Abstract
In genetics, the promoter is one of the most important regulatory elements controlling the spatiotemporal expression of a target gene. However, most studies have focused on core or proximal promoter regions, and information on regions that are more distant from the 5′-flanking region of the proximal promoter is often lacking. Here, approximately 4-kb of the sericin1 (Ser1) promoter was predicted to contain many potential transcriptional factor binding sites (TFBSs). Transgenic experiments have revealed that more TFBSs included in the promoter improved gene transcription. However, multi-copy proximal Ser1 promoter combinations did not improve gene expression at the transcriptional level. Instead, increasing the promoter copy number repressed transcription. Furthermore, a correlation analysis between two contiguous genes, firefly luciferase (FLuc) and EGFP, was conducted at the transcriptional level; a significant correlation was obtained regardless of the insertion site. The ELISA results also revealed a significant correlation between the transcriptional and translational EGFP levels. Therefore, the exogenous gene expression level can be predicted by simply detecting an adjacent EGFP. In conclusion, our results provide important insights for further investigations into the molecular mechanisms underlying promoter function. Additionally, a new approach was developed to quickly screen transgenic strains that highly express exogenous genes.
The development of transgenic technology constituted a crucial milestone in the history of life science, and is frequently used to alter biological traits and produce valuable foreign proteins in various organisms. Gene transcription is regulated by multiple cis-acting elements, such as promoters, enhancers, insulators, and silencers1,2,3. The promoter is the closest and most important element regulating target gene transcription. Universally, the promoter is a region of essentially contiguous DNA sequence in the immediate vicinity of the transcription start site (TSS), which initiates the transcription of a specific gene. However, the transcription of diverse genes uses distinct promoter types4, and there is no universal promoter that suits all types of genes. The core promoter5,6, the most important part of an active promoter, is a short sequence around the TSS. It acts as a platform for the assembly of transcription pre-initiation complexes (PICs) that include RNA polymerase II (pol II) and certain basal or general transcription factors (TFs), such as TFIIA, TFIIB, TFIID, TFIIE, TFIIF, and TFIIH7,8. Eventually, pol II accurately and efficiently directs transcription initiation9,10. The reason for the anchoring of PICs onto the core promoter is the existence of several core promoter elements, such as the classical TATA box11, the Initiator (Inr)12,13,14, and downstream core promoter element (DPE)15. In the current study, proximal promoters were selected to drive target gene transcription in vivo or in vitro. The proximal promoter controls gene transcription even though it may not include all regulatory elements4. Nevertheless, regulatory elements distant from the proximal promoters may be necessary to enhance the promoter's strength. We speculated that appropriately extending the length of the 5′-flanking sequence of a proximal promoter may improve gene transcription efficiency. We also speculated that combining multi-copy promoters may similarly contribute to enriching the PICs and enhance transcription efficiency.
Foreign genes integrate into the organism's genome and induce position effects. These events are influenced by the surrounding host chromatin and result in an exogenous gene that will demonstrate dramatically different expression patterns in various insertion sites16,17,18,19,20. In most transgenic studies, exogenous genes often randomly integrate into a host genome, and only a very low proportion of the integration events occur at favorable positions21. Therefore, the expression levels of foreign genes in different integration sites need to be assessed to identify the organisms with high foreign gene expression level potential, particularly in development of bioreactors to produce as much foreign protein as possible. This type of assessment is a time-consuming and labor-intensive process. A marker gene is generally used in transgenic experiments to screen positive individuals. Therefore, we reasoned that if the foreign- and marker genes are adjacent and in the same transcriptional direction in transgenic vectors, they may be similarly affected by host chromatin. As a result, there should be a significant linear correlation between the expression levels of these two genes, even in different insertion sites. Therefore, a promising method for predicting foreign gene expression levels may be detection of an adjacent marker gene, which can be more easily conducted.
The domestic silkworm, Bombyx mori, is an important economic insect that can synthesize and spin a large variety of proteins in silk glands to produce a cocoon before pupating. A method based on a piggyBac (PB) transposon-derived vector was successfully developed for stable germline transformation in silkworms22. Since its development, several studies have been performed using silk gland specific expression promoters, such as sericin (Ser) and fibroin light-chain (Fib-L), to drive exogenous gene expression in silk glands. These ideal bioreactors have helped express many valuable foreign proteins. However, most studies have indicated that foreign protein expression is still very low and accounts for less than 1% of a cocoon's weight23,24,25,26,27,28,29,30. It is noteworthy that the Ser and Fib-L promoters used in these studies are proximal or shorter promoters. Furthermore, positive individuals have to be selected from a large number of treated silkworms, rendering the process tedious and time-consuming. It is therefore necessary to develop a more efficient method for transgenic organism screening.
