Background: Unicellular cyanobacteria separate photosynthesis and nitrogen fixation using a day-night cycle.
Results: Cyanothece 51142 expresses a D1 protein exclusively in the dark period that assembles nonfunctional photosystem II.
Conclusion: The unusual D1 is named sentinel D1 because it prevents photosynthesis from occurring during nitrogen fixation.
Significance: Expression of sentinel D1 allows photosynthesis and nitrogen fixation to coexist in the cell.
Keywords: Enzyme Inactivation, Membrane Protein, Nitrogen Fixation, Photosynthesis, Photosystem II, D1 Protein
Abstract
Photosystem II, a large membrane-bound enzyme complex in cyanobacteria and chloroplasts, mediates light-induced oxidation of water to molecular oxygen. The D1 protein of PSII, encoded by the psbA gene, provides multiple ligands for cofactors crucial to this enzymatic reaction. Cyanobacteria contain multiple psbA genes that respond to various physiological cues and environmental factors. Certain unicellular cyanobacterial cells, such as Cyanothece sp. ATCC 51142, are capable of nitrogen fixation, a highly oxygen-sensitive process, by separating oxygen evolution from nitrogen fixation using a day-night cycle. We have shown that c-psbA4, one of the five psbA orthologs in this cyanobacterium, is exclusively expressed during nighttime. Remarkably, the corresponding D1 isoform has replacements of a number of amino acids that are essential ligands for the catalytic Mn4CaO5 metal center for water oxidation by PSII. At least 30 cyanobacterial strains, most of which are known to have nitrogen fixing abilities, have similar psbA orthologs. We expressed the c-psbA4 gene from Cyanothece 51142 in a 4E-3 mutant strain of the model non-nitrogen-fixing cyanobacterium Synechocystis sp. PCC 6803, which lacks any psbA gene. The resultant strain could not grow photoautotrophically. Moreover, these Synechocystis 6803 cells were incapable of PSII-mediated oxygen evolution. Based on our findings, we have named this physiologically relevant, unusual D1 isoform sentinel D1. Sentinel D1 represents a new class of D1 protein that, when incorporated in a PSII complex, ensures that PSII cannot mediate water oxidation, thus allowing oxygen-sensitive processes such as nitrogen fixation to occur in cyanobacterial cells.
Introduction
During oxygenic photosynthesis, the reaction centers of photosystem I and photosystem II (PSII),3 two pigment-protein complexes, convert light energy into chemical energy in the thylakoid membranes of cyanobacteria, plants, and eukaryotic algae. PSII is a large multiprotein, multicofactor enzyme complex that oxidizes water to molecular dioxygen with concomitant reduction of plastoquinones. The overall chemistry, as well as the organic and inorganic components of PSII, have remained remarkably conserved throughout the evolution of oxygenic photosynthetic organisms.
The structural details and the functional form of PSII have been elucidated during recent years (1, 2). The core of PSII is a heterodimer of the D1 and D2 proteins, which binds cofactors necessary for charge separation, namely P680, the special chlorophyll (3) pair, pheophytin, QA, QB, and YZ (4). D1 also provides the majority of the amino acid ligands for the Mn4CaO5 cluster, the catalytic site of water oxidation (4). These redox active residues render the D1 protein susceptible for oxidative damage and trigger the repair process that preferentially removes damaged D1 and inserts new D1 to maintain active PSII complexes in the cell (5). Thus PSII complexes are dynamic in nature (6, 7).
D1, encoded by the psbA gene, is initially synthesized in a precursor form, pD1 with a C-terminal extension typically of 16 amino acids in cyanobacteria (8) and 8–9 amino acid residues in eukaryotes (9, 10). pD1 is inserted into the thylakoid membrane as part of PSII precomplexes before it is processed by the protease CtpA to yield the mature form, D1 (11, 12). After this cleavage event, the Mn4CaO5 cluster assembles and the luminal extrinsic proteins bind the complex (13). At this point, PSII is capable of water oxidation.
All cyanobacterial strains sequenced to date have multiple copies of the psbA gene (14). The model cyanobacterium Synechocystis sp. PCC 6803 has three psbA genes. Among them, psbA2 (slr1311) and psbA3 (sll1867) encode two D1 isoforms with identical amino acids. psbA2 accounts for ∼90% of the psbA transcripts in the cell, and psbA3 accounts for the remaining 10% when cells are grown under standard laboratory conditions (15). However, upon deletion of psbA2, levels of psbA3 transcripts increase so that the total levels of psbA transcripts remain unchanged (15). Additional psbA3 transcription can be induced upon exposure to high light and UV-B (16, 17). psbA1 (slr1181) encodes a second isoform, D1′, which is expressed only under suboxic growth conditions (18). However, PSII with D1′ is functionally competent (19, 20). This multigene strategy presumably allows for adjustment of PSII activities according to environmental factors and is conserved in other cyanobacterial strains, including Synechococcus sp. PCC 7942 (21), Gloeobacter violaceus PCC 7421 (22), and Anabaena PCC 7120 (20).
Nitrogen-fixing cyanobacteria must balance the biochemical reactions of photosynthesis, which produces oxygen, with the requirements of nitrogenase, the enzyme that fixes atmospheric dinitrogen to ammonia (23). Nitrogenase is highly sensitive to oxygen. Filamentous cyanobacteria separate these reactions spatially by differentiation of 5–10% of the vegetative cells into specialized nitrogen-fixing cells called heterocysts (24). In contrast, unicellular diazotrophic cyanobacterial strains, such as Cyanothece sp. ATCC 51142, separate the processes temporally during a light/dark diel cycle (25). In these cells grown under a 12-h light/12-h dark cycle in medium lacking any nitrogen source, PSII activity peaks late in the light period, whereas nitrogenase activity exhibits a sharp peak of activity early in the dark phase (26). Whole genome transcriptomics analysis has shown that during the light period, 93% of genes associated with photosynthesis are up-regulated, and during the dark period, 100% of the genes associated with nitrogen fixation are up-regulated (26). Finally, concomitant with the high nitrogenase activity during the dark period, Cyanothece 51142 cells lose their capacity for PSII-mediated oxygen evolution (27, 28). The mechanism of this dark inactivation remains unknown. In contrast, in other cyanobacteria, such as the model organism Synechocystis sp. PCC 6803, PSII complexes are quite stable in the dark and retain 90% of their activity after 6 h of dark incubation (29).
A recent analysis of many sequenced cyanobacterial genomes has identified a new class of D1 proteins, each with a number of amino acid replacements crucial to D1 function, including residues critical for CtpA processing and binding of the catalytic manganese atoms (14). Of the five psbA genes in the Cyanothece 51142 genome (30), c-psbA4 encodes a member of this divergent D1 family. To characterize the function of the c-psbA4 gene product, we expressed it as the sole D1 protein in Synechocystis 6803. The PSII complexes in such a strain could not catalyze oxygen evolution. We suggest that the product of c-psbA4 is used in Cyanothece 51142 as a structural placeholder in PSII, preventing water oxidation during the nitrogen fixation stage of its diurnal cycle. We propose that this class of D1 isoform be referred to as sentinel D1 (sD1) to reflect its role in preventing water oxidation during the nitrogen-fixing period, protecting the oxygen-sensitive enzyme, nitrogenase.
EXPERIMENTAL PROCEDURES
Bacterial Strains and Culture Conditions
Cyanothece 51142 was grown in ASP2 media without NaNO3 (25) at 30 °C, with air bubbling, under alternating 50 μmol m−2 s−1 white light for 12 h with darkness for 12 h, as previously described (26). All Synechocystis sp. PCC 6803 (Synechocystis 6803) strains were maintained in BG11 medium (31) supplemented with 25 mm glucose and 10 μm DCMU at 30 °C under continuous 15 μmol m−2 s−1 of white light. The 4E-3 psbA triple deletion strain of Synechocystis 6803 was grown in the presence of 10 μm spectinomycin, 10 μm erythromycin, and 10 μm chloramphenicol. c-psbA1 4E-3 and c-psbA4 4E-3 strains were supplemented with 5 μm kanamycin. To compare growth rates, all strains were grown in triplicate in 12-well plates under 30 μmol m−2 s−1 of white light, and growth was monitored using OD730 nm on a μQuant Biotek plate reader (Bio-Tek Instruments).
Sequence Alignment and Phylogenetic Analysis
The five annotated psbA genes (c-psbA1 to c-psbA5) from Cyanothece 51142 were aligned with the reference sequence psbA2 from Synechocystis 6803 using CLUSTAL OMEGA (32, 33). To detect homologs of sD1 across 130 cyanobacterial genomes, NCBI BlastP was used to generate a list of sequences with E value of 0 (34). The list was manually curated to include entries that displayed similar sequence features as in sD1 of Cyanothece 51142. Typical D1 sequences for these species were chosen to use for phylogenetic tree analysis. D1 protein sequences from Synechocystis 6803 and Thermosynechococcus elongates BP-1 were chosen for comparison. These sequences were aligned in MEGA 6.06 (35) using CLUSTALW (32, 36). Phylogenetic trees were constructed in MEGA 6.06 (35) using the Jones-Taylor-Thornton model for amino acid substitution, and the tree was generated using neighbor joining method. Bootstrap values obtained after 1000 replications were attached to each node, and cutoff values of 50% were chosen for visualization on the tree.
