Abstract
Rhesus macaques (Macaca mulatta) are an important primate model species in several areas of biomedical research. The wide geographic distribution of this species has led to significant genetic differentiation among local and regional populations. These regional differences can be important factors in the selection of the most appropriate subjects for particular research studies, as animals from different populations can respond differently to the same experimental treatment. Consequently, it is valuable to confirm the ancestry of individual rhesus monkeys from geographically distinct populations. Using DNA samples obtained from rhesus macaques from six National Primate Research Centers, we tested a set of 384 potential ancestry informative single nucleotide polymorphisms (SNPs) and identified a final panel of 91 SNPs that can reliably distinguish Indian-origin from Chinese-origin rhesus monkeys. This genetic test can be used to determine the ancestral origin of animals and to detect individuals that are hybrids between these two regional populations. To demonstrate use of the SNP panel, we investigated the ancestry of 480 animals from the Yerkes NPRC (YNPRC) for which the colony records were insufficient to clearly establish ancestry. Three of the YNPRC animals tested were determined to be hybrids. This SNP ancestry tool will be useful to researchers, colony managers, and others who wish to evaluate the ancestral origin of individual rhesus macaques, and therefore will facilitate more effective and efficient use of these animals in biomedical research.
Keywords: ancestry, rhesus
Introduction
The rhesus macaque (Macaca mulatta) is the most widely used nonhuman primate species in biomedical research. Rhesus macaques provide critical models for studies of infectious disease (especially simian immunodeficiency virus infection as a model of HIV-AIDS), neurobiology, endocrinology, alcohol and drug addiction, metabolic disease and other aspects of human biology and health. This species has the widest geographic range of any nonhuman primate, extending from Pakistan and Afghanistan in the west across southern Asia to the northern parts of Thailand, Laos, Vietnam and north throughout China south of the Yangtze River [Groves, 2001]. Although fully capable of interbreeding, the animals from distant parts of this range are genetically distinct. Their anatomical and morphological diversity is reflected in taxonomy, with six subspecies recognized [Groves, 2001]. In U.S. research colonies, most rhesus macaques are derived from founding animals that were obtained from either India or China. While both Indian- and Chinese-origin animals range in color from dusty brown to auburn with reddish pink faces, adult Chinese-origin animals are generally heavier, with longer limbs and greater head-to-tail length than their Indian-origin conspecifics [Clarke and O’Neil, 1999]. This variation in size, external appearance and other traits [Champoux et al., 1997; Champoux et al., 1994] reflect underlying population genetic diversification dating to at least 160,000 years ago [Ferguson et al., 2007; Gibbs et al., 2007; Hernandez et al., 2007].
Importantly for biomedical research, the two geographic populations (Indian and Chinese) exhibit phenotypic differences that are directly relevant to their use in experimental studies. For example, the different responses to SIV infection in Indian and Chinese rhesus macaques lead to strong preferences for Indian-origin animals among researchers working on the rhesus model of SIV infection and AIDS [Cohen, 2002; Ling et al., 2002; Trichel et al., 2002]. Chinese-derived animals also have an array of MHC class I haplotypes that only partially overlaps with that found in Indian-derived animals, suggesting that immunological challenges may be processed by the immune systems of the two populations in different ways [Karl et al., 2013; Wiseman et al., 2009]. In addition, behavioral traits can differ between the two geographic populations as animals with Chinese ancestry exhibit increased behavioral reactivity compared to Indian origin subjects [Champoux et al., 1997; Champoux et al., 1994]. As a result of these and other biological differences between regional populations of this species, investigators frequently prefer to study rhesus macaques of one population or geographic ancestry than another.
However, the ancestry of individual rhesus macaques available for research is not always known with complete reliability. While most of the rhesus macaques in U.S. research and breeding colonies are Indian-origin animals, in recent years an increasing number of animals with Chinese ancestry have been imported into the US [2011]. Incomplete or inaccurate animal records can result in macaques of mixed Indian-Chinese ancestry being inadvertently incorporated into breeding colonies or experimental protocols. This admixture may not be recognized and could influence research results. An inexpensive and accurate genetic test for ancestry is needed to allow colony managers, veterinarians and researchers to verify the ancestry of individual rhesus macaques. The Genetics and Genomics Working Group (GGWG) of the NIH Nonhuman Primate Research Consortium has developed and validated such a genetic test, here called the Ancestry Informative Markers (AIMs) test. This assay uses single nucleotide polymorphisms (SNPs) that differ in allele frequencies between Indian-origin and Chinese-origin rhesus monkeys. The test can be performed in a short time using a small amount of DNA and either assigns an individual to one or the other regional population, or provides an estimate of admixture. We report here the methods used to develop and validate this test. Our intent is to make this assay and the associated analytical procedures available to the research community to facilitate the evaluation of specific animals for breeding programs and research protocols.
We also describe the use of this test for an extensive analysis of a single rhesus macaque breeding colony. The Yerkes National Primate Research Center (YNPRC) rhesus macaque colony is typical of those of the eight National Primate Research Centers (NPRCs); while all rhesus macaques currently housed at the YNPRC have been bred in captivity, the founders of that population were obtained through multiple sources. The YNPRC has acquired animals from other NPRCs and from commercial breeding facilities in an effort to increase genetic variation within this captive population. While these acquired animals were described as of Indian-origin, in many cases inadequate information was available to reliably document their ancestry. Therefore, as with other NPRCs, there is interest in validating the ancestry of the current YNPRC rhesus macaque colony.
Methods
Candidate SNPs with potential for use in this AIMs panel were identified from re-sequencing data as described in Ferguson et al. (2007) and Satkoski et al. (2008). A set of 384 SNPs was chosen for further evaluation based upon identified differences in minor allele frequencies (MAF) between Indian and Chinese rhesus macaque populations detected in these initial discovery studies [Ferguson et al., 2007; Kanthaswamy et al., 2009].