The objective of this study was to assess the correlations between different lengths and copy numbers of the Ser1 promoter and the foreign gene transcriptional level in silkworms. Additionally, a correlation analysis between two adjacent genes, a marker gene (EGFP) and foreign gene (FLuc), was conducted to elucidate their relationship.
Results
Architectural analysis of Ser1 promoter
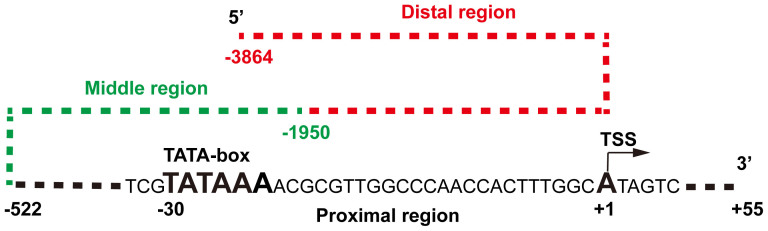
Promoters can be divided into two classes, conserved TATA box-enriched promoters and evolvable CpG-rich promoters4. The 4-kb long 5′-flanking Ser1 gene sequence, a highly tissue-specific gene that is only expressed in the middle silk gland31, was cloned and analyzed using bioinformatics methods. We confirmed the positions of the Ser1 gene TSS and the classical regulatory element TATA-box, which was located 30 bp upstream of the TSS (Fig. 1). In addition, we also predicted the potential TFBSs in three Ser1 promoter regions. The results suggested that the promoter's proximal, middle, and distal regions comprised 41, 87, and 105 potential TFBSs, respectively (Supplementary Table S1). Thus, we speculated that the 192 potential TFBSs located further away from the proximal promoter region may be beneficial for downstream gene transcription.
Figure 1. Schematic diagram of the Ser1 promoter sequence.
Black dashed line and capital letters represent the proximal promoter region; green and red dashed lines represent the middle and distal promoter regions respectively. TSS, transcription start site.
Ser1 promoter activity analysis
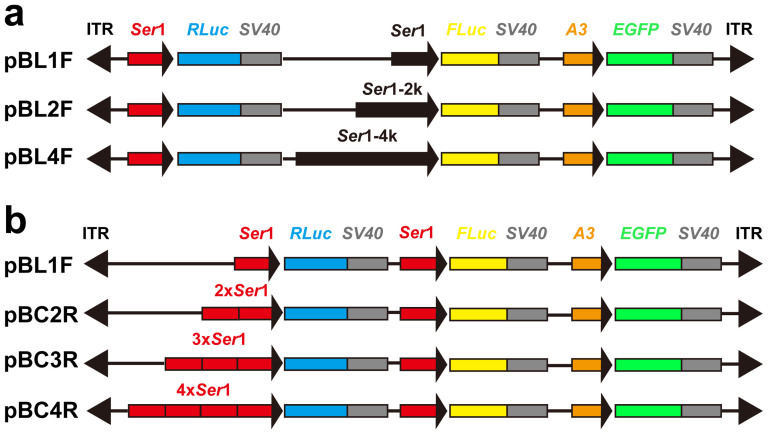
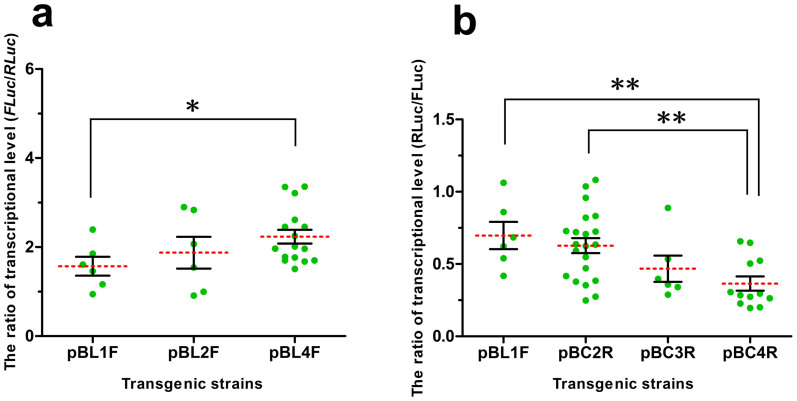
To accurately distinguish the different lengths of the Ser1 promoter that drive activity while eliminating the position effects as much as possible, we designed two expression frames in the same direction that were closely inserted in vectors (Fig. 2a). A reference gene (renilla luciferase, RLuc), driven by a proximal Ser1 promoter, was used in each vector. The ratio of FLuc/RLuc gene expression at the transcriptional level was calculated to determine the strength of the three different Ser1 promoters according to their lengths. Three types of pBL series-positive transgenic silkworms were obtained (Supplementary Table S2). Certain positive broods included multiple phenotypes, which mainly manifested in diverse patterns of EGFP expression resulting from PB integrating into different sites. This outcome is a common phenomenon in PB-mediated gene transposition studies. Therefore, silkworms with different EGFP expression patterns in the same brood were separated and defined as independent transgenic strains. We found that the highest level of FLuc transcription was in the pBL4F series transgenic silkworms, followed by the pBL2F series (Fig. 3a). To our surprise, the strength comparison among the four types of proximal promoter combinations (Fig. 2b) revealed that multi-copy proximal promoters did not enhance RLuc (FLuc served as an internal reference gene) transcription but repressed transcription (Fig. 3b). Indeed, RLuc transcription decreased as the promoter copy number increased.
