and APLP2. J. Cell Sci. 122, 368–377 [DOI] [PubMed] [Google Scholar]
- 33. Andersen O. M., Benhayon D., Curran T., Willnow T. E. (2003) Differential binding of ligands to the apolipoprotein e receptor 2. Biochemistry 42, 9355–9364 [DOI] [PubMed] [Google Scholar]
- 34. Jacobsen L., Madsen P., Jacobsen C., Nielsen M. S., Gliemann J., Petersen C. M. (2001) Activation and functional characterization of the mosaic receptor SorLA/LR11. J. Biol. Chem. 276, 22788–22796 [DOI] [PubMed] [Google Scholar]
- 35. Portelius E., Westman-Brinkmalm A., Zetterberg H., Blennow K. (2006) Determination of β-amyloid peptide signatures in cerebrospinal fluid using immunoprecipitation-mass spectrometry. J. Proteome Res. 5, 1010–1016 [DOI] [PubMed] [Google Scholar]
- 36. Spoelgen R., von Arnim C. A., Thomas A. V., Peltan I. D., Koker M., Deng A., Irizarry M. C., Andersen O. M., Willnow T. E., Hyman B. T. (2006) Interaction of the cytosolic domains of SorLA/LR11 with the amyloid precursor protein (APP) and β-secretase β-site APP-cleaving enzyme. J. Neurosci. 26, 418–428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Andersen O. M., Christensen L. L., Christensen P. A., Sørensen E. S., Jacobsen C., Moestrup S. K., Etzerodt M., Thogersen H. C. (2000) Identification of the minimal functional unit in the low density lipoprotein receptor-related protein for binding the receptor-associated protein (RAP). J. Biol. Chem. 275, 21017–21024 [DOI] [PubMed] [Google Scholar]
- 38. Andersen O. M., Schwarz F. P., Eisenstein E., Jacobsen C., Moestrup S. K., Etzerodt M., Thøgersen H. C. (2001) Dominant thermodynamic role of the third independent receptor binding site in the receptor-associated protein RAP. Biochemistry 40, 15408–15417 [DOI] [PubMed] [Google Scholar]
- 39. Jensen G. A., Andersen O. M., Bonvin A. M., Bjerrum-Bohr I., Etzerodt M., Thøgersen H. C., O'Shea C., Poulsen F. M., Kragelund B. B. (2006) Binding site structure of one LRP-RAP complex: implications for a common ligand-receptor binding motif. J. Mol. Biol. 362, 700–716 [DOI] [PubMed] [Google Scholar]
- 40. Fisher C., Beglova N., Blacklow S. C. (2006) Structure of an LDLR-RAP complex reveals a general mode for ligand recognition by lipoprotein receptors. Mol. Cell 22, 277–283 [DOI] [PubMed] [Google Scholar]
- 41. Verdaguer N., Fita I., Reithmayer M., Moser R., Blaas D. (2004) X-ray structure of a minor group human rhinovirus bound to a fragment of its cellular receptor protein. Nat. Struct. Mol. Biol. 11, 429–434 [DOI] [PubMed] [Google Scholar]
- 42. Yasui N., Nogi T., Kitao T., Nakano Y., Hattori M., Takagi J. (2007) Structure of a receptor-binding fragment of reelin and mutational analysis reveal a recognition mechanism similar to endocytic receptors. Proc. Natl. Acad. Sci. U.S.A. 104, 9988–9993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Yasui N., Nogi T., Takagi J. (2010) Structural basis for specific recognition of reelin by its receptors. Structure 18, 320–331 [DOI] [PubMed] [Google Scholar]
- 44. Beglov D., Lee C. J., De Biasio A., Kozakov D., Brenke R., Vajda S., Beglova N. (2009) Structural insights into recognition of b2-glycoprotein I by the lipoprotein receptors. Proteins 77, 940–949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Lee C. J., De Biasio A., Beglova N. (2010) Mode of interaction between b2GPI and lipoprotein receptors suggests mutually exclusive binding of b2GPI to the receptors and anionic phospholipids. Structure 18, 366–376 [DOI] [PubMed] [Google Scholar]