We next genotyped the 384 candidate SNPs in 341 rhesus macaques from six NPRCs. The sample set included animals presumed to be pure Indian or pure Chinese, based on purchase records, pedigree data or genotypes at short tandem repeat (STR) loci. DNA samples from unrelated animals, as well as 30 sets of trios (sire, dam and offspring), were included as test subjects. Finally, DNA samples from a set of 30 known hybrid animals from the California and Oregon NPRCs, including animals with 25%, 50% or 75% Indian ancestry, as inferred from pedigree data, were also analyzed. Table I lists the number of animals tested from the different centers.
Table I.
Source and reported geographic origin of the rhesus macaque samples used in the initial development of this SNP assay. The number of samples used for initial testing is listed first and the numbers of samples used for validating the AIMs SNP panel are in parentheses.
| Source | N | Origin |
|---|---|---|
| NENPRC | 50 (13) | Indian |
| WNPRC | 50 (6) | Indian |
| ONPRC | 19 (2) | Chinese |
| ONPRC | 50 (2) | Indian |
| ONPRC | 5 (0) | 50% Indian |
| ONPRC | 5 (1) | 75% Indian |
| TNPRC | 50 (6) | Indian |
| YNPRC | 50 (3) | Indian |
| CNPRC | 20 (4) | Indian |
| CPRC | 0 (20) | Indian |
| CNPRC | 5 (5) | 75% Indian |
| CNPRC | 10 (9) | 50% Indian |
| CNPRC | 5 (4) | 25% Indian |
| CNPRC | 22 (20) | Chinese |
| Total | 341 (95) |
Genotyping was performed at the Oregon Health Sciences University Gene Profiling Shared Resource using the Illumina, Inc. (San Diego, CA) BeadXpress Assay, BeadStudio Reader, and GoldenGate™ Platform. After initial analysis of the genotype data, we excluded individuals that had more than 10% missing genotypes. A total of 336 animals satisfied this criterion and were used in subsequent analyses. The genotype data were then used to identify the most informative SNPs for ancestry discrimination, using two criteria. First, the SNPs were ranked by their information value for discriminating the two ancestral populations, an approach that prioritizes SNPs having the largest differences in MAF between the two populations [Banks et al., 2003; Kanthaswamy et al., 2009]. Second, we considered SNP chromosomal positions to maximize the distribution of SNPs across the genome. Through this process we identified a subset of 96 candidate SNPs for inclusion in the AIMs panel (Supplemental Table I). The size of the SNP array was designed to achieve a balance between the need for accurate ancestry discrimination and the cost of each assay. Additional information on these SNPs, including chromosome coordinates and flanking sequences, is provided in the Supplemental Table I and II. Chromosomal locations were based on the Macaca mulatta genome assembly, rheMac2 (Gibbs et al 2007).
We further tested the reliability of the 96 SNP AIMS panel by re-genotyping a set of 95 samples consisting of 54 confirmed Indian-origin animals (including 20 samples from the Caribbean Primate Research Center (CPRC)), 22 confirmed Chinese-origin individuals, and 19 hybrid animals based on the initial round of genotyping (Table I). This round of SNP genotyping of the 95 samples was performed at a different genotyping facility, the UC Davis Genome Center, using the Illumina BeadExpress Assay, BeadStudio Reader and GoldenGate Platform as in the first round. We reviewed the genotypes to ensure GenCall and GenTrain quality scores were at least 0.60, and that the SNP profiles did not exhibit more than 5% missing information.
This AIMS assay was then applied to the YNPRC rhesus macaque colony as a case study and to demonstrate the utility of the test. The 480 YNPRC subject animals were selected based on the number of offspring each had produced, so as to represent the maximum proportion of the total genetic variance present within the YNPRC colony. Genotypes for each individual were assigned using Illumina’s GenomeStudio software (an upgrade of BeadStudio). Again, individual genotypes or SNPs with quality scores below 0.6 were eliminated from further analysis and only animals with at least 95% genotype calls were included in the analysis.
Genotype data from each of the above studies were analyzed using STRUCTURE 2.3.3 [Hubisz et al., 2009; Pritchard et al., 2000]. The program employs a Markov chain Monte Carlo (MCMC) method to compute P(X|K), the probability that the data fit the hypothesis of K genetically discrete clusters. STRUCTURE also calculates the fractional genetic composition of each animal derived from each inferred ancestral population (Q). In this analysis, it was assumed that all animals were members of one of two possible ancestral populations, i.e. Indian or Chinese (K = 2), or hybrids produced from those two populations, with Q defined as the proportion of Indian ancestry. STRUCTURE was run with 5 X 105 MCMC iterations after a burn-in period of 1 x 105 iterations to ensure that group assignments with the greatest probabilities were detected. It was assumed that allele frequencies are correlated among populations and that, despite prior assignment of an individual to a specific ancestral population, each individual could have ancestors that were derived from both populations. A discriminant analysis of principal components (DAPC), which provides a visual and quantitative method for identifying genetic clusters by partitioning within- and between-group variance [Jombart and Ahmed, 2011], was performed using the adegenet package in R [Jombart, 2008].
All research protocols reported in this manuscript were reviewed and approved by the appropriate institutional animal utilization and care committees and adhered to the legal and ethical requirements that regulate research with animals in the United States. The research also complied with the American Society of Primatologists (ASP) Principles for the Ethical Treatment of Non Human Primates as found at https://www.asp.org/society/resolutions/EthicalTreatmentOfNonHumanPrimates.cfm.