Figure 2. Structure diagram of transgenic vectors.
Ser1, Ser1-2 k, and Ser1-4 k indicate the ~0.5 kb, ~2 kb and ~4 kb of Ser1 gene 5′-flanking sequence, respectively. The 2×, 3× and 4× Ser1 represent 2, 3 and 4 copies of Ser1 proximal promoter, respectively. RLuc, renilla luciferase; FLuc, firefly luciferase; A3, Bombyx mori A3 cytoplasmic actin gene promoter; EGFP, enhanced green fluorescence protein; SV40, 3′-untranslated sequences; ITR, inverted terminal repeats of PB transposon.
Figure 3. Comparison of driving activities of the Ser1 promoters.
(a) Differences in FLuc transcription driven by different lengths of the Ser1 promoter. The final relative transcriptional level was calculated as FLuc relative expression level/RLuc relative expression level. (b) Differential transcription of RLuc driven by different copy numbers of proximal Ser1 promoter. Similarly, the final relative expression level was calculated as RLuc relative expression level/FLuc relative transcriptional level. *P < 0.05, **P < 0.01.
Integration site analysis in transgenic silkworms
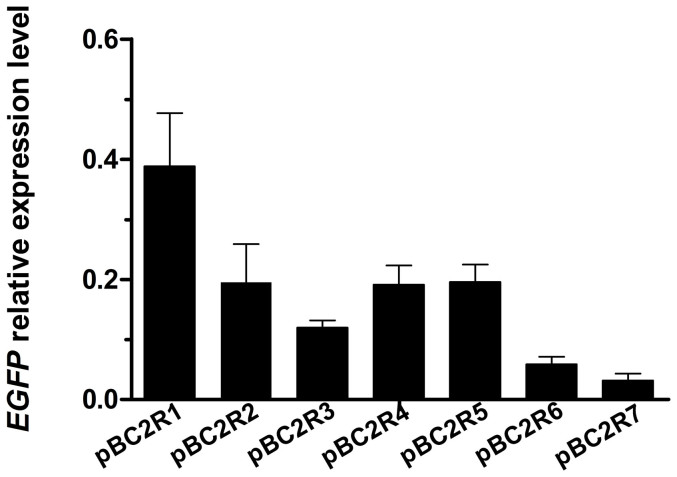
A foreign gene in different genomic loci demonstrates diverse expression levels due to position effects. An integration site analysis was conducted in the pBC2R series transgenic strains, which yielded the most positive individuals. As a representative sample, we selected seven transgenic strains, and each was found to have PB inserted at different genome positions (Supplementary Table S3). Meanwhile, the EGFP transcription levels were also determined in these strains, and huge differences were found among these levels; specifically, the transcription level of EGFP in pBC2R1 was 12 times higher than that in pBC2R7 (Fig. 4). These results further confirmed that PB-mediated transgenesis resulted in random insertions into the genome and that foreign gene transcription is dramatically influenced by the position effect.
Figure 4. Transcriptional level of EGFP in different integration sites.
The inverse PCR showed that the PB integration sites were different in the seven pBC2R transgenic strains (Supplementary Table S3). The EGFP mRNA levels differed among these strains due to position effects from the host chromatin. Data are presented as the mean ± SD of three separate experiments performed in triplicate.