Generation of Targeted Mutant Strains
To generate a c-psbA4 expression strain, we utilized a SLIC-based strategy similar to that described in Ref. 37. Using primers designed with 30-bp homology at the 5′ end, we amplified 500 bp upstream (including the promoter) of the psbA2 gene from Synechocystis 6803, c-psbA4 coding region from Cyanothece 51142, kanamycin resistance gene from pUC4K, 500 bp downstream sequence of the psbA2 coding region from Synechocystis 6803, and the origin of replication and ampicillin resistance gene from pCR2.1. Primer sequences are listed in Table 1. The resulting plasmid pSL2153 is diagrammed in Fig. 4A. pSL2153 was used for transformation of the 4E-3 strain to generate the strain c-psbA4 4E-3. A three-piece fusion PCR strategy was used to generate the c-psbA1 construct. psbA2us and KanR-psbA2ds fragments were PCR-amplified from pSL2153. The c-psbA1 coding region was PCR-amplified using Cyanothece 51142 genomic DNA. Primers used for amplification had 30-bp homology at their 5′ ends. All three PCR products were excised from an agarose gel, combined, and purified via the Invitrogen PureLink gel extraction kit. The eluate was used as a template for PCR using the A2usF and A2dsR primers (Table 1). The resulting 3-kb fusion psbA1 fusion PCR product was used for direct transformation of the 4E-3 strain to generate the strain c-psbA1 4E-3. Segregations of both the c-psbA4 and c-psbA1 mutations in the 4E-3 background were monitored by PCR with the A2usF and A2dsF primers (Table 1).
TABLE 1.
Primer sequences
All sequences are 5′ to 3′.
| Primer name | Primer sequence |
|---|---|
| A2usF | GTTCTTTCCTGCGTTATCCCCTGATTCTGTAATACATCCCCCTAAAAATATCAGAATCCT |
| A2usR | TTGGTTATAATTCCTTATGTATTTGTCGATGTTCAGATTGGAACTGACTAAACTTAGTCT |
| A4F | ATCGACAAATACATAAGGAATTATAACCAAATGACCAATGTATTTCCTCGCCAAAAAGAA |
| A4R | TTAACTGATCACTGTAGCCTTACCACTGGCTAAGGTTAAAGGAAAGTGATGAGTATTCGG |
| KanRF | GCCAGTGGTAAGGCTACAGTGATCAGTTAATCTGCCTCGTGAAGAAGGTGTTGCTGAC |
| KanRR | CGTTGTGTCTCAAAATCTCTGATGTTACATTGCACAAGATAAAAATATATCATCATGAAC |
| A2dsF | ATGTAACATCAGAGATTTTGAGACACAACGAAGAAAAAAAATGCCTCCTGGAGCATTGAA |
| A2dsR | TCTAGATAATCCTGGTCTTTGTAGCCATTACAAATGATTAGGGGCTTGGTATGCTTGTCC |
| OriCF | TAATGGCTACAAAGACCAGGATTATCTAGACGAGTTCTTCTGAATTGAAAAAGGAAGAGT |
| OriCR | ACAGAATCAGGGGATAACGCAGGAAAGAACATGTGAGCAAAAGGCCAGCA |
| A1F | ATCGACAAATACATAAGGAATTATAACCAAATGACTACTACCTTACAACAACGCGAGAGC |
| A1R | TTAACCATTGATAACAGGAGCAGATACAGGCTCAGCAGAAGCTAAGTCTAAGGGGAAGTT |
| A1KanRF | CCTGTATCTGCTCCTGTTATCAATGGTTAATCTGCCTCGTGAAGAAGGTGTTGCTGAC |
| 16SF | TGTAGCGGTGAAATGCGT |
| 16SR | AGGTTCTTCGCGTTGCAT |
FIGURE 4.
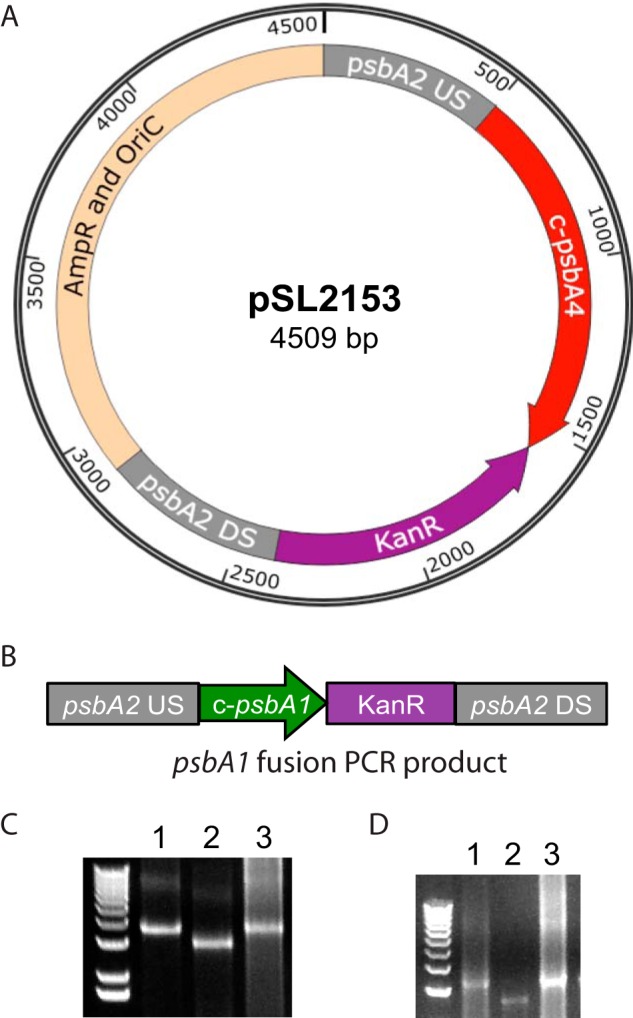
Construction of c-psbA 4E-3 strain. A, pSL2153 plasmid was constructed using a five-piece SLIC reaction, including 500 bp upstream sequence of Synechocystis 6803 psbA2, the entire coding region of c-psbA4 from Cyanothece 51142, the kanamycin resistance gene from pUC4K, 500 bp downstream sequence of Synechocystis 6803 psbA2, and the origin of replication and ampicillin resistance gene from pCR2.1. B, layout of c-psbA1 4E-3 construct created by fusion PCR including 500 bp upstream sequence of Synechocystis 6803 psbA2, the entire coding region of c-psbA1 from Cyanothece 51142, the kanamycin resistance gene from pUC4K, and 500 bp downstream sequence of Synechocystis 6803 psbA2. C, PCR showing complete segregation of c-psbA4 4E-3 construct. Lanes 1, 2, and 3 show c-psbA4 4E-3 genomic DNA, 4E-3 genomic DNA, and c-psbA4 plasmid DNA, respectively. D, PCR showing complete segregation of c-psbA1 4E-3 construct transformed into 4E-3. Lanes 1, 2, and 3 represent c-psbA1 4E-3 genomic DNA, 4E-3 genomic DNA control, and c-psbA1 fusion PCR product, respectively.
Semiquantitative RT-PCR
RNA isolation from Cyanothece 51142, 4E-3, c-psbA1 4E-3, and c-psbA4 4E-3 cells, and cDNA synthesis were carried out as described previously (38). Briefly, RNA was isolated using RNAWiz (Ambion), and cDNA was synthesized using SuperScript II with random primers (Invitrogen). Primer pairs A1F/A1R, A4F/A4R, and 16SF/16SR were used for PCR amplification of c-psbA1, c-psbA4, and the control 16 S rRNA genes, respectively. Primer sequences can be found in Table 1.
Membrane Preparation and Immunological Detection
Total membranes were isolated as previously described (39). Samples were fractionated by SDS-PAGE (16% acrylamide, 6 m urea), transferred to nitrocellulose, and incubated with specific antisera. Chemiluminescent signals were developed with Immobilon HRP reagent (Millipore) and visualized on an LAS-1000 Plus imager (Fujifilm).
Spectrophotometric Assays
After methanol extractions, Chl concentrations were measured on a DW2000 spectrophotometer (40).
Fluorescence Measurements
Fluorescence induction measurements were conducted using an FL-200 instrument (Photon System Instruments, Drásov, Czech Republic). Samples were diluted to 7 μg/ml Chl. Samples were dark incubated for 3 min prior to measurement, and 10 μm DCMU was used for DCMU treated samples. Low temperature (77 K) spectra of cyanobacterial cells were determined on a Fluoromax-2 fluorometer (Jobin Yvon, Longjumeau, France). Cells were adjusted to 5 μg/ml Chl for all measurements. Fluorescein diacetate was added to cells at final concentration 1 μm just prior to measurements. Samples were excited at 435 nm, and emission spectra were adjusted for same signal at the fluorescein peak at 522 nm. The fluorescein component from emission spectra was subtracted and normalized to the 721 nm emission band (41).
