Background: Neutrophils must generate well regulated forces to migrate within the confined, three-dimensional space of diseased tissues.
Results: Spatial confinement induces a switch to integrin-independent motility, but integrins regulate three-dimensional traction force generation.
Conclusion: Physical confinement is sufficient to induce integrin-independent motility.
Significance: Spatially confined double hydrogels are a biomimetic system that may allow for ex vivo testing of neutrophil-specific therapeutics.
Keywords: Cell Motility, Confocal Microscopy, Integrin, Leukocyte, Neutrophil
Abstract
Neutrophils are capable of switching from integrin-dependent motility on two-dimensional substrata to integrin-independent motion following entry into the confined three-dimensional matrix of an afflicted tissue. However, whether integrins still maintain a regulatory role for cell traction generation and cell locomotion under the physical confinement of the three-dimensional matrix is unknown, and this is challenging to deduce from motility studies alone. Using three-dimensional traction force microscopy and a double hydrogel sandwich system, we determined the three-dimensional spatiotemporal traction forces of motile neutrophils at unprecedented resolution and show, for the first time, that entry into a highly confined space (2.5D) is a sufficient trigger to convert to integrin-independent migration. We find that integrins exert a significant regulatory role in determining the magnitude and spatial distribution of tractions and cell speed on confined cells. We also find that 90% of neutrophil tractions are in the out-of-plane axis, and this may be a fundamental element of neutrophil traction force generation.
Introduction
An inflammatory response is initiated when circulating neutrophils enter the three-dimensional microenvironment of a damaged tissue or organ (1). A well regulated inflammatory response is rapid in onset, brief in duration, and sufficient to clear contaminating microbes and initiate healing. The consequence of an improperly regulated inflammatory response is medically significant. An inadequate response places the host at risk for the infection to spread. If the response is excessive, hyperactive neutrophils can cause damage to the tissues it is designed to protect (1, 2). In order to understand how an optimal inflammatory response occurs, it is important to identify factors within an infected or injured tissue that regulate neutrophil activity.
Similar to other mammalian cells, neutrophils generate and transmit forces to their surrounding environment in order to locomote; these are defined as cell tractions. On two-dimensional substrates, neutrophils, like most other eukaryotic cells, engage ligand-specific integrins to adhere and generate tractions (3–5). However, in vivo, as neutrophils pass through the blood vascular wall, they transit from a predominantly two-dimensional to a confined three-dimensional environment, where polymorphonuclear neutrophils reach migration speeds up to 20 μm/min (6, 7).
In 2008, Lämmermann et al. (8) reported that leukocytes devoid of integrins migrate with normal kinetics within a draining lymph node and in a confined fibrillar scaffold. This is consistent with an integrin-independent motility mechanism operative in three dimensions but not in two dimensions, in which surface contact forces produced by a forward expanding actin network are sufficient to drive cell motility (8–11). The biological relevance of the ability of a leukocyte to switch strategies of migration immediately upon entry into a three-dimensional extracellular microenvironment is that it allows these cells to function within any damaged or infected organ regardless of geometry or molecular composition. The physical components of a three-dimensional microenvironment include such factors as substrate stiffness, confinement, and matrix geometry, making it difficult to determine the contribution of each element in inducing the switch to integrin-independent migration in an extravasated leukocyte. Moreover, it is not clear whether integrins are entirely dispensable or exert a regulatory function in generating leukocyte traction in a three-dimensional setting.
To isolate the effect of physical confinement on the generation of traction, we combine three-dimensional traction force microscopy with a double hydrogel sandwich system (2.5D2 system) originally developed by Beningo et al. (12). The 2.5D terminology denotes the presence of confinement between two impenetrable gels without complete envelopment of the cell within the substratum. This experimental system overcomes the limitation of more widely used native gels, where changing confinement in the matrix structure coincidently affects matrix stiffness and ligand density, thereby coupling these physical parameters (13, 14). Furthermore, this system allows in situ temporal control of confinement and permits repeated measures of an individual cell as it transitions from a two-dimensional planar system to a 2.5D highly confined system.
Using three-dimensional traction force microscopy, we found that over 90% of neutrophil-generated forces are directed vertically against the extracellular matrix (out-of-plane direction). This disproportionate exertion of force in the out-of-plane direction by the neutrophil is unlike the distributions reported for adhesive cell types and for ameboid Dictyostelium cells, both of which produce both in-plane and out-of-plane forces but to equivalent extents (15–17). The magnitude of neutrophil traction increases significantly when the cells are placed on stiffer substrates or within a confined space, but the overall predominance of out-of-plane force generation is consistent and mechano-insensitive for stiffness and geometry. Furthermore, the relative spatiotemporal patterns of cell tractions remained unchanged upon entry into a highly confined space.
Function blocking of the β1β2 family integrins of confined neutrophils induced a significant change in the speed of cell migration and in the spatial distribution of cell tractions. Taken together, our data show that physical confinement itself is sufficient for triggering integrin-independent motility and that integrins continue to play a significant regulatory role for neutrophil force generation and migration speed.
EXPERIMENTAL PROCEDURES
Preparation and Mechanical Characterization of Polyacrylamide Substrates
Polyacrylamide gel substrates were prepared from acrylamide (40% (w/v); Bio-Rad) and N,N-methylene-bisacrylamide (2% (w/v); Bio-Rad) stock solutions as described previously (16, 17). The concentrations of acrylamide and N,N-methylene-bisacrylamide were varied to control the mechanical properties of the substrates and were chosen to be 8%/0.08% for stiffer substrates and 3%/0.2% for softer substrates. The chosen concentrations yielded Young's moduli of 8.3 ± 0.2 and 1.5 ± 0.1 kPa. All substrates contained 14% (w/v) fluorescent microspheres (0.5 μm in diameter, carboxylate-modified; Invitrogen). Polyacrylamide gel solution was vortexed for about 30 s, and 30 μl of polyacrylamide solution with green fluorescent microspheres were pipetted on the surface of an activated microscope slide (see below) for bottom substrate and sandwiched with a hydrophobic glass coverslip (22 mm in diameter), yielding a final thickness of around 60 μm. For the top substrate, 20 μl of polyacrylamide gel solution containing red fluorescent beads was deposited on top of an activated circular coverslide (30 mm in diameter) and sandwiched with a small hydrophobic coverslip (15 mm in diameter), yielding a final thickness of ∼60 μm.
Glass Coverslip and Microscope Slide Surface Modification
Rectangular (75 × 25 mm; Fisher) and circular (30 mm in diameter; Esslinger and Co., Mendota Heights, MN) microscope slides were chemically modified to allow covalent attachment of polyacrylamide substrates. Glass coverslips and slides were rinsed with ethanol and placed in a Petri dish containing a solution of 0.5% (v/v) 3-aminopropyltrimethoxysilane (Sigma-Aldrich) in ethanol for 5 min. Next, coverslips and microscope slides were washed with ethanol and submersed in a solution of 0.5% glutaraldehyde (Polysciences, Inc.) in deionized water for 30 min. Activated coverslips and slides were washed with deionized water and left to dry. To allow easier gel detachment, round glass coverslips of two different sizes (22 and 15 mm in diameter) were chemically modified to create hydrophobic surfaces. Briefly, glass coverslips were placed in a Petri dish containing 97% (v/v) hexane (Fisher), 2.5% (v/v) (tridecafluoro-1,1,2,2-tetrahydrooctyl)-triethoxysilane (Gelest, Morrisville, PA), and 0.5% (v/v) acetic acid (Sigma-Aldrich) for ∼1 min. Coverslips were then removed and left to dry.