Correlation analysis between marker- and target genes
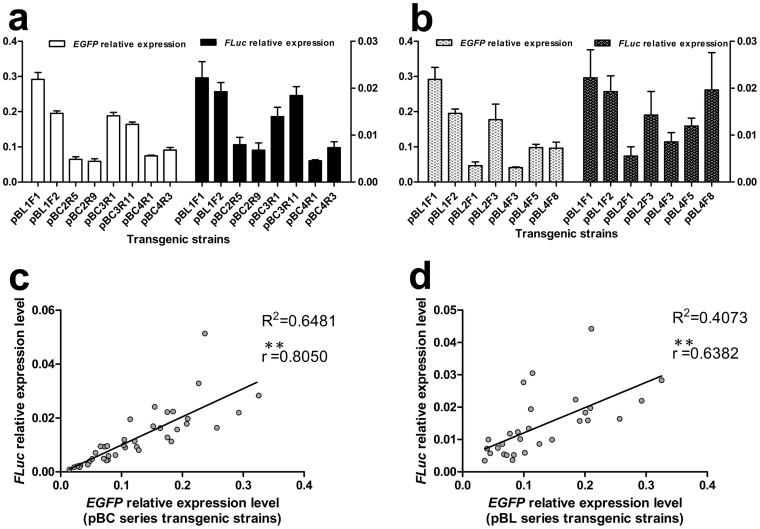
Two adjacent foreign genes should theoretically experience similar effects from the host chromatin. In our study, the marker gene was inserted close to the target gene with the same transcriptional orientation, e.g., EGFP and FLuc in Fig. 2. We speculated that the transcriptional levels of these two adjacent genes would demonstrate a linear correlation even if they were inserted into different genomic loci. Four series of transgenic silkworms, pBL1F, pBC2R, pBC3R, and pBC4R, shared a common feature, the promoter in front of FLuc and EGFP was unchanged (Fig. 2b). The qRT-PCR results indicated that the FLuc and EGFP transcriptional levels varied for the different integration sites, but the two adjacent genes exhibited similar expression patterns (Fig. 5a) with a significant correlation between their expression levels (Fig. 5c). However, the correlation among the pBL1F, pBL2F, and pBL4F series transgenic silkworms was lower compared to the previous four series of transgenic silkworms (Fig. 5b, d). This finding may be due to the different lengths of the Ser1 promoter in front of FLuc, which resulted in different Ser1 promoter strengths.
Figure 5. Correlation analysis between the marker (EGFP) and target (FLuc) genes at the transcriptional level.
(a, c) Transcription pattern comparison of EGFP and FLuc in the pBC (including pBL1F) series transgenic silkworms. A significant correlation was revealed between the EGFP and FLuc expression levels with the two expression frames assembled in close proximity. (b, d) However, the correlation between EGFP and FLuc in the pBL series transgenic silkworms was relatively lower; the Ser 1 promoters in front of FLuc were different (Fig. 2a), which affected the transcriptional level of FLuc. The qRT-PCR data are shown as the mean ± SD of three separate experiments performed in triplicate. **P < 0.01.
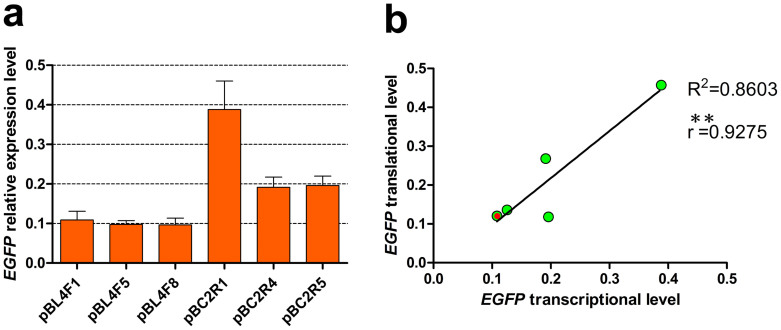
These findings suggested that FLuc expression was significantly correlated with EGFP expression at the transcriptional level. To determine whether the transcriptional and translational levels were also significantly correlated at the same developmental stage, we selected three strains with low and relatively high EGFP transcription from the pBL4F and pBC2R series transgenic silkworms, respectively (Fig. 6a). The ELISA results suggested that there was a highly significant correlation between the transcriptional and translational EGFP levels (Fig. 6b). Taken together, these results demonstrate that the expression level of a target gene (FLuc) in the host can be simply predicted by the expression level of an adjacent marker gene (EGFP).
Figure 6. Correlation analysis between the EGFP transcriptional and translational levels.
(a) Six strains with different EGFP transcription levels were selected for further analysis. Data are presented as the mean ± SD of three separate experiments performed in triplicate. (b) There was a highly significant correlation between the transcriptional and translational levels of EGFP. Each green dot represents one transgenic strain; a red dot is used to distinguish two overlapping dots. **P < 0.01.