- 46. Guttman M., Prieto J. H., Croy J. E., Komives E. A. (2010) Decoding of lipoprotein-receptor interactions: properties of ligand binding modules governing interactions with apolipoprotein E. Biochemistry 49, 1207–1216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Guttman M., Prieto J. H., Handel T. M., Domaille P. J., Komives E. A. (2010) Structure of the minimal interface between ApoE and LRP. J. Mol. Biol. 398, 306–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Beglova N., Blacklow S. C. (2005) The LDL receptor: how acid pulls the trigger. Trends Biochem. Sci. 30, 309–317 [DOI] [PubMed] [Google Scholar]
- 49. Huang S., Henry L., Ho Y. K., Pownall H. J., Rudenko G. (2010) Mechanism of LDL binding and release probed by structure-based mutagenesis of the LDL receptor. J. Lipid Res. 51, 297–308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. McFarlane I., Georgopoulou N., Coughlan C. M., Gillian A. M., Breen K. C. (1999) The role of the protein glycosylation state in the control of cellular transport of the amyloid β precursor protein. Neuroscience 90, 15–25 [DOI] [PubMed] [Google Scholar]
- 51. Perdivara I., Petrovich R., Allinquant B., Alliquant B., Deterding L. J., Tomer K. B., Przybylski M. (2009) Elucidation of O-glycosylation structures of the β-amyloid precursor protein by liquid chromatography-mass spectrometry using electron transfer dissociation and collision induced dissociation. J. Proteome Res. 8, 631–642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Halim A., Brinkmalm G., Rüetschi U., Westman-Brinkmalm A., Portelius E., Zetterberg H., Blennow K., Larson G., Nilsson J. (2011) Site-specific characterization of threonine, serine, and tyrosine glycosylations of amyloid precursor protein/amyloid β-peptides in human cerebrospinal fluid. Proc. Natl. Acad. Sci. U.S.A. 108, 11848–11853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Lane R. F., Steele J. W., Cai D., Ehrlich M. E., Attie A. D., Gandy S. (2013) Protein sorting motifs in the cytoplasmic tail of SorCS1 control generation of Alzheimer's amyloid-β peptide. J. Neurosci. 33, 7099–7107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Hermey G. (2009) The Vps10p domain receptor family. Cell. Mol. Life Sci. 66, 2677–2689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Kounnas M. Z., Moir R. D., Rebeck G. W., Bush A. I., Argraves W. S., Tanzi R. E., Hyman B. T., Strickland D. K. (1995) LDL receptor-related protein, a multifunctional ApoE receptor, binds secreted β-amyloid precursor protein and mediates its degradation. Cell 82, 331–340 [DOI] [PubMed] [Google Scholar]
- 56. Knauer M. F., Orlando R. A., Glabe C. G. (1996) Cell surface APP751 forms complexes with protease nexin 2 ligands and is internalized via the low density lipoprotein receptor-related protein (LRP). Brain Res. 740, 6–14 [DOI] [PubMed] [Google Scholar]
- 57. Cam J. A., Zerbinatti C. V., Knisely J. M., Hecimovic S., Li Y., Bu G. (2004) The LDL receptor-related protein 1B retains β-APP at the cell surface and reduces amyloid-β peptide production. J. Biol. Chem. 279, 29639–29646 [DOI] [PubMed] [Google Scholar]
- 58. Cam J. A., Zerbinatti C. V., Li Y., Bu G. (2005) Rapid endocytosis of the low density lipoprotein receptor-related protein modulates cell surface distribution and processing of the β-amyloid precursor protein. J. Biol. Chem. 280, 15464–15470 [DOI] [PubMed] [Google Scholar]
- 59. Pietrzik C. U., Yoon I. S., Jaeger S., Busse T., Weggen S., Koo E. H. (2004) FE65 constitutes the functional link between the low-density lipoprotein receptor-related protein and the amyloid precursor protein. J. Neurosci. 24, 4259–4265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Fuentealba R. A., Barría M. I., Lee J., Cam J., Araya C., Escudero C. A., Inestrosa N. C., Bronfman F. C., Bu G., Marzolo M. P. (2007) ApoER2 expression increases Aβ production while decreasing amyloid precursor protein (APP) endocytosis: possible role in the partitioning of APP into lipid rafts and in the regulation of gamma-secretase activity. Mol. Neurodegener. 