Oxygen Evolution Measurements
Steady state oxygen evolution from cyanobacterial cells was measured on a Clark type electrode. With Cyanothece 51142, whole chain oxygen evolution measurements were conducted under 8250 μmol m−2 s−1 of white light. With Synechocystis 6803, PSII-meditated oxygen evolution was measured in the presence of 0.5 mm 2,6-dichloro-p-benzoquinone (Eastman-Kodak) and 1 mm potassium ferricyanide K3FeCN6 under a range of light intensities. Light intensity was adjusted using neutral density filters. Synechocystis 6803 cells were diluted to 10 μg/ml Chl for all measurements.
RESULTS
Temporal Changes in PSII Activity in Cyanothece Cells
To examine the water oxidation ability of Cyanothece 51142 cells during a diel cycle, they were grown under a 12-h light/12-h dark regimen under nitrogen-fixing conditions. Cellular samples were collected 9 h after the onset of a light period (L9) and also 3 h after the onset of a dark period (D3) and examined for their light-induced PSII activities. As shown in Table 2, the D3 samples exhibited whole chain oxygen evolution activity that was negligible compared with that of the L9 samples. Measurements of Kautsky fluorescence induction showed a similar lack of PSII activity in the D3 sample (Fig. 1 and Table 2). Notably, there was no difference in fluorescence levels in the D3 samples in the presence or absence of the PSII acceptor side inhibitor DCMU. Also, the Fo levels were much higher in the D3 samples, and the Fv/Fm ratios were ∼25% of those observed in L9 samples (Table 2). This indicates that PSII complexes in Cyanothece 51142 at D3 were nonfunctional. Taken together, the oxygen evolution rates and kinetics of fluorescence induction indicated that PSII centers in Cyanothece 51142 were not photoactive during the dark period.
TABLE 2.
Parameters for fluorescence induction and oxygen evolution in whole cells (n = 3)
| Sample | Fo | Fv/Fm | Rate of oxygen evolution |
|---|---|---|---|
| μmol O2 mg Chl−1 h−1 | |||
| Cyanothece 51142 L9 | 0.98 ± 0.03 | 0.45 ± 0.06 | 220 ± 24 (whole chain) |
| Cyanothece 51142 D3 | 1.11 ± 0.002 | 0.12 ± 0.01 | 9 ± 3 (whole chain) |
| WT Synechocystis 6803 | 0.43 ± 0.02 | 0.62 ± 0.01 | 315 ± 0 (PSII-mediated) |
| 4E-3 | 1.71 ± 0.01 | 0.27 ± 0.001 | 67 ± 22 (PSII-mediated) |
| c-psbA4 4E-3 | 0.73 ± 0.01 | 0.30 ± 0.001 | 97 ± 46 (PSII-mediated) |
| c-psbA1 4E-3 | 0.52 ± 0.004 | 0.61 ± 0.02 | 263 ± 65 (PSII-mediated) |
FIGURE 1.
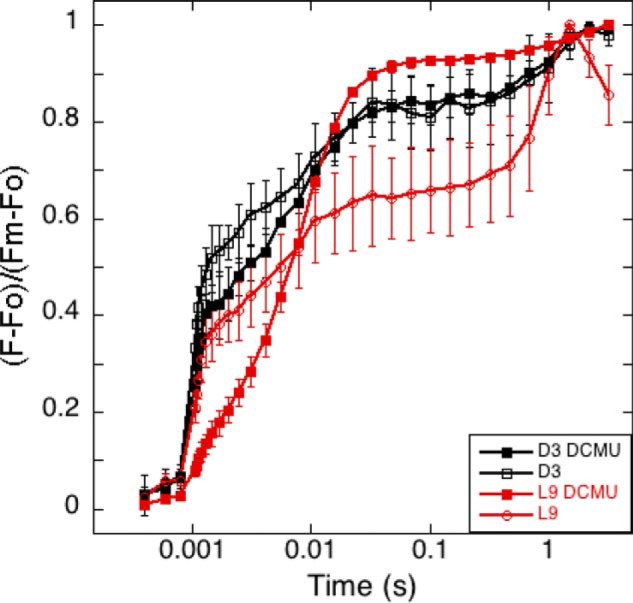
Fluorescence induction kinetics of Cyanothece 51142. Samples represent time points D3 (black squares) and L9 (red circles) in the absence (open shapes) and presence of DCMU (filled shapes). n = 3; error bars represent S.D.
psbA Genes of Cyanothece 51142 Exhibit Diverse Expression and Sequence Features
To account for the temporal inactivation of PSII during the dark period, we re-examined our previously published Cyanothece 51142 transcriptomics dataset at various times during the 12-h light/12-h dark cycle (26). In this earlier study, we determined that the majority of the transcripts for photosystem I, PSII, and the light-harvesting phycobillisome genes are up-regulated during the light (photosynthesis) period and are down-regulated during the dark (nitrogen fixation) period (26). Among the 26 annotated PSII transcripts, only one showed peak expression during the dark phase and down-regulation during the light period. This gene is c-psbA4 (cce_3477), one of the five psbA genes in the Cyanothece 51142 genome (26, 30). The expression levels of c-psbA1 and c-psbA4 genes are shown in Fig. 2. c-psbA4 displayed the highest transcript level at the D1 time point (26), whereas the level of the psbA1 transcript was highest at the L9 time point, consistent with its function during photosynthesis. Notably, a unique peptide corresponding to the sD1 isoform was observed in a global proteomic study of Cyanothece 51142, confirming that the sD1 protein is present in cells during the dark period (42).
FIGURE 2.
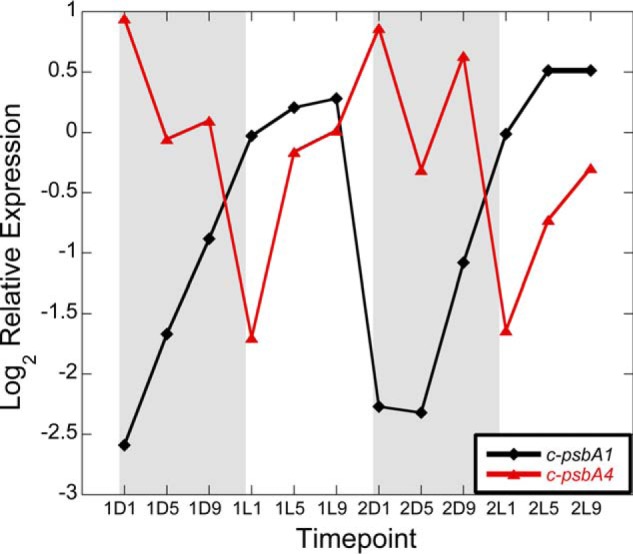
Transcript levels of c-psbA1 (32) and c-psbA4 (red) in Cyanothece 51142 during two consecutive diel cycles.
The five gene copies of psbA encode four D1 isoforms, as c-psbA1 (cce_3501) and c-psbA5 (cce_0636) encode D1 proteins with identical amino acid sequences, most similar (95% identical) to the product of the psbA2 gene of Synechocystis 6803. The D1 isoforms encoded by c-psbA3 (cce_0267) and c-psbA2 (cce_3411) are the second and third most conserved, 92 and 80% identical, respectively. The most divergent of the four Cyanothece 51142 D1 orthologs is the sD1 isoform encoded by c-psbA4 (cce_3477), with only 68% identity with the psbA2 gene product of Synechocystis 6803.