Discussion
It is important to develop high-efficiency bioreactors that meet societal demands. Identifying efficient cis-acting regulators, such as promoters and enhancers, is commonly used to improve foreign gene expression. Previous studies have demonstrated that TATA box-containing promoters are more constrained with slow evolution rates. The preferred distance between a TATA-box and TSS is 30 or 31 bp, and this promoter type generally has high tissue specificity in driving gene expression4. These findings are highly consistent with our results, and we further validated the characteristics of the TATA box-containing promoter. Promoter activity is closely related to its sequence length; longer promoters are superior regardless of the quantity and type of potential TFBSs (Supplementary Table S1). The 192 potential TFBSs in the middle and distal Ser1 promoter regions further enhanced promoter activity and facilitated downstream gene transcription. Therefore, we believe that the TFBSs are important parameters for improving transgene expression efficiency. However, lengthening the upstream regions of the Ser1 promoter did not demonstrate a pronounced improvement in the promoter activity; the promoter activity improved by approximately 1.5 times after using a 4-kb length Ser1 promoter. Currently, using efficient enhancers is a common method for enhancing the efficiency of exogenous gene expression in silkworms. However, expression efficiency remains very low. We believe that the precondition for improving transgene expression efficiency is to find a high-efficiency promoter. Therefore, promoter activity will be significantly amplified after the combined use of enhancers, and using the intrinsic functional elements of the promoter as much as possible is an efficient method for improving the level of exogenous gene expression.
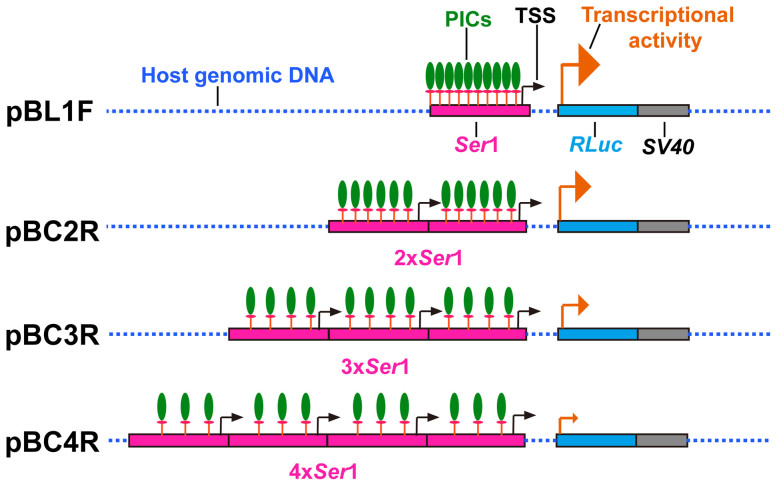
Surprisingly, the multi-copy promoter combinations had an adverse effect on gene transcription. The activity of weakened multi-copy promoter combinations may generate competition among the same promoters. This possibility is reflected in the simultaneous competition for the same TFs and pol II. We speculate that this phenomenon will induce limited substrates that dispersedly combine on promoters, with the promoter closest to the downstream gene serving as the main driving force of gene transcription (Fig. 7). As a result, the promoters function at a sub-optimal level due to a lack of PICs (composed of TFs and pol II) on the promoter closest to the gene. In addition, each proximal promoter contains a TSS (Fig. 7), implying another possibility that competition also exists among multiple TSSs. However, more detailed studies are necessary to investigate these two possible molecular mechanisms.
Figure 7. Schematic representation of the competition phenomenon among multi-copy promoters.
PICs, transcription pre-initiation complexes; TSS, transcription start site of the Ser1 promoter; orange arrows represent transcription activity and transcription orientation of RLuc; 2×, 3× and 4× of Ser1 represent 2, 3 and 4 copies of the Ser1 proximal promoter, respectively; RLuc, renilla luciferase; SV40, 3′-untranslated sequences.
Transgenic vectors often contain two parts, the marker gene and the exogenous gene expression cassettes. Our design approach was different from the standard approach, which does not take into consideration the special demands of transgenic vector construction. The marker gene has only been used to select transgenic animals in previous studies. However, another important function of the marker gene is to assist in determining the exogenous gene expression level, as described above. Our findings revealed that if the two expression cassettes are constructed close to one another with the same transcriptional orientation, then there is a significant linear correlation between their transcriptional levels, even if they are in different insertion sites. Furthermore, the transcriptional and translational levels of the marker genes were significantly correlated; therefore, it is possible to determine foreign gene expression levels by simply assessing the level of marker gene expression. Marker genes are widely used in life science studies and can be easily and conveniently measured using various methods. In conclusion, the assisted assessment of the expression level of an exogenous gene is a promising new method for the large-scale screening of transgenic strains characterized by a high-expression capacity for foreign proteins. Additionally, it can also be used for the real-time monitoring of gene expression patterns to more conveniently and thoroughly understand gene functions. Furthermore, it provides a foundation for further optimizing transgenesis technology.