To neutralize any effects due to complex lipopolysaccharides (LPS), 0.01% (v/v) of antibiotic polymyxin B (Invitrogen) was mixed into the gel solution. Cross-linking was initiated through the addition of ammonium persulfate (Sigma-Aldrich) and N,N,N,N-tetramethylethylenediamine (Invitrogen).
Mechanical Characterization of Polyacrylamide Gels
Characterization was based on our previously established testing protocols on a custom-built uniaxial compression device (16, 17). Gel samples were cast in circular nylon molds (16 mm in diameter and 10 mm in height). Following polymerization, the molds were removed, and the samples were submersed in deionized water. The custom-built compression device consisted of a centrally positioned linear actuator (Series A, Ultra Motion) equipped with a built-in linear encoder that provides displacement information with a displacement resolution of 1 μm. To measure the compressive forces acting on each sample, a 50-g linear force transducer (LCFA-50F, Omega Engineering Inc.) was attached to the end of the linear actuator.
To ensure uniaxial compression conditions, a ball point tip-platten top was placed on each gel sample, and all samples were carefully aligned along the central compression axis of the linear actuator. Nominal stress, σ, was computed by dividing the measured applied force by the circular contact area. Nominal strain, ϵ, was calculated by measuring the height change of each sample divided by the sample's original height.
Each sample was compressed at three different strain rates (10−3, 10−2, and 10−1) to capture any time dependence in the material behavior. Young's moduli were calculated from the slope of each linear stress-strain curve as E = σ/ϵ. The polyacrylamide gel can be described as neo-Hookean solid that has the same Young's modulus as the linear stress-strain relationship in the limit of small strains. The Poisson's ratio is taken to be 0.45, which is within the range of the typical values chosen for traction force microscopy studies. A set of representative material stress-strain curves used to determine the Young's modulus for each of the soft and stiff polyacrylamide gels is shown in Fig. 1.
FIGURE 1.
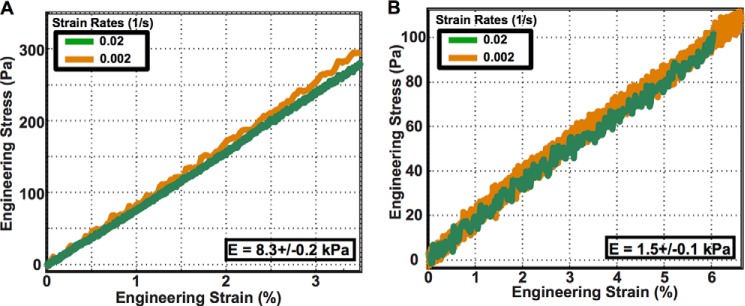
Stress-strain curves for an unconfined compression test of cylindrical polyacrylamide samples for stiffer (E = 8.3 kPa) substrates (A) and softer (E = 1.5 kPa) substrates (B).
Functionalization of Polyacrylamide Substrates
To promote cell attachment to polyacrylamide films, top and bottom substrates were functionalized with fibronectin using the bifunctional cross-linker, sulfo-SANPAH (Thermo Fisher) (16–18). Excess water was removed prior to deposition of 100 μl of sulfo-SANPAH (1 mg/ml) onto the surface of each film, followed by a 15-min exposure to UV light. The darkened sulfo-SANPAH solution was aspirated, and the procedure was repeated. The samples were thoroughly washed with deionized water and covered with a solution of 0.2 mg/ml fibronectin (Invitrogen) diluted in 50 mm HEPES (pH 8; Sigma-Aldrich) and left undisturbed at 4 °C overnight. Following overnight incubation, the substrates were rinsed three times with 1× PBS mixed with polymyxin B (0.01%, v/v) and sterilized with UV irradiation before depositing cells. Bifunctional cross-linker was also used for coating with polymer poly(l-lysine) with poly(ethylene glycol) chains (0.2 mg/ml final concentration; SuSoS AG).
Neutrophil Preparation
Neutrophils were isolated from healthy human volunteers into EDTA-containing Vacutainer tubes (BD Biosciences) according to Institutional Review Board procedures. Histopaque cell separation was followed by gravity sedimentation through 3% dextran. Hypotonic lysis to remove erythrocytes yielded a neutrophil purity of >95%. Neutrophils were suspended in Hanks' balanced salt solution (without Ca2+/Mg2+) on ice until use. All reagents contained <0.1 pg/ml endotoxin.
For traction measurements, ∼350,000 cells were resuspended in a vial containing 1 ml of phenol-free Leibovitz L-15 medium and 2 mg/ml glucose. Prior to imaging, formyl-methionyl-leucyl-phenylalanine (100 nm final concentration; Sigma-Aldrich) was added to the cell solution to activate and induce migration.
For integrin blocking experiments, we used a method described previously (4). For anti-β1 (clone 6S6; EMD Millipore) and anti-β2 (clone TS1/18; Thermo Fisher Scientific) integrin blockade, neutrophils were incubated in the 10 μg/ml concentration of antibody in Hanks' balanced salt solution(−/−) on ice for 20 min. Once incubation was complete, cells were transferred into L15 + glucose medium, where the antibody concentration was maintained throughout the experiment. For controls, neutrophils were pretreated with 20 μg/ml IgG1. Similar to blockade experiments, the concentration of 20 μg/ml was maintained throughout the whole experiment.
To inhibit actin polymerization in migrating neutrophils, cells were treated with latrunculin A (Invitrogen). At high concentrations of latrunculin A, treated neutrophils neither adhere to nor generate measurable forces on the substrates. Thus, a variety of the concentrations (0.5–0.01 μm) were tried during the experiment, and 0.2 μm was chosen as an optimal concentration for force measurements. Before each experiment, cells were incubated in the presence of latrunculin A on ice for 30 min. Once incubation was complete, cells were transferred into L15 + glucose medium, where the inhibitor concentration was maintained throughout the experiment.
To inhibit myosin II in migrating neutrophils, cells were pretreated with blebbistatin at 50 μm concentration on ice for 30 min. Similarly, cells were then transferred into L15 + glucose medium prior to the experiment, and the inhibitor concentration was maintained throughout the duration of the experiment. Due to sensitivity of blebbistatin to blue light (488-nm laser), both top and bottom substrates were impregnated with red fluorescent beads and excited with a green (561 nm) laser for all blebbistatin-treated cell experiments.
Live Cell Imaging
Three-dimensional image stacks were acquired using a Nikon A-1 confocal system mounted on a TI Eclipse inverted optical microscope controlled by NI-Elements Nikon Software. A ×40 plan fluor air objective mounted on a piezo objective positioner was used for all of the experiments, which allowed imaging at speeds up to 30 frames/s. Green fluorescent microspheres were used for the bottom substrate and excited with an argon (488-nm) laser. Red 0.5-μm fluorescent microspheres in the top substrate were excited with a diode 561-nm laser. 512 × 512 × z voxels (102 μm × 102 μm × z) confocal volume stacks were recorded every 15 s, with z ranging from 128 to 250 pixels (∼38–75 μm), depending on whether only the bottom (two-dimensional) or both the bottom and top (three-dimensional) substrates were imaged simultaneously. To ensure physiological imaging conditions within the imaging chamber, the temperature was controlled at 37 °C during time lapse recording as described previously (17). During the experiment, the cell outline was estimated from phase-contrast microscopy.