Results
Detailed review of the genotypes generated in the second round of testing identified 3 SNPs (CXCL123173, GREM13811 and ZAP1283210) that did not produce reliable genotype calls across the two different genotyping centers, and thus they were eliminated from the panel. Two X-linked markers (CD40LG:738G and MAOA:116G) were also eliminated from the panel. Therefore the final SNP set includes 91 SNP loci. The Supplemental Table II presents the chromosome coordinates, alleles, MAF for each ancestral population and ΔMAF values (differences in MAF between Indian- and Chinese-origin animals) for the 91 SNPs included in the AIMS panel. The ΔMAF values ranged from 0.09 (X34958656.8.D8YOWMI01A87RR) to 0.69 (FJX13286), with an average of 0.365 (±0.098).
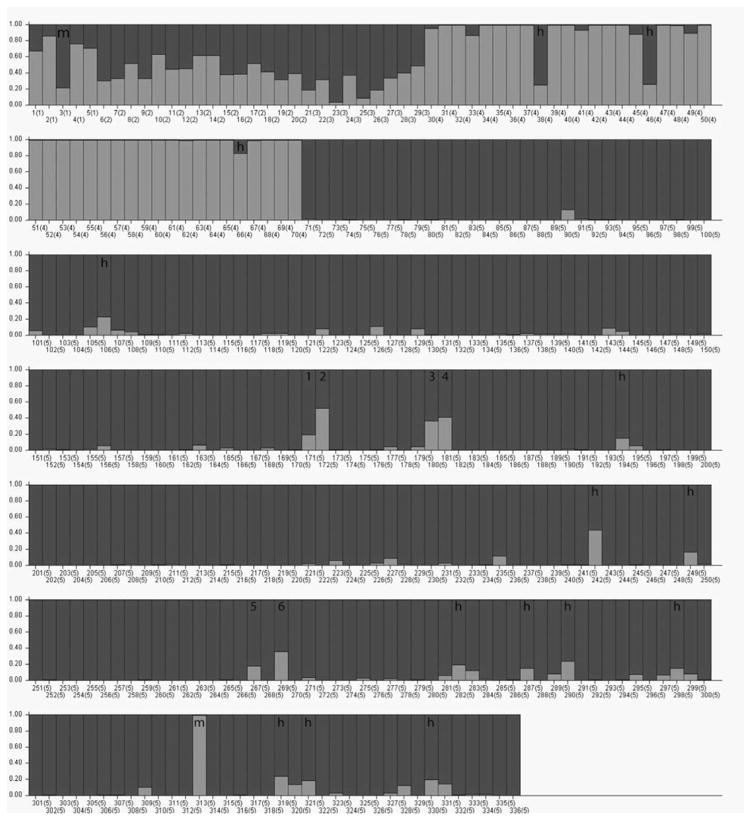
Overall, the SNP panel performed well in distinguishing animals from the two ancestral populations and for identifying hybrid individuals. Within the set of 336 rhesus macaques retained for analysis, the majority (93.5%) of animals reported as pure Chinese or pure Indian ancestry based on prior records were also classified as purebred using this ancestry assay (Fig. 1 and 2). Known hybrid animals exhibited mixed ancestry with Q values ranging from 0.171 to 0.846 (Figures 1 and 2).
Fig. 1.
The fractional genetic ancestry (Q) of each individual rhesus macaque (represented by a vertical bar) derived from the calculated Chinese (light grey) and Indian (dark grey) clusters, as estimated by STRUCTURE. Thirteen individuals predicted to have greater than 15% hybrid ancestry are indicated with an ‘h’. An additional three offspring/parent pairs (‘1/2’, ‘3/4’, ‘5/6’) with predicted hybrid ancestry are shown. A Chinese rhesus that was misclassified as an Indian rhesus and a 75% Indian animal that was misclassified as a 25% Indian rhesus are indicated with an ‘m’.
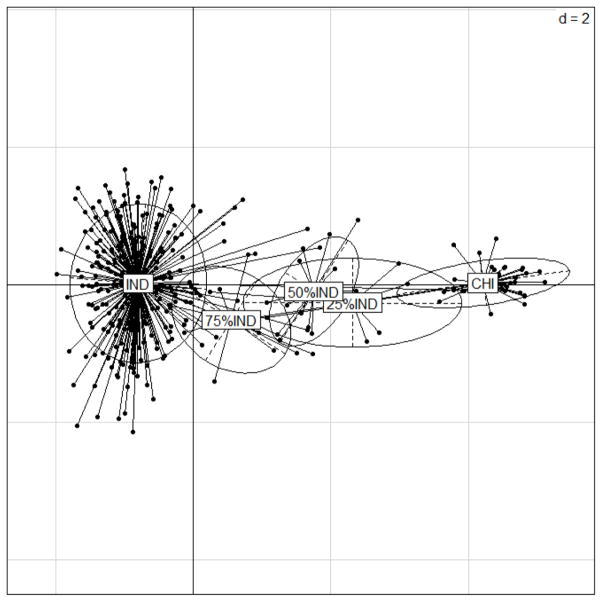
Fig. 2.
The first two PCA axes differentiating pure Chinese, Indian and hybrid rhesus macaques explain approximately 75% and 15% of the variation respectively.
STRUCTURE analysis provides not only a point estimate for the likelihood of membership in each cluster (population), but also the 90% credible interval (CI) for inclusion in each cluster. In our study, individuals that were estimated to have 15% or less admixture based on AIMS genotypes also had a 90% CI range that included 0% secondary ancestry contribution (purebred). Thus, the lower limit for the resolution of hybrid status using this assay was established as 15%.
Among the 336 NPRC animals, the AIMS assay identified 20 individuals whose genotypes did not correspond with predictions based on colony records or other prior information (Figure 1 and 2). These problematic cases provide clear demonstration of the need for and value of this SNP ancestry-testing panel. In one case, a DNA sample from an animal initially labeled as unmixed Indian-origin was assigned purebred (99.8 %) Chinese ancestry. This sample is labeled “m” in bottom row of Figure 1. It was subsequently discovered that the sample had been mislabeled as Indian and is actually of unmixed Chinese ancestry. Another sample initially assigned as 25% Indian ancestry (third sample, also labeled “m”, in the first row of Figure 1) was subsequently found to have incorrect sire and dam records, and is in fact 75% Indian ancestry. These cases provide both a cautionary tale and a series of real-world examples demonstrating that this panel will be useful.