Presence of sD1 Homologs in Other Nitrogen-fixing Cyanobacteria
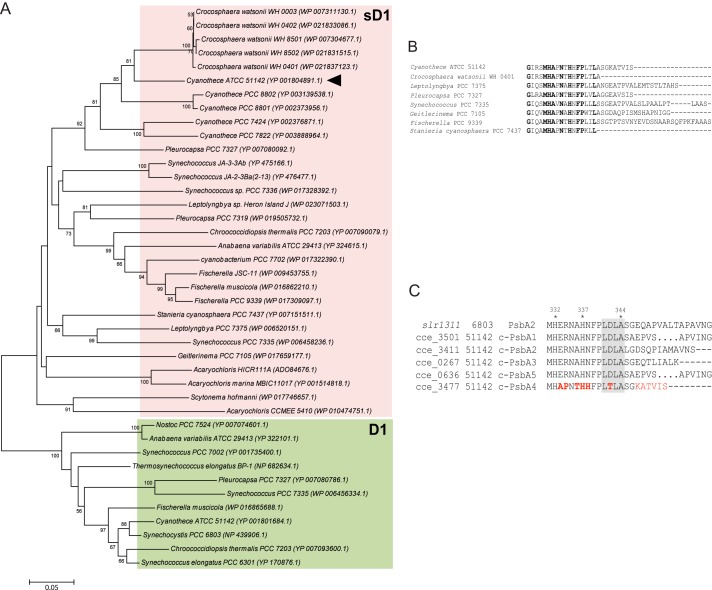
We identified 29 other cyanobacterial strains with sequences homologous to sD1 of Cyanothece 51142 (Fig. 3A). These include the 10 strains previously described in Ref. 14. Newly identified strains in the current study include organisms whose genomes were sequenced more recently (43–45). Most organisms in this group are diazotrophic nitrogen fixers except Acaryochloris marina MBIC11017, which contains vestigial nif genes in its genome (46). In this context, it is noteworthy that the recently sequenced Acaryochloris sp. HICR111A has active genes for nitrogen fixation (3, 47). Among the diazotrophs, Nostoc sp. PCC 7524 lacks an sD1 isoform. As is evident, during evolution, sD1 sequences diverged earlier from D1 sequences to form a separate clade (Fig. 3A). Among the sD1 homologs, we observed several variations in the amino acid sequence, and a consensus sequence for sD1 was not observed. However, all sD1 sequences had critical amino acids replacements in the C-terminal extension. Cyanothece 54112 and the Crocosphaera watsonii species were grouped together in the tree and bear high degrees of similarity in amino acid replacements in the C-terminal extension, as well as in residues required for binding the proteins of the lumenal oxygen-evolving complex. sD1 from Synechococcus PCC 7335 with an unusual C-terminal sequence (14) was found to have sequence similarity with sD1 proteins from Leptolyngbya PCC 7375 and Leptolyngbya Heron Island J. sD1 from Fischerella PCC 9339 was observed to have the longest C-terminal tail with 24 amino acids after Ala-344, the C-terminal residue of the mature D1 protein. Fig. 3B shows the amino acid sequence alignment of the C terminus of sD1 sequences emphasizing the variation of amino acids residues in this region. In all 30 organisms identified with sD1, the conserved recognition sequence LDLA-SGE for the enzyme CtpA was modified, suggesting the inability of this protein to undergo cleavage of its C-terminal extension. In addition to the C terminus, residues at the oxygen-evolving complex, for example, Asp-61, Asp-160, Glu-189, His-332, Glu-333, Asp-342, and Ala-344, were consistently different in all sD1 sequences. These residue changes suggest that PSII complexes with sD1 should not be able to assemble a functional oxygen-evolving complex.
FIGURE 3.
Phylogenetic analysis and sequence alignment of PsbA4 from Cyanothece 51142. A, phylogenetic tree showing newly identified sD1 homologs across 130 cyanobacteria with sequenced genomes. sD1 and normal D1 sequences are highlighted in pink and green boxes, respectively. The scale bar indicates number of amino acid substitutions per site. B, amino acid sequence alignment of the C-terminal regions of sD1 protein from selected cyanobacterial species (using CLUSTAL OMEGA). Alignment illustrates the variability of amino acid sequence after Ala-344 between the different sD1 homologs. Conserved residues are highlighted in bold. C, alignment of amino acid sequences for PsbA2 from Synechocystis 6803 and all five c-PsbAs from Cyanothece 51142. C-terminal regions are shown to indicate the CtpA recognition sequences (gray-shaded area). The amino acid replacements in c-PsbA4 are highlighted in red.
Amino acid sequence alignment at the C terminus of all five c-PsbA (D1) proteins in Cyanothece 51142 to that of the PsbA1 protein in Synechocystis 6803 is shown in Fig. 3C. CtpA cleaves the C-terminal extension, transforming pD1 into the mature, functional D1 (for a review of D1 processing, see Ref. 48). Processing of pD1 by CtpA to yield D1 is a critical step in PSII assembly and is a requirement for the binding of the catalytic Mn4CaO5 cluster, the site of water oxidation (13). The mutations observed in c-psbA4 suggest that the sD1 protein would not be processed by CtpA.
Expression of c-PsbA4 as the Sole D1 Protein in Synechocystis 6803
Cyanothece 51142 is recalcitrant to targeted genetic modification. Hence, we exploited the model cyanobacterium Synechocystis 6803 to study the function of the c-PsbA4 protein. The 4E-3 strain of Synechocystis 6803 is a triple D1 deletion mutant in which the majority of the coding regions of psbA1, psbA2, and psbA3 genes have been replaced with resistance genes for spectinomycin, erythromycin, and chloramphenicol, respectively (49). This 4E-3 strain was transformed with two constructs to express either c-psbA4 or the control c-psbA1 from Cyanothece 51142 (Fig. 4). (psbA genes from Cyanothece 51142 are referred to as c-psbA to differentiate them from Synechocystis 6803 psbA genes.) In both the psbA4 and psbA1 constructs, the Cyanothece psbA (c-psbA) genes were targeted to the psbA2 locus in Synechocystis 6803, driven by the native Synechocystis 6803 psbA2 promoter and contain a kanamycin resistance (KanR) gene (Fig. 4, A and B). Complete segregation of the mutant lines was confirmed via PCR using primers corresponding to the psbA2us and psbA2ds regions of the psbA2 locus (Fig. 4, C and D, and Table 1).
Transcript levels, as determined by semiquantitative RT-PCR, showed that the expression of c-psbA4 in the c-psbA4 4E-3 strain was stronger than that in Cyanothece 51142 (Fig. 5A). This increased expression was likely due to the strong psbA2 promoter in Synechocystis 6803. In contrast, the control strain c-psbA1 4E-3 displayed c-psbA1 expression levels similar to those in Cyanothece 51142 (Fig. 5B). Western blots using anti-D1 antibodies showed that the c-PsbA4 D1 isoform (sD1) was produced in the c-psbA4 4E-3 line (Fig. 5C), and the c-PsbA1 D1 isoform was produced in the psbA1 4E-3 line (Fig. 5D). Western blots using anti-CP43 and anti-CP47 antibodies also demonstrated that these proteins were present in the c-psbA4 4E-3, as well as in the 4E-3 strains (Fig. 5C). Decreased steady state levels of D1 in the c-psbA4 4E-3 could be due to differences in the antibody reactivity, because the amino acid sequence for c-PsbA4 has several residues mutated from the traditional PsbA sequence. Another plausible explanation is that because of the unusual C-terminal extension sequence, CtpA is unable to process the C terminus, thereby making it susceptible for degradation.
FIGURE 5.
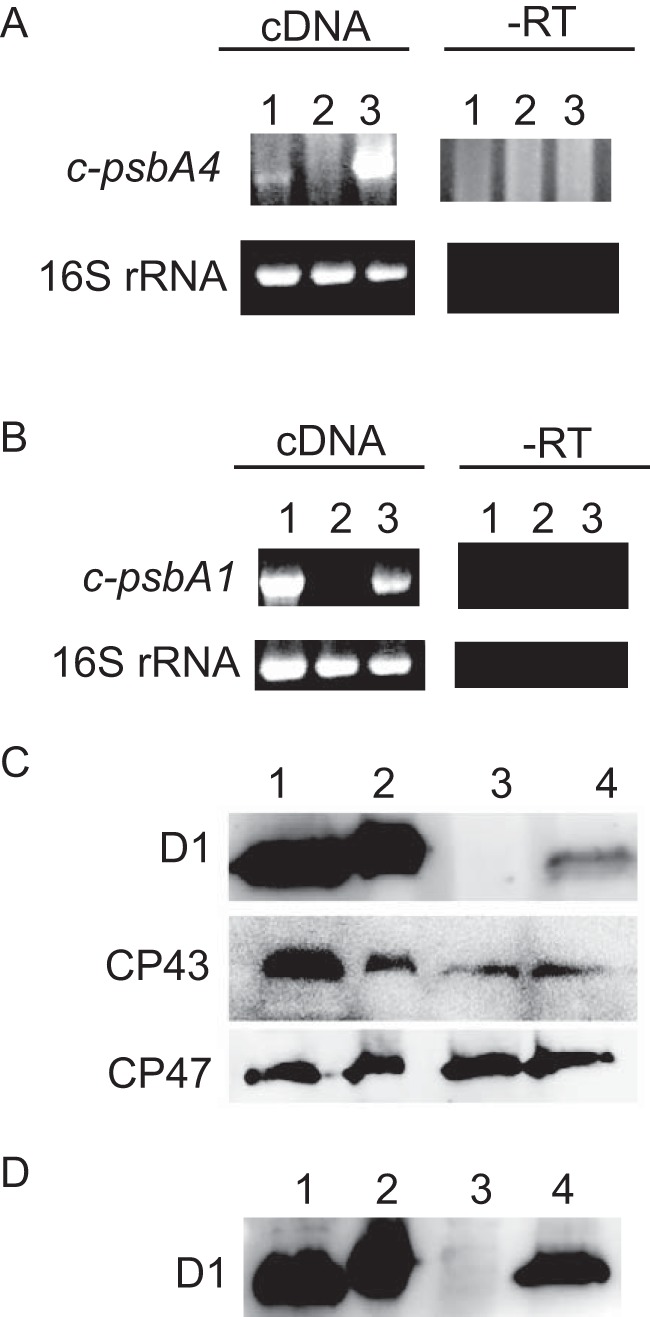
Expression levels of psbA. A, transcript levels of c-psbA4 and the control 16 S rRNA in Cyanothece 51142, 4E-3, and c-psbA4 4E-3 cells shown in lanes 1, 2, and 3, respectively. B, transcript levels of c-psbA1 and the control 16 S rRNA in Cyanothece 51142, 4E-3, and c-psbA1 4E-3 cells shown in lanes 1, 2, and 3, respectively. C, D1, CP43, and CP47 protein levels in isolated membranes of Synechocystis 6803 WT, ΔctpA, 4E-3, and c-psbA4 4E-3 shown in lanes 1, 2, 3, and 4, respectively. D, D1 protein levels in isolated membranes from Synechocystis 6803 WT, ΔctpA, 4E-3, and c-psbA1 4E-3 cells, respectively. Samples for Western blots were loaded at 5 μg Chl per lane. -RT, no reverse transcriptase used as a control.