Calculation of Displacements and Three-dimensional Tractions
Three-dimensional image stacks of the motion of the fluorescent beads were recorded by using a Nikon A-1 confocal system mounted on a TI Eclipse inverted optical microscope controlled by NI-Elements Nikon software. Cell-generated full-field displacements and tractions were determined using our recently developed large deformation three-dimensional traction force microscopy method (19). Briefly, three-dimensional time-lapsed volumetric images of fluorescent beads embedded in polyacrylamide substrates were recorded using laser scanning confocal microscopy. A new fast iterative digital volume correlation algorithm was used to track the motion of the embedded fluorescent beads in all three dimensions between time increments (20). This newly developed fast iterative digital volume correlation technique provides significantly higher spatial resolution, signal/noise, and faster computation times than our previous digital volume correlation algorithm (21). When quantifying neutrophil-induced substrate deformations, we find that the displacement gradients are non-negligible, which requires stress and traction analysis within a finite, rather than a linear, continuum framework. As shown previously, polyacrylamide gels undergoing large deformations can be constitutively described as a neo-Hookean material (22). Thus, cell-generated substrate stresses, σ, can be calculated as follows,
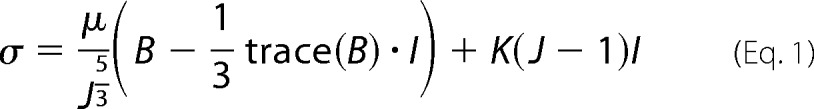 |
where μ and K represent the shear and bulk moduli of the polyacrylamide gels. Both of the quantities are related to the Young's modulus of a material by the following:
 |
 |
The quantity B = F·FT is the left Cauchy-Green tensor, and J is the Jacobian of deformation gradient tensor F. In both of these relations, the deformation gradient tensor F is calculated from the full-field displacements using an 11-tap discrete differentiation kernel described by Farid et al. (23), which we showed to provide optimal spatial accuracy in the presence of noise (20).
Finally, the three-dimensional neutrophil surface tractions can be calculated using the well known Cauchy relation,
where n is the surface normal vector. The surface normal vectors were determined directly from the raw laser scanning confocal microscopy images. In brief, we take advantage of the spatial locations of the embedded fluorescent beads in the raw laser scanning confocal microscopy images. A scattered data representation of the surface is built by finding a bead's maximum z location within a window that slides in the x–y plane. After least-squared fitting and surface gradient smoothing, the surface normals are calculated from a Delaunay triangulation of the surface (24). The subsequent surfaces in the time lapse are computed by translating the surface points in the reference configuration by the displacement calculated via our fast iterative digital volume correlation algorithm. The magnitude of the three-dimensional traction vector in the global coordinate system is then calculated as follows,
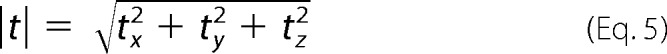 |
where tx and ty are the in-plane traction force components under the cell and tz corresponds to the out-of-plane component. We calculate the total force exerted by the cell on its substrate by integrating the magnitude of all surface tractions over the total cell area S as follows.
 |
Similarly, the traction vector was resolved into components acting in plane and out of plane to the surface (25).
In-plane and out-of-plane forces were then calculated as follows.
 |
 |
The volume integrals in this study were numerically carried out using an eight-point Gaussian quadrature of hexahedral elements with nodes centered at the grid point locations of the stress-strain values. All computations were performed in MATLAB 2013B (MathWorks, Natick, MA).
The resolution of our fast iterative digital volume correlation technique was determined by measuring the noise floor outside of the projected cell area for each experimentally obtained cell deformation field. This was done for all of the data sets for bottom and top substrates separately. Based on laser scanning confocal microscopy, this technique yields a displacement noise floor of 0.08 μm in the x and y (in-plane) and 0.15 μm in the z (out-of-plane) directions. The calculated noise floor for the in-plane and out-of-plane tractions and total forces are summarized in Table 1. Any traction data with a signal/noise ratio less than 2 was excluded from analysis.
TABLE 1.
Noise floor estimates for the in-plane and out-of-plane tractions (Pa) and total forces (nN)
|
E = 1.5 kPa (soft) |
E = 8.3 kPa (stiff) |
||||
|---|---|---|---|---|---|
| Bottom | Top | Bottom | Top | ||
| In-plane tractions, t | 40 Pa | 50 Pa | 200 Pa | 250 Pa | |
| Out-of-plane tractions, t | 200 Pa | 200 Pa | 700 Pa | 800 Pa | |
| In-plane force, F | 0.4 nN | 0.4 nN | 1.7 nN | 2.2 nN | |
| Out-of-plane force, F | 1.7 nN | 1.7 nN | 6.2 nN | 7.1 nN | |
Cell Speed Calculations
To calculate polymorphonuclear neutrophil speed during migration in both two- and three-dimensional systems, cell boundaries from captured phase images were recorded, and cell centroids were computed during each imaging increment using a custom-written Matlab algorithm. Using this information, cell speed per time increment can be calculated as follows.
 |
It should be noted that because the cell speed information is based on centroid calculations derived from phase images, only the in-plane (x, y) velocity components could be detected.
Global Force and Moment Balance
We computed the sum of all forces and moments acting on any given control volume inside each polyacrylamide gel at every time point to verify that static force and moment equilibrium are satisfied. The overall procedure is identical to our previously published methodology (17). Under all experimental conditions and imaging time points, we found static equilibrium satisfied with force and moment magnitudes on the order of 10−10 to 10−9 newtons (forces) and 10−15 to 10−14 newtons·m (moments), respectively. These numbers are consistent with our noise floor measurements.
Statistical Analysis
Data were subjected to the χ2 goodness of fit test to determine whether a data sample came from a normal distribution. The Kruskal-Wallis non-parametric test was used to compare the medians of the groups of data to determine whether the samples came from the same population. The Wilcoxon rank-sum test was used for comparison of the medians of the data and their interactions.
RESULTS
Neutrophil Force Generation as a Function of Spatial Confinement
To investigate how confinement affects the traction signatures of moving neutrophils, cells were seeded between two compliant fibronectin-coated polyacrylamide substrates separated by an adjustable gap distance to switch from a two-dimensional to a highly confined (2.5D) microenvironment. In order to control confinement of the neutrophil gel system, a custom-designed micromanipulator system was built. Fig. 2 presents a schematic of the micromanipulator and neutrophil gel system (Fig. 2A) as well as the orientation of the surface normal vectors used in the three-dimensional traction calculations (Fig. 2B).
FIGURE 2.
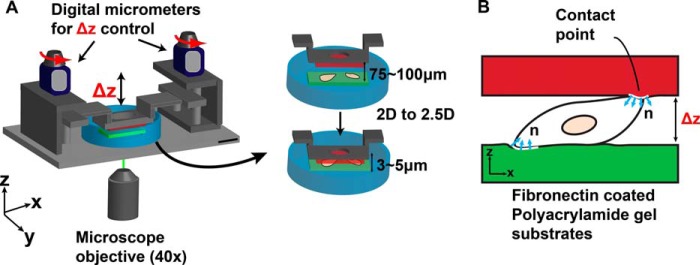
A, schematic overview of a custom-built micromanipulator to switch from a two-dimensional to a confined 2.5D neutrophil gel system. B, a schematic of the cell surface contact points and definition of the surface normal on the top (red) and bottom (green) substrates.