In all cases in which animals were initially labeled as purebred Indian, but were indicated by the SNP testing to be hybrids, subsequent analysis of colony records showed that either a parent or grandparent of the predicted hybrid was obtained from breeding facilities outside the NPRC system. These acquired breeders were labeled as being purebred Indian origin by their suppliers, but our genetic test suggests that this was probably not the case. Thus, genetic ancestry testing was immediately useful in these specific cases. In the future, this approach can be used to validate the ancestry of individuals prior to their introduction into NPRC breeding or research populations.
In the test of the reproducibility of the 91 SNP assay using the 95 individuals selected for re-genotyping, five DNA samples failed for all 91 SNPs and were thus omitted from further analysis. The Q values of all of the unmixed samples were consistent with pure ancestry of their assumed countries of origin. All eight of the remaining predicted hybrids identified in Round One of our assays that were re-genotyped in Round Two were reconfirmed as hybrids (Figure 3). Their Q values range from 0.517 to 0.730. This testing also confirmed that one animal, originally thought to be ~25% Indian ancestry, was in fact ~75% Indian in both the first pass and confirmatory analysis (animal m in Figure 3).
Fig. 3.
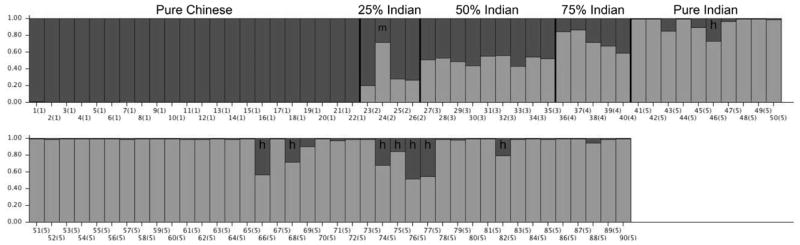
The probability of assignment or fractional ancestry (Q) of each individual rhesus macaque in the retest panel (represented by a vertical bar) derived from the putative Chinese (dark grey) and putative Indian (light grey) samples, as estimated by STRUCTURE. Eight individuals are labeled with ‘h’ to indicate that they have greater than 15% hybrid ancestry. A 75% Indian animal that was misclassified as a 25% Indian rhesus in Figure 1 is indicated with an ‘m’ in this figure.
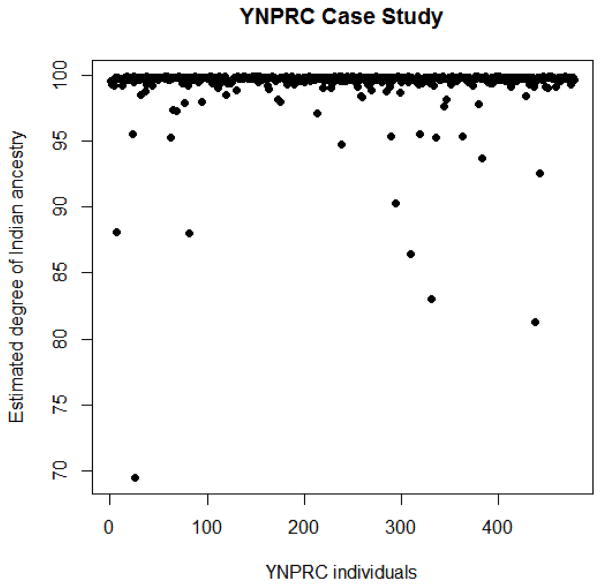
To demonstrate use of the SNP ancestry assay for extensive analysis of a single breeding center, a set of 480 rhesus macaques born at the YNPRC were genotyped using the 91 SNP set (Figure 4). The 75 rhesus macaques whose ancestries were re-confirmed in the replication study, and the 20 new CPRC animals, were used as a reference panel for STRUCTURE analysis in this study. All but three of the YNPRC rhesus macaques were unambiguously assigned to the pure Indian subpopulation, having at least 85% probability of Indian ancestry. Each of these three animals was subsequently removed from the breeding population. In tracing the origin of these animals it was discovered that one of three, as well as a common ancestor of the other two, were acquired from a common outside vendor, again highlighting the importance of ancestry testing on newly acquired animals.
Fig. 4.
With the exception of three individuals, all animals among the YNPRC sample were assigned to 85% or greater Indian ancestry and were considered pure Indian ancestry.
Discussion
The genetic and physiological differences among rhesus macaques from different parts of the species’ natural geographic range are significant enough to influence results in some types of experimental studies. The difference in response to SIV infection is the most widely cited example (Ling et al. 2002; Cohen 2002; Trichel et al. 2002), but others have also been described (Champoux et al. 1994, 1997). Ancestry records for specific animals may be incomplete or in doubt, making it useful in some circumstances to confirm the ancestry of individual rhesus macaques using a genetic test.
The SNP-based test described here is capable of distinguishing purebred Chinese-origin from purebred Indian-origin rhesus macaques with great reliability. This AIMs test is also capable of detecting hybrid ancestry when the proportion of admixture is at least 15%. Though no wild-caught animal samples were available for use as controls in this test, the overwhelming agreement between the reported animal ancestry and the ancestry predicted by this SNP panel across hundreds of subjects provides confidence that the assay correctly distinguishes the two ancestral populations of Indian and Chinese rhesus macaques.