2, 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. He X., Cooley K., Chung C. H., Dashti N., Tang J. (2007) Apolipoprotein receptor 2 and X11a/b mediate apolipoprotein E-induced endocytosis of amyloid-β precursor protein and β-secretase, leading to amyloid-β production. J. Neurosci. 27, 4052–4060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Casey J. R., Grinstein S., Orlowski J. (2010) Sensors and regulators of intracellular pH. Nat. Rev. Mol. Cell Biol. 11, 50–61 [DOI] [PubMed] [Google Scholar]
- 63. Hampe W., Riedel I. B., Lintzel J., Bader C. O., Franke I., Schaller H. C. (2000) Ectodomain shedding, translocation and synthesis of SorLA are stimulated by its ligand head activator. J. Cell Sci. 113, 4475–4485 [DOI] [PubMed] [Google Scholar]
- 64. Hermey G., Sjøgaard S. S., Petersen C. M., Nykjaer A., Gliemann J. (2006) Tumour necrosis factor α-converting enzyme mediates ectodomain shedding of Vps10p domain receptor family members. Biochem. J. 395, 285–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Nyborg A. C., Ladd T. B., Zwizinski C. W., Lah J. J., Golde T. E. (2006) Sortilin, SorCS1b, and SorLA Vps10p sorting receptors, are novel γ-secretase substrates. Mol. Neurodegener. 10.1186/1750-1326-1-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Böhm C., Seibel N. M., Henkel B., Steiner H., Haass C., Hampe W. (2006) SorLA signaling by regulated intramembrane proteolysis. J. Biol. Chem. 281, 14547–14553 [DOI] [PubMed] [Google Scholar]
- 67. Hemming M. L., Elias J. E., Gygi S. P., Selkoe D. J. (2009) Identification of β-secretase (BACE1) substrates using quantitative proteomics. PLoS ONE 4, e8477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Lane R. F., Gatson J. W., Small S. A., Ehrlich M. E., Gandy S. (2010) Protein kinase C and rho activated coiled coil protein kinase 2 (ROCK2) modulate Alzheimer's APP metabolism and phosphorylation of the Vps10 domain protein, SorL1. Mol. Neurodegener. 5, 62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Dunphy W. G., Rothman J. E. (1985) Compartmental organization of the Golgi stack. Cell 42, 13–21 [DOI] [PubMed] [Google Scholar]
- 70. Påhlsson P., Shakin-Eshleman S. H., Spitalnik S. L. (1992) N-Linked glycosylation of β-amyloid precursor protein. Biochem. Biophys. Res. Commun. 189, 1667–1673 [DOI] [PubMed] [Google Scholar]
- 71. Graebert K. S., Popp G. M., Kehle T., Herzog V. (1995) Regulated O-glycosylation of the Alzheimer β-A4 amyloid precursor protein in thyrocytes. Eur. J. Cell Biol. 66, 39–46 [PubMed] [Google Scholar]
- 72. Georgopoulou N., McLaughlin M., McFarlane I., Breen K. C. (2001) The role of post-translational modification in beta-amyloid precursor protein processing. Biochem. Soc. Symp. 67, 23–36 [DOI] [PubMed] [Google Scholar]
- 73. Tomita S., Kirino Y., Suzuki T. (1998) Cleavage of Alzheimer's amyloid precursor protein (APP) by secretases occurs after O-glycosylation of APP in the protein secretory pathway. Identification of intracellular compartments in which APP cleavage occurs without using toxic agents that interfere with protein metabolism. J. Biol. Chem. 273, 6277–6284 [DOI] [PubMed] [Google Scholar]
- 74. Ullrich S., Münch A., Neumann S., Kremmer E., Tatzelt J., Lichtenthaler S. F. (2010) The novel membrane protein TMEM59 modulates complex glycosylation, cell surface expression, and secretion of the amyloid precursor protein. J. Biol. Chem. 285, 20664–20674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Xue Y., Lee S., Wang Y., Ha Y. (2011) Crystal structure of the E2 domain of amyloid precursor protein-like protein 1 in complex with sucrose octasulfate. J. Biol. Chem. 286, 29748–29757 [DOI] [PMC free article] [PubMed] [Google Scholar]