To determine dynamic cell tractions, neutrophil-generated substrate deformation fields were recorded and analyzed using our recently developed high resolution three-dimensional traction force microscopy technique (19). Although the basic working principle is similar to our previous three-dimensional traction force microscopy technique (16, 17), this new technique features significantly improved spatial resolution, signal/noise, and computational efficiency. To determine the influence of substrate stiffness on neutrophil force generation and speed as the matrix changes from an unconfined two-dimensional to a confined 2.5D system, two substrate stiffnesses were chosen: E = 1.5 kPa to mimic soft tissues, such as endothelial or lung tissue, and E = 8.3 kPa to mimic stiffer tissues, like muscle (Fig. 1).
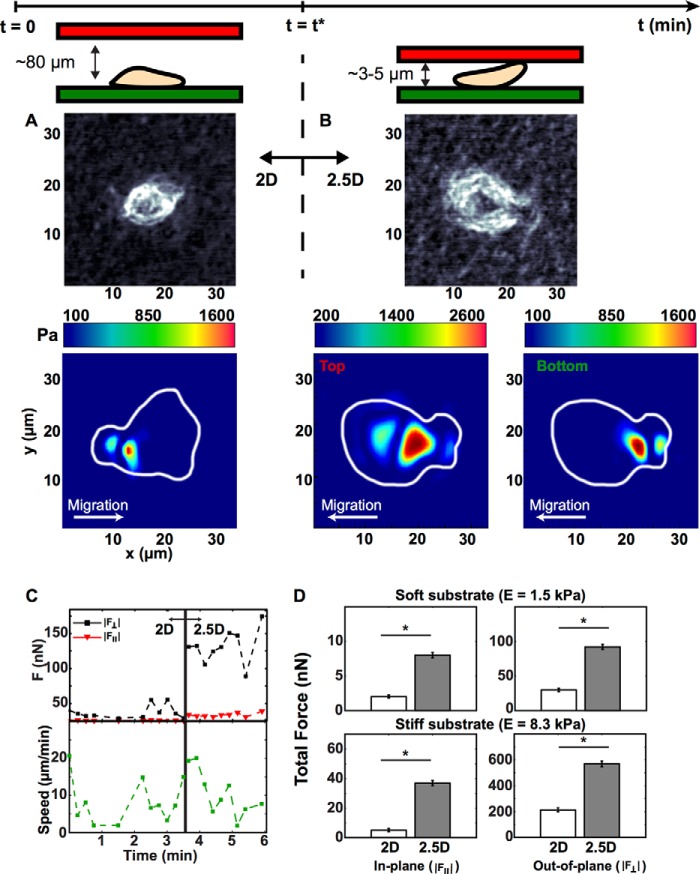
Initially, polymorphonuclear neutrophils were seeded on the bottom gel (Fig. 2A) while maintaining a gap height of ∼80 μm, which is significantly larger than the average spherical cell diameter (8 μm (26)), thus simulating a two-dimensional environment. Once adhered to the bottom gel, time lapse videos of cellular motion and cell-generated substrate deformations were recorded every 15 s for a total of 10 min via confocal microscopy. This time period is sufficient to recapitulate cellular morphologies and trajectories when compared with previous two-dimensional studies (3–5). Fig. 3 shows a representative map of cellular tractions as a single neutrophil switched from a two-dimensional to a confined environment (gel modulus: E = 1.5 kPa). Fig. 3A depicts a snapshot of the neutrophil morphology and a color contour map of the magnitude of the three-dimensional traction vector during chemokinesis in two dimensions (gap height ∼80 μm). The magnitude of the three-dimensional traction vector peaked around 1300 Pa at the rear of the cell relative to its direction of motion. The observation of asymmetric traction profiles is consistent with previously reported two-dimensional studies on neutrophil motility (3, 5).
FIGURE 3.
A and B, phase-contrast images of a neutrophil and its associated cell traction profiles in a two-dimensional system (A) and a 2.5D system (B). The color contour map displays the magnitude of the three-dimensional traction vector. C, dynamic out-of-plane (black) and in-plane (red) total forces and cell speed (green) for a neutrophil during dimensional switch from a two-dimensional to a 2.5D system. D, total out-of-plane (right column) and in-plane (left column) force components in our two-dimensional (white) and 2.5D (gray) systems on soft and stiff substrates (top and bottom rows). Quantitative data reflect the median and S.E. (*, p < 0.01 versus two-dimensional, Kruskal-Wallis test; n > 100).
Following the initial two-dimensional acquisition period (t = t*), the top substrate was lowered to an effective substrate-to-substrate gap size of ∼3–5 μm, which is slightly less than the average spherical neutrophil cell diameter (26). This change in gap size generated a confined 2.5D space, allowing for neutrophil attachment to the top and bottom surface. Cell position and substrate deformations were recorded for another 10 min at 15-s intervals. Fig. 3B is representative of the typically observed neutrophil morphology and the magnitude of the three-dimensional traction vectors for both bottom and top substrates. The bottom contour plot shows cellular traction generation with magnitudes similar to that shown in Fig. 3A, whereas the top substrate shows roughly a doubling of tractions when compared with the two-dimensional case. The asymmetric distribution of tractions seen in two dimensions was maintained in confinement in both the upper and lower gels. Although these data represent only single time points during time lapse, they represent the typical behavior observed for most neutrophils in two-dimensional and confined 2.5D environments.
Even more striking is the breakdown of the total force into its in-plane and out-of-plane components during the transition to confinement. Fig. 3C illustrates strong up-regulation of the out-of-plane force component as the neutrophil switched from a two-dimensional to a confined 2.5D environment with little attenuation in its in-plane component. Similarly striking is the observation that the average neutrophil speed remained unattenuated despite the significant increase in total traction force. Dynamic oscillations in force and speed magnitudes observed in Fig. 3C are similar to the force undulations observed in previous two-dimensional studies, where these oscillations are generated by variations in the amoeboid cell protrusion, contraction, adhesion, and relaxation force transmission cycle (27, 28).
To account for the donor dependence and variance among individual neutrophils, we computed the total in-plane and out-of-plane forces for all recorded cells (n > 100; summarized in Fig. 3D, represented by their median and S.E. For both substrate stiffnesses, the in-plane and out-of-plane force components are approximately 3 times larger in the confined system as compared with two dimensions, although force magnitudes scale with substrate stiffness. The ratios of out-of-plane to in-plane force components are significantly higher than previous measurements on other anchorage-dependent cells (17, 29). Higher out-of-plane force measurements support previous theories of a fast peeling mechanism during cellular detachment for amoeboid-type locomoting cells (30–32).