There are at least two circumstances in which this genetic test will be valuable to colony managers or researchers. First, it can be used to confirm the genetic background of new animals introduced into a colony for breeding. If a colony consists of purebred animals of one origin, it may be appropriate to test new potential breeders to maintain a population with unmixed ancestry. The potential value of this test was unexpectedly demonstrated during these development studies, and again in the survey of the YNPRC breeding group, by the identification of several hybrid animals originally thought to be unmixed. Second, the test will be appropriate in cases where the background of a set of existing research animals, based on available records, is in some doubt and documentation from an independent assay is desired.
At this time, it is beyond the power of this test to reliably detect low (less than 15%) levels of admixture. Thus, it is important to bear in mind that when the result of this test for a given animal is either “Indian-origin” or “Chinese-origin,” this actually means that the animal shows no evidence of > 15% admixture. Thus, an animal with one great-grandparent (out of eight) with a geographic (genetic) origin different from the other seven may be indistinguishable from a true purebred individual using this test. In addition, this test cannot determine which parent or grandparent contributed the contrasting genetic heritage, though direct testing of each of those parents or ancestors would probably provide this information.
While the current panel of SNPs is effective and reliable for the intended purpose, this assay could be improved or expanded in the future. Expansion of the AIMs panel by incorporating additional informative SNPs would provide increased statistical power that might facilitate detection of admixture below 15%. Improved information concerning allele frequencies in different parts of the natural range of rhesus macaques would help identify other subpopulations with potentially differing allele distributions. The establishment of a well-characterized reference panel of animals of known and fully confirmed ancestry would also ensure the consistency of results across studies.
Finally, as we learn more about the genetic basis of the phenotypic traits that differ between Indian and Chinese rhesus macaques, such as their differential response to SIV or differences in temperament and behavior, it should be possible to develop a panel of genetic tests that includes some of those functionally significant variants. A SNP panel including a series of functional variants that drive phenotypic variation might provide more useful predictions about the response of specific individuals to certain experimental protocols, independent of their overall ancestry. In the long run, researchers may also consider targeted genetic testing for specific genetic variants that influence specific phenotypes of interest, as compared to testing for geographic origin, which measures underlying, but currently unidentified functional genetic differences between populations. Finally, we note that the same type of test could be developed for cynomolgus macaques (M. fascicularis), to distinguish among animals born or with ancestry in Indonesia, Malaysia, Indochina, the Philippines or Mauritius, and among whom genetic differentiation exceeds that among regional populations of rhesus macaques [Kanthaswamy et al., 2008; Smith et al., 2007]. This type of genetic test could, in principle, also be developed for different regional populations of baboons, African green monkeys, squirrel monkeys or any other primate species that exhibits substantial genetic variation among its natural regional populations.
Supplementary Material
Table II.
Chromosomal locations for the 96 SNPs originally identified for the development of the AIMs panel. The genomic coordinates are based on the rheMac2 assembly. Minor allele frequencies and delta values for the 91 SNPs used for ancestry determination are provided below.
| Indian | Chinese | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| SNP | Chromosome | Position | Change | ΔMAF | Minor Allele | Major Allele | MAF | Minor Allele | Major Allele | MAF |
| X28654019.1.D8YOWMI02G4FKQ | 1 | 28654019 | G/T | 0.34 | T | G | 0.19 | G | T | 0.46 |
| X35009123.1.D8YOWMI02GFCBO | 1 | 35009123 | A/G | 0.25 | A | G | 0.28 | A | G | 0.04 |
| LRP8:647C | 1 | 56125158 | C/T | 0.27 | C | T | 0.29 | C | T | 0.02 |
