Background: Zelda initiates widespread transcription of the zygotic genome during embryogenesis.
Results: Zelda binds DNA using C-terminal zinc fingers and activates transcription through a low-complexity domain.
Conclusion: Transcriptional activation by Zelda is conserved in insects and uses domains we have identified to bind cis-regulatory regions and drive gene expression.
Significance: We provide the first insights into the functional domains of the essential activator Zelda.
Keywords: DNA Binding Protein, Drosophila, Embryo, Transcription, Zinc Finger, Maternal-to-Zygotic Transition, Zygotic Genome Activation
Abstract
Delayed transcriptional activation of the zygotic genome is a nearly universal phenomenon in metazoans. Immediately following fertilization, development is controlled by maternally deposited products, and it is not until later stages that widespread activation of the zygotic genome occurs. Although the mechanisms driving this genome activation are currently unknown, the transcriptional activator Zelda (ZLD) has been shown to be instrumental in driving this process in Drosophila melanogaster. Here we define functional domains of ZLD required for both DNA binding and transcriptional activation. We show that the C-terminal cluster of four zinc fingers mediates binding to TAGteam DNA elements in the promoters of early expressed genes. All four zinc fingers are required for this activity, and splice isoforms lacking three of the four zinc fingers fail to activate transcription. These truncated splice isoforms dominantly suppress activation by the full-length, embryonically expressed isoform. We map the transcriptional activation domain of ZLD to a central region characterized by low complexity. Despite relatively little sequence conservation within this domain, ZLD orthologs from Drosophila virilis, Anopheles gambiae, and Nasonia vitripennis activate transcription in D. melanogaster cells. Transcriptional activation by these ZLD orthologs suggests that ZLD functions through conserved interactions with a protein cofactor(s). We have identified distinct DNA-binding and activation domains within the critical transcription factor ZLD that controls the initial activation of the zygotic genome.
Introduction
During the first stages of embryogenesis, the zygotic genome is transcriptionally quiescent, and development is controlled by maternally deposited mRNAs and proteins (1, 2). Only at later cell cycles, after the cells have become totipotent, does the embryo begin to control its own development with the initiation of widespread transcription. This critical stage is termed the maternal-to-zygotic transition (MZT)2 and is characterized by a monumental reorganization of the transcriptional profile of the embryo (2). This process is required for future development, and failure to undergo this transition is lethal to the embryo. Despite the fact that the MZT is nearly universal among metazoans, the mechanisms governing this process and how they relate to the coordinated establishment of a totipotent state remain unknown.
The transcription factor Zelda (ZLD) was recently identified in the Drosophila melanogaster embryo as the first known global activator of zygotic gene expression (3). Maternally deposited zld mRNA is necessary for the transcriptional activation of hundreds of genes at the MZT, and ZLD is required for early embryonic development (3). Embryos lacking maternally contributed zld die at the MZT. ZLD binds to a set of related heptameric DNA sequence motifs, termed TAGteam elements (3–6), and these elements are enriched in the promoters and enhancers of genes expressed during the MZT (7–9). TAGteam elements are essential for early gene expression, and the number of TAGteam sites correlates with the levels and timing of gene expression (7, 9). Through its sequence-specific DNA-binding activity, ZLD localizes to the enhancers and promoters of thousands of genes activated over the MZT (5, 6). ZLD occupancy in vivo is governed largely by sequence, with 64% of the canonical TAGteam sequence, CAGGTAG, being occupied by ZLD in the early embryo (5). Therefore, in vivo ZLD occupancy is determined largely by sequence, and it is this sequence-specific binding that drives transcriptional activation of the zygotic genome. However, the mechanism by which ZLD mediates widespread transcriptional activation is not well understood.
In early embryos, ZLD is a 1596-amino acid protein with six C2H2 zinc fingers and stretches of highly repetitive amino acid sequences (low-complexity regions) (Fig. 1A) (3, 10). There are no other identifiable protein domains that might suggest an enzymatic activity. Although it has been demonstrated that a region containing the cluster of four C-terminal zinc fingers can bind DNA in vitro (11), it remains to be determined which, if any, of these zinc fingers are required for sequence-specific DNA binding. Therefore, the primary sequence of this large protein offers few clues as to how it mediates genome activation. Additional splice isoforms of zld have also been identified, and at least one is expressed both in larvae and in the central nervous system of late-stage embryos (12–14). The proteins resulting from translation of these isoforms, ZLD-PD and ZLD-PF (Fig. 1A), lack three of the four C-terminal zinc fingers, suggesting that they may have altered DNA-binding properties. The function of these additional splice isoforms and whether they are translated in vivo remains unclear.
FIGURE 1.
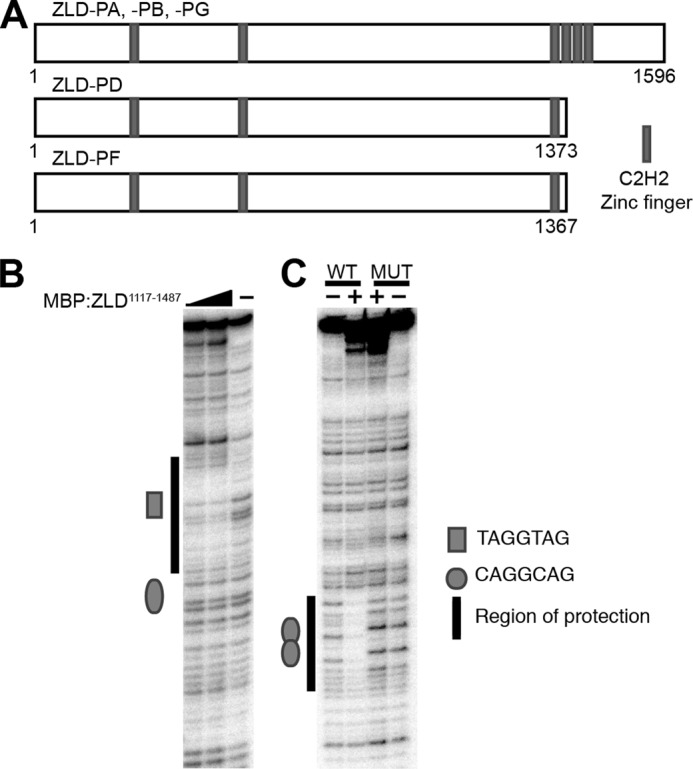
Amino acids 1117–1487 of ZLD, containing a cluster of four zinc fingers, provide sequence-specific DNA-binding activity. A, ZLD polypeptides (ZLD-PA, ZLD-PB, ZLD-PD, ZLD-PF, and ZLD-PG) generated from different splice isoforms. B, DNase I protection of a zen ventral repression element DNA fragment by increasing amounts of MBP:ZLD1117–1487. C, DNase I protection of a fragment of Sxlpe containing either WT or mutated (MUT) overlapping CAGGCAG elements by MBP:ZLD1117–1487.
By establishing a cell-culture based system to assay for ZLD-mediated transcriptional activation, we defined functional domains in this large and poorly characterized protein. We show here that all of the four C-terminally clustered zinc fingers are required for ZLD to bind DNA and that splice isoforms containing only one of the four zinc fingers fail to activate transcription. Furthermore, these truncated ZLD isoforms dominantly suppress the ability of the 1596-amino acid form to drive transcription. We mapped the activation domain of ZLD to an internal region of ∼400 amino acids containing a number of low-complexity domains that are poorly conserved in other insects. Nonetheless, insect orthologs of ZLD activated transcription in Drosophila cells, suggesting that ZLD regulates transcription through conserved interactions with a protein cofactor(s). Together these data specifically identify the DNA-binding and transcriptional activation domains of this essential activator of the zygotic genome and demonstrate that ZLD activation is mediated through a process that has been conserved over ∼300 million years of evolution.