Methods
Silkworm strains
Lan10, the multivoltine with diapause ability silkworm strain, and Qiufeng, a high silk-yield strain, were used. All experimental silkworms were reared on fresh mulberry leaves under the standard conditions (25°C, 80% R.H).
Bioinformatics analysis of Ser1 promoter
The preselected ~4-kb length 5′-flanking Ser1 gene sequence was analyzed using the TFSEARCH (version 1.3) software32 to identify putative TATA-box and TFBSs (threshold value: 85.0). Moreover, the TSS location was also analyzed using the Neural Network Promoter Prediction software33.
Transgenic vector construction
Genomic DNA was extracted from Qiufeng silk glands using a DNA extraction kit (Sangon, Shanghai, China). Then, different Ser1 promoter lengths, 0.5 kb (proximal region, 577 bp), 2 kb (proximal and middle region, 2005 bp) and 4 kb (proximal, middle, and distal region, 3919 bp), were cloned using high fidelity PrimeSTAR HS DNA Polymerase (TAKARA BIO INC., Otsu, Shiga, Japan). The promoters were sequenced after cloning into the pMD19-T vector (TAKARA BIO INC., Otsu, Shiga, Japan). Three transgenic vectors, pBL1F, pBL2F, and pBL4F, were constructed based on the bi-fluorescence plasmid pA3RLucFLuc (conserved in our lab) and the PB transposon plasmid pBA3EGFP to compare the driving activities of the three different Ser1 promoters. EGFP served as a marker gene used for positive individual screening, and RLuc was an internal reference gene to standardize the FLuc transcription level. Furthermore, combinations of two, three, and four copies of Ser1 proximal promoters were constructed in the pBC2R, pBC3R, and pBC4R vectors, respectively, and their driving activities were compared. Similarly, FLuc was used as an internal reference gene to standardize the RLuc transcription level. All data were analyzed by multiple comparisons (LSD) after one-way analysis of variance (ANOVA).
Embryo microinjection and positive individual screening
The detailed methodologies for embryo microinjection and positive animal screening have been previously described in detail22,34. Briefly, the fertilized eggs were timely collected and microinjection was performed within 8 h of oviposition. The injection plasmids consisted of transposition- and helper plasmids, at a concentration ratio of 1:0.5, and a total injection volume of approximately 10 nL for each egg (the total concentration of DNA was 0.4 μg/μL) was used. After microinjection, G0 generation zygotes were cultured under standard conditions to mature moths. Each moth was mated with native Lan10 silkworms to produce G1 generations. Finally, positive individuals were selected from each G1 brood using an Olympus SZX16 fluorescence microscope (Olympus, Tokyo, Japan).
Total RNA Extraction and Quantitative RT-PCR Analysis
Certain procedures were performed as previously described35. Briefly, total RNA was extracted from fresh middle silk glands collected from day 3 fifth instar larvae using TRIzol reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer's instructions. The cDNA was obtained using the PrimeScript RT reagent kit with gDNA Eraser (Perfect Real Time) (TAKARA BIO INC., Otsu, Shiga, Japan). The qRT-PCR was performed by a LightCycler480 instrument (Roche Diagnostics, Rotkreuz, Switzerland) in a 20-μL volume reaction that included 50 ng cDNA, 10 μL of SYBR Premix Ex Taq™ (2X) (TAKARA BIO INC., Otsu, Shiga, Japan), and 4 μM each of forward and reverse primer. The amplification program began with 30 s denaturation at 95°C, followed by 40 cycles of 5 s at 95°C, 20 s at 60°C, and 15 s at 72°C. The relative quantification analyses of the mRNA levels of target genes were based on the delta Ct value normalized with a reference gene, Rp49 gene (accession number: NM_001098282). Each sample had three independent replicates, and the gene-specific primers (Supplementary Table S4) were designed using the Primer Premier 5.0 software (Premier Biosoft International, Palo Alto, CA).
Inverse PCR analysis
Genomic DNA was isolated from the positive transgenic silkworms as described above. The inverse PCR analysis was conducted as previously described22, 1 μg of each genomic DNA was digested with Sau3A I at 37°C for 2 h and then self-circularized overnight at 16°C using T4 DNA ligase (TAKARA BIO INC., Otsu, Shiga, Japan). The ligated products (approximately 50 ng DNA) were amplified using EX Taq polymerase (TAKARA BIO INC., Otsu, Shiga, Japan) and the designed primers (Supplementary Table S4) with 4 min denaturing cycle at 94°C, followed by 40 cycles of 30 s at 94°C, 30 s at 58°C, and 2 min at 72°C, and a final extension at 72°C for 10 min. Amplified products were sequenced after cloning into pMD19-T to determine the insertion sites.