Expression of psbA4 in Synechocystis 6803 Does Not Restore Photoautotrophy
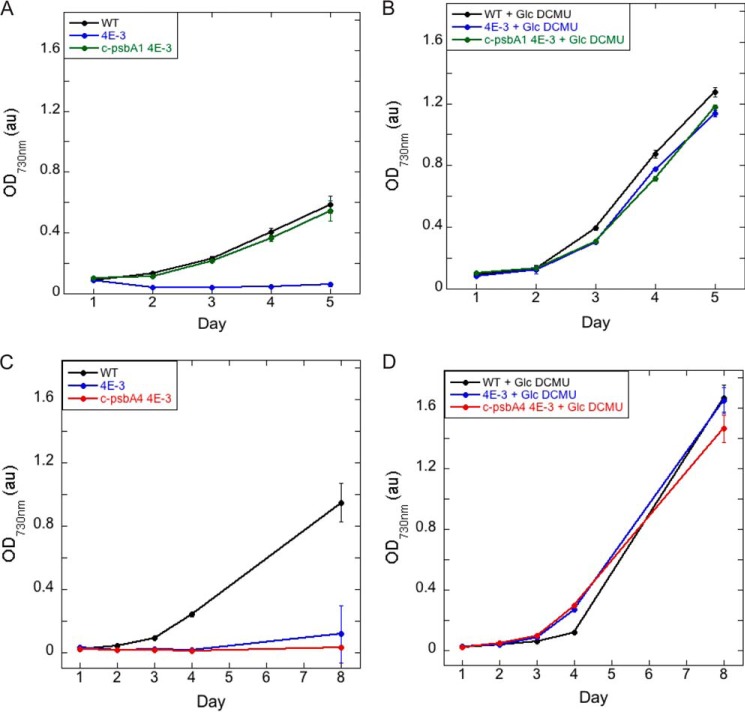
The background psbA deletion strain 4E-3 was not capable of autotrophic growth and required the presence of glucose and DCMU for growth (Fig. 6, A and B). The addition of c-psbA1 to 4E-3 restored photoautotrophic growth (Fig. 6A), confirming that this c-psbA gene from Cyanothece 51142 can restore D1 function in Synechocystis 6803 cells. In contrast, c-psbA4 4E-3 could not grow in the absence of glucose and DCMU (Fig. 6C) but could grow with those supplements at rates similar to those found in WT Synechocystis 6803 (Fig. 6D). Thus incorporation of sD1 as the D1 protein in PSII did not allow photosynthetic growth of Synechocystis 6803 cells.
FIGURE 6.
Growth of Synechocystis WT, 4E-3, c-psbA1 4E-3, and c-psbA4 4E-3 cells. A, growth of Synechocystis WT, 4E-3, and c-psbA1 4E-3 strains in BG11. B, growth of Synechocystis WT, 4E-3, and c-psbA1 4E-3 strains in the presence of 25 mm glucose and 10 μm DCMU. C, growth of Synechocystis WT, 4E-3, and c-psbA4 4E-3 strains in BG11. D, growth of Synechocystis WT, 4E-3, and c-psbA4 4E-3 strains in the presence of 25 mm glucose and 10 μm DCMU. n = 3; error bars represent S.D.
sD1 Isoform Does Not Restore PSII Activity in 4E-3
To determine the basis for the photoautotrophic defect of c-psbA4 4E-3, PSII in these cells was evaluated. As shown in the 77 K fluorescence spectra of different cyanobacterial strains (Fig. 7), WT Synechocystis 6803 cells as well as c-psbA1 4E-3 had emission peaks at 685 and 695 nm from PSII and 720 nm from photosystem I. The 4E-3 cells, in contrast, had only a 683 peak in this region. In the presence of c-psbA4, the emission peaks were at 685 and 693 nm, suggesting the presence of assembled PSII complexes. CP47 and CP43 were both present in the PSII complexes purified from c-psbA4 4E-3 (data not shown). Therefore, PSII with sD1 isoform assembles into a holocomplex consisting of both CP47 and CP43 (6).
FIGURE 7.
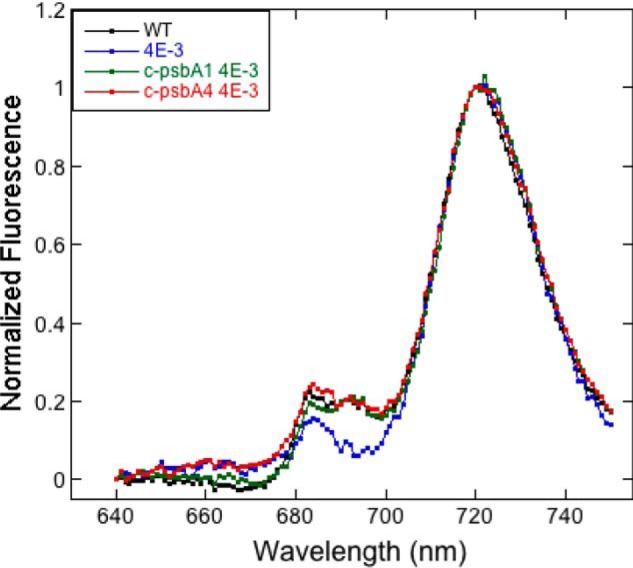
Low temperature (77 K) fluorescence emission spectra of Synechocystis 6803 WT, 4E-3, c-psbA1 4E-3, and c-psbA4 4E-3 cells with excitation at 435 nm. Samples were adjusted to 5 μg/ml Chl. Fluorescein diacetate was used as an internal standard to adjust to the same signal intensity. Fluorescein component was subtracted from emission spectra and subsequently normalized at 721 nm.
Next, PSII-mediated oxygen evolution was assayed over a range of light intensities (Fig. 8A). The c-psbA1 4E-3 cells had oxygen evolution rates similar to that in wild type cells. In contrast, c-psbA4 4E-3 exhibited negligible oxygen evolution activity, similar to that in 4E-3. Hence, the sD1 isoform is not functionally competent for PSII to catalyze water oxidation. Fluorescence induction kinetics were also investigated for all four strains in both the absence (Fig. 8B) and the presence (Fig. 8C) of DCMU. 4E-3 and c-psbA4 4E-3 exhibited little to no variable fluorescence and displayed no difference in kinetics in the presence of DCMU. As expected, wild type and c-psbA1 4E-3 cells displayed normal kinetics. Taken together, the lack of oxygen evolution and variable fluorescence indicated that PSII complexes with the sD1 protein were inactive.
FIGURE 8.