EXPERIMENTAL PROCEDURES
Antibodies and Plasmids
The scute promoter, including the four TAGteam motifs (described in Ref. 15) was cloned upstream of firefly luciferase in pGL3-Basic (Promega). The reporter cassette was subcloned into pCo_PURO (Addgene plasmid 17533) (16) for the generation of stable cell lines. Reporters with mutated TAGteam sites contained changes described in ten Bosch et al. (9). Protein coding regions were cloned into pAc5.1 (Invitrogen) for protein expression in S2 tissue culture cells and pMAL (New England Biolabs) for the production of recombinant MBP fusion protein in Escherichia coli. Site-directed mutagenesis was used to introduce a thymine in the zld open reading frame that results in a stop codon after the fourth amino acid. Actin:Renilla and GAL4Rho:firefly were provided by Robert Tjian (17). The antibodies used for immunoblots were rabbit anti-ZLD antibodies at 1:750 (4) and M2 conjugated to horseradish peroxidase at 1:500 (Sigma).
Cloning ZLD Orthologs
BLAST searches were used to identify possible zld orthologs in Drosophila virilis, Anopheles gambiae, and Nasonia vitripennis. Analysis of genomic sequences confirmed that, as in D. melanogaster, zld was encoded by a single coding exon in these species (18) and that both a highly conserved N-terminal zinc finger region and a conserved cluster of four zinc fingers were contained within a single open reading frame. An intron was predicted in the N. vitripennis zld by the Nv2RefSeq, but this predicted intron removed a stretch of highly conserved sequence, suggesting that this predicted intron is unlikely to be spliced. PCR was used to amplify the open reading frames of predicted ZLD orthologs from genomic DNA obtained from D. virilis, A. gambiae, and N. vitripennis (provided by Sean Carroll, Brian Lazzaro, and Jack Werren, respectively). Amplified products of the predicted size were cloned for expression into pAc5.1 (Invitrogen).
Cell Culture, Generation of Stable Cell Lines, and Luciferase Assays
Drosophila S2 cells were cultured at 25 °C in Schneider's medium (Invitrogen) supplemented with 10% fetal bovine serum (Omega Scientific) and antibiotic/antimycotic (Invitrogen). Transfections were performed in triplicate in 24-well dishes with a total of 300 ng of DNA using Effectene transfection reagent (Qiagen). S2 cell lines with the scute:firefly luciferase reporter stably integrated were made by transfecting cells and subsequently selecting with 2 μg/ml of puromycin (16). Luciferase assays were performed using the Dual-Luciferase assay system (Promega). -Fold activation was determined by comparison with transfections using a plasmid containing the actin promoter but no expression sequence. Representative data sets are shown, with error bars indicating the standard deviation.
Protein Expression and Purification
MBP-ZLD1117–1487 was purified from E. coli in column buffer (20 mm Hepes (pH 7.6), 0.2 m NaCl, 1 mm EDTA, 10 mm β-mercaptoethanol, and 2 mm PMSF) using amylose resin (New England Biolabs). After washing, protein was eluted with 20 mm maltose in PBS and dialyzed against an excess of PBS to remove the maltose.
Baculovirus Infection and Immunoprecipitation
Baculovirus expressing FLAG-tagged ZLD has been described previously (4), and the HA-tagged version was generated in a similar manner. Infection of Hi5 cells was performed as described in Harrison et al. (4), with equal volumes of virus expressing FLAG:ZLD and HA:ZLD. Cells were harvested 48 h post-infection, and cytoplasmic extract was prepared as in Ilves et al. (19). Briefly, cells were lysed in hypotonic lysis buffer and by physical disruption. The extract was divided and used for immunoprecipitation with either anti-FLAG (M2, Sigma) or anti-HA (HA-7, Sigma) resin. The resin was washed extensively under gentle conditions (25 mm Hepes (pH 7.6), 150 mm KCl, 0.02% Tween, 10% glycerol, 1 mm EGTA, 0.4 mm PMSF, 2 mm β-mercaptoethanol, and 0.01 mm ZnCl2). Protein was eluted by boiling in SDS-containing sample buffer and detected by immunoblot analysis.
S2 cells were lysed (25 mm Hepes (pH 7.6), 100 mm KCl, 10% glycerol, 1% Triton X-100, 1 mm EDTA, and 0.4 mm PMSF), and the lysate was cleared by centrifugation. The lysate was immunoprecipitated by incubation with anti-FLAG (M2, Sigma) resin. The resin was washed under gentle conditions (25 mm Hepes (pH 7.6), 150 mm KCl, 20% glycerol, 0.01% Nonidet P-40, and 1 mm EDTA). Protein was eluted by boiling in SDS-containing sample buffer and detected by immunoblotting.
DNase I Protection Assays
DNase I protection assays were performed essentially as described in Jones et al. (20). DNA fragments of the Sxl or zen regulatory regions containing TAGteam elements were 5′ end-labeled with 32P and used as probes. The position of the protected sites within the DNA sequences was identified by sequencing each probe using fmol DNA cycle sequencing (Promega). These sequencing reactions were run on TBE urea gels in parallel with the DNase I protection assays and have been published previously (see supplemental Fig. 1 in Harrison et al. (4)).
Electromobility Shift Assays
EMSAs were performed essentially as described in Harrison et al. (4), except that, instead of using radioactively labeled oligonucleotides, Cy5 5′ end-labeled oligonucleotides were used to detect nucleic acid mobility. In addition, samples were electrophoresed for 30 min at 150 V and 4 °C in 4% polyacrylamide gels (29:1). 40 fmol of oligonucleotide probe and between 0.5–4 pmol of recombinant protein were used in each reaction. For the EMSAs with MBP-ZLD1117–1487 containing mutated zinc fingers, 4 pmol of protein was used. EMSAs were performed in triplicate, and a single example is shown. The sequence of the oligonucleotides probes that were Cy5 labeled were as follows: GAGAGAG ACTACCTGTGGCTCACT (wild type) and GAGA GAGAGTAGTTCTGGCTCACT (mutant).
RESULTS
ZLD Binds DNA via a C-terminal Cluster of Four Zinc Fingers
ZLD is expressed in early embryos as a 1596-amino acid protein encoded by a single exon (ZLD-PA, Fig. 1A) (3, 10, 14). We have demonstrated previously that recombinant ZLD-PA binds to TAGteam elements (4), and this activity was mapped to a large C-terminal region of the protein (amino acids 1239–1596) (3). To specifically define the DNA-binding domain, we tested the ability of a ZLD fragment containing amino acids 1117–1487 to bind specifically to TAGteam elements. ZLD1117–1487 was expressed and purified as a fusion to maltose binding protein (MBP-ZLD1117–1487) and used in DNase I protection assays with DNA fragments derived from the zen ventral repression element and in the Sxl establishment promoter (Sxlpe), two regions known to be bound and regulated by ZLD in vivo (3–6, 9, 21–23). MBP-ZLD1117–1487 bound to TAGteam elements in both the zen ventral repression element and Sxlpe (Fig. 1, B and C). The binding site in the zen ventral repression element included the TAGGTAG TAGteam element but not the CAGGCAG site, and the extent of protection provided by MBP-ZLD1117–1487 is identical to the full-length protein (4). Binding to Sxlpe occurred on the overlapping CAGGCAG elements and specifically required these TAGteam motifs because mutation of the CAGGCAG abrogated binding (Fig. 1C). Furthermore, because the protection provided by amino acids 1117–1487 of ZLD spanned the same residues as the full-length protein, our DNase I protection assays suggested that this region of ZLD comprises the entirety of the DNA-binding domain. In agreement with our DNase I protection assays, a recombinant protein containing only amino acids 1240–1470 has been shown to specifically bind DNA using electromobility shift assays (11). Together, these data narrowed the DNA-binding domain to a region containing the cluster of four zinc fingers.