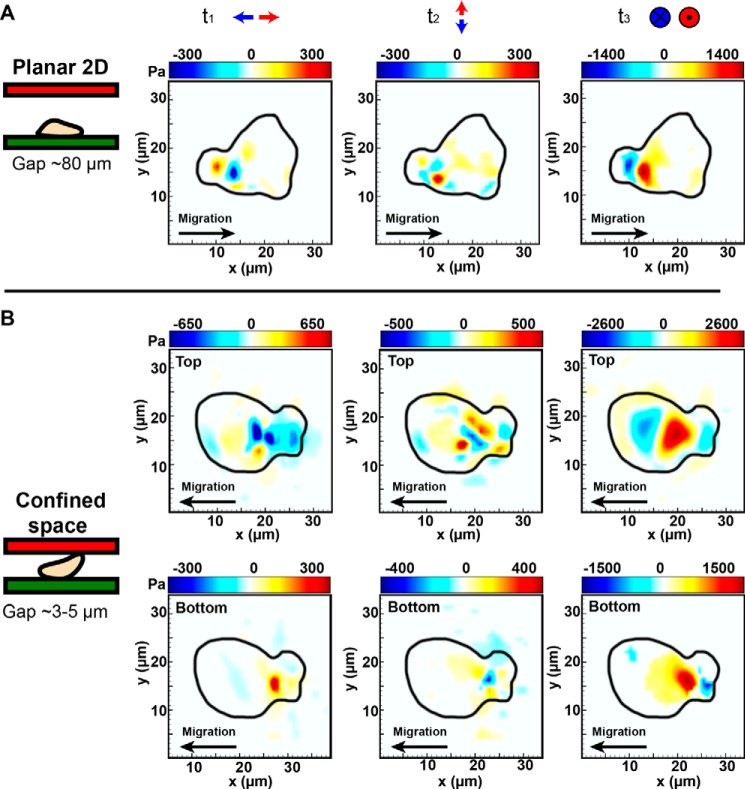
Although the spatial distribution of surface tractions shows strong similarities in the bottom gels for both the two-dimensional and 2.5D cases, the intriguing distribution of tractions in the upper gels prompted closer examination of each traction component individually plotted in Fig. 4. As can been seen in Fig. 4B, the out-of-plane (t3) traction component is the major contributor of all surface tractions generated in the upper gel. Spatial localization of the t3 traction component in the upper gel visually coincides with the nuclear region observed in the phase images, suggesting a significant contribution of the nuclear region to the measured upper gel surface tractions during locomotion.
FIGURE 4.
Decomposition of traction forces shown in Fig. 3 (A and B) into their t1,t2 (in-plane) and t3 (out-of-plane) traction components in two dimensions (A) and 2.5D (B). All of the contour maps and color bars represent tractions in Pa. Positive and negative t3 traction are defined using the local, positive outward normal vector for each upper and lower gel as illustrated in Fig. 2B. Comparison of the two-dimensional and 2.5D traction profiles reveals similar spatiotemporal patterns with significant traction generation in the uropod of the cell. Traction profiles in 2.5D show a significant contribution from the central, nuclear region of the neutrophil against the top surface.
Neutrophil Forces and Speed Are Integrin-independent in Confined 2.5D Environments
On two-dimensional substrates, neutrophils engage ligand-specific integrins to adhere and generate forces (4, 5). However, it has been reported that integrins are dispensable for migration in three-dimensional spaces (10, 33). A recently proposed three-dimensional leukocyte migration model suggests that there might be two different modes of force generation. The dominant mechanism in two dimensions relies on forming adhesive ligand-integrin contacts and is, therefore, adhesion-based. Locomotion in three dimensions is characterized by pressure-induced substrate deformations, which do not rely on the use of integrins and so are integrin-independent (9, 10).
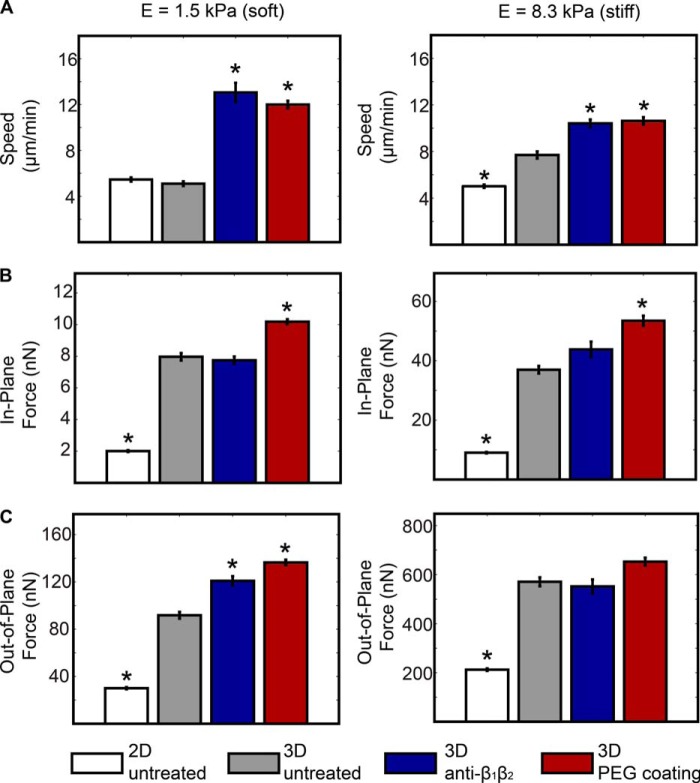
Our studies isolated spatial confinement as an experimental variable to determine whether confinement is sufficient for the acquisition of integrin-independent neutrophil motility as would be encountered in a complex three-dimensional setting. Measures of cell speed, force generation, and the spatial distribution of tractions of a given cell were determined on fibronectin-conjugated hydrogels of different stiffnesses beginning in two dimensions with repeated measures taken over time as confinement (2.5D) was imposed. Speed, traction, and force measures were also determined in the presence or absence of function-blocking integrin antibodies (Fig. 5). Integrin blockade was performed to cover all β1 and β2 integrins (such as CD11b/CD18), and cells were also assayed on PEGylated surface providing complete absence of matrix ligands. Under conditions of pan-β1 and β2 inhibition on soft and stiff gels, neutrophil speed and force generation was either unchanged or increased, demonstrating a lack of dependence on integrins for traction force generation. Cells treated with nonspecific IgG at identical concentrations were not different from untreated control cells (not shown).
FIGURE 5.
Neutrophils migrate and generate forces in the absence of integrin-engaged adhesions. Bar plots present the average cell speed (A) and in-plane (B) and out-of-plane forces (C) on soft (left) and stiff (right) substrates in two-dimensional and 2.5D systems. All of the data are presented for untreated and treated (anti-β1β2 (10 μg/ml each)) neutrophils on fibronectin and untreated neutrophils on PEG-coated substrates. All quantitative data are represented by their medians and S.E. (error bars). Statistical significance is shown against the untreated cells in 2.5D (*, p < 0.01 versus 2.5D, untreated, Kruskal-Wallis test; n > 100).
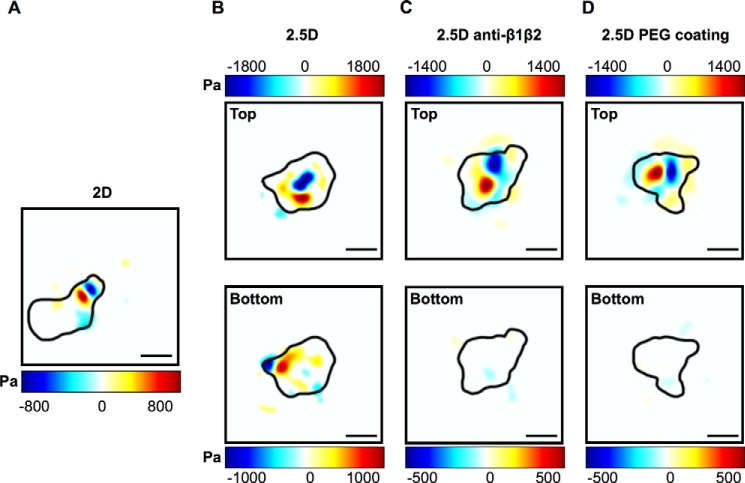
Although these calculations (Fig. 5) show that upon imposition of confinement, the total force magnitudes and speeds produced by neutrophils are not reliant on integrins, they do not determine whether integrins play a subtler regulatory role in the traction generation under confinement. Fig. 6 presents traction patterns generated by cells as a function of confinement and integrin utilization. Findings shown are for the predominating out-of-plane traction component for neutrophils moving on planar two-dimensional and, subsequently, under highly confined 2.5D conditions. The application of confinement does not alter traction distribution. Pan-inhibition of β1 and β2 integrins or the absence of any ligand (PEG surface) significantly alters traction patterns during locomotion (Fig. 6, C and D). Here, instead of the typically featured traction dipole pattern (Figs. 3 and 6 (A and B)), which is characteristic of an adhesion-based motility mode (27), the primary traction contribution probably stems from surface contact pressures between the nuclear lobes of the cell with the top surface. Although the bottom surfaces for both Fig. 6, C and D, do not show any significant tractions above our measurement noise, integration of our resolution limit (Table 1) across the bottom surface yields a force equal in magnitude and opposite to the neutrophil's force imposed against the top surface. This implies that the type of motility featured in Fig. 6, C and D, does not engage the surface at focal points but rather distributes its contact forces across the entire surface of the cell. Thus, these data show that integrins continue to be utilized by a locomoting neutrophil to affect traction distribution even after entry into a highly confined space.