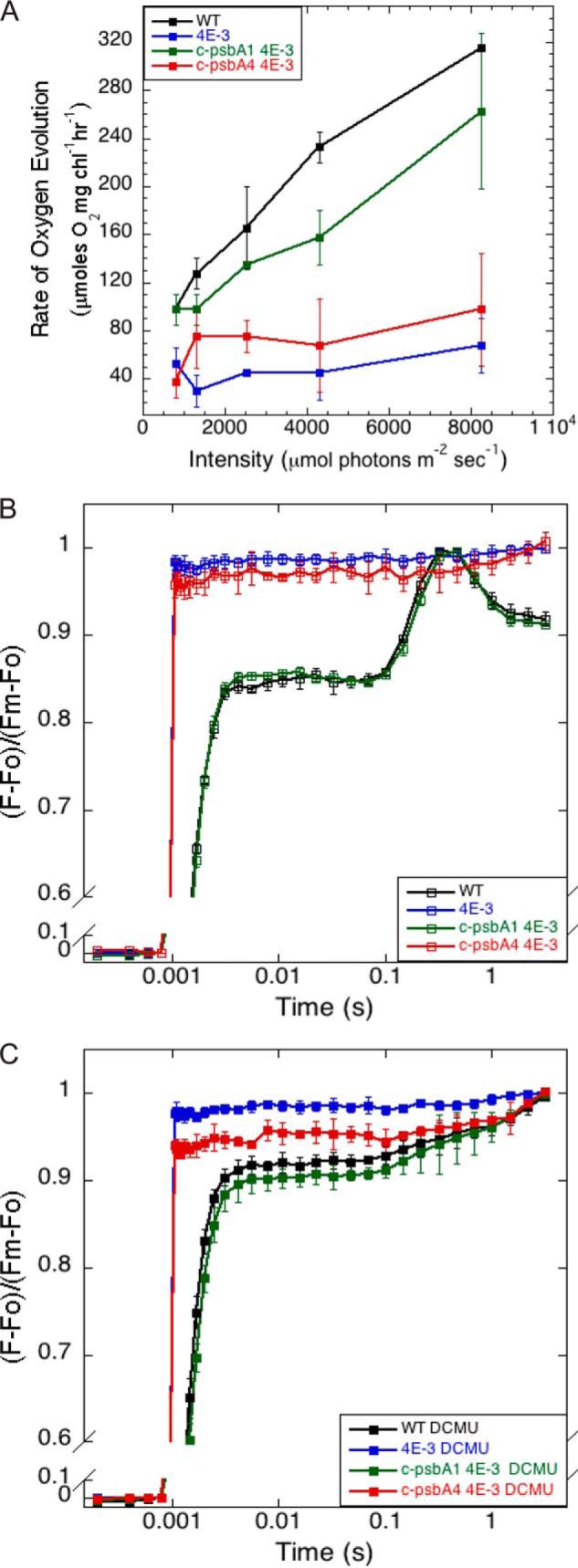
PSII activities in Synechocystis WT, 4E-3, c-psbA1 4E-3, and c-psbA4 4E-3 cells. A, PSII-mediated oxygen evolution, samples at 10 μg/ml Chl. n = 3; error bars represent S.D. B, fluorescence induction without DCMU (open symbols). C, fluorescence induction with DCMU (closed symbols), samples at 7 μg/ml Chl. n = 3; error bars represent S.D.
DISCUSSION
Nitrogen fixation and photosynthesis are contrasting processes, one being oxygen-sensitive and the other an oxygen-evolving process. Certain cyanobacteria separate these opposing processes spatially by sequestering them in different specific cell types like heterocysts. Cyanothece 51142 and other unicellular diazotrophs separate the processes temporally by limiting photosynthesis to the light period and nitrogen fixation to the dark period. We found that Cyanothece 51142 is incapable of water oxidation in the dark phase of growth under a 12 h light-dark cycle, as shown by the lack of variable fluorescence and oxygen evolution activity (Fig. 1 and Table 2) at time point D3. This is in agreement with previously published data that Cyanothece 51142 is not capable of oxygen evolution while fixing nitrogen during the dark phase of growth (27, 28).
This temporal inactivation of PSII activity could be achieved by either disassembly of the PSII complex or by altering the composition of the complex to render it inactive. Energetically, it would be less costly for the cell to make more subtle alterations to PSII, as opposed to total disassembly and de novo complex assembly. The D1 protein, with its central role in PSII photochemistry, is a likely candidate for PSII inactivation. The observation that c-psbA4 is the only PSII gene that exhibits an increase in transcription in the dark period (26) lends supports to the hypothesis that an alteration in D1 could account for the temporal inactivation in Cyanothece 51142 (27, 28). Sequence comparisons to more traditional D1 isoforms indicate several key amino acid replacements in the sD1 protein encoded by c-psbA4. One of the predicted consequences is that the precursor form of sD1 cannot be processed by CtpA (14). This processing step is required to bind the catalytic Mn4CaO5 cluster and represents a critical checkpoint in PSII assembly (13).
In this study, we report the functional characterization of the unusual D1 isoform by expression of c-psbA4 in the model cyanobacterium Synechocystis 6803. We utilized a triple psbA deletion mutant, 4E-3, of Synechocystis 6803 to express either c-psbA4 or c-psbA1. Expression of c-psbA4 as the sole D1 isoform in 4E-3 is not sufficient to restore photoautotrophic growth, oxygen evolution activity, and variable fluorescence to 4E-3 (Figs. 6 and 8 and Table 2). Despite substantial expression of c-psbA4 detected at transcript and protein levels (Fig. 5, A and C), the c-psbA4 4E-3 strain lacks PSII activity. In comparison, c-PsbA1 of Cyanothece 51142, the D1 homolog most similar to Synechocystis 6803 PsbA2, was able to rescue the 4E-3 phenotypes (Figs. 6 and 8 and Table 2). The fluorescence induction kinetics of 4E-3 and c-psbA4 4E-3 cells (Fig. 8, B and C) exhibited little to no variable fluorescence and displayed no difference in kinetics in the presence and absence of DCMU, similar to that observed in measurements of Cyanothece 51142 D3 samples (Fig. 1 and Table 2). However, the kinetics observed in WT Synechocystis 6803 and c-psbA1 4E-3 (Fig. 8, B and C) were similar to that seen Cyanothece 51142 L9 samples (Fig. 1 and Table 2). Low (77 K) temperature emission spectra indicate that c-psbA4 4E-3 assembles PSII complexes (Fig. 7). Taken together, these results imply that PSII complexes in c-psbA4 4E-3 are inactive because of the presence of sD1.
We identified 30 cyanobacterial species with c-PsbA4 orthologs (Fig. 3A). These represent a divergent class of D1 with most species possessing nitrogen fixation ability. Thus Cyanothece 51142 and other unicellular diazotrophs maintain these genes in their genomes and expend the energy to transcribe and translate these nonfunctional genes to form inactive PSII assemblies. In filamentous diazotrophic cyanobacteria, nitrogen fixation is confined to heterocysts where PSII is inactive. Earlier studies have shown the presence of D1 in heterocysts (51). Moreover, monomeric PSII complexes from heterocysts are incapable of oxygen evolution (52).
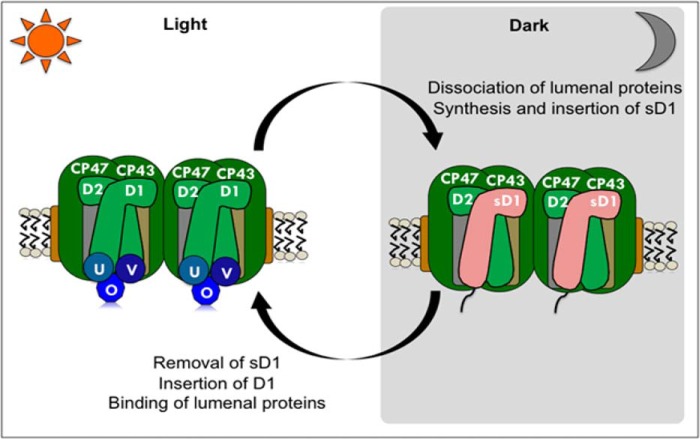
Sentinel D1, a divergent class of D1, protects nitrogenase activity. The effects of oxygen on nitrogen fixation are numerous. The catalytic Fe4S4 centers of the nitrogenase are irreversibly damaged by oxygen in vitro (53). Oxygen also represses transcription of nitrogenase genes (54). Furthermore, any reactive oxygen species generated by photosynthesis also affects the integrity of nitrogenase (55). We propose that in unicellular diazotrophs like Cyanothece 51142, nonfunctional sD1 serves as a safety mechanism to prevent oxygen evolution during the oxygen-sensitive nitrogen-fixing period. Fig. 9 describes a schematic model for the alteration in the PSII complexes during light-dark transitions. During the dark phase, with the increase in expression of sD1, the functional D1 protein is removed, and the sD1 protein is inserted to form an inactive PSII complex. With the transition into the light phase, sD1 is removed and replaced with a processed functional copy of D1 to assemble a fully functional PSII. This model best explains our findings and emphasizes the protective function of the sD1 isoform.
FIGURE 9.
A schematic model for changes in PSII in unicellular nitrogen-fixing cyanobacteria during light-dark transitions. During the dark period, D1 protein in PSII is replaced by sD1. PSII complexes with sD1 are inactive. A transition into the light phase results in the replacement of sD1 by D1, followed by binding of other lumenal proteins to form a functional PSII complex.
Although moonlight and starlight provide less irradiance than laboratory growth conditions, the natural condition under which Cyanothece 51142 and other unicellular diazotrophs have evolved is not always absolute dark during nighttime. Maximal estimates for moonlight and starlight are 5 nmol photons m−2 s−1 (56) and 1 fmol photons m−2 s−1 (57), respectively. Whereas physiological measurements indicate that 2 μmol m−2 s−1 is the lowest irradiance observed to support survival of photoautotrophic cells (58), it is estimated that 10 nmol photons m−2 s−1 is the minimal irradiance required to support oxygenic photoautotrophic growth (59). Thus it is conceivable that some oxygen evolution and the generation of reactive oxygen species could occur even during nighttime.