To test the functional importance of this DNA-binding domain on ZLD activity, we established a cell culture-based assay system. Previous data have shown that TAGteam sites in the promoter of the sex determination factor scute are required for proper gene expression (9, 15), and we have demonstrated that ZLD is bound to these sites in the early embryo (5). Therefore, we created a reporter of ZLD-mediated activation by cloning a region of the scute promoter, including the four TAGteam sites, upstream of the coding sequence for firefly luciferase (Fig. 2A). Because ZLD is not endogenously expressed in Drosophila S2 cells, we cotransfected the reporter with a plasmid constitutively expressing ZLD, resulting in robust activation (Fig. 2B). ZLD expression failed to activate transcription from a reporter in which all four TAGteam sites in the scute promoter were mutated, demonstrating that activation is dependent on specific DNA binding by ZLD (Fig. 2B). A reporter driven by Sxlpe demonstrated a similar dependence on TAGteam sites for transcriptional activation by ZLD (data not shown). Finally, expression of zld containing a premature stop codon failed to activate transcription, indicating that expression from the scute promoter was not mediated by a feature of ZLD mRNA (Fig. 2B). Therefore, we established a highly specific and responsive system to monitor ZLD- and TAGteam-mediated gene expression.
FIGURE 2.

The C-terminal zinc fingers recruit ZLD to DNA. A, firefly luciferase reporter driven by a wild-type scute promoter (top sequence) or a promoter with mutated ZLD-binding sites (bottom sequence). Dots indicate identity between the two reporters. B, -fold luciferase activation of either wild-type or mutant scute reporters by full-length ZLD or a form of ZLD truncated after four amino acids (ZLDSTOP). C, activation of the scute reporter by either full-length FLAG-tagged ZLD or a truncation lacking the final 299 amino acids (FLAG:ZLD1–1297). Protein levels were determined by immunoblot analysis for the FLAG epitope. For all activation assays, n = 3, mean ± S.D.
We exploited this transcriptional assay system to determine the functional importance of specific domains in ZLD. We first assessed the role of the DNA-binding domain identified in our in vitro protection assays. Deletion of the C-terminal 299 amino acids (ZLD1–1297), which removes the cluster of four zinc fingers and their associated DNA-binding activity, dramatically impaired transcriptional activation by ZLD compared with the full-length protein (Fig. 2C). Immunoblotting determined that both the full-length and truncated proteins were expressed at relatively similar levels, demonstrating that the lower activity of the ZLD truncation was not the result of decreased protein expression or stability. Therefore, our biochemical data, along with our cell-based transcription assays, showed that the cluster of four zinc fingers in the C terminus of ZLD provide the DNA-binding activity necessary for transcriptional activation.
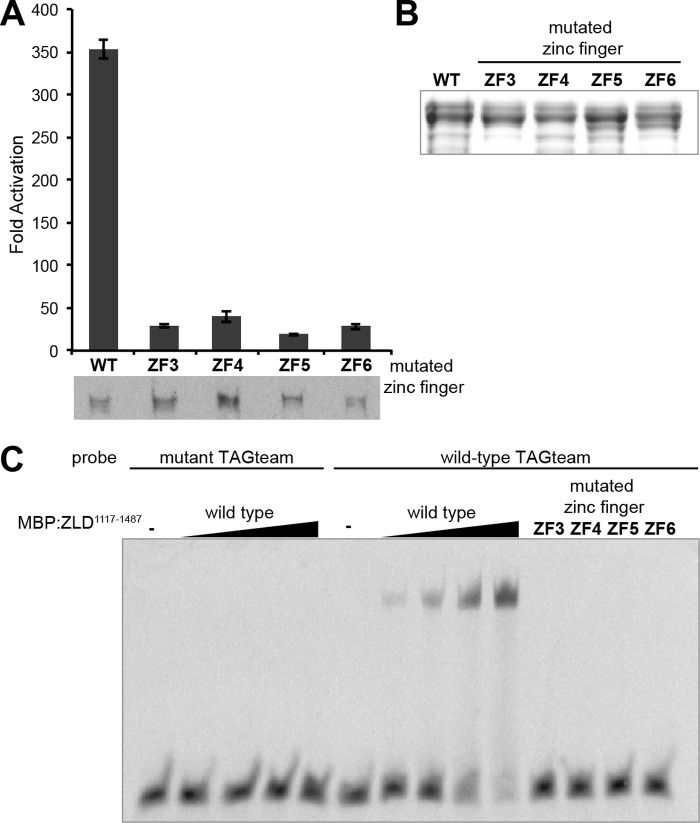
Because the zinc fingers are each predicted to bind three base pairs and because the canonical binding site for ZLD is only seven base pairs, it was possible that only a subset of the four zinc fingers in the mapped DNA-binding domain was required for binding. To determine whether all four zinc fingers or only a subset thereof are required for DNA binding, we mutated the two metal-binding cysteines to serines in each of the four zinc fingers (zinc fingers 3–6) individually and tested the ability of the resulting proteins to activate transcription. Mutations in any single zinc finger almost completely abrogated TAGteam-dependent transcriptional activation (Fig. 3A and data not shown). Approximately equal amounts of wild-type and mutant proteins were detected by immunoblot analysis, indicating that the lack of activity is not due to the absence of a stable polypeptide.
FIGURE 3.
The four C-terminal zinc-fingers of ZLD are required for DNA binding and transcriptional activity. A, -fold activation mediated by wild-type ZLD or by ZLD mutants in which individual zinc fingers in the DNA-binding domain (zinc fingers (ZF) 3–6) were mutated. Protein levels were assayed by immunoblot analysis for ZLD. B, Coomassie-stained gel of purified wild-type MBP:ZLD1117–1487 or the same protein with each of the individual zinc fingers mutated. C, electromobility shift assays using increasing concentrations of wild-type MBP:ZLD1117–1487 (0, 25, 50, 100, or 200 nm) or maximal amounts of individual zinc finger mutants (200 nm). The sequence of the probes corresponds to either a wild-type or mutant version of a CAGGTAG element from the scute promoter.
We next tested whether each individual zinc finger is similarly required for DNA binding. We purified recombinant MBP-ZLD1117–1487 containing wild-type zinc fingers or with individual zinc fingers mutated and used these proteins for EMSAs (Fig. 3, B and C). Wild-type MBP-ZLD1117–1487 bound to oligonucleotide probes corresponding to a CAGGTAG element from the scute promoter but did not bind when this site was mutated (Fig. 3C). These data demonstrated that our EMSAs detect sequence-specific DNA binding by the cluster of four zinc fingers in ZLD, similar to what we have shown previously for full-length ZLD (4). This domain alone binds with a Kd of ∼75 nm. This is in remarkable agreement with an estimated Kd of ∼15 nm for the full-length protein based on our previously published EMSAs (4). This suggests that the majority of the DNA-binding activity for ZLD is localized in the zinc-finger domain, but we cannot exclude minor contributions from other regions. Mutation of any individual zinc finger ablated DNA binding in EMSAs (Fig. 3C), even when we used protein amounts corresponding to the maximum amount of wild-type MBP-ZLD1117–1487 used. Therefore, these data show that the cluster of four zinc fingers comprises the DNA-binding domain of ZLD and that all four zinc fingers are necessary for DNA binding and transcriptional activation.
ZLD Splice Isoforms Lacking Three of the Four C-terminal Zinc Fingers Are Dominant Negative Inhibitors of ZLD-mediated Activation
At least one of two nearly identical zld splice isoforms that lack the coding region for three of the four clustered zinc fingers is expressed in larvae and late-stage embryos (12, 13). The predicted proteins resulting from these two different splice isoforms differ by the number and sequence of amino acids following the single remaining zinc finger: 22 amino acids in ZLD-PD and 17 amino acids in ZLD-PF (Fig. 1A). Given our data showing that all four zinc fingers are required for DNA binding, we used our cell culture-based transcriptional activation assay to determine whether these truncated proteins could bind to TAGteam elements to activate transcription. Neither of these splice isoforms activated transcription from the scute promoter in a TAGteam-dependent manner, suggesting that the single remaining zinc finger does not provide sufficient DNA-binding activity to activate transcription (Fig. 4A and data not shown).
FIGURE 4.