FIGURE 6.
Color contour maps of the out-of-plane component of the three-dimensional traction vector highlighting integrin-dependent traction generation in unconfined two-dimensional (A) and confined 2.5D (B) systems as well as integrin independence upon anti-β1β2 blockade (C) and in the absence of matrix ligand on PEG-coated substrates (D). Integrin-dependent traction generation shows a distinct localized out-of-plane traction distribution in the uropod of the cell (A and B) similar to the results shown in Fig. 4. In the absence of integrins, neutrophils generate most of their surface tractions using contact pressure of their nuclear region (C and D). All of the color bars represent tractions in Pa. Positive and negative t3 traction are defined using the local, positive outward normal vector for each upper and lower gel as illustrated in Fig. 2B. Scale bar, 10 μm.
We examined the temporal evolution of the total force and speed patterns of untreated and antibody-treated neutrophils to determine whether integrin blockade would introduce significant differences in their temporal profiles (Fig. 7). We did not find any significant differences in the temporal distribution of neutrophil speed and force patterns except for changes in their mean levels (Fig. 7, dashed line), which are reflected in Fig. 5. The general oscillations in the neutrophil-generated forces shown in Fig. 7 are similar to the force undulations observed in previous two-dimensional studies during chemokineses, where these oscillations are generated by variations in the amoeboid cell protrusion, contraction, adhesion, and relaxation force transmission cycle (27, 28). Experiments were conducted to investigate whether changes in cell-generated forces correlate to changes in cell speed for control and integrin-blocked cells. Dynamic oscillations between speed and in-plane and out-of-plane forces do not statistically correlate. This result is in agreement with the work of Meili et al. (28), where it is shown that migrating leukocytes attain their peak forces during the contraction phase, whereas the highest speeds are reached during the protrusion step.
FIGURE 7.
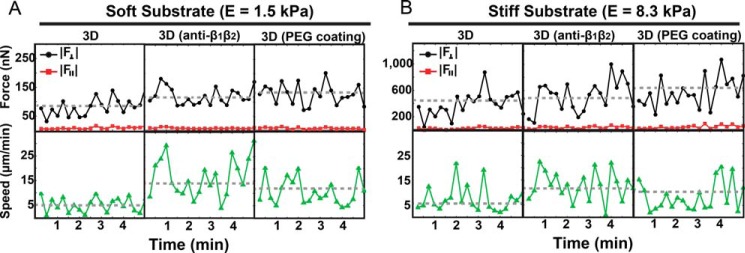
Temporal evolution of the out-of-plane (black) and in-plane (red) neutrophil-generated total forces and the corresponding cell speeds (green) in a 2.5D system on soft (A) and stiff (B) substrates. The data are presented in 2.5D for untreated neutrophils and pan-β1β2 (10 μg/ml of each) antibody-blocked cells migrating on fibronectin-coated substrates as well as untreated neutrophils on PEG-coated substrates.
The Role of Actin-based Protrusive and Myosin-based Contractile Forces during Confinement
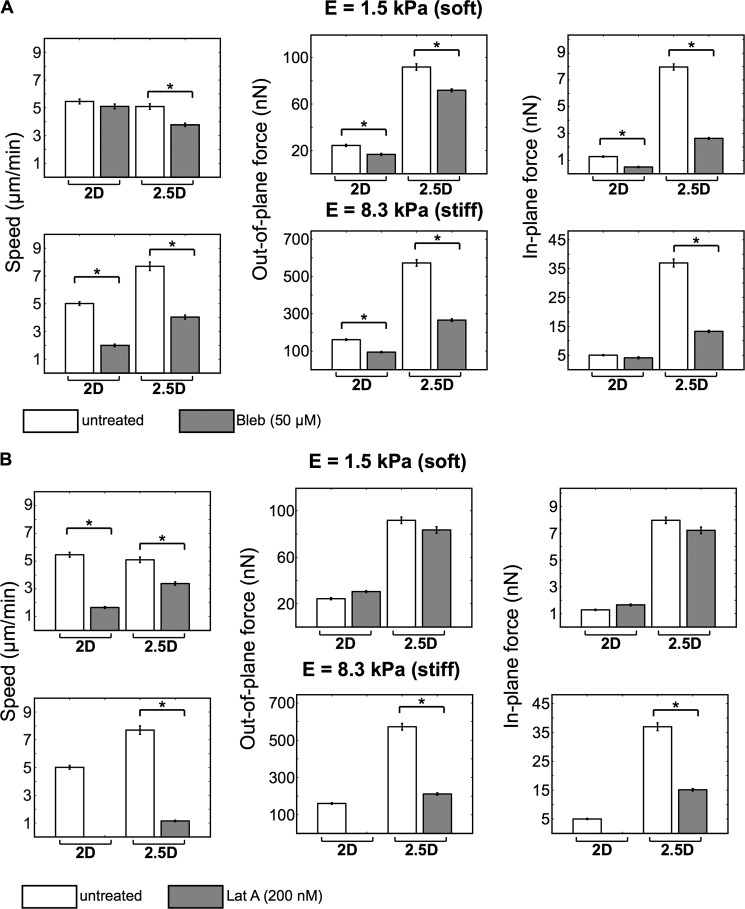
To determine the effect of confinement on the cytoskeletal force-motility machinery of the neutrophil, actin and myosin were differentially blocked with latrunculin A and blebbistatin. In confined 2.5D environments, cell speed was dependent on contributions from both actin- and myosin-based machineries and is not a function of matrix stiffness (Fig. 8). The role of myosin in cell speed and force generation, other than a modest effect of blebbistatin on soft substrates, is significant regardless of confinement and substrate stiffness (Fig. 8A). Of speed, in-plane, and out-of-plane forces, myosin inhibition had the most pronounced effect on reducing the in-plane shear forces of confined neutrophils irrespective of substrate stiffness.
FIGURE 8.
Neutrophil motility and force generation decrease with inhibition of myosin-based contractile (A) and actin-based protrusive (B) forces. Bar plots present the average cell speed and in-plane and out-of-plane forces on soft (top) and stiff (bottom) substrates in our two-dimensional and 2.5D systems. All of the data are presented for untreated and treated (50 μm blebbistatin and 200 nm latrunculin A, respectively) neutrophils on fibronectin-coated substrates. All quantitative data are represented by their medians and S.E. (error bars). Statistical significance is shown against the untreated cells (*, p < 0.01 versus untreated, Kruskal-Wallis test; n > 100).