Interestingly, other alternate native D1 isoforms in various cyanobacteria investigated to date with varying expression patterns are capable of supporting PSII activity (16, 18, 22, 60, 61). In contrast, the sD1 isoform, identified in at least 30 cyanobacterial species, renders PSII inactive to protect the nitrogenase enzyme. The previously given name for these D1 isoforms, “rogue D1” (14), suggests a negative connotation and does not reflect its physiological function. Zhang and Sherman (50) has raised similar objections about this name and proposed an alternate name psbA4 (dd) (for dark diazotrophic) for this isoform. This naming designation is also inadequate because of the specificity of the gene number in the name (which varies among different species. Therefore, we have renamed this isoform sentinel D1 (sD1), because this name reflects the protective function of this protein isoform in the cyanobacterial cell.
Acknowledgments
We thank Dr. Robert Burnap and Dr. Richard Debus for the kind gift of the 4E-3 strain of Synechocystis 6803 and all members of the Pakrasi lab for collegial discussions.
This work was supported by National Science Foundation Grants MCB 0745611 and MCB 1331194 (to H. B. P.).
- PSII
- photosystem II
- sD1
- sentinel D1
- DCMU
- 3,4-dichlorophenyl)-1,1-dimethylurea
- Chl
- chlorophyll
- Ln
- time n hours after the onset of a light period
- Dn
- time n hours after the onset of a dark period.
REFERENCES
- 1. Ferreira K. N., Iverson T. M., Maghlaoui K., Barber J., Iwata S. (2004) Architecture of the photosynthetic oxygen-evolving center. Science 303, 1831–1838 [DOI] [PubMed] [Google Scholar]
- 2. Loll B., Kern J., Saenger W., Zouni A., Biesiadka J. (2005) Towards complete cofactor arrangement in the 3.0 Å resolution structure of photosystem II. Nature 438, 1040–1044 [DOI] [PubMed] [Google Scholar]
- 3. Mohr R., Voss B., Schliep M., Kurz T., Maldener I., Adams D. G., Larkum A. D., Chen M., Hess W. R. (2010) A new chlorophyll d-containing cyanobacterium: evidence for niche adaptation in the genus Acaryochloris. ISME J 4, 1456–1469 [DOI] [PubMed] [Google Scholar]
- 4. Umena Y., Kawakami K., Shen J. R., Kamiya N. (2011) Crystal structure of oxygen-evolving photosystem II at a resolution of 1.9 A. Nature 473, 55–60 [DOI] [PubMed] [Google Scholar]
- 5. Nagarajan A., Burnap R. L. (2014) Parallel expression of alternate forms of psbA2 gene provides evidence for the existence of a targeted D1 repair mechanism in Synechocystis sp. PCC 6803. Biochem. Biophys. Acta 1837, 1417–1426 [DOI] [PubMed] [Google Scholar]
- 6. Nixon P. J., Michoux F., Yu J., Boehm M., Komenda J. (2010) Recent advances in understanding the assembly and repair of photosystem II. Ann. Bot. 106, 1–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Komenda J., Sobotka R., Nixon P. J. (2012) Assembling and maintaining the Photosystem II complex in chloroplasts and cyanobacteria. Curr. Opin. Plant Biol. 15, 245–251 [DOI] [PubMed] [Google Scholar]
- 8. Nixon P. J., Trost J. T., Diner B. A. (1992) Role of the carboxy terminus of polypeptide D1 in the assembly of a functional water-oxidizing manganese cluster in photosystem II of the cyanobacterium Synechocystis sp. PCC 6803: assembly requires a free carboxyl group at C-terminal position 344. Biochemistry 31, 10859–10871 [DOI] [PubMed] [Google Scholar]
- 9. Takahashi Y., Nakane H., Kojima H., Satoh K. (1990) Chromatographic purification and determination of the carboxy-terminal sequences of photosystem II reaction center protein, Dl and D2. Plant Cell Physiol. 31, 273–280 [Google Scholar]
- 10. Takahashi M., Shiraishi T., Asada K. (1988) COOH-terminal residues of Dl and 44-kDa CPa-2 at spinach photosystem II core complex. FEBS Lett. 240, 6–8 [DOI] [PubMed] [Google Scholar]
- 11. Anbudurai P. R., Mor T. S., Ohad I., Shestakov S. V., Pakrasi H. B. (1994) The ctpA gene encodes the C-terminal processing protease for the D1 protein of the photosystem II reaction center complex. Proc. Natl. Acad. Sci. U.S.A. 91, 8082–8086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Diner B. A., Ries D. F., Cohen B. N., Metz J. G. (1988) COOH-terminal processing of polypeptide D1 of the photosystem II reaction center of Scenedesmus obliquus is necessary for the assembly of the oxygen-evolving complex. J. Biol. Chem. 263, 8972–8980 [PubMed] [Google Scholar]
- 13. Roose J. L., Pakrasi H. B. (2004) Evidence that D1 processing is required for manganese binding and extrinsic protein assembly into photosystem II. J. Biol. Chem. 279, 45417–45422 [DOI] [PubMed] [Google Scholar]
- 14. Murray J. W. (2012) Sequence variation at the oxygen-evolving centre of photosystem II: a new class of “rogue” cyanobacterial D1 proteins. Photosynth. Res. 110, 177–184 [DOI] [PubMed] [Google Scholar]
- 15. Mohamed A., Eriksson J., Osiewacz H. D., Jansson C. (1993) Differential expression of the psbA genes in the cyanobacterium Synechocystis 6803. Mol. Gen. Genet. 238, 161–168 [DOI] [PubMed] [Google Scholar]
- 16. Kós P. B., Deák Z., Cheregi O., Vass I. (2008) Differential regulation of psbA and psbD gene expression, and the role of the different D1 protein copies in the cyanobacterium Thermosynechococcus elongatus BP-1. Biochim. Biophys. Acta 1777, 74–83 [DOI] [PubMed] [Google Scholar]
- 17. Máté Z., Sass L., Szekeres M., Vass I., Nagy F. (1998) UV-B-induced differential transcription of psbA genes encoding the D1 Protein of photosystem II in the cyanobacterium Synechocystis 6803. J. Biol. Chem. 273, 17439–17444 [DOI] [PubMed] [Google Scholar]
- 18. Summerfield T. C., Toepel J., Sherman L. A. (2008) Low-oxygen induction of normally cryptic psbA genes in cyanobacteria. Biochemistry 47, 12939–12941 [DOI] [PubMed] [Google Scholar]
- 19. Salih G. F., Jansson C. (1997) Activation of the silent psbA1 gene in the cyanobacterium Synechocystis sp strain 6803 produces a novel and functional D1 Protein. Plant Cell 9, 869–878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sicora C., Wiklund R., Jansson C., Vass I. (2004) Charge stabilization and recombination in Photosystem II containing the D1′ protein product of the psbA1 gene in Synechocystis 6803. Phys. Chem. Chem. Phys. 6, 4832–4837 [Google Scholar]
- 21. Campbell D., Clarke A. K., Gustafsson P., Öquist G. (1999) Oxygen-dependent electron flow influences photosystem II function and psbA gene expression in the cyanobacterium Synechococcus sp. PCC 7942. Physiol. Plant. 105, 746–755 [Google Scholar]
- 22. Sicora C. I., Brown C. M., Cheregi O., Vass I., Campbell D. A. (2008) The psbA gene family responds differentially to light and UVB stress in Gloeobacter violaceus PCC 7421, a deeply divergent cyanobacterium. Biochim. Biophys. Acta 1777, 130–139 [DOI] [PubMed] [Google Scholar]
- 23. Berman-Frank I., Lundgren P., Falkowski P. (2003) Nitrogen fixation and photosynthetic oxygen evolution in cyanobacteria. Res. Microbiol. 154, 157–164 [DOI] [PubMed] [Google Scholar]
- 24. Meeks J. C., Elhai J., Thiel T., Potts M., Larimer F., Lamerdin J., Predki P., Atlas R. (2001) An overview of the genome of Nostoc punctiforme, a multicellular, symbiotic cyanobacterium. Photosynth. Res. 70, 85–106 [DOI] [PubMed] [Google Scholar]
- 25. Reddy K. J., Haskell J. B., Sherman D. M., Sherman L. A. (1993) Unicellular, aerobic nitrogen-fixing cyanobacteria of the genus Cyanothece. J. Bacteriol. 175, 1284–1292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Stöckel J., Welsh E. A., Liberton M., Kunnvakkam R., Aurora R., Pakrasi H. B. (2008) Global transcriptomic analysis of Cyanothece 51142 reveals robust diurnal oscillation of central metabolic processes. Proc. Natl. Acad. Sci. U.S.A. 105, 6156–6161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sherman L. A., Meunier P., Colon-Lopez M. S. (1998) Diurnal rhythms in metabolism: a day in the life of a unicellular, diazotrophic cyanobacterium. Photosynth. Res. 58, 25–42 [Google Scholar]