A truncated splice isoform (ZLD-PD) can dominantly inhibit ZLD-PA-mediated transcriptional activation. A, -fold luciferase activation of either a WT or mutant (MUT) scute reporter by ZLD-PA or the splice isoform, ZLD-PD, which lacks three of the four C-terminal zinc fingers. Protein levels were assayed by immunoblot analysis for ZLD. B, -fold activation by ZLD-PA and ZLD-PD expressed individually or simultaneously. (Data for ZLD-PA and ZLD-PD are the same as in A.) For all activation assays, n = 3, mean ± S.D. C, HA or FLAG immunoprecipitation (IP) from cells coinfected with baculovirus expressing HA:ZLD-PA and FLAG:ZLD-PA. Immunoblots (IB) for FLAG and HA are indicated on the left. D, FLAG immunoprecipitation from cells coexpressing FLAG:ZLD-PA and ZLD-PD. Also shown are immunoblots for ZLD. IN, 1% of input; IP, 33% of immunoprecipitation.
To better understand a potential function for these truncated isoforms, we coexpressed ZLD-PD or ZLD-PF along with the full 1596-amino acid ZLD-PA in the transcriptional activation assay. Coexpression of ZLD-PD (or ZLD-PF) with ZLD-PA significantly reduced gene expression, demonstrating that ZLD-PD and ZLD-PF act dominantly to suppress ZLD-mediated transcriptional activation (Fig. 4B and data not shown). This dominant-negative activity could be explained if ZLD dimerizes and if a heterodimer containing ZLD-PD and ZLD-PA is inactive. To test this possibility, we assessed the ability of ZLD to dimerize by coexpressing differentially tagged versions of the protein. We coinfected Hi5 cells with baculovirus encoding either an N-terminally FLAG-tagged ZLD or HA-tagged ZLD. Lysates were immunoprecipitated for either the FLAG or HA tag and probed for these tags by immunoblot analysis. The FLAG-tagged ZLD was clearly detected when immunoprecipitated with anti-FLAG antibodies, but the HA-tagged version was not coprecipitated. The reverse was also true when the HA-tagged ZLD was purified. Therefore, ZLD failed to form dimers in our assay, regardless of the epitope tag used for immunoprecipitation (Fig. 4C). Additionally, in S2 cells, immunoprecipitation of FLAG-tagged ZLD-PA did not precipitate the truncated ZLD-PD isoform (Fig. 4D). Given the lack of evidence for dimerization, we instead propose that the dominant-negative functionality results from the N-terminal region of ZLD interacting with a cofactor(s) required to mediate transcriptional activation and that the truncated splice isoform competes with ZLD-PA for their binding.
Amino Acids 904–1297 Comprise the Transcriptional Activation Domain of ZLD
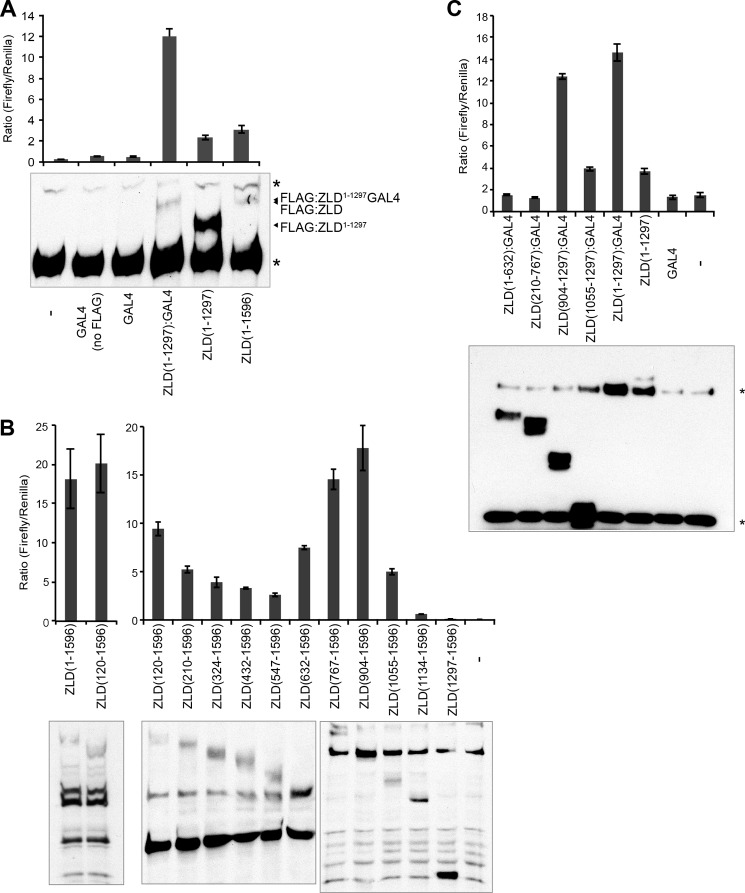
Because ZLD-PD and ZLD-PF likely bind a cofactor necessary for mediating transcriptional activation, this interaction domain must exist N-terminally to the DNA-binding domain we identified. To test whether only this region was sufficient to drive transcription, we replaced the C-terminal 299 amino acids of ZLD, including the DNA-binding domain, with the GAL4 DNA-binding domain. A firefly luciferase reporter with upstream GAL4-binding sites was used to assay for transcriptional activation by this chimeric protein. Neither full-length ZLD nor ZLD1–1297 could drive significant levels of transcription from this reporter (Fig. 5A). By contrast, ZLD1–1297 fused to the GAL4 DNA-binding domain could drive robust transcriptional activation, even though immunoblotting showed that it was expressed at low levels compared with the protein lacking the DNA-binding domain (Fig. 5A). Although the C-terminal zinc fingers mediate DNA binding (Figs. 1–3), these data demonstrate that the first 1297 amino acids of ZLD are sufficient for activating transcription.
FIGURE 5.
Amino acids 904–1297 constitute a minimal domain required to drive ZLD-mediated transcriptional activation. A, FLAG-tagged constructs (except where noted) were expressed. Ratio of a firefly luciferase reporter containing upstream GAL4 DNA-binding sites to a transfection control (Renilla luciferase) is shown. Protein expression was verified by immunoblot analysis against the FLAG epitope. B, transcriptional activation by a series of N-terminally truncated ZLD proteins. Shown is the ratio of scute:firefly luciferase reporter expression to a transfection control. Amino acids included in the truncated protein are shown. Protein levels were determined by immunoblot analysis with anti-ZLD antibodies. The graphs show representative assays performed in a single experiment. (A number of cross-reactive bands are evident and vary depending on the percentage of acrylamide in the PAGE gel.). C, FLAG-tagged ZLD truncations were expressed as GAL4 fusions in the presence of a GAL4 DNA-binding site reporter construct. Ratio of the firefly luciferase reporter to a transfection control is shown. Protein expression was verified by immunoblot analysis with anti-FLAG antibodies. For activation assays, n = 3, mean ± S.D. The asterisks indicate nonspecific immunoreactivity.
We next used a series of N-terminal truncations in our assay system to determine the residues necessary for activity. All of the truncations activated gene expression to varying levels above background, except for the truncation that contained only amino acids 1134–1596. Immunoblots confirmed the expression of all truncated proteins at approximately equivalent levels (Fig. 5B). ZLD767–1596 and ZLD904–1596 were the most potent activators, whereas there was a severe reduction in activity when amino acids 904–1134 were removed, suggesting that these residues were required for transcriptional activity. There was also an increase in activity when amino acids 632–904 were truncated, allowing for the possibility that this region may contain an inhibitory activity.
To determine whether amino acids 904–1297 were sufficient to activate transcription, we generated a series of proteins in which a portion of ZLD was fused to the GAL4 DNA-binding domain. These fusions were then assayed for their ability to activate transcription from a reporter containing GAL4 binding sites. Although amino acids 1–632, 210–767, and 1055–1297 did not activate transcription in this assay, robust activation was detected when the amino acids between 904–1297 were fused to the GAL4 DNA-binding domain (Fig. 5C). Together, these data demonstrate that the 393 amino acids spanning residues 904–1297 are necessary and sufficient for ZLD-mediated transcriptional activation.