Like myosin, actin plays an essential role in generating cell speed under confinement and irrespective of stiffness as noted by sensitivity to latrunculin A (Fig. 8B). Sensititivity to latrunculin A with regard to traction force production is mechanosensitive under confinement, affecting both in-plane and out-of-plane forces on stiff, but not soft, gels (Fig. 8). These results are consistent with the notion that cells generate larger tractions during locomotion on stiffer substrates, which requires significant actin polymerization and F-actin assembly to sustain the increase in cell tractions. Whereas in 2.5D, neutrophils can generate significant contact pressures against the confining upper and lower surfaces, cells in two dimensions rely exclusively on actomyosin force generation, which requires myosin recruitment to F-actin. Without the capacity to polymerize enough F-actin, actomyosin force generation is unsuccessful, leading to complete abrogation of force and speed generation, as shown in Fig. 8B.
A similar and consistent picture emerges when examining the temporal evolution of neutrophil total force and speed patterns in the presence of differential actomyosin blockade (Fig. 9). Of note is the importance of both actin polymerization and myosin II activity to achieve large temporal changes in speed and force production indicated by the pronounced undulations in the force and speed profiles for untreated cells versus blebbistatin- and latrunculin-treated cells.
FIGURE 9.
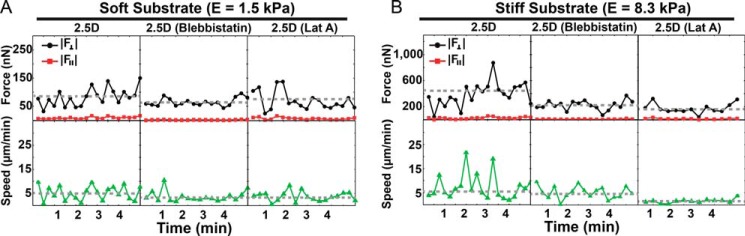
Temporal evolution of the out-of-plane (black) and in-plane (red) neutrophil-generated total forces and corresponding speeds (green) in a 2.5D system on soft (A) and stiff (B) substrates. The data are presented for neutrophils with and without respective blebbistatin and latrunculin A treatment.
DISCUSSION
In this study, we utilized a tunable, double-hydrogel system that allowed neutrophils to be studied under conditions that more closely model a tissue microenvironment. Using three-dimensional traction force microscopy, both in-plane (x,y) and out-of-plane (z) traction force measurements were obtained for neutrophils in two dimensions and under highly confined 2.5D conditions. We find that the traction force signature of the neutrophil is highly three-dimensional, with the strongest traction force contribution stemming from the out-of-plane traction component regardless of confinement or substrate stiffness. This suggests that the predominance of out-of-plane tractions is a generalized mechanism by which neutrophils exert forces upon their substratum, whether planar or confined, and under integrin-dependent or -independent conditions.
Whereas inhibition of ligand-specific integrins prohibits cell attachment and force generation in two dimensions, we show that integrin blockade does not limit force generation or cell speed in a highly confined system. We show that in the presence of integrin function, neutrophil-generated traction patterns are up-regulated in 2.5D as compared with two-dimensional planar surfaces. Moreover, integrin blockade of cells in 2.5D resulted in a distinct change in cellular traction patterns and increase in cell speeds. Taken together, these findings show that a change in physical confinement is sufficient to switch neutrophils to an integrin-independent motility mode, but the magnitude of traction and the speed of migration are integrin-regulated. The 2.5D system also showed that the cell need not be completely enveloped by a three-dimensional matrix in order to generate in-plane and out-of-plane force without integrins. The observation of significant out-of-plane tractions under such conditions can be explained by a general friction-based motility mechanism, which had been previously put forth theoretically but never quantified experimentally at the level of traction resolution presented here (34, 35). Adhesion strength of bonds formed between cells and substrate is a direct function of traction angle. A traction angle of 90° (i.e. out of plane) correlates with the weakest adhesion and shortest bond lifetime, and formation of such bonds may correlate with rapid motility (36, 37). This model of rapid motility is in contrast to the situation for the adhesion of mesenchymal cells, which form a relatively even distribution of in-plane and out-of-plane tractions but adhere more tightly and move more slowly (16, 38).
Surprisingly, for neutrophils placed under hydrogel confinement, integrin blockade up-regulated cell speed on both soft and stiff matrices, suggesting that integrins may serve as a regulatory brake for motility as a function of confinement. The biological impact of regulating the migration speed of extravasated neutrophils by integrins cannot be discerned by an ex vivo system, but our findings indicate that, upon entry into a confined microenvironment, integrins may continue to be consequential to functions relevant to host defense. For example, integrins have been shown to regulate neutrophil apoptosis, respiratory burst, phagocytosis, and NETosis (2, 39). Whether integrins continue to regulate these functions within a three-dimensional microenvironment warrants further study.
We investigated the role of actin and myosin II activity during force-dependent motility and found that neutrophil speed and force generation in 2.5D decreased upon inhibition of myosin II and actin polymerization but was not decreased by the absence of integrin engagement (Figs. 5–7). To our knowledge, these are the first quantitative three-dimensional force measurements for human neutrophils across different substrate dimensions and stiffnesses following inhibition of their cellular force machinery. Similar observations have been documented previously for dendritic cells confined in a three-dimensional collagen network, where the inhibition of actin polymerization and myosin II contraction reduces cell migration speeds, but integrin blockade was without effect (9).
In sum, we demonstrate that confinement is a sufficient condition to switch neutrophil motility from integrin dependence to integrin independence but that integrins continue to operate in a regulatory capacity. The sensitivity to both geometric and mechanical cues encountered by neutrophils in the three-dimensional microenvironment should be considered in the future development of anti-inflammatory drugs.
Acknowledgments
We thank Xian O'Brien and Angel Byrd for helpful discussions.
This work was supported, in whole or in part, by National Institutes of Health Grants GM066194 (to J. S. R.) and AI101469 (to J. S. R. and C. F.). This work was also supported by a Brown University seed grant (to J. S. R. and C. F.).

This article contains supplemental Figs. S1–S3.
- 2.5D
- combination of three-dimensional traction force microscopy with double hydrogel sandwich
- Pa
- pascals.