| X74753921.1.D8YOWMI01A81XP | 1 | 74753921 | G/T | 0.39 | T | G | 0.42 | T | G | 0.02 |
| AK53266 | 1 | 80406471 | A/G | 0.33 | G | A | 0.36 | G | A | 0.04 |
| X133878311.1.D8YOWMI02H3OT4 | 1 | 133878311 | A/T | 0.33 | A | T | 0.01 | A | T | 0.34 |
| TLR5:389T | 1 | 147143765 | C/T | 0.31 | T | C | 0.39 | T | C | 0.09 |
| CR2193 | 1 | 162907848 | A/G | 0.27 | A | G | 0.01 | A | G | 0.28 |
| GLUL3670 | 1 | 188288891 | A/T | 0.47 | T | A | 0.05 | A | T | 0.49 |
| X189341999.1.D8YOWMI01ALA6J | 1 | 189341999 | C/T | 0.28 | T | C | 0.32 | C | T | 0.40 |
| XCL1:320C | 1 | 198313836 | C/T | 0.24 | C | T | 0.26 | C | T | 0.02 |
| CHRM32590 | 1 | 217281301 | C/T | 0.39 | T | C | 0.44 | T | C | 0.05 |
| TARSH3441 | 2 | 20931497 | C/T | 0.43 | T | 0.00 | C | T | 0.43 | |
| X27827510.2.D8YOWMI01BI0G3 | 2 | 27827510 | A/G | 0.39 | A | G | 0.42 | G | A | 0.19 |
| CD471555 | 2 | 28185807 | C/T | 0.32 | T | C | 0.16 | T | C | 0.49 |
| TTLL33375 | 2 | 51156234 | C/T | 0.25 | C | T | 0.26 | C | T | 0.01 |
| BCHE:76G | 2 | 121586169 | C/G | 0.25 | G | C | 0.28 | G | C | 0.04 |
| X135860354.2.D8YOWMI01BBML4 | 2 | 135860354 | C/T | 0.32 | C | T | 0.24 | T | C | 0.44 |
| X138464766.2.D8YOWMI02GMF29 | 2 | 138464766 | C/T | 0.40 | T | C | 0.35 | C | T | 0.24 |
| X149899985.2.D8YOWMI01A7OV1 | 2 | 149899985 | A/G | 0.19 | A | G | 0.41 | A | G | 0.22 |
| CCR4:420C | 2 | 175430090 | A/C | 0.24 | C | A | 0.01 | C | A | 0.26 |
| ADIPOQ3282 | 2 | 179211380 | C/G | 0.38 | G | C | 0.23 | C | G | 0.39 |
| ITGB2:413G | 3 | 1814133 | A/G | 0.53 | G | A | 0.09 | A | G | 0.38 |
| X86532773.3.D8YOWMI02JDS52 | 3 | 86532773 | A/G | 0.32 | G | A | 0.31 | A | G | 0.37 |
| IL62404 | 3 | 103513696 | A/G | 0.55 | A | G | 0.41 | G | A | 0.04 |
| X138181005.3.D8YOWMI01BKZ5G | 3 | 138181005 | A/T | 0.37 | T | A | 0.45 | T | A | 0.09 |
| X167559647.3.D8YOWMI02H5ZKN | 3 | 167559647 | A/G | 0.36 | G | A | 0.39 | G | A | 0.04 |
| ALDH5A12101 | 4 | 24520863 | A/C | 0.42 | A | C | 0.07 | A | C | 0.49 |
| CDKN1A1416 | 4 | 36438276 | A/G | 0.31 | A | G | 0.34 | A | G | 0.02 |
| X60611841.4.D8YOWMI01D27HH | 4 | 60611841 | A/C | 0.33 | A | C | 0.35 | A | C | 0.02 |
| WFS12271 | 5 | 2511912 | A/G | 0.30 | A | G | 0.31 | A | G | 0.01 |
| CHRNA93449 | 5 | 35384836 | A/G | 0.39 | G | A | 0.33 | A | G | 0.28 |
| X123375995.5.D8YOWMI02IVQDH | 5 | 123375995 | A/G | 0.25 | A | G | 0.27 | A | G | 0.01 |
| ABCE12593 | 5 | 137306033 | A/T | 0.32 | T | A | 0.02 | T | A | 0.34 |
| X11634432.6.D8YOWMI01CE37I | 6 | 11634432 | C/T | 0.39 | T | C | 0.19 | C | T | 0.41 |
| X19209816.6.D8YOWMI01EIA7V | 6 | 19209816 | A/G | 0.35 | G | A | 0.04 | G | A | 0.39 |
| X119189982.6.D8YOWMI01EM0A3 | 6 | 119189982 | A/G | 0.52 | G | A | 0.34 | A | G | 0.15 |
| X145283831.6.D8YOWMI02I6ST3 | 6 | 145283831 | G/T | 0.22 | G | T | 0.17 | G | T | 0.39 |
| CD74358 | 6 | 146876592 | C/T | 0.29 | C | T | 0.30 | C | T | 0.01 |
| CHRM5144 | 7 | 12206127 | C/T | 0.25 | T | C | 0.29 | T | C | 0.04 |
| X47713241.7.D8YOWMI01A7MWI | 7 | 47713241 | A/G | 0.27 | A | G | 0.30 | A | G | 0.04 |
| X52102599.7.D8YOWMI01CWETE | 7 | 52102599 | C/G | 0.39 | C | G | 0.41 | C | G | 0.02 |
| X73733494.7.D8YOWMI02IY81A | 7 | 73733494 | A/G | 0.32 | G | A | 0.42 | A | G | 0.26 |
| APEX11216 | 7 | 83159550 | A/G | 0.34 | A | G | 0.39 | A | G | 0.04 |
| TRAF31704 | 7 | 166139737 | C/G | 0.32 | G | C | 0.40 | G | C | 0.07 |
| X34958656.8.D8YOWMI01A87RR | 8 | 34958656 | A/G | 0.09 | A | G | 0.38 | A | G | 0.29 |
| STAR:522G | 8 | 38598828 | G/T | 0.40 | G | T | 0.41 | G | T | 0.01 |
| X62326623.8.D8YOWMI01BDLZK | 8 | 62326623 | A/G | 0.39 | G | A | 0.31 | A | G | 0.29 |
| X82509260.9.D8YOWMI02GPQPE | 9 | 82509260 | A/G | 0.29 | A | G | 0.33 | G | A | 0.37 |
| X101742820.9.D8YOWMI01BH8Y8 | 9 | 101742820 | C/G | 0.57 | G | C | 0.39 | C | G | 0.04 |
| X122398406.9.D8YOWMI01EH54Y | 9 | 122398406 | C/T | 0.31 | T | C | 0.01 | T | C | 0.32 |
| SPO113501 | 10 | 7190042 | C/G | 0.45 | G | C | 0.30 | C | G | 0.26 |
| X41420257.10.D8YOWMI01DBC16 | 10 | 41420257 | A/T | 0.40 | A | T | 0.46 | A | T | 0.06 |
| SNAP252454 | 10 | 43536876 | A/G | 0.26 | G | A | 0.29 | G | A | 0.04 |
| X2063897.11.D8YOWMI02IKCMS | 11 | 2063897 | A/G | 0.29 | A | G | 0.09 | A | G | 0.38 |
| CD43558 | 11 | 7001554 | C/T | 0.58 | C | T | 0.34 | T | C | 0.07 |
| STK38L3112 | 11 | 27621826 | C/T | 0.28 | C | T | 0.35 | C | T | 0.07 |
| STK38L3391 | 11 | 27622105 | A/G | 0.29 | A | G | 0.34 | A | G | 0.05 |