Our data and data from others have suggested that ZLD may, in part, mediate transcriptional activation through changes in the chromatin structure (5, 24). Therefore, we tested whether the DNA-binding and activation domains that we identified using our transient transfection-based cell culture assay system were necessary and sufficient to activate transcription of a reporter in the context of chromatin. We created a stable S2 cell line with the scute reporter integrated into the genome. ZLD-mediated transcriptional activation was again assayed on this chromatinized reporter. Identical to what was shown in our previous assays, wild-type and FLAG-tagged ZLD-PA activated transcription (Fig. 6). The DNA-binding domain, containing amino acids 1297–1596, was required for transcriptional activation. Furthermore, amino acids 904–1596, containing both the DNA-binding domain and the minimal activation domain, were sufficient to drive transcription (Fig. 6). Therefore, the C-terminal 692 amino acids provide the activities necessary for ZLD to mediate transcriptional activation.
FIGURE 6.
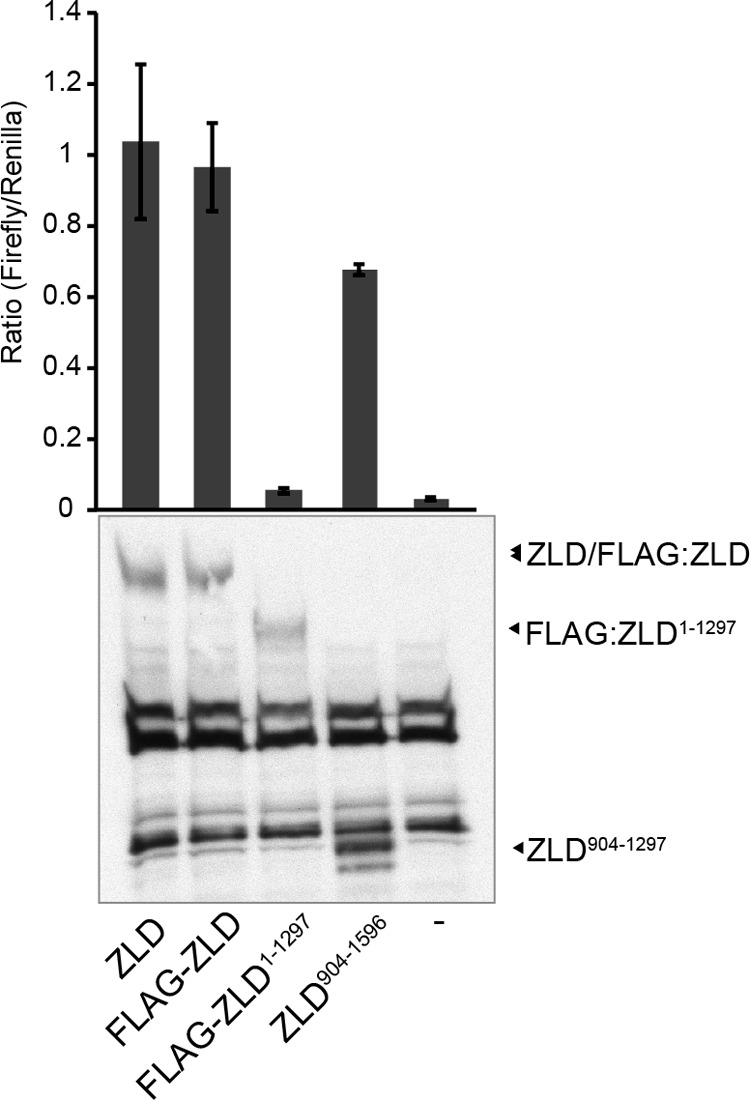
Amino acids 904–1596 are sufficient to activate transcription from an integrated reporter. Constructs expressing ZLD or portions of ZLD were cotransfected along with a transfection control (Renilla luciferase) into cells with the scute:firefly luciferase reporter integrated. Ratios of firefly luciferase to Renilla luciferase expression are shown. Protein levels were determined by immunoblot analysis with anti-ZLD antibodies (n = 3, mean ± S.D.). (A number of cross-reactive bands are evident.)
ZLD Orthologs Can Drive Transcription of a ZLD Target Gene
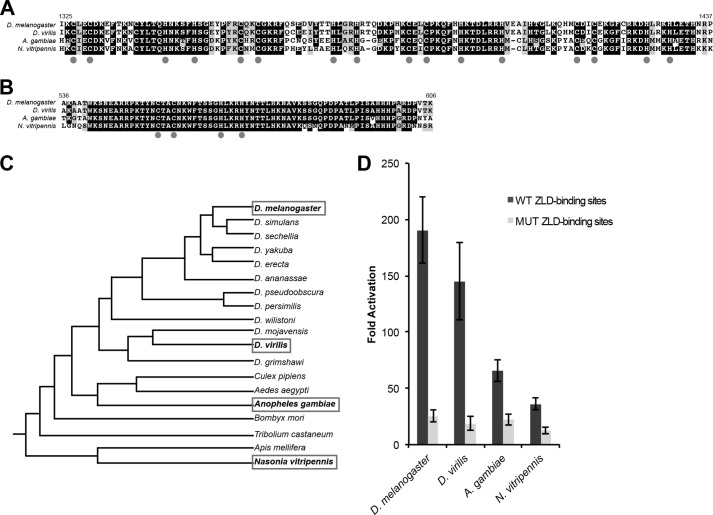
ZLD orthologs were identified in other insect species, and conservation was most strongly evident in the DNA-binding domain (Fig. 7A) and in a region surrounding the second of the N-terminal zinc fingers (Fig. 7B). The conservation of the DNA-binding domain suggests that these orthologs may be able to bind to sequences similar to the TAGteam elements. However, because the conserved N-terminal region is outside of our identified activation domain, it was not clear whether these orthologs would be able to activate transcription in our cell culture assay system. To test this, we cloned zld open reading frames from three additional insect species that show a varying range of identity with D. melanogaster ZLD: D. virilis (56% identity), A. gambiae (32% identity), and N. vitripennis (25% identity) (Fig. 7C). Because ZLD-PA from D. melanogaster is expressed from a single coding exon, we identified a similar single open reading frame in these other species that included both the N-terminal zinc finger and the DNA-binding domain. We amplified the presumed zld coding sequences and cloned them into an expression vector for use in our assay system. ZLD orthologs from all three species activated transcription from the D. melanogaster scute promoter to varying degrees (Fig. 7D). This activation required the presence of TAGteam elements because gene expression is reduced from the mutated reporter (Fig. 7D). These data suggest that ZLD orthologs bind to sequences similar to the TAGteam elements and drive transcriptional activation by interacting with a cofactor(s) conserved in D. melanogaster.
FIGURE 7.
ZLD orthologs from other insects activate TAGteam-dependent transcription. A, alignment of the DNA-binding domain of ZLD with sequence from three additional insect species (D. virilis, A. gambiae, and N. vitripennis). Gray dots indicate the cysteines and histidines in the four C2H2 zinc fingers. B, alignment of a highly conserved N-terminal zinc finger. C, phylogeny showing the relation of the four insect species analyzed (25, 26). D, -fold activation driven by expression of ZLD orthologs of the scute:firefly reporter with either WT or mutant (MUT) ZLD-binding sites. For activation assays, n = 4, mean ± S.D., and p ≤ 0.0002 between activation of wild-type and mutant reporters for all orthologs (Student's t test).