- 28. Meunier P. C., Colon-Lopez M. S., Sherman L. A. (1998) Photosystem II cyclic heterogeneity and photoactivation in the diazotrophic, unicellular cyanobacterium Cyanothece species ATCC 51142. Plant Physiol. 116, 1551–1562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Burnap R. L., Qian M., Pierce C. (1996) The manganese stabilizing protein of photosystem II modifies the in vivo deactivation and photoactivation kinetics of the H2O oxidation complex in Synechocystis sp. PCC 6803. Biochemistry 35, 874–882 [DOI] [PubMed] [Google Scholar]
- 30. Welsh E. A., Liberton M., Stöckel J., Loh T., Elvitigala T., Wang C., Wollam A., Fulton R. S., Clifton S. W., Jacobs J. M., Aurora R., Ghosh B. K., Sherman L. A., Smith R. D., Wilson R. K., Pakrasi H. B. (2008) The genome of Cyanothece 51142, a unicellular diazotrophic cyanobacterium important in the marine nitrogen cycle. Proc. Natl. Acad. Sci. U.S.A. 105, 15094–15099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Allen M. M. (1968) Simple conditions for growth of unicellular blue-green algae on plates. J. Phycol. 4, 1–4 [DOI] [PubMed] [Google Scholar]
- 32. Larkin M. A., Blackshields G., Brown N. P., Chenna R., McGettigan P. A., McWilliam H., Valentin F., Wallace I. M., Wilm A., Lopez R., Thompson J. D., Gibson T. J., Higgins D. G. (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948 [DOI] [PubMed] [Google Scholar]
- 33. Goujon M., McWilliam H., Li W., Valentin F., Squizzato S., Paern J., Lopez R. (2010) A new bioinformatics analysis tools framework at EMBL-EBI. Nucleic Acids Res. 38, W695–W699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. (1990) Basic local alignment search tool. J. Mol. Biol. 215, 403–410 [DOI] [PubMed] [Google Scholar]
- 35. Tamura K., Stecher G., Peterson D., Filipski A., Kumar S. (2013) MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Goujon M., McWilliam H., Li W., Valentin F., Squizzato S., Paern J., Lopez R. (2010) A new bioinformatics analysis tools framework at EMBL-EBI. Nucleic Acids Res. 38, W695–W699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Li M. Z., Elledge S. J. (2007) Harnessing homologous recombination in vitro to generate recombinant DNA via SLIC. Nat. Methods 4, 251–256 [DOI] [PubMed] [Google Scholar]
- 38. Bandyopadhyay A., Stöckel J., Min H., Sherman L. A., Pakrasi H. B. (2010) High rates of photobiological H2 production by a cyanobacterium under aerobic conditions. Nat. Commun. 1, 139. [DOI] [PubMed] [Google Scholar]
- 39. Norling B., Zak E., Andersson B., Pakrasi H. (1998) 2D-isolation of pure plasma and thylakoid membranes from the cyanobacterium Synechocystis sp. PCC 6803. FEBS Lett. 436, 189–192 [DOI] [PubMed] [Google Scholar]
- 40. Mannan R. M., Pakrasi H. B. (1993) Dark heterotrophic growth conditions result in an increase in the content of photosystem II units in the filamentous cyanobacterium Anabaena variabilis ATCC 29413. Plant Physiol. 103, 971–977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Rakhimberdieva M. G., Vavilin D. V., Vermaas W. F., Elanskaya I. V., Karapetyan N. V. (2007) Phycobilin/chlorophyll excitation equilibration upon carotenoid-induced non-photochemical fluorescence quenching in phycobilisomes of the cyanobacterium Synechocystis sp. PCC 6803. Biochim. Biophys. Acta 1767, 757–765 [DOI] [PubMed] [Google Scholar]
- 42. Stöckel J., Jacobs J. M., Elvitigala T. R., Liberton M., Welsh E. A., Polpitiya A. D., Gritsenko M. A., Nicora C. D., Koppenaal D. W., Smith R. D., Pakrasi H. B. (2011) Diurnal rhythms result in significant changes in the cellular protein complement in the cyanobacterium Cyanothece 51142. PLoS One 6, e16680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Bench S. R., Heller P., Frank I., Arciniega M., Shilova I. N., Zehr J. P. (2013) Whole genome comparison of six Crocosphaera watsonii strains with differing phenotypes. J. Phycol. 49, 786–801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Shih P. M., Wu D., Latifi A., Axen S. D., Fewer D. P., Talla E., Calteau A., Cai F., Tandeau de Marsac N., Rippka R., Herdman M., Sivonen K., Coursin T., Laurent T., Goodwin L., Nolan M., Davenport K. W., Han C. S., Rubin E. M., Eisen J. A., Woyke T., Gugger M., Kerfeld C. A. (2013) Improving the coverage of the cyanobacterial phylum using diversity-driven genome sequencing. Proc. Natl. Acad. Sci. U.S.A. 110, 1053–1058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Dagan T., Roettger M., Stucken K., Landan G., Koch R., Major P., Gould S. B., Goremykin V. V., Rippka R., Tandeau de Marsac N., Gugger M., Lockhart P. J., Allen J. F., Brune I., Maus I., Pühler A., Martin W. F. (2013) Genomes of stigonematalean cyanobacteria (subsection V) and the evolution of oxygenic photosynthesis from prokaryotes to plastids. Genome Biol. Evol. 5, 31–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Bandyopadhyay A., Elvitigala T., Welsh E. A., Stockel J., Liberton M., Min H., Sherman L. A., Pakrasi H. B. (2011) Novel metabolic attributes of the genus cyanothece, comprising a group of unicellular nitrogen-fixing Cyanothece. mBio. 2, e00214–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Pfreundt U, Stal L. J., Voss B., Hess W. R. (2012) Dinitrogen fixation in a unicellular chlorophyll d-containing cyanobacterium. ISME J 6, 1367–1377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Satoh K., Yamamoto Y. (2007) The carboxyl-terminal processing of precursor D1 protein of the photosystem II reaction center. Photosynth. Res. 94, 203–215 [DOI] [PubMed] [Google Scholar]
- 49. Debus R. J., Nguyen A. P., Conway A. B. (1990) Identification of ligands to manganese and calcium in photosystem II by site-directed mutagenesis. Curr. Res. Photosynth. 1, 829–832 [Google Scholar]
- 50. Zhang X., Sherman L. A. (2012) Alternate copies of D1 are used by cyanobacteria under different environmental conditions. Photosynth. Res. 114, 133–135 [DOI] [PubMed] [Google Scholar]
- 51. Thiel T., Hartnett T., Jr., Pakrasi H. B. (1990) Examination of photosystem II in heterocysts of the cyanobacterium Nostoc sp. ATCC 29150. Curr. Res. Photosynth. 1, 291–294 [Google Scholar]
- 52. Cardona T., Battchikova N., Zhang P., Stensjö K., Aro E. M., Lindblad P., Magnuson A. (2009) Electron transfer protein complexes in the thylakoid membranes of heterocysts from the cyanobacterium Nostoc punctiforme. Biochim. Biophys. Acta 1787, 252–263 [DOI] [PubMed] [Google Scholar]
- 53. Postgate J. R. (1998) Nitrogen Fixation, Cambridge University, Cambridge, UK [Google Scholar]
- 54. Gallon J. R. (1992) Reconciling the incompatible: N2 fixation and O2. New Phytol. 122, 571–609 [Google Scholar]
- 55. Williams R. J. P., da Silva J. J. R. F. (1997) The Natural Selection of the Chemical Elements: The Environment and Life's Chemistry, Clarendon Press, Wotton-under-Edge, Gloucestershire UK [Google Scholar]
- 56. Gorbunov M. Y., Falkowski P. G. (2002) Photoreceptors in the cnidarian host allow symbiotic corals to sense blue moonlight. Limnol. Oceanogr. 47, 309–315 [Google Scholar]
- 57. Munz F. W., McFarland W. N. (1973) The significance of spectral position in the rhodopsins of tropical marine fishes. Vision Res. 13, 1829–1874 [DOI] [PubMed] [Google Scholar]
- 58. Overmann J., Garcia-Pichel F. (2013) The phototrophic way of life. In The Prokaryotes (Rosenberg E., DeLong E., Lory S., Stackebrandt E., Thompson F. eds), pp. 203–257, Springer Berlin, Heidelberg [Google Scholar]
- 59. Raven J. A., Cockell C. S. (2006) Influence on photosynthesis of starlight, moonlight, planetlight, and light pollution (reflections on photosynthetically active radiation in the universe). Astrobiology 6, 668–675 [DOI] [PubMed] [Google Scholar]
- 60. Kulkarni R. D., Golden S. S. (1994) Adaptation to high light intensity in Synechococcus sp. strain PCC 7942: regulation of three psbA genes and two forms of the D1 protein. J. Bacteriol. 176, 959–965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Gan F., Zhang S., Rockwell N. C., Martin S. S., Lagarias J. C., Bryant D. A. (2014) Extensive remodeling of a cyanobacterial photosynthetic apparatus in far-red light. Science 345, 1312–1317 [DOI] [PubMed] [Google Scholar]