| X122225487.11.D8YOWMI02G3O7L | 11 | 122225487 | C/G | 0.26 | G | C | 0.29 | G | C | 0.02 |
| ITGA4:321G | 12 | 45134560 | A/G | 0.53 | G | A | 0.02 | A | G | 0.45 |
| X31733062.13.D8YOWMI02JXLV2 | 13 | 31733062 | A/G | 0.41 | A | G | 0.44 | A | G | 0.02 |
| X98825742.13.D8YOWMI01D4G8Q | 13 | 98825742 | C/T | 0.26 | T | C | 0.00 | T | C | 0.27 |
| INHBB:131T | 13 | 126574228 | C/T | 0.27 | T | C | 0.33 | T | C | 0.06 |
| FJX13286 | 14 | 36208486 | C/T | 0.69 | C | T | 0.01 | T | C | 0.29 |
| CD44:471C | 14 | 36608996 | C/T | 0.29 | C | T | 0.01 | C | T | 0.30 |
| X65705967.14.D8YOWMI01DDMKZ | 14 | 65705967 | A/G | 0.36 | A | G | 0.39 | A | G | 0.02 |
| X100225126.14.D8YOWMI02JGR1F | 14 | 100225126 | C/T | 0.39 | C | T | 0.40 | C | T | 0.01 |
| X7707542.15.D8YOWMI02GFL0I | 15 | 7707542 | C/T | 0.34 | T | C | 0.37 | T | C | 0.04 |
| TLR4:703C | 15 | 18363421 | C/T | 0.33 | C | T | 0.01 | C | T | 0.34 |
| ASTN23424 | 15 | 19658319 | A/G | 0.31 | G | A | 0.01 | G | A | 0.32 |
| X24174624.15.D8YOWMI02G1031 | 15 | 24174624 | A/G | 0.29 | A | G | 0.31 | A | G | 0.02 |
| DDX581136 | 15 | 44683588 | A/G | 0.33 | G | A | 0.01 | G | A | 0.33 |
| X64630215.15.D8YOWMI02FJRC7 | 15 | 64630215 | A/G | 0.43 | A | G | 0.16 | G | A | 0.41 |
| X80445623.15.D8YOWMI02I3FAC | 15 | 80445623 | A/G | 0.31 | A | G | 0.33 | A | G | 0.02 |
| FBXW103174 | 16 | 15297924 | A/G | 0.38 | A | G | 0.40 | A | G | 0.02 |
| CCL8:516A | 16 | 29606160 | A/G | 0.43 | A | G | 0.46 | A | G | 0.02 |
| CCL5:690C | 16 | 31144359 | A/C | 0.33 | C | A | 0.01 | C | A | 0.34 |
| X50387595.16.D8YOWMI01AOJ7S | 16 | 50387595 | A/G | 0.50 | A | G | 0.04 | G | A | 0.46 |
| PYY:151C | 16 | 54029444 | C/T | 0.25 | C | T | 0.27 | C | T | 0.02 |
| X21413256.17.D8YOWMI02G62LU | 17 | 21413256 | A/T | 0.43 | T | A | 0.29 | A | T | 0.28 |
| HTR2A1425 | 17 | 25714351 | C/G | 0.56 | C | G | 0.42 | G | C | 0.03 |
| HTR2A1120 | 17 | 25714656 | A/G | 0.60 | A | G | 0.04 | G | A | 0.36 |
| X8046984.18.D8YOWMI02G8JPN | 18 | 8046984 | C/T | 0.38 | T | C | 0.07 | T | C | 0.45 |
| X15112147.18.D8YOWMI02H9T1B | 18 | 15112147 | C/T | 0.34 | T | C | 0.37 | T | C | 0.03 |
| X29194447.18.D8YOWMI02I1NC9 | 18 | 29194447 | A/G | 0.32 | A | G | 0.35 | A | G | 0.02 |
| X37863883.18.D8YOWMI02GJMMQ | 18 | 37863883 | A/C | 0.34 | C | A | 0.25 | A | C | 0.41 |
| X59111711.18.D8YOWMI01CKKVH | 18 | 59111711 | C/T | 0.41 | T | C | 0.37 | C | T | 0.22 |
| X39470934.19.D8YOWMI02H8KAL | 19 | 39470934 | A/G | 0.25 | A | G | 0.26 | A | G | 0.01 |
| SLC6A22495 | 20 | 53920793 | A/C | 0.38 | A | C | 0.42 | A | C | 0.04 |
| X63419598.20.D8YOWMI01CXU1A | 20 | 63419598 | A/G | 0.31 | G | A | 0.24 | A | G | 0.45 |
| GAN2690 | 20 | 79486479 | C/G | 0.33 | C | G | 0.05 | C | G | 0.38 |
| SNPs that were eliminated from the AIMs panel | ||||||||||
| CD40LG:738G | X | 286214 | ||||||||
| GREM13811 | 7 | 170922 | ||||||||
| ZAP1283210 | 7 | 136681838 | ||||||||
| MAOA:116G | X | 811542 | ||||||||
| CXCL123173 | 9 | 1392538 | ||||||||
Acknowledgments
The genetic test reported here developed from discussions within the Genetics and Genomics Working Group of the NIH Nonhuman Primate Research Consortium. The authors wish to thank the other members of that group (A. Vinson, J. Ha, L. Cox, D. O’Connor, R. Palermo, J. Nylander, J.P. Capitanio, C.J. Dubay, C.E. Hotchkiss, L.S. Kean, S. Lank and G.M. Miller) for their comments and suggestions during the development of this assay. Funding for the development of this SNP assay was provided by the Office of Research Infrastructure Programs, within Office of the Director, NIH and received through the following grants: P51-OD011107 for CNPRC and P51-OD011092 for ONPRC. We also wish to express our thanks to Dr. John Harding for his consistent and valuable support of the GGWG and its activities. All of the work described in this manuscript was carried out in compliance with the legal and ethical requirements for animal care and utilization.
Footnotes
Conflict of Interest Statement:
Two authors (SK and DGS) have a relationship with Primate Products Inc. in which the company markets and manages a service for genetic testing to assess geographic ancestry in macaques. Primate Products, Inc. receives biological samples from commercial clients, and sends those samples as well as financial support for supplies and technical staff to the laboratory of SK and DGS, where SK and DGS oversee the genetic testing and return results back to Primate Products, Inc. No other authors have conflicts of interest.