DISCUSSION
Embryonic gene expression undergoes a dramatic reorganization at the MZT as the embryo switches from utilizing maternally provided mRNAs and proteins to widespread activation of its own genome. ZLD was the first identified global activator of zygotic transcription at the MZT in any organism (3). Nonetheless, the mechanism by which ZLD activates gene expression remains unclear. Here we developed a cell-based transcriptional activation assay to probe the functional domains of ZLD. We defined the DNA-binding domain of D. melanogaster ZLD as a C-terminal cluster of four zinc fingers that specifically binds to TAGteam sequences within the regulatory regions of early expressed genes. This cluster of zinc fingers is conserved in ZLD orthologs from evolutionarily distant insect species, and these orthologs retain the ability to activate transcription from TAGteam-containing promoters. Additionally, we mapped the activation domain to a poorly conserved, low-complexity protein domain just N-terminal to the DNA-binding domain. Finally, we showed that splice isoforms lacking three of the four zinc fingers in the DNA-binding domain fail to drive gene expression and can dominantly inhibit the ability of the 1596-amino acid isoform to activate transcription. Together, these data define the functional domains of this essential activator of the zygotic genome and support a model in which ZLD interacts with a conserved protein cofactor(s) to drive transcription in the early embryo.
Truncated Splice Isoforms of ZLD May Regulate Gene Expression by Competing with ZLD-PA for a Cofactor
Using DNase I protection assays, we demonstrated that a region containing the C-terminal cluster of four zinc fingers bound the same DNA sequence as the full-length protein. Using our cell-based transcription assay, we further showed that these zinc fingers were each individually required to activate gene expression, and EMSAs demonstrated that all four zinc fingers were necessary for DNA binding. Furthermore, expression of ZLD variants in which the C-terminal 299 amino acids, including the zinc fingers, were replaced by the GAL4 DNA-binding domain activated transcription from a GAL4-responsive promoter, suggesting that the primary function of these zinc fingers is DNA binding and not transcriptional activation. Thus, we used our assay system to demonstrate that all four zinc fingers in the C terminus are both necessary and sufficient for TAGteam-specific DNA binding and constitute the minimal DNA-binding domain. The zinc fingers are each predicted to bind approximately three base pairs. The requirement of all four zinc fingers for DNA binding provides evidence that sequences outside of the canonical seven-base pair TAGteam element may help direct ZLD specificity. Alternatively, a subset of zinc fingers may play a structural role and may not contribute to sequence-specific DNA binding.
Previous work has shown that multiple ZLD splice isoforms are expressed in late-stage embryos and in larvae. These isoforms are predicted to produce protein products of either 1596 (ZLD-PA) or 1373 (ZLD-PD) amino acids. (The 1367 amino acid isoform ZLD-PF is likely not expressed (12, 13).) Although ZLD-PA, which contains all four zinc fingers of the DNA-binding domain, activated robust gene expression, neither of the truncated isoforms, ZLD-PD or ZLD-PF, activated transcription. These data are consistent with our mutagenic analysis of the DNA-binding domain of ZLD-PA, demonstrating that all four zinc fingers are required for DNA binding. This suggests that the failure of ZLD-PD and ZLD-PF to activate transcription is due to these truncated proteins failing to bind DNA. Surprisingly, we demonstrated that these truncated splice isoforms can dominantly suppress the ability of ZLD-PA to activate transcription. This dominant inhibition could be explained by either the formation of inactive ZLD-PA/ZLD-PD multimers or by competition of these two isoforms for interaction with cofactors required to activate transcription. However, we have seen no evidence for the formation of stable ZLD multimers.
Instead, our data support the hypothesis that, when coexpressed, ZLD-PD is competing with ZLD-PA for a binding partner required for activity. In this scenario, limiting amounts of a necessary transcriptional coactivator would be titrated away from the activation-competent ZLD-PA by the truncated isoform. This would provide a mechanism whereby ZLD binding could be established in the early embryo, when ZLD-PA is the primary splice isoform and the genome may be in a relatively accessible conformation. This binding established early in embryogenesis would then be maintained throughout development. ZLD-regulated genes could then be activated or repressed on the basis of the levels of ZLD-PD generated. Both isoforms are expressed in the embryo, although it remains to be determined whether these isoforms are simultaneously expressed in a single cell. If they are coexpressed, this competition could mediate repression of ZLD-activated genes without changing levels of ZLD-PA occupancy at promoters and enhancers.
Conservation of ZLD and TAGteam Sites in Driving Genome Activation
The coding region for ZLD is conserved in other insects, and zld expression in late-stage embryos of multiple Drosophila species has been observed (13). Here we tested the ability of orthologs from three additional insect species (D. virilis, A. gambiae, and N. vitripennis) to activate transcription in a TAGteam-dependent manner in D. melanogaster cells. D. melanogaster and D. virilis are relatively closely related because they are estimated to have diverged 40 million years ago (25, 26). By contrast, A. gambiae and N. vitripennis diverged from D. melanogaster about 250 and 300 million years ago, respectively (27–29). The zinc-finger DNA-binding domain we identified is highly conserved in these species, suggesting that these orthologs may bind to TAGteam-related sequences in these insects. However, outside of the six zinc-finger domains, there is little conservation at the sequence level between D. melanogaster ZLD and its orthologs. Despite this limited sequence conservation, ZLD from these divergent insect species activated transcription in a TAGteam-dependent manner, although, as might be expected, the more divergent ZLD orthologs activated to a lesser degree. These data indicate that the capacity of ZLD to bind related sequence motifs and activate transcription has been conserved over 300 million years of evolution.
Supporting this conserved role of ZLD in driving genome activation in other insects, sequences similar to TAGteam elements (VBRGGTA, V = A/C/G, B = C/G/T, r = A/G) are enriched in the 400-base pair regions upstream of the set of genes activated at the MZT in the mosquito Aedes aegypti (30). Comparison of position weight matrices for the A. aegypti sequence and D. melanogaster TAGteam elements suggests that these two sequence elements may be homologous, and, like the D. melanogaster TAGteam sites, a region containing this sequence is capable of driving early zygotic transcription (9, 30). Together, these data suggest that both the DNA-binding domain of ZLD and the sequence to which it binds are conserved in divergent species. Therefore, the ability of ZLD to drive widespread gene activation at the MZT is likely to be a conserved feature.
Further evidence for a conserved role of ZLD within Drosophilids comes from comparative studies of transcription factor binding sites in multiple Drosophila species. Transcription factor binding sites have been defined in multiple Drosophila species using chromatin immunopreciptation coupled with microarrays and with high-throughput sequencing (ChIP-chip and ChIP-seq) (8, 25, 31–33). The canonical TAGteam sequence CAGGTAG is highly enriched in the regions bound by multiple different transcription factors in D. melanogaster (8, 32, 33), and this is similarly true for the other Drosophila species examined (25, 31). Furthermore, analysis suggests that changes in the binding sites of these transcription factors between different species correlates with changes in ZLD-binding sites, suggesting that ZLD binding may be a driving force in the evolution of these transcription factor binding sites (25, 31). Therefore, our functional data combined with the evolutionary analysis of the conservation of ZLD-binding sites supports a role for ZLD in genome activation in a broad range of insects.
Low-complexity Domains Mediate Transcriptional Activation by ZLD
The activation domain we defined does not contain any identifiable domains and is comprised of a low-complexity sequence. Low-complexity regions contain repeated stretches of single or low numbers of amino acids and are characteristically unstructured (34). It is this feature of being relatively unstructured that may allow such regions to interact with a large number of different binding partners (35). Low-complexity regions are often found within transcription factors and can function as transcriptional activation domains (35, 36). Recent evidence suggests that at least some low-complexity domains polymerize to form amyloid-like fibers, and it is this capacity that mediates transcriptional activation (36–38). Therefore, a mechanistic understanding of how ZLD activates transcription may help further define the functions of this large class of transcription factors.
Because ZLD orthologs contain low-complexity regions and similarly activate transcription, it is likely that this shared feature provides the capacity to activate transcription. The poor conservation of the activation domain at the sequence level, the absence of any recognizable catalytic domains, and our data showing that ZLD-PD can dominantly suppress activation mediated by ZLD-PA cumulatively suggest that ZLD is activating transcription by interacting with a protein cofactor(s) through these low-complexity domains. Future studies identifying these cofactors and their function will be vital to understanding how ZLD mediates activation of the zygotic genome.