REFERENCES
- 1. Nathan C. (2006) Neutrophils and immunity: challenges and opportunities. Nat. Rev. Immunol. 6, 173–182 [DOI] [PubMed] [Google Scholar]
- 2. Mayadas T. N., Cullere X. (2005) Neutrophil β2 integrins: moderators of life or death decisions. Trends Immunol. 26, 388–395 [DOI] [PubMed] [Google Scholar]
- 3. Smith L. A., Aranda-Espinoza H., Haun J. B., Dembo M., Hammer D. A. (2007) Neutrophil traction stresses are concentrated in the uropod during migration. Biophys. J. 92, L58–L60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Oakes P. W., Patel D. C., Morin N. A., Zitterbart D. P., Fabry B., Reichner J. S., Tang J. X. (2009) Neutrophil morphology and migration are affected by substrate elasticity. Blood 114, 1387–1395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Jannat R. A., Dembo M., Hammer D. A. (2011) Traction forces of neutrophils migrating on compliant substrates. Biophys. J. 101, 575–584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Stroka K. M., Aranda-Espinoza H. (2009) Neutrophils display biphasic relationship between migration and substrate stiffness. Cell Motil. Cytoskeleton 66, 328–341 [DOI] [PubMed] [Google Scholar]
- 7. Butler K. L., Ambravaneswaran V., Agrawal N., Bilodeau M., Toner M., Tompkins R. G., Fagan S., Irimia D. (2010) Burn injury reduces neutrophil directional migration speed in microfluidic devices. PLoS One 5, e11921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lämmermann T., Bader B. L., Monkley S. J., Worbs T., Wedlich-Söldner R., Hirsch K., Keller M., Förster R., Critchley D. R., Fässler R., Sixt M. (2008) Rapid leukocyte migration by integrin-independent flowing and squeezing. Nature 453, 51–55 [DOI] [PubMed] [Google Scholar]
- 9. Lämmermann T., Sixt M. (2009) Mechanical modes of “amoeboid” cell migration. Curr. Opin. Cell Biol. 21, 636–644 [DOI] [PubMed] [Google Scholar]
- 10. Renkawitz J., Sixt M. (2010) Mechanisms of force generation and force transmission during interstitial leukocyte migration. EMBO Rep. 11, 744–750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lämmermann T., Germain R. N. (2014) The multiple faces of leukocyte interstitial migration. Semin. Immunopathol. 36, 227–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Beningo K. A., Dembo M., Wang Y.-L. (2004) Responses of fibroblasts to anchorage of dorsal extracellular matrix receptors. Proc. Natl. Acad. Sci. U.S.A. 101, 18024–18029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Miron-Mendoza M., Seemann J., Grinnell F. (2010) The differential regulation of cell motile activity through matrix stiffness and porosity in three dimensional collagen matrices. Biomaterials 31, 6425–6435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Carey S. P., Kraning-Rush C. M., Williams R. M., Reinhart-King C. A. (2012) Biophysical control of invasive tumor cell behavior by extracellular matrix microarchitecture. Biomaterials 33, 4157–4165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Delanoë-Ayari, Rieu J. P., Sano M. (2010) 4D traction force microscopy reveals asymmetric cortical forces in migrating Dictyostelium cells. Phys. Rev. Lett. 105, 248103. [DOI] [PubMed] [Google Scholar]
- 16. Maskarinec S. A., Franck C., Tirrell D. A., Ravichandran G. (2009) Quantifying cellular traction forces in three dimensions. Proc. Natl. Acad. Sci. U.S.A. 106, 22108–22113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Franck C., Maskarinec S. A., Tirrell D. A., Ravichandran G. (2011) Three-dimensional traction force microscopy: a new tool for quantifying cell-matrix interactions. PLoS One 6, e17833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Dembo M., Wang Y.-L. (1999) Stresses at the cell-to-substrate interface during locomotion of fibroblasts. Biophys. J. 76, 2307–2316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Toyjanova J., Bar-Kochba E., López-Fagundo C., Reichner J., Hoffman-Kim D., Franck C. (2014) High resolution, large deformation 3D traction force microscopy. PLoS One 9, e90976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bar-Kochba E., Toyjanova J., Andrews E., Kim K-S., Franck C. (2014) A fast iterative digital volume correlation algorithm for large deformations. Exp. Mech. 10.1007/s11340-014-9874-2 [DOI] [Google Scholar]
- 21. Franck C., Hong S., Maskarinec S. A., Tirrell D. A., Ravichandran G. (2007) Three-dimensional full-field measurements of large deformations in soft materials using confocal microscopy and digital volume correlation. Exp. Mech. 10.1007/s11340-007-9037-9 [DOI] [Google Scholar]
- 22. Long R., Hall M. S., Wu M., Hui C.-Y. (2011) Effects of gel thickness on microscopic indentation measurements of gel modulus. Biophys. J. 101, 643–650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Farid H., Simoncelli E. P. (2004) Differentiation of discrete multi-dimensional signals. IEEE Trans. Image Process. 13, 496–508 [DOI] [PubMed] [Google Scholar]
- 24. D'Errico J. (2005) Surface fitting using gridfit. MATLAB Central File Exchange [Google Scholar]
- 25. Bower A. F. (2010) Applied Mechanics of Solids, pp. 100–102, CRC Press, Boca Raton, FL [Google Scholar]
- 26. Ting-Beall H. P., Needham D., Hochmuth R. M. (1993) Volume and osmotic properties of human neutrophils. Blood 81, 2774–2780 [PubMed] [Google Scholar]
- 27. Shin M. E., He Y., Li D., Na S., Chowdhury F., Poh Y. C., Collin O., Su P., de Lanerolle P., Schwartz M. A., Wang N., Wang F. (2010) Spatiotemporal organization, regulation, and functions of tractions during neutrophil chemotaxis. Blood 116, 3297–3310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Meili R., Alonso-Latorre B., del Alamo J. C., Firtel R. A., Lasheras J. C. (2010) Myosin II is essential for the spatiotemporal organization of traction forces during cell motility. Mol. Biol. Cell 21, 405–417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hur S. S., Zhao. Y., Li Y.-S., Botvinick E., Chien S. (2009) Live cells exert 3-dimensional traction forces on their substrata. Cell Mol. Bioeng. 2, 425–436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sundd P., Gutierrez E., Pospieszalska M. K., Zhang H., Groisman A., Ley K. (2010) Quantitative dynamic footprinting microscopy reveals mechanisms of neutrophil rolling. Nat. Methods 7, 821–824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Pospieszalska M. K., Lasiecka I., Ley K. (2011) Cell protrusions and tethers: a unified approach. Biophys. J 100, 1697–1707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lin Y. (2010) A model of cell motility leading to biphasic dependence of transport speed on adhesive strength. J. Mech. Phys. Solids 10.1016/j.jmps.2010.01.010 [DOI] [Google Scholar]
- 33. Brown A. F. (1982) Neutrophil granulocytes: adhesion and locomotion on collagen substrata and in collagen matrices. J. Cell Sci. 58, 455–467 [DOI] [PubMed] [Google Scholar]
- 34. Aubry D., Thiam H., Piel M., Allena R. (2014) A computational mechanics approach to assess the link between cell morphology and forces during confined migration. Biomech. Model. Mechanobiol. 14, 143–157 [DOI] [PubMed] [Google Scholar]
- 35. Hawkins R. J., Piel M., Faure-Andre G., Lennon-Dumenil A. M., Joanny J. F., Prost J., Voituriez R. (2009) Pushing off the walls: a mechanism of cell motility in confinement. Phys. Rev. Lett. 102, 058103 [DOI] [PubMed] [Google Scholar]
- 36. Qian J., Wang J., Lin Y., Gao H. (2009) Lifetime and strength of Periodic bond clusters between elastic media under inclined loading. Biophys. J. 97, 2438–2445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ward M. D., Hammer D. A. (1993) A theoretical analysis for the effect of focal contact information on cell-substrate attachment strength. Biophys. J. 64, 936–959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. López-Fagundo C., Bar-Kochba E., Livi L. L., Hoffman-Kim D., Franck C. (2014) Three-dimensional traction forces of Schwann cells on compliant substrates. J. R. Soc. Interface 11, 20140247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Byrd A. S., O'Brien X. M., Johnson C. M., Lavigne L. M., Reichner J. S. (2013) An extracellular matrix-based mechanism of rapid neutrophil extracellular trap formation in response to Candida albicans. J. Immunol. 190, 4136–4148 [DOI] [PMC free article] [PubMed] [Google Scholar]