References
- 1.Animal Research in a Global Environment: Meeting the Challenges: Proceedings of the November 2008 International Workshop; Washington DC. [PubMed] [Google Scholar]
- 2.Banks MA, Eichert W, Olsen JB. Which genetic loci have greater population assignment power? Bioinformatics. 2003;19(11):1436–1438. doi: 10.1093/bioinformatics/btg172. [DOI] [PubMed] [Google Scholar]
- 3.Champoux M, Higley JD, Suomi SJ. Behavioral and physiological characteristics of Indian and Chinese-Indian hybrid rhesus macaque infants. Dev Psychobiol. 1997;31(1):49–63. [PubMed] [Google Scholar]
- 4.Champoux M, Suomi SJ, Schneider ML. Temperament differences between captive Indian and Chinese-Indian hybrid rhesus macaque neonates. Lab Anim Sci. 1994;44(4):351–7. [PubMed] [Google Scholar]
- 5.Clarke MR, O’Neil JA. Morphometric comparison of Chinese-origin and Indian-derived rhesus monkeys (Macaca mulatta) Am J Primatol. 1999;47(4):335–46. doi: 10.1002/(SICI)1098-2345(1999)47:4<335::AID-AJP5>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 6.Cohen J. Vaccine studies stymied by shortage of animals. Science. 2002:959–960. doi: 10.1126/science.287.5455.959. [DOI] [PubMed] [Google Scholar]
- 7.Ferguson B, Street SL, Wright H, Pearson C, Jia Y, Thompson SL, Allibone P, Dubay CJ, Spindel E, Norgren RB. Single nucleotide polymorphisms (SNPs) distinguish Indian-origin and Chinese-origin rhesus macaques (Macaca mulatta) Bmc Genomics. 2007:8. doi: 10.1186/1471-2164-8-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gibbs RA, Rogers J, Katze MG, Bumgarner R, Weinstock GM, Mardis ER, Remington KA, Strausberg RL, Venter JC, Wilson RK, et al. Evolutionary and biomedical insights from the rhesus macaque genome. Science. 2007;316(5822):222–34. doi: 10.1126/science.1139247. [DOI] [PubMed] [Google Scholar]
- 9.Groves C. Primate Taxonomy. Washington, DC: Smithsonian Institution Press; 2001. p. 350. [Google Scholar]
- 10.Hernandez RD, Hubisz MJ, Wheeler DA, Smith DG, Ferguson B, Rogers J, Nazareth L, Indap A, Bourquin T, McPherson J, et al. Demographic histories and patterns of linkage disequilibrium in Chinese and Indian rhesus macaques. Science. 2007;316(5822):240–3. doi: 10.1126/science.1140462. [DOI] [PubMed] [Google Scholar]
- 11.Hubisz M, Falush D, Stephens M, Pritchard J. Inferring weak population structure with the assistance of sample group information. Molecular Ecology Resources. 2009;9(5):1322–1332. doi: 10.1111/j.1755-0998.2009.02591.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jombart T. adegenet: a R package for the multivariate analysis of genetic markers. Bioinformatics. 2008;24(11):1403–5. doi: 10.1093/bioinformatics/btn129. [DOI] [PubMed] [Google Scholar]
- 13.Jombart T, Ahmed I. adegenet 1.3-1: new tools for the analysis of genome-wide SNP data. Bioinformatics. 2011;27(21):3070–1. doi: 10.1093/bioinformatics/btr521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kanthaswamy S, Capitanio JP, Dubay CJ, Ferguson B, Folks T, Ha JC, Hotchkiss CE, Johnson ZP, Katze MG, Kean LS, et al. Resources for genetic management and genomics research on non-human primates at the National Primate Research Centers (NPRCs) J Med Primatol. 2009;38(Suppl 1):17–23. doi: 10.1111/j.1600-0684.2009.00371.x. [DOI] [PubMed] [Google Scholar]
- 15.Kanthaswamy S, Satkoski J, George D, Kou A, Erickson BJ, Smith DG. Interspecies Hybridization and the Stratification of Nuclear Genetic Variation of Rhesus (Macaca Mulatta) and Long-Tailed Macaques (Macaca Fascicularis) Int J Primatol. 2008;29(5):1295–1311. doi: 10.1007/s10764-008-9295-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Karl JA, Bohn PS, Wiseman RW, Nimityongskul FA, Lank SM, Starrett GJ, O’Connor DH. Major histocompatibility complex class I haplotype diversity in Chinese rhesus macaques. G3 (Bethesda) 2013;3(7):1195–201. doi: 10.1534/g3.113.006254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ling B, Veazey RS, Luckay A, Penedo C, Xu K, Lifson JD, Marx PA. SIV(mac) pathogenesis in rhesus macaques of Chinese and Indian origin compared with primary HIV infections in humans. AIDS. 2002;16(11):1489–96. doi: 10.1097/00002030-200207260-00005. [DOI] [PubMed] [Google Scholar]
- 18.Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Smith DG, McDonough JW, George DA. Mitochondrial DNA variation within and among regional populations of longtail macaques (Macaca fascicularis) in relation to other species of the fascicularis group of macaques. Am J Primatol. 2007;69(2):182–98. doi: 10.1002/ajp.20337. [DOI] [PubMed] [Google Scholar]
- 20.Trichel AM, Rajakumar PA, Murphey-Corb M. Species-specific variation in SIV disease progression between Chinese and Indian subspecies of rhesus macaque. J Med Primatol. 2002;31(4–5):171–8. doi: 10.1034/j.1600-0684.2002.02003.x. [DOI] [PubMed] [Google Scholar]
- 21.Wiseman RW, Karl JA, Bimber BN, O’Leary CE, Lank SM, Tuscher JJ, Detmer AM, Bouffard P, Levenkova N, Turcotte CL, et al. Major histocompatibility complex genotyping with massively parallel pyrosequencing. Nat Med. 2009;15(11):1322–6. doi: 10.1038/nm.2038. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.