It has been shown recently that, in addition to their well defined role as canonical pluripotency factors, Pou5f1, Sox2, and Nanog function in a manner similar to ZLD to drive global activation of the zygotic genome during the initial stages of zebrafish embryonic development (39, 40). These data indicate a connection between zygotic genome activation and the establishment of the pluripotent state. Although these proteins are not conserved in Drosophila at the sequence level, the general features governing the activation of the zygotic genome and the cofactors required possibly represent a common mode of zygotic genome activation. Therefore, our dissection of the functional domains of ZLD required for transcriptional activation is likely to provide important insights into the connection between genome activation and pluripotency.
Acknowledgments
We thank Jack Werren, Brian Lazzaro, Sean Carroll, Francis Castellino, and Robert Tjian for reagents. We also thank Andrew Mehle, John Denu, and Richard Harrison for helpful discussions and critical reading of the manuscript.
This work was supported by a Basil O'Connor Starter Scholar Research Award from the March of Dimes (to M. M. H.) and by a National Science Foundation predoctoral fellowship (to D. C. H.).
- MZT
- maternal-to-zygotic transition
- ZLD
- Zelda
- MBP
- maltose-binding protein.
REFERENCES
- 1. Newport J., Kirschner M. (1982) A major developmental transition in early Xenopus embryos: II. Control of the onset of transcription. Cell 30, 687–696 [DOI] [PubMed] [Google Scholar]
- 2. Tadros W., Lipshitz H. D. (2009) The maternal-to-zygotic transition: a play in two acts. Development 136, 3033–3042 [DOI] [PubMed] [Google Scholar]
- 3. Liang H. L., Nien C. Y., Liu H. Y., Metzstein M. M., Kirov N., Rushlow C. (2008) The zinc-finger protein Zelda is a key activator of the early zygotic genome in Drosophila. Nature 456, 400–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Harrison M. M., Botchan M. R., Cline T. W. (2010) Grainyhead and Zelda compete for binding to the promoters of the earliest-expressed Drosophila genes. Dev. Biol. 345, 248–255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Harrison M. M., Li X. Y., Kaplan T., Botchan M. R., Eisen M. B. (2011) Zelda binding in the early Drosophila melanogaster embryo marks regions subsequently activated at the maternal-to-zygotic transition. PLoS Genet. 7, e1002266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Nien C. Y., Liang H. L., Butcher S., Sun Y., Fu S., Gocha T., Kirov N., Manak J. R., Rushlow C. (2011) Temporal coordination of gene networks by Zelda in the early Drosophila embryo. PLoS Genet. 7, e1002339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. De Renzis S., Elemento O., Tavazoie S., Wieschaus E. F. (2007) Unmasking activation of the zygotic genome using chromosomal deletions in the Drosophila embryo. PLoS Biol. 5, e117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Li X. Y., MacArthur S., Bourgon R., Nix D., Pollard D. A., Iyer V. N., Hechmer A., Simirenko L., Stapleton M., Luengo Hendriks C. L., Chu H. C., Ogawa N., Inwood W., Sementchenko V., Beaton A., Weiszmann R., Celniker S. E., Knowles D. W., Gingeras T., Speed T. P., Eisen M. B., Biggin M. D. (2008) Transcription factors bind thousands of active and inactive regions in the Drosophila blastoderm. PLoS Biol. 6, e27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. ten Bosch J. R., Benavides J. A., Cline T. W. (2006) The TAGteam DNA motif controls the timing of Drosophila pre-blastoderm transcription. Development 133, 1967–1977 [DOI] [PubMed] [Google Scholar]
- 10. Staudt N., Fellert S., Chung H. R., Jäckle H., Vorbrüggen G. (2006) Mutations of the Drosophila zinc finger-encoding gene vielfaltig impair mitotic cell divisions and cause improper chromosome segregation. Mol. Biol. Cell 17, 2356–2365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Struffi P., Corado M., Kaplan L., Yu D., Rushlow C., Small S. (2011) Combinatorial activation and concentration-dependent repression of the Drosophila even skipped stripe 3+7 enhancer. Development 138, 4291–4299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Giannios P., Tsitilou S. G. (2013) The embryonic transcription factor Zelda of Drosophila melanogaster is also expressed in larvae and may regulate developmentally important genes. Biochem. Biophys. Res. Commun. 438, 329–333 [DOI] [PubMed] [Google Scholar]
- 13. Pearson J. C., Watson J. D., Crews S. T. (2012) Drosophila melanogaster Zelda and Single-minded collaborate to regulate an evolutionarily dynamic CNS midline cell enhancer. Dev. Biol. 366, 420–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. St Pierre S. E., Ponting L., Stefancsik R., McQuilton P., FlyBase C. (2014) FlyBase 102: advanced approaches to interrogating FlyBase. Nucleic Acids Res. 42, D780–D788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wrischnik L. A., Timmer J. R., Megna L. A., Cline T. W. (2003) Recruitment of the proneural gene scute to the Drosophila sex-determination pathway. Genetics 165, 2007–2027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Iwaki T., Figuera M., Ploplis V. A., Castellino F. J. (2003) Rapid selection of Drosophila S2 cells with the puromycin resistance gene. BioTechniques 35, 482-484, 486 [DOI] [PubMed] [Google Scholar]
- 17. Wright K. J., Marr M. T., 2nd, Tjian R. (2006) TAF4 nucleates a core subcomplex of TFIID and mediates activated transcription from a TATA-less promoter. Proc. Natl. Acad. Sci. U.S.A. 103, 12347–12352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. (1990) Basic local alignment search tool. J. Mol. Biol. 215, 403–410 [DOI] [PubMed] [Google Scholar]
- 19. Ilves I., Petojevic T., Pesavento J. J., Botchan M. R. (2010) Activation of the MCM2–7 helicase by association with Cdc45 and GINS proteins. Mol. Cell 37, 247–258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Jones K. A., Yamamoto K. R., Tjian R. (1985) Two distinct transcription factors bind to the HSV thymidine kinase promoter in vitro. Cell 42, 559–572 [DOI] [PubMed] [Google Scholar]
- 21. Jiang J., Rushlow C. A., Zhou Q., Small S., Levine M. (1992) Individual dorsal morphogen binding sites mediate activation and repression in the Drosophila embryo. EMBO J. 11, 3147–3154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Keyes L. N., Cline T. W., Schedl P. (1992) The primary sex determination signal of Drosophila acts at the level of transcription. Cell 68, 933–943 [DOI] [PubMed] [Google Scholar]
- 23. Kirov N., Zhelnin L., Shah J., Rushlow C. (1993) Conversion of a silencer into an enhancer: evidence for a co-repressor in dorsal-mediated repression in Drosophila. EMBO J. 12, 3193–3199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Foo S. M., Sun Y., Lim B., Ziukaite R., O'Brien K., Nien C. Y., Kirov N., Shvartsman S. Y., Rushlow C. A. (2014) Zelda potentiates morphogen activity by increasing chromatin accessibility. Curr. Biol. 24, 1341–1346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Paris M., Kaplan T., Li X. Y., Villalta J. E., Lott S. E., Eisen M. B. (2013) Extensive divergence of transcription factor binding in Drosophila embryos with highly conserved gene expression. PLoS Genet. 9, e1003748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Russo C. A., Takezaki N., Nei M. (1995) Molecular phylogeny and divergence times of drosophilid species. Mol. Biol. Evol. 12, 391–404 [DOI] [PubMed] [Google Scholar]