Enzyme-linked immunosorbent assay (ELISA)
The middle silk glands were dissected from the day 3 fifth instar larvae of transgenic silkworms and were thoroughly grinded in liquid nitrogen after adding ice-cold 2× cell lysis buffer (Cell Signaling Technology, Inc., Danvers, MA, USA) at a ratio of 100 mg of tissue to 400 μL buffer. Next, the samples were incubated on ice for 30 min with vortexing several times, centrifuged at 15,000 × g at 4°C for 10 min, and then the supernatant was collected in a new tube. The protein concentration was determined using a DC protein assay kit (Bio-Rad, California, USA). The tissue lysates were stored at −80°C in single-use aliquots. Each tissue lysate was diluted to the same concentration (10 mg/mL), and 1 mg of total protein was used for the ELISA assay. EGFP was detected using the PathScan® Total GFP Sandwich ELISA Kit (Cell Signaling Technology, Inc., Danvers, MA, USA) according to the manufacturer's instructions. Finally, the absorbance readings at 450 nm were measured using a microplate reader (PerkinElmer, Massachusetts, USA).
Author Contributions
L.Y. designed and conducted experiments, performed data analysis and wrote the paper. Q.Q. conducted experiments and performed data analysis. Y.Z. and Z.Y. conducted experiments. J.C. and J.S. reared and harvested all the samples. B.Z. conceived, designed and conducted experiments, performed data analysis, and revised the manuscript. All authors read and revised manuscript before submission.
Supplementary Material
Supplementary information
Acknowledgments
We thank Dr. Toshiki Tamura (National Institute of Sericultural and Entomological Science, Tsukuba, Ibaraki, Japan) for providing the piggyBac transposon pPIGA3EGFP and helper plasmid. This work is supported by the grants from the National Basic Research Program of China (No. 2012CB114601), Projects of Zhejiang Provincial Science and Technology Plans (No. 2013C32048, No. 2012C12910).
References
- Blackwood E. M. & Kadonaga J. T. Going the distance: a current view of enhancer action. Science 281, 60–63 (1998). [DOI] [PubMed] [Google Scholar]
- West A. G., Gaszner M. & Felsenfeld G. Insulators: many functions, many mechanisms. Genes Dev 16, 271–288 (2002). [DOI] [PubMed] [Google Scholar]
- Bulger M. & Groudine M. Looping versus linking: toward a model for long-distance gene activation. Genes Dev 13, 2465–2477 (1999). [DOI] [PubMed] [Google Scholar]
- Carninci P. et al. Genome-wide analysis of mammalian promoter architecture and evolution. Nat Genet 38, 626–635 (2006). [DOI] [PubMed] [Google Scholar]
- Smale S. T. & Kadonaga J. T. The RNA polymerase II core promoter. Annu Rev Biochem 72, 449–479 (2003). [DOI] [PubMed] [Google Scholar]
- Thomas M. C. & Chiang C. M. The general transcription machinery and general cofactors. Crit Rev Biochem Mol Biol 41, 105–178 (2006). [DOI] [PubMed] [Google Scholar]
- Matsui T., Segall J., Weil P. A. & Roeder R. G. Multiple factors required for accurate initiation of transcription by purified RNA polymerase II. J Biol Chem 255, 11992–11996 (1980). [PubMed] [Google Scholar]
- Zawel L. & Reinberg D. Initiation of transcription by RNA polymerase II: a multi-step process. Prog Nucleic Acid Res Mol Biol 44, 67–108 (1993). [DOI] [PubMed] [Google Scholar]
- Struhl K. Promoters, activator proteins, and the mechanism of transcriptional initiation in yeast. Cell 49, 295–297 (1987). [DOI] [PubMed] [Google Scholar]
- Weis L. & Reinberg D. Transcription by RNA polymerase II: initiator-directed formation of transcription-competent complexes. FASEB J 6, 3300–3309 (1992). [DOI] [PubMed] [Google Scholar]
- Breathnach R. & Chambon P. Organization and expression of eucaryotic split genes coding for proteins. Annu Rev Biochem 50, 349–383 (1981). [DOI] [PubMed] [Google Scholar]
- Ohler U., Liao G. C., Niemann H. & Rubin G. M. Computational analysis of core promoters in the Drosophila genome. Genome Biol 3, RESEARCH0087 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- FitzGerald P. C., Sturgill D., Shyakhtenko A., Oliver B. & Vinson C. Comparative genomics of Drosophila and human core promoters. Genome Biol 7, R53 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gershenzon N. I., Trifonov E. N. & Ioshikhes I. P. The features of Drosophila core promoters revealed by statistical analysis. BMC Genomics 7, 161 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke T. W. & Kadonaga J. T. The downstream core promoter element, DPE, is conserved from Drosophila to humans and is recognized by TAFII60 of Drosophila. Genes Dev 11, 3020–3031 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spradling A. C. & Rubin G. M. The effect of chromosomal position on the expression of the Drosophila xanthine dehydrogenase gene. Cell 34, 47–57 (1983). [DOI] [PubMed] [Google Scholar]