- 27. Gaunt M. W., Miles M. A. (2002) An insect molecular clock dates the origin of the insects and accords with palaeontological and biogeographic landmarks. Mol. Biol. Evol. 19, 748–761 [DOI] [PubMed] [Google Scholar]
- 28. Trautwein M. D., Wiegmann B. M., Beutel R., Kjer K. M., Yeates D. K. (2012) Advances in insect phylogeny at the dawn of the postgenomic era. Annu. Rev. Entomol. 57, 449–468 [DOI] [PubMed] [Google Scholar]
- 29. Werren J. H., Richards S., Desjardins C. A., Niehuis O., Gadau J., Colbourne J. K., Nasonia Genome Working Group, Werren J. H., Richards S., Desjardins C. A., Niehuis O., Gadau J., Colbourne J. K., Beukeboom L. W., Desplan C., Elsik C. G., Grimmelikhuijzen C. J., Kitts P., Lynch J. A., Murphy T., Oliveira D. C., Smith C. D., van de Zande L., Worley K. C., Zdobnov E. M., Aerts M., Albert S., Anaya V. H., Anzola J. M., Barchuk A. R., Behura S. K., Bera A. N., Berenbaum M. R., Bertossa R. C., Bitondi M. M., Bordenstein S. R., Bork P., Bornberg-Bauer E., Brunain M., Cazzamali G., Chaboub L., Chacko J., Chavez D., Childers C. P., Choi J. H., Clark M. E., Claudianos C., Clinton R. A., Cree A. G., Cristino A. S., Dang P. M., Darby A. C., de Graaf D. C., Devreese B., Dinh H. H., Edwards R., Elango N., Elhaik E., Ermolaeva O., Evans J. D., Foret S., Fowler G. R., Gerlach D., Gibson J. D., Gilbert D. G., Graur D., Grunder S., Hagen D. E., Han Y., Hauser F., Hultmark D., Hunter H. C. t., Hurst G. D., Jhangian S. N., Jiang H., Johnson R. M., Jones A. K., Junier T., Kadowaki T., Kamping A., Kapustin Y., Kechavarzi B., Kim J., Kim J., Kiryutin B., Koevoets T., Kovar C. L., Kriventseva E. V., Kucharski R., Lee H., Lee S. L., Lees K., Lewis L. R., Loehlin D. W., Logsdon J. M., Jr., Lopez J. A., Lozado R. J., Maglott D., Maleszka R., Mayampurath A., Mazur D. J., McClure M. A., Moore A. D., Morgan M. B., Muller J., Munoz-Torres M. C., Muzny D. M., Nazareth L. V., Neupert S., Nguyen N. B., Nunes F. M., Oakeshott J. G., Okwuonu G. O., Pannebakker B. A., Pejaver V. R., Peng Z., Pratt S. C., Predel R., Pu L. L., Ranson H., Raychoudhury R., Rechtsteiner A., Reese J. T., Reid J. G., Riddle M., Robertson H. M., Romero-Severson J., Rosenberg M., Sackton T. B., Sattelle D. B., Schluns H., Schmitt T., Schneider M., Schuler A., Schurko A. M., Shuker D. M., Simoes Z. L., Sinha S., Smith Z., Solovyev V., Souvorov A., Springauf A., Stafflinger E., Stage D. E., Stanke M., Tanaka Y., Telschow A., Trent C., Vattathil S., Verhulst E. C., Viljakainen L., Wanner K. W., Waterhouse R. M., Whitfield J. B., Wilkes T. E., Williamson M., Willis J. H., Wolschin F., Wyder S., Yamada T., Yi S. V., Zecher C. N., Zhang L., Gibbs R. A. (2010) Functional and evolutionary insights from the genomes of three parasitoid Nasonia species. Science 327, 343–348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Biedler J. K., Hu W., Tae H., Tu Z. (2012) Identification of early zygotic genes in the yellow fever mosquito Aedes aegypti and discovery of a motif involved in early zygotic genome activation. PloS ONE 7, e33933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bradley R. K., Li X. Y., Trapnell C., Davidson S., Pachter L., Chu H. C., Tonkin L. A., Biggin M. D., Eisen M. B. (2010) Binding site turnover produces pervasive quantitative changes in transcription factor binding between closely related Drosophila species. PLoS Biol. 8, e1000343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. MacArthur S., Li X. Y., Li J., Brown J. B., Chu H. C., Zeng L., Grondona B. P., Hechmer A., Simirenko L., Keränen S. V., Knowles D. W., Stapleton M., Bickel P., Biggin M. D., Eisen M. B. (2009) Developmental roles of 21 Drosophila transcription factors are determined by quantitative differences in binding to an overlapping set of thousands of genomic regions. Genome Biol. 10, R80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. modENCODE Consortium, Roy S., Ernst J., Kharchenko P. V., Kheradpour P., Negre N., Eaton M. L., Landolin J. M., Bristow C. A., Ma L., Lin M. F., Washietl S., Arshinoff B. I., Ay F., Meyer P. E., Robine N., Washington N. L., Di Stefano L., Berezikov E., Brown C. D., Candeias R., Carlson J. W., Carr A., Jungreis I., Marbach D., Sealfon R., Tolstorukov M. Y., Will S., Alekseyenko A. A., Artieri C., Booth B. W., Brooks A. N., Dai Q., Davis C. A., Duff M. O., Feng X., Gorchakov A. A., Gu T., Henikoff J. G., Kapranov P., Li R., MacAlpine H. K., Malone J., Minoda A., Nordman J., Okamura K., Perry M., Powell S. K., Riddle N. C., Sakai A., Samsonova A., Sandler J. E., Schwartz Y. B., Sher N., Spokony R., Sturgill D., van Baren M., Wan K. H., Yang L., Yu C., Feingold E., Good P., Guyer M., Lowdon R., Ahmad K., Andrews J., Berger B., Brenner S. E., Brent M. R., Cherbas L., Elgin S. C., Gingeras T. R., Grossman R., Hoskins R. A., Kaufman T. C., Kent W., Kuroda M. I., Orr-Weaver T., Perrimon N., Pirrotta V., Posakony J. W., Ren B., Russell S., Cherbas P., Graveley B. R., Lewis S., Micklem G., Oliver B., Park P. J., Celniker S. E., Henikoff S., Karpen G. H., Lai E. C., MacAlpine D. M., Stein L. D., White K. P., Kellis M. (2010) Identification of functional elements and regulatory circuits by Drosophila modENCODE. Science 330, 1787–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Huntley M. A., Golding G. B. (2002) Simple sequences are rare in the Protein Data Bank. Proteins 48, 134–140 [DOI] [PubMed] [Google Scholar]
- 35. Coletta A., Pinney J. W., Solís D. Y., Marsh J., Pettifer S. R., Attwood T. K. (2010) Low-complexity regions within protein sequences have position-dependent roles. BMC Syst. Biol. 4, 43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kwon I., Kato M., Xiang S., Wu L., Theodoropoulos P., Mirzaei H., Han T., Xie S., Corden J. L., McKnight S. L. (2013) Phosphorylation-regulated binding of RNA polymerase II to fibrous polymers of low-complexity domains. Cell 155, 1049–1060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Han T. W., Kato M., Xie S., Wu L. C., Mirzaei H., Pei J., Chen M., Xie Y., Allen J., Xiao G., McKnight S. L. (2012) Cell-free formation of RNA granules: bound RNAs identify features and components of cellular assemblies. Cell 149, 768–779 [DOI] [PubMed] [Google Scholar]
- 38. Kato M., Han T. W., Xie S., Shi K., Du X., Wu L. C., Mirzaei H., Goldsmith E. J., Longgood J., Pei J., Grishin N. V., Frantz D. E., Schneider J. W., Chen S., Li L., Sawaya M. R., Eisenberg D., Tycko R., McKnight S. L. (2012) Cell-free formation of RNA granules: low complexity sequence domains form dynamic fibers within hydrogels. Cell 149, 753–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lee M. T., Bonneau A. R., Takacs C. M., Bazzini A. A., DiVito K. R., Fleming E. S., Giraldez A. J. (2013) Nanog, Pou5f1 and SoxB1 activate zygotic gene expression during the maternal-to-zygotic transition. Nature 503, 360–364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Leichsenring M., Maes J., Mössner R., Driever W., Onichtchouk D. (2013) Pou5f1 transcription factor controls zygotic gene activation in vertebrates. Science 341, 1005–1009 [DOI] [PubMed] [Google Scholar]