- al-Shawi R., Kinnaird J., Burke J. & Bishop J. O. Expression of a foreign gene in a line of transgenic mice is modulated by a chromosomal position effect. Mol Cell Biol 10, 1192–1198 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markstein M., Pitsouli C., Villalta C., Celniker S. E. & Perrimon N. Exploiting position effects and the gypsy retrovirus insulator to engineer precisely expressed transgenes. Nat Genet 40, 476–483 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis E. B. The phenomenon of position effect. Adv Genet 3, 73–115 (1950). [DOI] [PubMed] [Google Scholar]
- Levis R., Hazelrigg T. & Rubin G. M. Effects of genomic position on the expression of transduced copies of the white gene of Drosophila. Science 229, 558–561 (1985). [DOI] [PubMed] [Google Scholar]
- Rubin G. M. & Spradling A. C. Genetic transformation of Drosophila with transposable element vectors. Science 218, 348–353 (1982). [DOI] [PubMed] [Google Scholar]
- Tamura T. et al. Germline transformation of the silkworm Bombyx mori L. using a piggyBac transposon-derived vector. Nat Biotechnol 18, 81–84 (2000). [DOI] [PubMed] [Google Scholar]
- Tomita M. et al. Transgenic silkworms produce recombinant human type III procollagen in cocoons. Nat Biotechnol 21, 52–56 (2003). [DOI] [PubMed] [Google Scholar]
- Adachi T., Tomita M., Shimizu K., Ogawa S. & Yoshizato K. Generation of hybrid transgenic silkworms that express Bombyx mori prolyl-hydroxylase alpha-subunits and human collagens in posterior silk glands: Production of cocoons that contained collagens with hydroxylated proline residues. J Biotechnol 126, 205–219 (2006). [DOI] [PubMed] [Google Scholar]
- Hino R., Tomita M. & Yoshizato K. The generation of germline transgenic silkworms for the production of biologically active recombinant fusion proteins of fibroin and human basic fibroblast growth factor. Biomaterials 27, 5715–5724 (2006). [DOI] [PubMed] [Google Scholar]
- Ogawa S., Tomita M., Shimizu K. & Yoshizato K. Generation of a transgenic silkworm that secretes recombinant proteins in the sericin layer of cocoon: production of recombinant human serum albumin. J Biotechnol 128, 531–544 (2007). [DOI] [PubMed] [Google Scholar]
- Yanagisawa S. et al. Improving cell-adhesive properties of recombinant Bombyx mori silk by incorporation of collagen or fibronectin derived peptides produced by transgenic silkworms. Biomacromolecules 8, 3487–3492 (2007). [DOI] [PubMed] [Google Scholar]
- Iizuka M. et al. Production of a recombinant mouse monoclonal antibody in transgenic silkworm cocoons. FEBS J 276, 5806–5820 (2009). [DOI] [PubMed] [Google Scholar]
- Tateno M. et al. Production and characterization of the recombinant human mu-opioid receptor from transgenic silkworms. J Biochem 145, 37–42 (2009). [DOI] [PubMed] [Google Scholar]
- Wen H. et al. Transgenic silkworms (Bombyx mori) produce recombinant spider dragline silk in cocoons. Mol Biol Rep 37, 1815–1821 (2010). [DOI] [PubMed] [Google Scholar]
- Okamoto H., Ishikawa E. & Suzuki Y. Structural analysis of sericin genes. Homologies with fibroin gene in the 5′ flanking nucleotide sequences. J Biol Chem 257, 15192–15199 (1982). [PubMed] [Google Scholar]
- Heinemeyer T. et al. Databases on transcriptional regulation: TRANSFAC, TRRD and COMPEL. Nucleic Acids Res 26, 362–367 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reese M. G. Application of a time-delay neural network to promoter annotation in the Drosophila melanogaster genome. Comput Chem 26, 51–56 (2001). [DOI] [PubMed] [Google Scholar]
- Zhong B., Li J., Chen J., Ye J. & Yu S. Comparison of transformation efficiency of piggyBac transposon among three different silkworm Bombyx mori Strains. Acta Biochim Biophys Sin (Shanghai) 39, 117–122 (2007). [DOI] [PubMed] [Google Scholar]
- Wang S. H. et al. Quantitative proteomic and transcriptomic analyses of molecular mechanisms associated with low silk production in silkworm Bombyx mori. J Proteome Res 13, 735–751 (2014). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary information