Key Points
Using an in vivo model for primary MLL-rearranged infant ALL, we identified phenotypically and functionally distinct LICs and HSCs.
In MLL ALL patient samples, molecules differentially expressed between LICs and HSCs including CD9, CD32, and CD24 were identified.
Abstract
Translocation of the mixed-lineage leukemia (MLL) gene with AF4, AF9, or ENL results in acute leukemia with both lymphoid and myeloid involvement. We characterized leukemia-initiating cells (LICs) in primary infant MLL-rearranged leukemia using a xenotransplantation model. In MLL-AF4 patients, CD34+CD38+CD19+ and CD34−CD19+ cells initiated leukemia, and in MLL-AF9 patients, CD34−CD19+ cells were LICs. In MLL-ENL patients, either CD34+ or CD34− cells were LICs, depending on the pattern of CD34 expression. In contrast, in patients with these MLL translocations, CD34+CD38−CD19−CD33− cells were enriched for normal hematopoietic stem cells (HSCs) with in vivo long-term multilineage hematopoietic repopulation capacity. Although LICs developed leukemic cells with clonal immunoglobulin heavy-chain (IGH) rearrangement in vivo, CD34+CD38−CD19−CD33− cells repopulated recipient bone marrow and spleen with B cells, showing broad polyclonal IGH rearrangement and recipient thymus with CD4+ single positive (SP), CD8+ SP, and CD4+CD8+ double-positive (DP) T cells. Global gene expression profiling revealed that CD9, CD32, and CD24 were over-represented in MLL-AF4, MLL-AF9, and MLL-ENL LICs compared with normal HSCs. In patient samples, these molecules were expressed in CD34+CD38+ and CD34− LICs but not in CD34+CD38−CD19−CD33− HSCs. Identification of LICs and LIC-specific molecules in primary human MLL-rearranged acute lymphoblastic leukemia may lead to improved therapeutic strategies for MLL-rearranged leukemia.
Introduction
The mixed-lineage leukemia (MLL) gene, located on 11q23, is a mammalian homolog of the Drosophila melanogaster trithorax gene and serves as a component of the proteins associated with SET1 (COMPASS)-like complex.1 COMPASS-like complexes methylate Lys4 on histone H3 (H3K4) through their conserved SET domain, and MLL associates with cofactors such as menin, a tumor suppressor, for chromatin localization and H3K4 trimethylation of genes including HOX genes.2-4 In addition, MLL may regulate epigenetic inheritance by promoting transcriptional reactivation following mitotic chromosome condensation through a H3K4 trimethylation-independent mechanism.5 In mouse development, Mll is necessary for establishment of definitive hematopoiesis and expansion of hematopoietic progenitors, whereas in adult hematopoiesis, it maintains hematopoietic stem cell (HSC) quiescence and promotes progenitor proliferation.6-8 In Mll knockout mice, the defect in hematopoietic progenitor expansion is reversible on re-expression of Hox genes, demonstrating that Hox gene expression, as regulated by Mll, is critical in normal hematopoiesis.
MLL translocations are associated with pathogenesis of multiple types of leukemia, identified in up to 10% of de novo acute lymphoblastic leukemia (ALL) and acute myeloid leukemia (AML).9,10 In ALL, t(4;11) is the most frequent translocation, whereas t(9;11)(p21;q23) is most commonly associated with AML and myelodysplastic syndrome (MDS)/secondary leukemia, respectively. In MLL-rearranged leukemia, translocation of MLL with a variety of translocation partner genes, such as AF4, AF9, and ENL, results in the loss of the catalytic SET domain. Instead, the products of fusion partner genes confer the ability to recruit a histone methyltransferase, DOT1L, to target genes determined by specific recognition elements in the remaining portion of MLL.11,12 DOT1L, when recruited to MLL target genes, aberrantly methylates genes such as HOXA9 and MEIS1, enhancing their expression.13-16 This may result in leukemogenesis, because ectopic expression of Hox genes such as Hoxb8, Hoxa7, and Hoxa9 have been shown to induce leukemia in mice, and t(7;11)(p15;p15) HOXA9-NUP98 fusion is associated with some human AML.17-19 MLL rearrangement may also lead to leukemogenesis via deregulation of cell cycle and proliferation in myeloid or lymphoid lineages, through binding of rearranged MLL with c-Myc, and ATR and upregulation of the Wnt/β-catenin pathway.20-22
ALL in infants is biologically distinct from ALL in older children, with 70% to 80% of infant cases associated with MLL translocations.23 Although long-term event-free survival rates of ∼80% are reported in older children with ALL, prognosis for infants is at ∼40%, and prognosis for infants diagnosed with MLL-rearranged leukemia is significantly poorer than in MLL-nonrearranged cases.23-25
In some malignancies such as adult AML, malignant stem cells may play important roles both in the initiation of disease and in disease relapse. The high failure rate in the treatment of infant MLL-rearranged leukemia is associated with disease relapse, suggesting that leukemia-initiating cell (LIC)-targeted therapy may improve patient outcomes. Unlike childhood B-cell lineage ALL in which CD34, CD10, and CD19 have been reported as potential markers for LICs,26 MLL-rearranged ALL is characterized by co-expression of B-cell and myeloid lineage antigens, suggesting that malignant transformation may have occurred in earlier stages of hematopoiesis. Characterizing MLL-rearranged ALL LICs and understanding the developmental origin and hierarchy in MLL ALL may lead to identification of mechanisms for disease relapse and development of effective therapeutic strategies.
Transplantation of murine or human HSCs/hematopoietic progenitor cells (HPCs) expressing MLL fusion proteins such as MLL-AF9 and MLL-ENL has provided highly informative in vivo models of MLL-rearranged leukemia. Although MLL-ENL–expressing murine HSCs/HPCs induce myeloid leukemia, MLL-ENL–expressing human lineage-negative cells induce lymphoid leukemia exclusively.27,28 In patients, MLL-ENL is associated more frequently in ALL than AML. MLL-AF9–expressing human cord blood (CB) CD34+ HSC/HPCs develop either myeloid or lymphoid leukemia in vivo depending on the expression of human cytokines in the mouse microenvironment.29 These studies illustrate the complexity of developmental hierarchy and lineage determination in MLL-rearranged leukemia.
In the present study, we identified phenotypically and functionally distinct populations of MLL-rearranged ALL LICs in MLL-AF4, MLL-AF9, and MLL-ENL patient samples in an in vivo xenotransplantation model. In addition, we demonstrated the presence of normal HSC-enriched MLL translocation-negative population in MLL-rearranged ALL patient samples, which had the capacity to repopulate normal human hematopoiesis in vivo. Finally, we identified genes differentially expressed between MLL LICs and normal HSCs including cell surface molecules that may serve as therapeutic targets. Our data, through direct analysis of primary MLL-rearranged leukemia in vivo, give insights into hierarchy of leukemogenesis in infant MLL-rearranged leukemia and identify potential therapeutic targets in MLL LICs.
Materials and methods
Patient samples
Patient samples were collected with written informed consent from parents/guardians of infant ALL patients in accordance with the Declaration of Helsinki and under approval of the Institutional Review Boards at each participating institution. All experiments were performed according to research protocol associated with the Japan Infant Leukemia Study Group protocol MLL96 and the Japanese Pediatric Leukemia/Lymphoma Study Group (JPLSG) protocol MLL-10 (UMIN Clinical Trials Registry number UMIN000004801; research protocol approval number 016). Samples were obtained fresh and mononuclear cells were isolated using density-gradient centrifugation before analysis and/or sorting. Normal CB and bone marrow (BM) mononuclear cells were purchased from Cambrex (Walkerville, MD).
Mice
NOD.Cg-PrkdcscidIl2rgtmlWjl/Sz (NOD/SCID/IL2rgnull [NSG]) mice developed at the Jackson Laboratory were used for in vivo transplantation experiments. Mice were bred and maintained under defined flora at the animal facility of RIKEN and at The Jackson Laboratory according to guidelines established by the Institutional Animal Committees at each institution.
Flow cytometry and fluorescence-activated cell sorting
For flow cytometric analysis and fluorescence-activated cell sorting (FACS), patient-derived mononuclear cells and recipient BM, spleen, liver, testis, and peripheral blood (PB) cells were labeled with monoclonal antibodies as listed in supplemental Table 8 available on the Blood Web site. The purity of sorted cells was >98%.
Xenotransplantation
Newborn NSG mice received 150 cGy total body irradiation followed by intravenous injection of sorted cells. To evaluate in vivo leukemia initiating capacity, 102 to 105 sorted patient BM or PB cells were injected per recipient. The sorting strategy and phenotypes of transplanted cells for each transplantation experiment are described in Results and indicated in the figures.
Morphological analysis
Cytospin preparations were made using Shandon Cytospin 4 cytocentrifuge (Thermo Electric, Waltham, MA). May-Grunwald-Giemsa staining was performed using standard procedures. Light microscopy was performed using Zeiss Axiovert 200 (Carl Zeiss).
Fluorescence in situ hybridization
Cytospin specimens were fixed with methanol and glacial acid for 15 minutes on ice. After denaturation with 70% formamide and dehydration with 70% ethanol, the cells were stained with a probe detecting MLL fusion (LSI MLL Dual color Break Apart Rearrangement Probe; Abbott) at 37°C overnight. Nuclei were counterstained with 4,6 diamidino-2-phenylindole.
Histological studies
Tissues were fixed with 4% paraformaldehyde. Hematoxylin-eosin staining was performed using standard methods. Confocal microscopy was performed using Zeiss LSM710 (Carl Zeiss). Light microscopy was performed using Zeiss Axiovert 200 (Carl Zeiss).
B-cell clonality assay
DNA samples derived from MLL-rearranged infant ALL were subjected to B-cell clonality assay based on rearrangement patterns of immunoglobulin heavy chain (IGH) gene by using the IGH Gene Clonality Assay (Invivoscribe Technologies; Cat No. 1-101-0061). Polymerase chain reaction products were sized on an ABI 3130xl DNA sequencer (Life Technologies).
Gene expression profiling
To identify molecules expressed in MLL LICs, CD34+CD38+CD19+ cells, and CD34− CD19+ cells derived from 12 patients and normal HSC samples were subjected to microarray analysis. From 20 ng total RNA extracted from FACS-purified cells using TRIzol (Invitrogen), cDNA was amplified using Ovation RNA Amplification System V2 Kit (NuGEN). The amplified cDNA was fragmented and labeled for Human Genome U133 plus 2.0 GeneChip (Affymetrix) expression array analysis using the FL-Ovation cDNA Biotin Module V2 kit. Microarray data were analyzed using the Bioconductor package (http://www.bioconductor.org/). The signal intensities of the probe sets were normalized using the GeneChip Robust Multiarray Averaging (GC-RMA) program.30 RankProd was used to select differentially expressed genes between LICs and HSCs with a cutoff P value of .01 and an estimated false-positive rate of 0.05.31 Limma was used to selected differentially expressed genes among MLL-AF4, MLL-ENL, and MLL-AF9 with a cutoff P value of .01 and 1.5-fold change (log2).32 Gene annotation data were obtained from the RefDIC database.33
Statistical analysis
Numerical data are presented as means ± standard error of the mean. The differences were examined using a 2-tailed t test (GraphPad Prism; GraphPad).
Accession numbers
Affymetrix gene expression data will be deposited in RefDIC database under the following accession numbers: RSM08287, RSM08288, RSM08289, RSM08292, RSM08293, RSM08294, RSM08295, RSM08395, RSM09150, RSM09151, RSM09152, RSM09189, RSM09190, RSM09536, RSM09537, RSM13162, RSM13489, RSM13490, and RSM13491.
Results
Distinct expression patterns of CD34 and CD38 in MLL-rearranged ALL
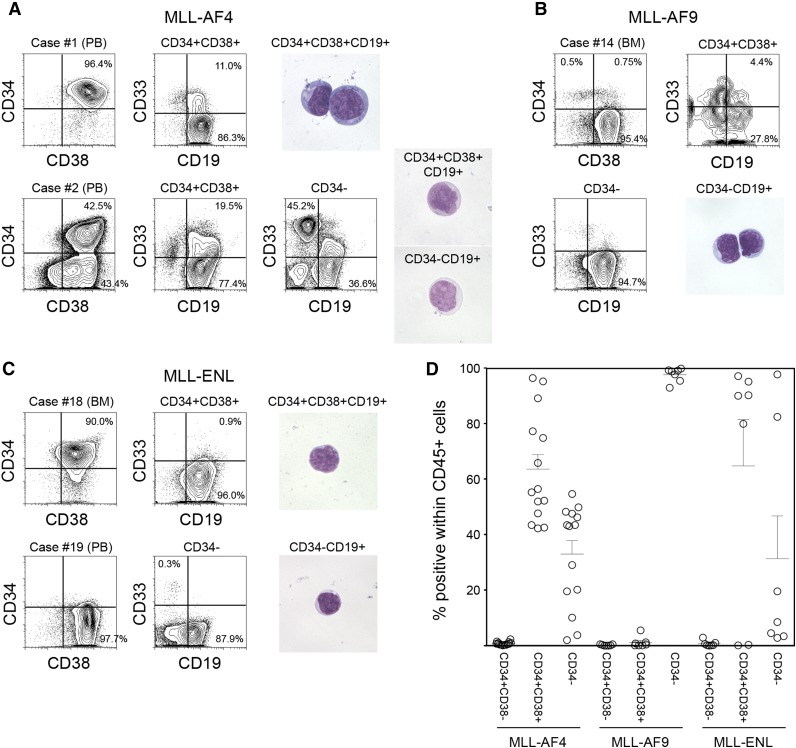
We analyzed expression of CD34 and CD38 in samples obtained from infants with MLL-rearranged ALL associated with 3 distinct translocations: MLL-AF4 (n = 14), MLL-AF9 (n = 7), and MLL-ENL (n = 7). Patient characteristics and frequency of cell populations as determined by surface expression of CD45, CD34, CD38, CD19, and CD33 are shown in Table 1. Expression patterns of CD34 and CD38 varied depending on MLL fusion partner. In MLL-AF4 cases, we found both CD34+CD38+CD19+ and CD34−CD38+CD19+ cell populations (63.6 ± 5.2% and 32.9 ± 4.9% within human CD45+ cells, respectively). In one case (case 1), 96.4% of total leukocytes were CD34+CD38+ cells (Figure 1A, upper), whereas in 13 of 14 cases, distinct CD34+CD38+CD19+ and CD34−CD19+ populations were observed (case 2 shown as an example in Figure 1A, lower). In contrast, in all MLL-AF9 cases, CD45+ leukocytes were mostly CD34− cells (97.6 ± 1.0%, n = 7; case 14 shown as an example in Figure 1B). In 4 of 7 MLL-ENL cases, CD45+ leukocytes were largely CD34+ (CD34+: 94.3 ± 1.4%, CD34−: 4.8 ± 1.3%), whereas in 2 cases, they were mainly CD34− cells (CD34+: 1.5 ± 0.1%, CD34−: 97.1 ± 0.3%) (cases 18 and 19 shown as examples in Figure 1C). CD34+CD38− cells accounted for 0.8 ± 0.2%, 0.2 ± 0.1%, and 0.7 ± 0.4% within human CD45+ cells in MLL-AF4, MLL-AF9, and MLL-ENL patient samples, respectively. CD34 and CD38 expression patterns in CD45+ leukocytes obtained from MLL-rearranged ALL patients are summarized in Figure 1D. Based on these findings, we went on to determine the significance of CD34 and CD38 expression by evaluating in vivo leukemia initiation capacity of CD34+CD38+CD19+ and CD34−CD19+ cells derived from MLL-AF4+, MLL-AF9+, and MLL-ENL+ ALL.
Table 1.
Patient characteristics
| Translocation/rearrangement | Patient ID | Age at diagnosis (months) | Gender | WBC (×103/µL) | Extramedullary involvement | Specimen type | % in human CD45+ | % in human CD34+CD38+ | % in human CD34− | % in human CD34+38− | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CNS | Testis | Skin | LN | Hepato- megaly (cm) | Spleno- megaly (cm) | CD34+CD38− | CD34+CD38+ | CD34−CD38+ | CD19+CD33− | CD19+CD33+ | CD19−CD33+ | CD19+CD33− | CD19+CD33+ | CD19−CD33+ | CD19+CD33− | CD19+CD33+ | CD19−CD33+ | CD19−CD33− | ||||||
| t(4;11)/MLL-AF4 | 1 | 6 | Female | 262.0 | (−) | (−) | (−) | (−) | 3 | 5 | PB | 1.0 | 96.4 | 2.0 | 86.3 | 11.0 | 1.7 | 62.2 | 1.1 | 17.2 | 65.1 | 4.3 | 3.5 | 27.2 |
| 2 | 5 | Female | 59.0 | (−) | (−) | (−) | (−) | 4 | 3 | PB | 0.3 | 42.5 | 43.4 | 77.4 | 19.5 | 1.5 | 36.6 | 9.2 | 45.2 | 42.8 | 1.0 | 17.3 | 38.9 | |
| 3 | 4 | Female | 176.7 | (−) | (−) | (−) | (−) | 2 | 2 | PB | 0.3 | 55.2 | 43.4 | 75.8 | 22.0 | 1.0 | 30.8 | 44.3 | 12.0 | 26.0 | 0.4 | 20.3 | 53.3 | |
| 4 | 4 | Female | 403.5 | (−) | (−) | (−) | (−) | 3 | 3 | PB | 0.6 | 43.4 | 54.6 | 96.2 | 2.8 | 0.1 | 94.0 | 2.7 | 1.0 | 63.8 | 0.6 | 3.5 | 32.2 | |
| 5 | 1 | Female | 398.0 | + | (−) | (−) | (−) | 4 | 6 | PB | 0.3 | 89.1 | 10.1 | 94.9 | 0.6 | 0.1 | 87.6 | 3.9 | 2.1 | 47.1 | 0.0 | 1.3 | 51.6 | |
| 6 | 1 | Male | 328.8 | (−) | (−) | + | (−) | 5 | 5 | PB | 0.5 | 65.8 | 29.0 | 62.2 | 12.8 | 0.7 | 50.6 | 10.9 | 17.8 | 16.8 | 0.3 | 40.3 | 42.6 | |
| 7 | 8 | Female | 166.4 | + | (−) | (−) | (−) | 5 | 5 | PB | 2.3 | 42.2 | 48.2 | 70.7 | 28.1 | 0.7 | 45.9 | 51.5 | 0.7 | 37.1 | 35.5 | 14.4 | 13.0 | |
| 8 | 4 | Male | 1290.0 | (−) | (−) | (−) | (−) | 6 | 7 | BM | 0.4 | 56.4 | 43.0 | 83.5 | 8.7 | 0.1 | 86.7 | 9.9 | 0.6 | 71.4 | 9.5 | 9.5 | 9.5 | |
| 9 | 3 | Female | 223.5 | + | (−) | (−) | (−) | 5 | 8 | BM | 1.0 | 51.9 | 46.2 | 13.4 | 85.9 | 0.6 | 13.5 | 50.3 | 35.8 | 84.0 | 9.0 | 2.0 | 5.0 | |
| 10 | 3 | Female | 294.0 | + | (−) | (−) | + | 7 | 4 | PB | 0.1 | 52.2 | 47.5 | 97.5 | 1.7 | 0.3 | 91.8 | 6.6 | 1.1 | 27.5 | 0.0 | 5.0 | 67.5 | |
| 11 | 6 | Female | 161.2 | (−) | (−) | (−) | (−) | 2 | 5 | BM | 0.7 | 74.8 | 20.1 | 87.1 | 12.8 | 0.1 | 71.8 | 24.5 | 0.8 | 29.4 | 0.0 | 0.0 | 70.6 | |
| 12 | 5 | Male | 832.8 | (−) | (−) | (−) | (−) | 10 | 10 | PB | 0.7 | 95.2 | 3.8 | 87.0 | 4.5 | 2.1 | 51.4 | 4.7 | 28.6 | 44.1 | 1.2 | 12.4 | 42.2 | |
| 13 | 2 | Male | 257.4 | + | (−) | (−) | (−) | 4 | 6 | PB | 1.3 | 47.6 | 49.8 | 89.9 | 7.5 | 0.6 | 66.4 | 19.4 | 6.0 | 75.1 | 1.8 | 6.7 | 16.4 | |
| 25 | 6 | Female | 67.6 | (−) | (−) | (−) | (−) | 1 | 1 | PB | 1.5 | 77.2 | 19.5 | 87.6 | 4.2 | 2.6 | 61.5 | 3.8 | 25.1 | 10.5 | 1.7 | 51.9 | 1.7 | |
| t(9;11)/MLL-AF9 | 14 | 7 | Male | 142.4 | (−) | + | + | + | 5 | 4 | BM | 0.5 | 0.8 | 95.4 | 27.8 | 4.4 | 22.7 | 94.7 | 0.2 | 0.2 | 0.7 | 0.2 | 2.6 | 96.5 |
| 15 | 9 | Female | 97.4 | (−) | (−) | + | (−) | 2 | 0 | PB | 0.3 | 0.4 | 98.8 | n.a. | n.a. | n.a. | 72.9 | 23.6 | 1.7 | 0.0 | 0.2 | 34.9 | 64.9 | |
| 16 | 1 | Male | 473.5 | + | (−) | + | (−) | 4 | 0 | BM | 0.5 | 1.2 | 97.7 | 67.5 | 9.3 | 1.1 | 97.9 | 0.3 | 0.3 | 3.2 | 0.5 | 0.5 | 95.8 | |
| 17 | 6 | Male | 33.0 | (−) | (−) | (−) | (−) | 0 | 0 | BM | 0.1 | 0.1 | 99.2 | n.a. | n.a. | n.a. | 77.1 | 8.6 | 0.7 | 1.5 | 3.0 | 0.0 | 95.5 | |
| 26 | 5 | Male | 2.4 | (−) | (−) | (−) | + | 3 | 2 | BM | 0.0 | 5.5 | 92.9 | 11.2 | 24.1 | 61.2 | 73.7 | 12.9 | 4.0 | n.a. | n.a. | n.a. | n.a. | |
| 27 | 2 | Female | 270.0 | (−) | (−) | + | + | 4 | 3 | BM | 0.0 | 0.1 | 99.8 | n.a. | n.a. | n.a. | 95.1 | 4.1 | 0.3 | n.a. | n.a. | n.a. | n.a. | |
| 28 | 5 | Female | 1.7 | (−) | (−) | (−) | (−) | 1 | 0 | BM | 0.0 | 0.1 | 99.1 | n.a. | n.a. | n.a. | 89.6 | 0.4 | 0.0 | n.a. | n.a. | n.a. | n.a. | |
| t(11;19)/MLL-ENL | 18 | 12 | Female | 499.5 | (−) | (−) | (−) | (−) | 2 | 2 | BM | 1.0 | 90.0 | 8.5 | 96.0 | 0.9 | 0.0 | 90.0 | 1.6 | 0.1 | 87.5 | 0.9 | 0.2 | 11.5 |
| 19 | 5 | Male | 261.6 | (−) | (−) | (−) | (−) | 7 | 5 | PB | 0.1 | 0.1 | 97.7 | n.a. | n.a. | n.a. | 87.9 | 0.0 | 0.3 | 0.0 | 0.0 | 0.0 | 100.0 | |
| 20 | 4 | Female | 60.4 | (−) | (−) | + | + | 5 | 5 | BM | 0.2 | 0.3 | 82.4 | 30.0 | 0.2 | 1.0 | 94.5 | 0.1 | 0.1 | 2.0 | 0.2 | 0.0 | 97.8 | |
| 21 | 3 | Female | 215.1 | (−) | (−) | (−) | (−) | 6 | 4 | BM | 2.9 | 90.2 | 4.5 | 90.9 | 8.1 | 0.5 | 45.4 | 8.4 | 32.9 | 82.4 | 0.5 | 2.3 | 14.8 | |
| 22 | 6 | Male | 993.0 | (−) | + | + | + | 3 | 8 | BM | 0.2 | 79.9 | 19.5 | 98.7 | 0.7 | 0.1 | 94.9 | 0.2 | 0.6 | 52.8 | 0.0 | 5.6 | 41.7 | |
| 23 | 1 | Female | 1750.0 | + | (−) | (−) | (−) | 7 | 7 | PB | 0.1 | 97.1 | 2.8 | 39.9 | 40.4 | 5.3 | 8.2 | 24.3 | 53.0 | 28.6 | 14.3 | 0.0 | 57.1 | |
| 24 | 5 | Female | 123.1 | (−) | (−) | (−) | (−) | 4 | 5 | PB | 0.6 | 95.1 | 3.4 | 80.4 | 11.5 | 0.2 | 75.7 | 7.7 | 4.2 | 56.5 | 3.2 | 0.0 | 40.3 | |
| Normal CB MNCs | 1 | 0 | CB | 0.0 | 0.1 | 60.1 | 39.9 | |||||||||||||||||
| 2 | 0 | CB | 0.0 | 0.5 | 66.4 | 33.1 | ||||||||||||||||||
| 3 | 0 | CB | 0.0 | 0.0 | 29.2 | 70.8 | ||||||||||||||||||
CNS, central nervous system; LN, lymph node; n.a., frequency not available due to low frequency of parent population; WBC, white blood cell count; (−), absence of extramedullary disease; +, presence of extramedullary disease.
Figure 1.
Phenotypic characteristics of MLL-AF4, MLL-AF9, and MLL-ENL ALL cells. (A-C) Representative flow cytometry plots showing expression of CD34, CD38, CD19, and CD33 in cells derived from infant ALL patients with (A) MLL-AF4, (B) MLL-AF9, and (C) MLL-ENL translocations. Morphology of leukemic cells derived from representative cases is shown. (D) Frequencies of CD34+CD38−, CD34+CD38+, and CD34− cells in MLL-rearranged infant leukemia (MLL-AF4, n = 14; MLL-AF9, n = 7; MLL-ENL, n = 7).
LICs in MLL-AF4+ ALL
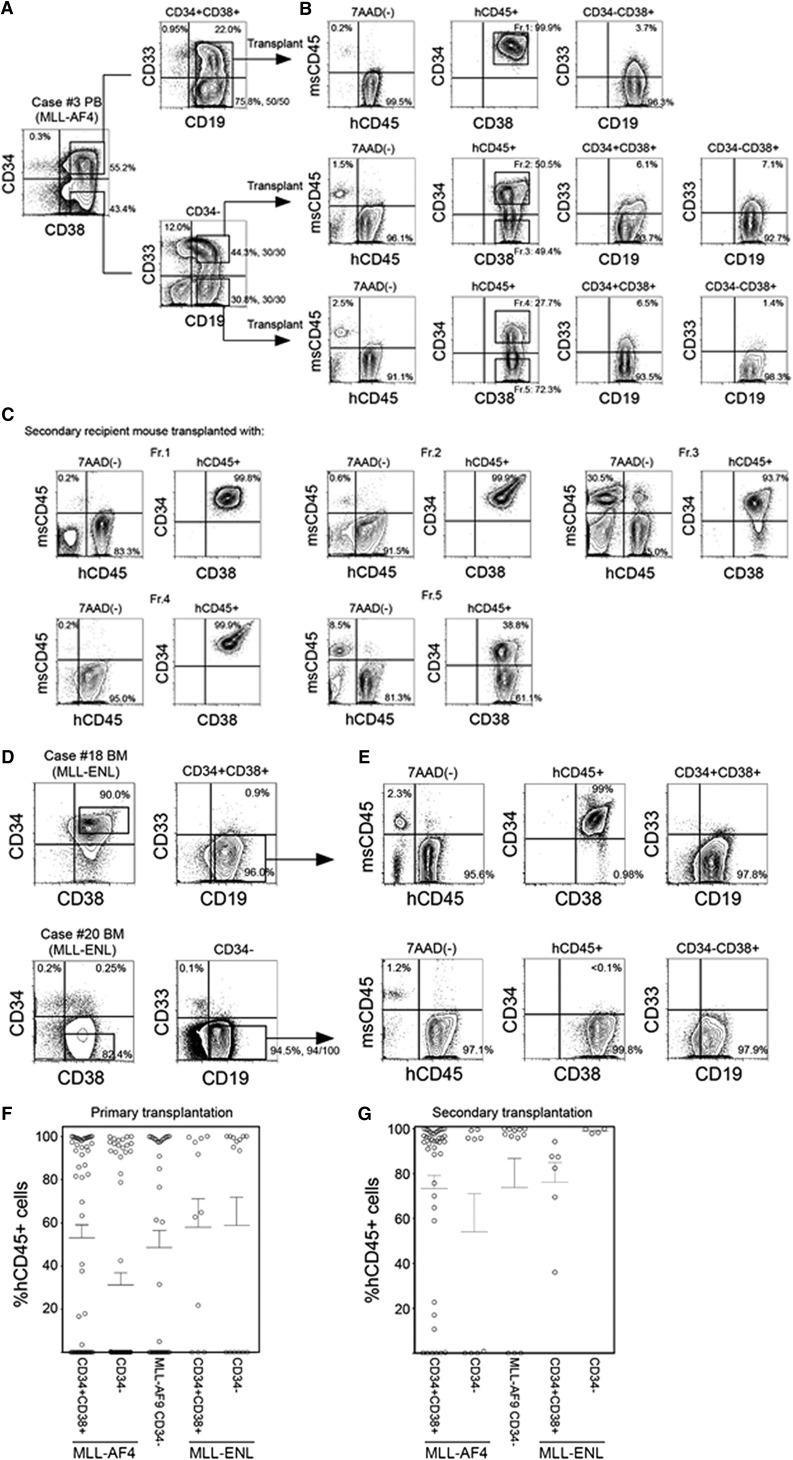
We transplanted CD34+CD38+CD19+ and CD34−CD19+ cells isolated from MLL-AF4 patients into NSG newborns. As shown in supplemental Table 1, 102 to 104 CD34+CD38+CD19+ cells or 102 to 104 CD34−CD19+ cells were capable of achieving high levels of engraftment in NSG recipient BM. We then examined the functional significance of CD33 expression in MLL-AF4+ ALL. From MLL-AF4 case 3, we isolated and transplanted 1000 CD34−CD19+CD33+ or CD34−CD19+CD33− cells into NSG newborns and found leukemia initiation from both CD33+ and CD33− fractions (Figure 2A-B, middle and bottom; supplemental Table 1). Transplantation of CD34+CD38+CD19+ cells derived from the same patient also resulted in high levels of human B-ALL engraftment in recipient mice regardless of the expression of CD33 (Figure 2A-B, top; supplemental Table 1). Among 11 cases of MLL-AF4+ ALL examined, in vivo ALL-initiating cells were contained in the CD34+CD38+ population in 10 cases and in the CD34− population in 8 cases.
Figure 2.
In vivo leukemia initiation by CD34+CD38+CD19+ and CD34−CD19+ cells with MLL translocations. (A-C) Representative flow cytometry plots showing identification of LIC populations in MLL-AF4 case 3. (A) Using sort gates indicated by rectangles, 1000 CD34+CD38+CD19+, CD34−CD19+CD33+, and CD34−CD19+CD33− cells were FACS purified and transplanted into newborn NSG recipients. (B) Leukemia initiation was confirmed by the presence of engrafted human CD45+ cells in the BM of recipients. Expression of CD34, CD38, CD19, and CD33 was analyzed in the engrafted human CD45+ cells. Five cell fractions (indicated by rectangles labeled Fr. 1-Fr. 5) were isolated and transplanted into secondary recipients. (C) Engraftment of human CD45+ cells and expression patterns of CD34 and CD38 in the secondary NSG recipients. (D) Representative flow cytometry plots showing identification of LIC populations in 2 MLL-ENL cases: one with dominant CD34+ cell population (case 18) and another with dominant CD34− cell population (case 20). Using sort gates indicated by rectangles, CD34−CD19+ or CD34+CD38+CD19+ cells were FACS purified and transplanted into NSG recipients. (E) Leukemia initiation was confirmed by the presence of engrafted human CD45+ cells. Expression of CD34, CD38, CD19, and CD33 in engrafted human CD45+ cells are shown. In A-E, frequencies of each population is indicated and the number of MLL translocation-positive cells out of total number of cells examined are shown as fractions beside sort gates. (F,G) Engraftment levels of human CD45+ cells in (F) primary and (G) secondary recipient BM. Primary transplantation: MLL-AF4 CD34+CD38+ recipients, n = 54; CD34-CD19+ recipients, n = 60, from 11 patients; MLL-AF9 CD34−19+ recipients, n = 35 from 6 patients; MLL-ENL CD34+CD38+ recipients, n = 11; CD34−CD19+ recipients, n = 15, from 4 patients. Secondary transplantation: MLL-AF4 CD34+CD38+ recipients, n = 38; CD34−CD19+ recipients, n = 14, from 6 patients; MLL-AF9 CD34−19+ recipients, n = 12, from 4 patients; MLL-ENL CD34+CD38+ recipients, n = 6, from 2 patients; CD34−CD19+ recipients, n = 4, from 2 patients.
In vivo, both primary MLL-AF4+ CD34+CD38+CD19+ and CD34−CD19+ LICs generated cells with phenotypes different from the respective initial populations in variable frequencies. In recipients engrafted with MLL-AF4+ CD34+CD38+CD19+ cells, the majority of hCD45+ cells were CD34+CD38+CD19+ cells (BM: 93.7 ± 2.1%, spleen: 86.8 ± 3.3%; n = 33; supplemental Table 1). Engraftment of primary MLL-AF4+ CD34−CD19+ cells resulted in emergence of both CD34+CD38+ (69.7 ± 7.0%) and CD34− (29.7 ± 6.9%) cells (n = 20), and the frequency of engrafted CD34− cells was higher compared with CD34+CD38+CD19+ cell recipients (P = .0002; supplemental Table 1). In these recipients, 99.6 ± 0.2% of CD34+CD38+ and 99.7 ± 0.2% of CD34− cells expressed CD19 (n = 19; case 3 recipient shown as an example in Figure 2B). Overall, we found CD34+CD38+CD19+ cells in the BM of 18 of 20 recipients engrafted with CD34−CD19+ cells from 8 cases of MLL-AF4+ infant ALL.
Human CD45+ ALL cell infiltration was observed in the liver and kidney of recipients transplanted with CD34+CD38+ and CD34− MLL-AF4+ cells (CD34+CD38+ recipients, n = 30: liver 90.5 ± 2.3%, kidney 80.1 ± 11.3%; CD34− recipients, n = 18: liver 80.1 ± 11.3%, kidney 60.0 ± 9.1%; (supplemental Figure 1; supplemental Table 1). CD34−CD19+ MLL-AF4+ cells gave rise to both CD34− and CD34+CD38+ cells in the liver and kidney, as seen in the BM of CD34+38+19+ and CD34−CD19+ recipients (supplemental Table 1).
To demonstrate secondary repopulating capacity, we performed serial transplantation of CD34+CD38+CD19+ and CD34−CD19+ populations from the BM of primary recipients transplanted with either CD34+CD38+CD19+ cells or CD34−CD19+ cells (supplemental Table 1). From primary CD34+CD38+CD19+ cell recipient BM, 1000 CD34+CD38+CD19+ cells sorted from fraction 1 (Fr.1; Figure 2B) initiated leukemia in secondary recipients, demonstrated by engraftment in the PB, BM, and spleen. Similarly, 1000 CD34+CD38+CD19+ cells in fraction 2 (Fr.2) and 1000 CD34−CD19+ cells in fraction 3 (Fr.3) of primary CD34−CD19+CD33+ cell recipient BM initiated leukemia in secondary recipients (supplemental Table 1; case 3 shown as an example in Figure 2C). From primary CD34−CD19+CD33− cell recipient BM, both CD34+CD38+CD19+ cells in fraction 4 (Fr.4) and CD34−CD19+ cells in fraction 5 (Fr.5) initiated leukemia in secondary recipients (supplemental Table 1; case 3 shown as an example in Figure 2C). In the BM of secondary recipients engrafted with fraction 1, fraction 2, and fraction 4 (CD34+CD38+CD19+) cells, CD34+CD38+CD19+ ALL cells were predominant (supplemental Table 1; case 3 shown as an example in Figure 2C). In contrast, in the BM of the recipient of fraction 3 and fraction 5 (CD34−CD19+) cells, CD34+CD38+CD19+ cells were generated, as well as CD34−CD19+ cells (supplemental Table 1; case 3 shown as an example in Figure 2C). These findings indicate the presence of both CD34+CD38+CD19+ and CD34−CD19+ LICs with secondary-reconstituting capacity in MLL-AF4+ infant ALL.
LICs in MLL-AF9+ and MLL-ENL+ MLL-ALL
We went on to identify LICs in MLL-AF9+ and MLL-ENL+ MLL-ALL. First, we isolated CD34+CD38+CD19+ and CD34−CD19+ cells from 6 MLL-AF9+ patients for xenotransplantation into NSG newborns. The majority of CD19+ cells in MLL-AF9+ patients were CD34 negative, with rare CD34+CD38+ cells (0.5 ± 0.4%). We matched the numbers of transplanted CD34−CD19+ and CD34+CD38+ cells whenever possible (supplemental Table 2; case 14 shown as an example in supplemental Figure 2). Twenty of 35 recipients transplanted with MLL-AF9+ CD34−CD19+ cells developed ALL as demonstrated by the engraftment of human CD45+CD19+ cells (supplemental Table 2; case 14 shown as an example in supplemental Figure 2B, lower). In these recipients, the majority of human CD45+ cells in the BM and spleen were CD34 negative (BM: 97.3 ± 1.3%, spleen: 95.7 ± 2.1%; n = 16; supplemental Table 2). Male recipients transplanted with 1000 or 10 000 CD34−CD19+ cells from patient 14, with testicular disease at diagnosis, showed testicular infiltration by human ALL cells (n = 4; supplemental Table 2). In contrast, of 8 primary recipients transplanted with MLL-AF9+ CD34+CD38+CD19+ cells, 1 recipient showed human CD45+ engraftment, which consisted of mature human CD10−CD20+ B cells but not ALL.
We then performed secondary transplantation from primary recipients of MLL-AF9+ CD34−CD19+ cells from 4 patients. Transplantation of 1000 to 16 000 CD45+CD34−CD19+ cells resulted in leukemia initiation in 9 of 12 secondary recipients (supplemental Table 2; case 14 shown as an example in supplemental Figure 2C), confirming CD34−CD19+ MLL-AF9+ cells are self-renewing LICs.
Next, we examined the leukemia initiation capacity of cells derived from 4 MLL-ENL+ cases. In MLL-ENL+ patient samples, there were 2 distinct CD34 expression patterns (cases 18 and 20 shown as examples in Figure 2D). In cases 18 and 21, the major cell population is CD34+CD38+. In cases 19 and 20, the major population is CD34 negative. In all 4 cases, the major cell population (whether CD34+CD38+ or CD34−) contained self-renewing LICs that initiate leukemia in primary and secondary recipients (supplemental Table 3; cases 18 and 20 shown as examples in Figure 2E). Cell surface phenotype of engrafted cells was similar to that of injected LICs; that is, CD34+CD38+CD19+ cells (cases 18 and 21) engrafted with CD34+CD38+CD19+ cells and CD34−CD19+ cells (cases 19 and 20) engrafted with CD34−CD19+ cells.
The levels of in vivo human CD45+ ALL engraftment in primary and secondary recipients of MLL-AF4+, MLL-AF9+, and MLL-ENL+ LICs are summarized in Figure 2F-G. These findings demonstrate that both CD34+CD38+CD19+ and CD34−CD19+ populations may contain self-renewing LICs in MLL-AF4+, MLL-AF9+, and MLL-ENL+ patients. In all MLL-AF9+ cases and 2 of 4 MLL-ENL+ cases examined, CD34−CD19+ population contains self-renewing LICs that generate mainly CD34−CD19+ ALL cells. In contrast, in MLL-AF4+ cases and in 2 of 4 MLL-ENL+ cases examined, the CD34+CD38+CD19+ population contains self-renewing LICs that generate mainly CD34+CD38+CD19+ ALL cells. In addition, in MLL-AF4+ patients, the CD34−CD19+ population also contains self-renewing LICs that generate both CD34+CD38+CD19+ and CD34−CD19+ cells.
Analysis of primitive CD34+CD38− cells in MLL-rearranged infant ALL
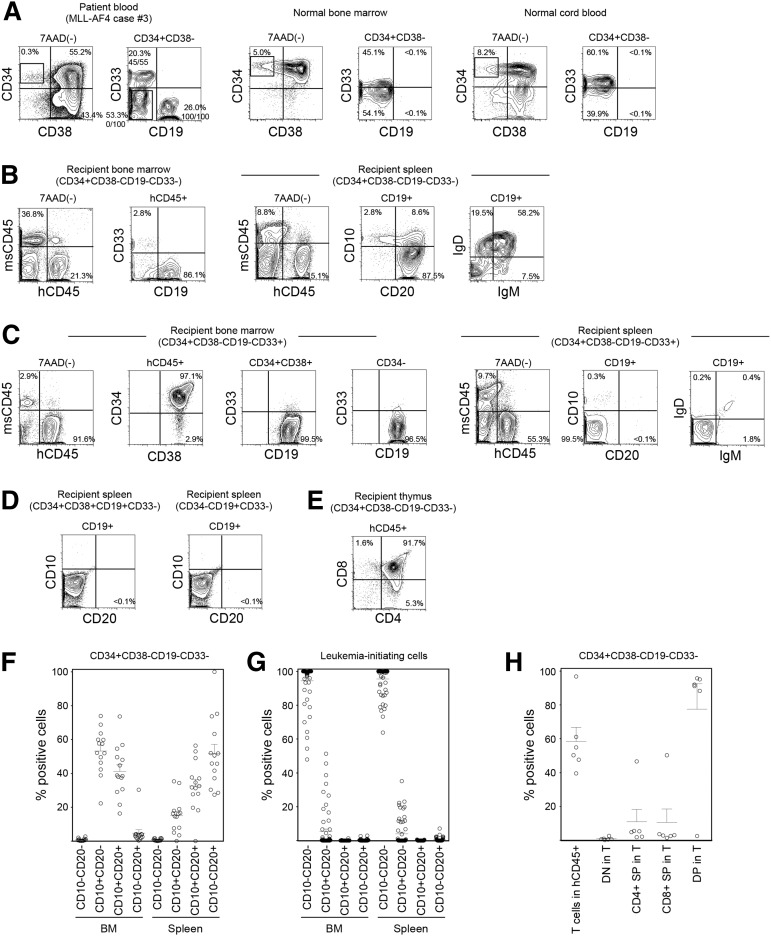
The CD34+CD38− population is enriched for HSCs in normal hematopoiesis and for leukemia stem cells in adult AML, suggesting that malignant transformation may occur within this population. To determine at which stage normal HSCs acquire MLL rearrangement and become malignant, we examined subpopulations of CD34+CD38− cells for MLL translocation in MLL-AF4+, MLL-AF9+, and MLL-ENL+ infant ALL patient samples by fluorescence in situ hybridization (FISH). In MLL-AF4+ case 3, CD19+CD33−, CD19−CD33+, and CD19−CD33− subpopulations were present within the CD34+CD38− population, whereas normal BM and CB CD34+CD38− cells do not express CD19 (Figure 3A; Table 1). In MLL-AF4+ case 4, CD19+(CD33+/−) and CD19−CD33− subpopulations were present within the CD34+CD38− population. In both cases 3 and 4, CD34+CD38−CD19−CD33− cells did not possess rearrangements involving MLL (0 of 100 cells examined for each). In contrast, MLL translocation was identified in 100 of 100 CD34+CD38−CD19+CD33− cells and 45 of 55 CD34+CD38−CD19−CD33+ cells in MLL-AF4+ case 3 and in 15 of 20 CD34+CD38−CD19+(CD33+/−) cells in MLL-AF4+ case 4. In MLL-AF9, CD34+CD38− populations from 3 cases were nearly uniformly CD19−CD33−, and these cells were FISH negative for MLL translocation (supplemental Figure 3; case 16 shown as an example in supplemental Figure 4). In 2 cases of MLL-ENL, 41 of 43 and 98 of 100 CD34+CD38−CD19−CD33− cells, respectively, were FISH negative for MLL translocation (supplemental Figure 3).
Figure 3.
CD34+CD38−CD19−CD33− cells in MLL ALL are enriched for normal hematopoietic stem/progenitor cells. (A-E) Representative flow cytometry plots showing the identification of HSC/HPC population in MLL-AF4 case 3. (A) Within the CD34+CD38− population, CD34+CD38−CD19−CD33−, CD34+CD38−CD19+CD33−, and CD34+CD38−CD19−CD33+ cells were sorted according to rectangular gates indicated and analyzed for MLL rearrangement by FISH. Numbers of MLL translocation-positive cells in total number of cells examined are shown as fractions. Sorted CD34+CD38−CD19−CD33− cells were transplanted into NSG recipients. In normal human BM and CB almost all CD34+CD38− cells do not express CD19. (B) Differentiation and maturation of human CD45+CD19+ B cells engrafted in recipient spleen examined through the expression of human CD10, CD20, IgM, and IgD. (C) BM and spleen of CD34+CD38−CD19−CD33+ cells were analyzed for human CD45+ cell engraftment and expression of CD34, CD38, CD19, CD33, CD10, CD20, IgD, and IgM. (D) Human CD10 and CD20 were not expressed by human CD19+ leukemia cells in the spleen of recipients transplanted with CD34+CD38+CD19+CD33− and CD34−CD19+CD33− cells. (E) Thymic repopulation by the CD34+CD38−CD19−CD33− cells indicated by the engraftment of CD4+CD8+ DP cells, CD4 SP cells, and CD8 SP cells. (F,G) B-cell maturation in the BM and spleen of recipients transplanted with (F) CD34+CD38−CD19−CD33− cells (MLL-AF4, n = 5; MLL-AF9, n = 7; MLL-ENL, n = 2) and those transplanted with (G) CD34+CD38+CD19+ or CD34−CD19+ LICs (MLL-AF4, n = 39; MLL-AF9, n = 16; MLL-ENL, n = 7) is summarized. (H) Frequencies of human CD3+ T cells within human CD45+ cells and the frequencies of double negative, DP, and single positive cells within human CD45+CD3+ cells in CD34+CD38−CD19−CD33− recipient thymus are summarized (MLL-AF4, n = 3; MLL-AF9, n = 2; MLL-ENL, n = 1).
Next we examined functional characteristics of MLL rearrangement-negative CD34+CD38−CD19−CD33− cells through newborn NSG transplantation. At 3 to 5 months after transplantation of CD34+CD38−CD19−CD33− cells isolated from 2 MLL-AF4 patient samples (cases 3 and 6), 4 MLL-AF9 patient samples (cases 14-17) and 3 MLL-ENL patient samples (cases 19-21), we confirmed human hematopoietic engraftment and multilineage repopulation in the BM and spleen of 14 of 19 recipients (case 3 shown as an example in Figure 3B; supplemental Table 4). In the spleen, 51.7 ± 5.5% of CD19+ B-lineage cells were CD10−CD20+ (MLL-AF4, n = 5; MLL-AF9, n = 7; MLL-ENL, n = 2; total, n = 14) and expressed immunoglobulin (Ig)M or IgD on their surface (Figure 3B,F). In nearly all CD34+CD38−CD19−CD33− recipient CD19+ B cells and CD33+ myeloid cells examined, MLL rearrangement was not detected (except 1 CD19+ cell of a case 17 recipient), demonstrating that CD34+CD38−CD19−CD33− MLL-ALL cells give rise to normal B- and myeloid lineage cells in vivo (supplemental Figure 3). In contrast, when MLL-AF4+ CD34+CD38−CD19−CD33+ cells were transplanted, the CD19+ population in the recipient spleen consisted of CD10−CD20− cells, similar to the phenotype of patient CD34+CD38+ and CD34− leukemia cells and that of engrafted leukemia cells in the recipients of CD34+CD38+ and CD34− LICs (Figure 3C,D,G). Furthermore, in the MLL-AF4 CD34+CD38−CD19−CD33− recipient thymus, DP T cells predominated, whereas CD4+ SP and CD8+ SP cells were more infrequent (case 3 shown as an example in Figure 3E). In addition, MLL-AF9 CD34+CD38−CD19−CD33− cells generated CD10−CD20+ mature B cells without MLL rearrangement in the spleen (0 of 50 cells examined) and DP and SP T cells in the thymus (case 16 shown as an example in supplemental Figure 4). Thymic repopulation by CD34+CD38−CD19−CD33− cells is summarized in Figure 3H. These findings indicate that CD34+CD38−CD19−CD33− cells in infant MLL-AF4, -AF9, and -ENL ALL are highly enriched for normal hematopoietic stem/progenitor cells.
IGH rearrangements in infant MLL ALL LICs
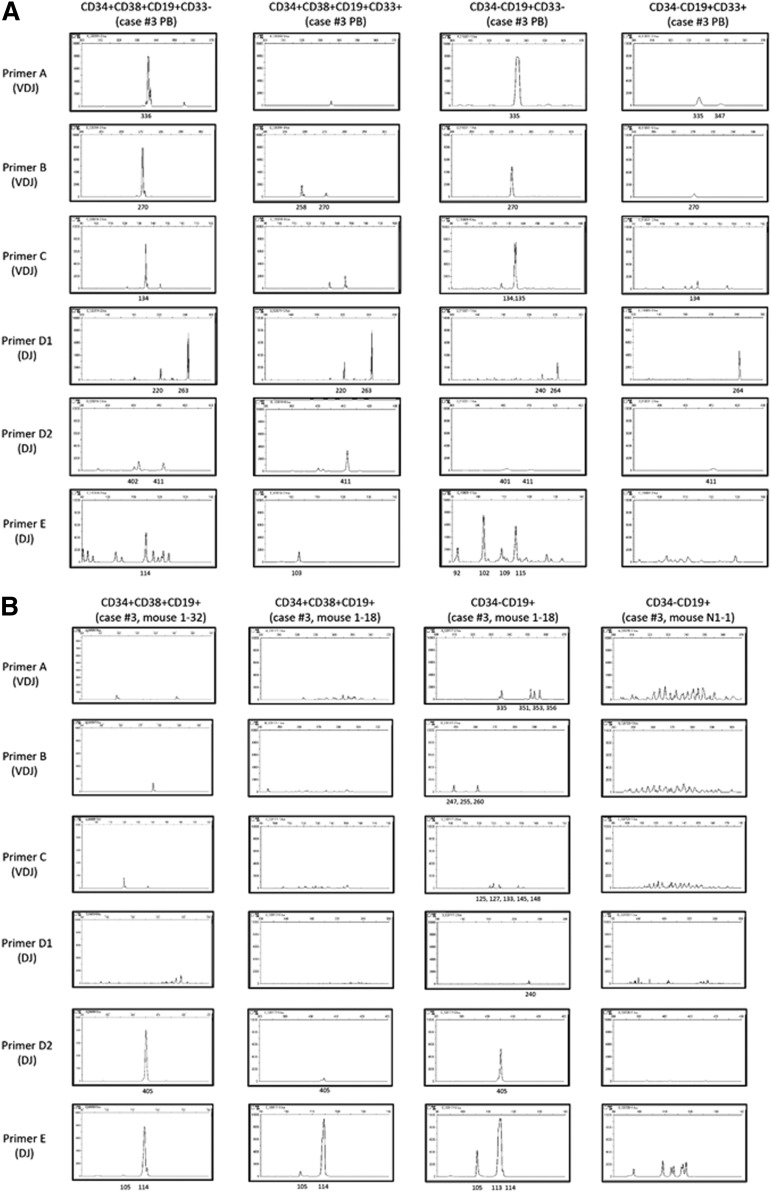
We proceeded to determine whether dominant leukemia clones existed in patients and recipient mice by examining IGH rearrangement. VDJ and DJ rearrangements were identified in CD34+CD38+CD19+ cells and CD34−CD19+ cells derived from 6 MLL-AF4 patients by polymerase chain reaction using 5 different primer sets (primer sets A, B, and C for VDJ and primer sets D and E for DJ; supplemental Table 5).34 In individual patients, CD34+CD38+CD19+CD33+, CD34+CD38+CD19+CD33−, CD34−CD19+CD33−, and CD34−CD19+CD33+ cells shared clonal populations as evidenced by IGH rearrangement patterns (supplemental Table 5; shared peaks at 270, 134, and 411 bp using primer set B, C, and D-2 in case 3 shown as examples in Figure 4A). In addition, there were dominant clones present among MLL-AF4 patients as evidence by shared IGH rearrangement peaks (indicated in blue in supplemental Table 5). MLL-AF4 patient CD34+CD38−CD19+CD33− cells, which were found to contain the MLL rearrangement, also showed clonal bands (supplemental Table 5).
Figure 4.
IGH rearrangement analysis shows oligoclonal expansion from LICs and polyclonal expansion from CD34+CD38−CD19−CD33− cells. (A) In MLL-AF4 case 3, PCR for IGH rearrangements showed the presence of a dominant clone present in CD34+ and CD34− cells, regardless of CD33 expression. (B) IGH rearrangement patterns of 4 engrafted cell populations from recipients of CD34+CD38−CD19−CD33+ LICs (left column of panels), CD34−CD19+ LICs (second and third columns of panels), and CD34+CD38−CD19−CD33− cells (right column of panels) isolated from MLL-AF4 case 3. Shared dominant clones were detected in (left) CD34+CD38+CD19+ cells from recipient 1-32 engrafted with CD34+CD38−CD19−CD33+ LICs, and (second from left) CD34+ and (third from left) CD34− cells from recipient 1-18 engrafted with CD34−CD19+ LICs. In contrast, CD19+CD33− B cells in recipient N1-1 engrafted with CD34+CD38−CD19−CD33− cells from MLL-AF4 case 3 (right column) show polyclonal pattern of IGH rearrangement.
We next investigated whether dominant IGH clones detected in MLL ALL patients persisted in the recipients. In MLL-AF4+ ALL, dominant IGH clones detected in the donor (patient) cells were also present in the recipient BM CD34+CD38+CD19+ and CD34−CD19+ cells (indicated in red in supplemental Table 5). Additional IGH rearrangements were also detected in the recipients (supplemental Table 5). Shared IGH clones were also detected in CD34+CD38+CD19+ cells and CD34−CD19+ cells of recipients (supplemental Table 5; case 3 shown as an example in Figure 4B). In a recipient engrafted with CD34+CD38−CD19−CD33+ cells (case 3 recipient 1-32), we did not detect shared VDJ-rearranged leukemic clones, but found DJ-rearranged clones (405 in primer set D and 114 in primer set E) that were shared by all recipients of CD34+CD38+ and CD34− LICs. Similarly, in both MLL-AF9 and -ENL ALL, there were shared IGH clones in CD34−CD19+ cells among patients or between patients and recipients (supplemental Table 5; case 17 shown as an example in supplemental Figure 5).
In contrast, human CD19+ B cells in recipients of CD34+CD38−CD19−CD33− cells (case 3 recipient N-1/N-2, case 6 recipient N-2, case 15 recipient N-3, and case 17 recipient N-1/N-2 in supplemental Table 5) showed polyclonal IGH rearrangement patterns comparable to those of human B cells in normal CB HSC recipients (CB HSC-engrafted recipient-1/2 in supplemental Table 5) that exhibit physiological diversity of IGH rearrangements (case 3 shown as an example in Figure 4B).
Gene expression signatures in MLL LICs and HSCs
We performed global transcriptome analyses of LICs from MLL-rearranged infant ALL patient samples (supplemental Figure 6). First, genes specifically up- and downregulated in LICs isolated from MLL-AF4, MLL-AF9, and MLL-ENL patient samples were identified through 3-way comparison (supplemental Figure 6A; supplemental Table 6).
Genes differentially expressed in MLL-AF4 LICs compared with MLL-AF9 and MLL-ENL LICs included HOXA clusters (downregulated) and TET2 and DR1 (upregulated). TET2 encodes a methylcytosine dioxygenase, a member of the Ten-Eleven-Translocation (TET) family of proteins, which mediates the process of active DNA demethylation.35-37 Loss-of-function mutation in TET2 is frequently found in adult AML and is associated with adverse prognosis.38-40 DR1 encodes a transcriptional repressor of TATA box-binding protein (TBP) promoter complexes. TBP-related factors are required for normal early development and hematopoiesis.41
ZNF521 was among the genes upregulated in MLL-AF9 LICs compared with MLL-AF4 and MLL-ENL LICs. ZNF521 encodes a zinc finger protein that inhibits early B-cell factor 1 (EBF1) which is required for normal B-cell development and differentiation. In Ebf1 knockout mice, accumulation of early lymphoid progenitors that lack B-lineage priming but have T- and myeloid lineage potential occurs while mature B cells are completely absent.42,43 Mice transgenic for Zfp521, a murine homolog for ZNF521, were also reported to develop B-lineage ALL.44
Genes upregulated in MLL-ENL LICs compared with MLL-AF4 and MLL-AF9 LICs included SIRPA and SATB1. SIRPA encodes an Ig superfamily inhibitory immunoreceptor, signal regulatory protein-α (SIRPα), expressed on macrophages, dendritic cells, and neurons.45-48 SIRPα binding with its ligand CD47 results in decreased phagocytosis by macrophages.49 SIRPα-CD47 interaction may be one of the mechanisms by which malignant cells evade innate immune responses, as suggested by reports that CD47 blocking antibodies result in elimination of tumor cells in various models of human leukemia and solid tumors.50-54 SATB1 encodes a matrix protein that recruits chromatin-remodeling factors by binding nuclear matrix and scaffold-associating DNAs, thereby regulating chromatin structure and gene expression. Special AT-rich sequence-binding protein 1 (SATB1) expression is associated with multiple solid tumors including colorectal cancer, breast cancer, malignant melanoma, gastric cancer, bladder cancer, and prostate cancer. SATB1 is also involved in normal T cell immunity. It is known to inhibit the expression of Forkhead box P3 transcription factor (FoxP3), a key determinant of normal regulatory T-cell development and function, and is required for Th2 cytokine expression.55,56 Some of the genes were commonly upregulated in our study and in previously reported MLL translocation-specific gene expression signatures (supplemental Figure 6B).57
Next, we compared gene expression profiles of CD34+CD38+CD19+ LICs and CD34−CD19+ LICs derived from 5 MLL-AF4 patient samples. We found that expression profiles clustered by patient not by surface phenotype, indicating that LIC gene expression signatures are relatively patient specific (supplemental Figure 6C). However, some genes such as DNTT, CCND2, and BAALC showed differential expression between CD34+CD38+CD19+ and CD34−CD19+ LIC populations (supplemental Figure 6D). CCND2 (cyclin D2) and BAALC expression have prognostic relevance in leukemia and other malignancies.58,59
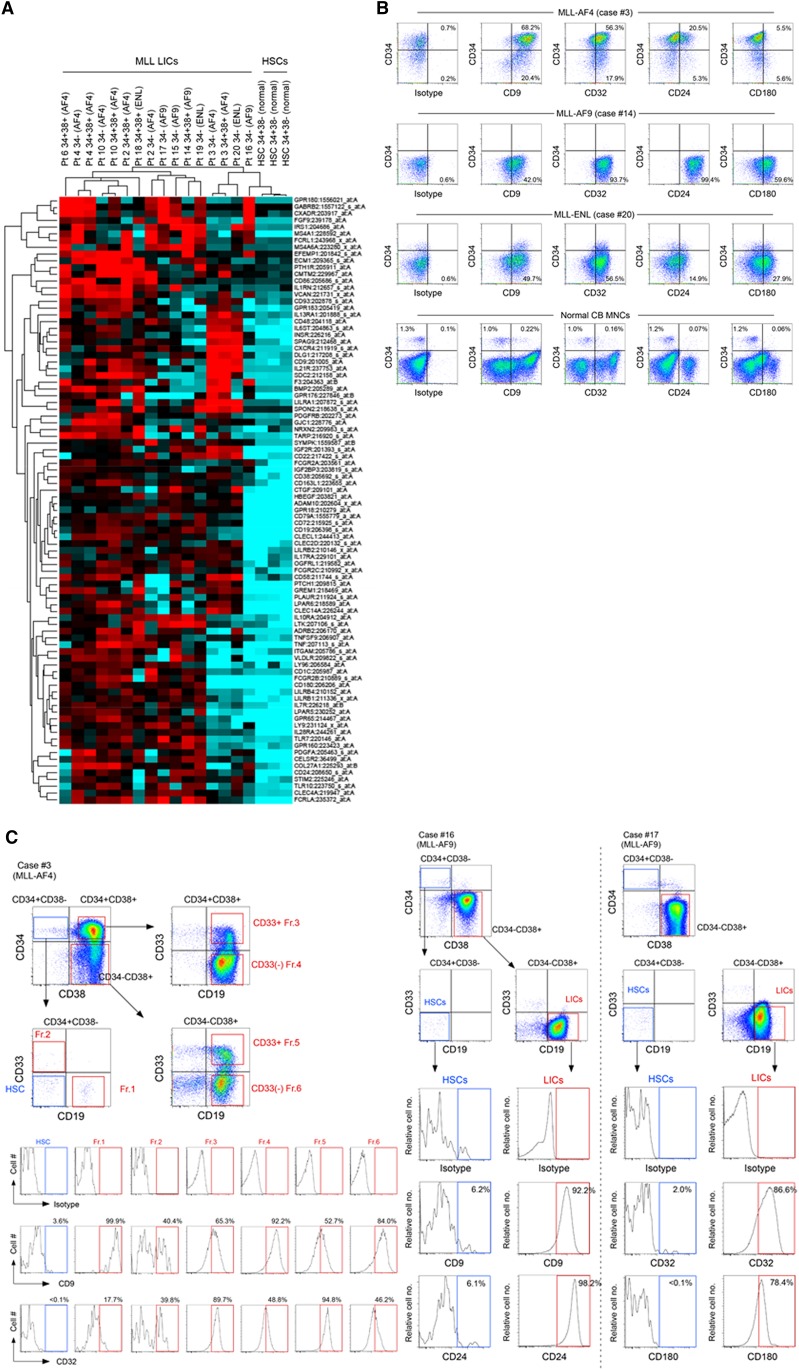
In addition, we compared gene expression profiles of 5 MLL-AF4 cases, 4 MLL-AF9 cases, and 3 MLL-ENL cases with those of normal human BM CD34+CD38− HSCs isolated from 3 healthy donors. We found relative overexpression of cell surface molecules CD9, CD24, CD32, CD127, and CD180b in MLL-AF4 CD34+CD38+CD19+ cells, MLL-AF4 CD34− cells, MLL-AF9 CD34− cells, MLL-ENL CD34+CD38+CD19+ cells, and MLL-ENL CD34−CD19+ cells (Figure 5A). We validated the expression of these molecules in MLL-ALL LICs (4 cases of MLL-AF4, 6 cases of MLL-AF9, and 4 cases of MLL-ENL) and in normal human CB mononuclear cells (MNCs; n = 3) by flow cytometry (Figure 5B; supplemental Table 7). In MLL-AF4, CD9 and CD32 were highly expressed in CD34+CD38+CD19+ cells and CD34−CD19+ cells in all 4 cases examined. CD24 was highly expressed by both CD34+CD38+CD19+ and CD34−CD19+ MLL-AF4 cases in 3 of 4 cases examined. In MLL-AF9 CD34−CD19+ LICs, CD9, CD32, and CD24 showed high expression in 5, 5, and 4 of 6 cases examined, respectively. In MLL-ENL, CD9 was highly expressed by CD34+CD38+CD19+ LICs and CD34−CD19+ LICs. In normal CB, the frequency of normal CD34+CD38− hematopoietic stem/progenitor cells expressing these molecules was low, whereas CD34−CD19+ B cells express these antigens.
Figure 5.
Global gene expression profiling identifies genes expressed in MLL-ALL LICs but not in HSC/HPCs. (A) Transcripts of cell surface molecules over-represented in MLL-AF4 (cases 2, 3, 4, 6, and 10), MLL-AF9 (cases 14-17), and MLL-ENL (cases 18-20) LICs compared with 3 independent normal human BM HSCs. (B) Representative flow cytometry plots showing expression of CD9, CD32, CD24, and CD180 in human CD45+ ALL cells (MLL-AF4 case 3, MLL-AF9 case 14, and MLL-ENL case 20) and normal CB MNCs. Frequencies of cells expressing the markers are indicated. (C) Expression of CD9 and CD32 (MLL-AF4+ case 3), CD9 and CD24 (MLL-AF9+ case 16), and CD32 and CD180 (MLL-AF9+ case 17) in the HSC-enriched CD34+CD38−CD19−CD33− population and LIC populations was analyzed by flow cytometry.
Finally, we examined the expression of these surface molecules in MLL-ALL patient cells that we determined to be HSCs (CD34+CD38−CD19−CD33− cells) or leukemic cells (CD34+CD38+CD19+, CD34−CD19+, CD34+CD38−CD19+CD33+, CD34+CD38−CD19+CD33−, and CD34−CD19+CD33− cells), both functionally (via xenotransplantation) and genetically (by FISH for MLL translocation). In MLL-AF4 (case 3), all 5 leukemia cell populations were found to express CD9 or CD32, whereas the normal HSC-enriched CD34+CD38−CD19−CD33− population did not (Figure 5C). In MLL-AF9 cases 16 and 17, CD9 and CD24, and CD32 and CD180, respectively, were highly expressed by CD34−CD19+CD33− LICs, but not by CD34+CD38−CD19−CD33− cells (Figure 5C). In MLL-AF9 case 14, CD24 was differentially expressed between HSC/HPC and LIC populations (supplemental Figure 7A). Finally, in MLL-ENL case 21, CD34+CD38+CD19+CD33+ LICs and CD34+CD38+CD19+CD33− leukemia cells expressed CD9 and CD32 at high levels and CD34+CD38−CD19+ cells expressed CD9, whereas CD34+CD38−CD19−CD33− HSCs/HPCs did not express either CD9 or CD32 (supplemental Figure 7B).
Discussion
Despite significant improvement in overall clinical outcomes of pediatric ALL in the last few decades, the prognosis in MLL-rearranged ALL has remained poor. To understand the pathogenesis of this disease entity and to develop effective therapeutic strategies, mouse syngeneic and xenogeneic transplantation of MLL-AF9– or -ENL–expressing mouse or human BM cells have been developed. These systems have provided important insights into the pathogenesis of MLL-rearranged ALL and have led to identification of potential therapeutic targets (eg, Rac signaling or DOT1L). In addition, direct evaluation of primary patient cells in vivo may also facilitate translation of research findings into medicine. In the present study, we examined the biology of primary LICs in infant MLL-rearranged ALL using the newborn NSG xenogeneic transplantation system.
The NSG xenogeneic transplantation system supports long-term engraftment and multilineage differentiation of normal HSCs.60,61 The NSG xenograft system also allows the identification of LICs and the analysis of engrafted human leukemia cells in vivo.62 In samples obtained from infants with MLL ALL, we identified LICs with distinct surface expression patterns of CD34, CD38, CD19, and CD33 according to fusion partners. Our findings do not suggest large differences in LIC frequency among leukemia-initiating populations in MLL-ALL patient samples examined, although formal limiting dilution transplantation experiments are required to determine LIC frequencies rigorously.
MLL leukemia is characterized by surface expression of B-cell antigens (eg, CD19), as well as myeloid cell antigens (eg, CD33, CD15, and CD13). Although CD19 expression is considered an important marker for B-ALL initiating cells,63 B-ALL initiation by CD19-negative cells was confirmed in NOD/SCID repopulating assay in several reports.26,64 In this study, we found no significant difference in leukemia-initiating capacity between CD34−CD19+CD33+ and CD34−CD19+CD33− cells in the NSG xenograft system. Interestingly, in MLL-AF4 and -AF9, CD34+CD38−CD19−CD33− cells did not carry the MLL rearrangement nor did they initiate B-ALL in vivo. Instead, CD34+CD38−CD19−CD33− cells repopulated recipients with normal human multilineage hematopoiesis. In addition, B-lineage progeny of these cells do not share dominant IGH clones or mono- or oligoclonal IGH rearrangement pattern found in leukemic progeny of LICs, indicating that in MLL-AF4 and -AF9, CD34+CD38−CD19−CD33− cells are highly enriched for normal HSCs. Furthermore, in 1 case of MLL-AF4, we found that CD34+CD38−CD19−CD33+ cells initiated B-ALL in vivo, suggesting that malignant transformation may occur in parallel with the acquisition of CD33 in AF4 disease.
These findings provide some insight regarding the hierarchy of malignant transformation in infant MLL-ALL. In both AF4- and AF9-rearranged ALL, more primitive CD34+CD38−CD19−CD33− populations contain normal HSCs that have not undergone chromosomal translocation or malignant transformation. In AF4 disease, malignant transformation appears to occur as CD34+CD38−CD19− early hematopoietic progenitors differentiate, and as a result, both CD34+ and more differentiated CD34− cells contain LICs. In contrast, in AF9 disease, malignant transformation appears to occur in more differentiated B-cell progenitors that have lost CD34 expression, as CD34+ cells do not initiate leukemia in vivo but CD34−CD19+ cells do. Interestingly, in ENL disease, we found that there are 2 groups of patients. In some MLL-ENL ALL patients, leukemic transformation may have occurred earlier in hematopoietic differentiation (CD34+ LICs), whereas in others, it occurred later during B-cell development (CD34− LICs). Of note, the frequency of CD34+CD38− cells in MLL-ALL patient samples appears to be higher than expected, possibly due to mobilization of primitive hematopoietic stem/progenitor cells analogous to that occurring in myeloid leukemia. In MLL-AF9 and MLL-ENL, additional transplantation experiments using more patient samples are required to definitively conclude that CD34+CD38− population in MLL-ALL does not contain LICs.
To determine potential therapeutic targets in infant MLL-ALL, we examined gene expression profiles in primary MLL ALL LICs. We found differential expression of some genes or groups of genes according to type of MLL translocation (eg, HOXA clusters and related genes) and phenotype of LICs (eg, DNTT, CCND2, and BAALC), but we also found patient-specific differences in LIC signature. By comparing expression profiles of primary MLL ALL LICs with those of HSCs, we identified CD9, CD32, and CD24 as cell surface molecules that more specifically mark LICs and not HSCs. Of note, CD32 shows higher levels of expression compared with CD9, CD24, or CD180 in CB-derived CD34+CD38− cells, consistent with previously reported expression of CD32 in CD34+CD38−CD133− hematopoietic progenitors and not in CD34+CD38−CD133+ HSCs.65 By using agents that target surface molecules with selective expression in LICs compared with HSCs, it may be possible to minimize treatment-related hematopoietic adverse effects only targeting mature hematopoietic subsets such as normal human B cells. Knockout mice for these molecules have not shown serious defects in hematopoiesis, giving some support to the speculation.66-68 CD9, CD32, and CD24 are reported to be expressed by non–MLL-rearranged hematologic malignancies and solid tumors.65,69-71 Targeting CD24 with antibody resulted in the significant reduction of immortalized lung tumor cells in a xenograft model.72 Recent studies have reported significantly improved outcomes in B-lymphoid malignancies by use of antibody drug and chimeric-antigen receptor T cells (CART).73,74 Development of new therapeutic agents targeting molecules identified in the present study using CART technology may prove beneficial to MLL-rearranged ALL, the majority which are infants. Therapeutic strategies that combine antibody drug/CART with drugs that target key molecules in MLL-induced leukemogenesis may provide us with a better chance for improved outcomes in patients with MLL-rearranged ALL. Whether LIC-specific surface markers identified are suitable for such use awaits further study.
Acknowledgments
This research has been done as an affiliated research program for JPLSG infant leukemia clinical study MLL-10. The authors thank all the participating pediatricians for providing patient samples for this research. The authors also thank all the JPLSG infant leukemia committee members and data managers of NPO-OSCR data center for supporting this research.
This research was supported in part by Grants-in-Aid for Cancer Research from the Ministry of Health, Labor, and Welfare Japan (H23-H22-11) and National Institutes of Health, National Cancer Institute Core grants CA034196 and CA171983.
Footnotes
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: Y.A., Y.S., O.O., L.D.S., S.M., and F.I. designed this study; Y.A., T.W., N.S., Y.K., A.H., S.F., R.O., K.S., and A.K. performed experiments and analyzed data; M.T., D.T., M.E., M.E.-I., K.K., E.I., and S.M. analyzed the data; Y.A., Y.S., O.O., L.D.S., S.M., and F.I. wrote the manuscript; and all authors read and approved the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Fumihiko Ishikawa, Laboratory for Human Disease Models, Center for Integrative Medical Sciences, RIKEN Yokohama Institute North Research Building N305, 1-7-22 Suehiro-cho, Tsurumi-ku, Yokohama 230-0045 Japan; e-mail: f_ishika@rcai.riken.jp.
References
- 1.Schuettengruber B, Martinez AM, Iovino N, Cavalli G. Trithorax group proteins: switching genes on and keeping them active. Nat Rev Mol Cell Biol. 2011;12(12):799–814. doi: 10.1038/nrm3230. [DOI] [PubMed] [Google Scholar]
- 2.Wu M, Wang PF, Lee JS, et al. Molecular regulation of H3K4 trimethylation by Wdr82, a component of human Set1/COMPASS. Mol Cell Biol. 2008;28(24):7337–7344. doi: 10.1128/MCB.00976-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yang Y, Hua X. In search of tumor suppressing functions of menin. Mol Cell Endocrinol. 2007;265-266:34–41. doi: 10.1016/j.mce.2006.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yokoyama A, Cleary ML. Menin critically links MLL proteins with LEDGF on cancer-associated target genes. Cancer Cell. 2008;14(1):36–46. doi: 10.1016/j.ccr.2008.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blobel GA, Kadauke S, Wang E, et al. A reconfigured pattern of MLL occupancy within mitotic chromatin promotes rapid transcriptional reactivation following mitotic exit. Mol Cell. 2009;36(6):970–983. doi: 10.1016/j.molcel.2009.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ernst P, Fisher JK, Avery W, Wade S, Foy D, Korsmeyer SJ. Definitive hematopoiesis requires the mixed-lineage leukemia gene. Dev Cell. 2004;6(3):437–443. doi: 10.1016/s1534-5807(04)00061-9. [DOI] [PubMed] [Google Scholar]
- 7.Ernst P, Mabon M, Davidson AJ, Zon LI, Korsmeyer SJ. An Mll-dependent Hox program drives hematopoietic progenitor expansion. Curr Biol. 2004;14(22):2063–2069. doi: 10.1016/j.cub.2004.11.012. [DOI] [PubMed] [Google Scholar]
- 8.Jude CD, Climer L, Xu D, Artinger E, Fisher JK, Ernst P. Unique and independent roles for MLL in adult hematopoietic stem cells and progenitors. Cell Stem Cell. 2007;1(3):324–337. doi: 10.1016/j.stem.2007.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pui CH, Campana D. Age-related differences in leukemia biology and prognosis: the paradigm of MLL-AF4-positive acute lymphoblastic leukemia. Leukemia. 2007;21(4):593–594. doi: 10.1038/sj.leu.2404598. [DOI] [PubMed] [Google Scholar]
- 10.Rowley JD, Olney HJ. International workshop on the relationship of prior therapy to balanced chromosome aberrations in therapy-related myelodysplastic syndromes and acute leukemia: overview report. Genes Chromosomes Cancer. 2002;33(4):331–345. doi: 10.1002/gcc.10040. [DOI] [PubMed] [Google Scholar]
- 11.Liu H, Cheng EH, Hsieh JJ. MLL fusions: pathways to leukemia. Cancer Biol Ther. 2009;8(13):1204–1211. doi: 10.4161/cbt.8.13.8924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jo SY, Granowicz EM, Maillard I, Thomas D, Hess JL. Requirement for Dot1l in murine postnatal hematopoiesis and leukemogenesis by MLL translocation. Blood. 2011;117(18):4759–4768. doi: 10.1182/blood-2010-12-327668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Krivtsov AV, Feng Z, Lemieux ME, et al. H3K79 methylation profiles define murine and human MLL-AF4 leukemias. Cancer Cell. 2008;14(5):355–368. doi: 10.1016/j.ccr.2008.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nguyen AT, Taranova O, He J, Zhang Y. DOT1L, the H3K79 methyltransferase, is required for MLL-AF9-mediated leukemogenesis. Blood. 2011;117(25):6912–6922. doi: 10.1182/blood-2011-02-334359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Okada Y, Feng Q, Lin Y, et al. hDOT1L links histone methylation to leukemogenesis. Cell. 2005;121(2):167–178. doi: 10.1016/j.cell.2005.02.020. [DOI] [PubMed] [Google Scholar]
- 16.Thiel AT, Blessington P, Zou T, et al. MLL-AF9-induced leukemogenesis requires coexpression of the wild-type Mll allele. Cancer Cell. 2010;17(2):148–159. doi: 10.1016/j.ccr.2009.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nakamura T, Largaespada DA, Lee MP, et al. Fusion of the nucleoporin gene NUP98 to HOXA9 by the chromosome translocation t(7;11)(p15;p15) in human myeloid leukaemia. Nat Genet. 1996;12(2):154–158. doi: 10.1038/ng0296-154. [DOI] [PubMed] [Google Scholar]
- 18.Nakamura T, Largaespada DA, Shaughnessy JD, Jr, Jenkins NA, Copeland NG. Cooperative activation of Hoxa and Pbx1-related genes in murine myeloid leukaemias. Nat Genet. 1996;12(2):149–153. doi: 10.1038/ng0296-149. [DOI] [PubMed] [Google Scholar]
- 19.Perkins A, Kongsuwan K, Visvader J, Adams JM, Cory S. Homeobox gene expression plus autocrine growth factor production elicits myeloid leukemia. Proc Natl Acad Sci USA. 1990;87(21):8398–8402. doi: 10.1073/pnas.87.21.8398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jin S, Zhao H, Yi Y, Nakata Y, Kalota A, Gewirtz AM. c-Myb binds MLL through menin in human leukemia cells and is an important driver of MLL-associated leukemogenesis. J Clin Invest. 2010;120(2):593–606. doi: 10.1172/JCI38030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu H, Takeda S, Kumar R, et al. Phosphorylation of MLL by ATR is required for execution of mammalian S-phase checkpoint. Nature. 2010;467(7313):343–346. doi: 10.1038/nature09350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yeung J, Esposito MT, Gandillet A, et al. β-Catenin mediates the establishment and drug resistance of MLL leukemic stem cells. Cancer Cell. 2010;18(6):606–618. doi: 10.1016/j.ccr.2010.10.032. [DOI] [PubMed] [Google Scholar]
- 23.Hilden JM, Dinndorf PA, Meerbaum SO, et al. Children’s Oncology Group. Analysis of prognostic factors of acute lymphoblastic leukemia in infants: report on CCG 1953 from the Children’s Oncology Group. Blood. 2006;108(2):441–451. doi: 10.1182/blood-2005-07-3011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pieters R, Schrappe M, De Lorenzo P, et al. A treatment protocol for infants younger than 1 year with acute lymphoblastic leukaemia (Interfant-99): an observational study and a multicentre randomised trial. Lancet. 2007;370(9583):240–250. doi: 10.1016/S0140-6736(07)61126-X. [DOI] [PubMed] [Google Scholar]
- 25.Tomizawa D, Koh K, Sato T, et al. Outcome of risk-based therapy for infant acute lymphoblastic leukemia with or without an MLL gene rearrangement, with emphasis on late effects: a final report of two consecutive studies, MLL96 and MLL98, of the Japan Infant Leukemia Study Group. Leukemia. 2007;21(11):2258–2263. doi: 10.1038/sj.leu.2404903. [DOI] [PubMed] [Google Scholar]
- 26.le Viseur C, Hotfilder M, Bomken S, et al. In childhood acute lymphoblastic leukemia, blasts at different stages of immunophenotypic maturation have stem cell properties. Cancer Cell. 2008;14(1):47–58. doi: 10.1016/j.ccr.2008.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Barabé F, Kennedy JA, Hope KJ, Dick JE. Modeling the initiation and progression of human acute leukemia in mice. Science. 2007;316(5824):600–604. doi: 10.1126/science.1139851. [DOI] [PubMed] [Google Scholar]
- 28.Lavau C, Szilvassy SJ, Slany R, Cleary ML. Immortalization and leukemic transformation of a myelomonocytic precursor by retrovirally transduced HRX-ENL. EMBO J. 1997;16(14):4226–4237. doi: 10.1093/emboj/16.14.4226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wei J, Wunderlich M, Fox C, et al. Microenvironment determines lineage fate in a human model of MLL-AF9 leukemia. Cancer Cell. 2008;13(6):483–495. doi: 10.1016/j.ccr.2008.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wu ZJ, Irizarry RA, Gentleman R, Martinez-Murillo F, Spencer F. A model-based background adjustment for oligonucleotide expression arrays. J Am Stat Assoc. 2004;99(468):909–917. [Google Scholar]
- 31.Hong F, Breitling R, McEntee CW, Wittner BS, Nemhauser JL, Chory J. RankProd: a bioconductor package for detecting differentially expressed genes in meta-analysis. Bioinformatics. 2006;22(22):2825–2827. doi: 10.1093/bioinformatics/btl476. [DOI] [PubMed] [Google Scholar]
- 32.Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3(1):1–25. doi: 10.2202/1544-6115.1027. [DOI] [PubMed] [Google Scholar]
- 33.Hijikata A, Kitamura H, Kimura Y, et al. Construction of an open-access database that integrates cross-reference information from the transcriptome and proteome of immune cells. Bioinformatics. 2007;23(21):2934–2941. doi: 10.1093/bioinformatics/btm430. [DOI] [PubMed] [Google Scholar]
- 34.van Dongen JJ, Langerak AW, Brüggemann M, et al. Design and standardization of PCR primers and protocols for detection of clonal immunoglobulin and T-cell receptor gene recombinations in suspect lymphoproliferations: report of the BIOMED-2 Concerted Action BMH4-CT98-3936. Leukemia. 2003;17(12):2257–2317. doi: 10.1038/sj.leu.2403202. [DOI] [PubMed] [Google Scholar]
- 35.He YF, Li BZ, Li Z, et al. Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science. 2011;333(6047):1303–1307. doi: 10.1126/science.1210944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ito S, Shen L, Dai Q, et al. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science. 2011;333(6047):1300–1303. doi: 10.1126/science.1210597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tahiliani M, Koh KP, Shen Y, et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324(5929):930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Patel JP, Gönen M, Figueroa ME, et al. Prognostic relevance of integrated genetic profiling in acute myeloid leukemia. N Engl J Med. 2012;366(12):1079–1089. doi: 10.1056/NEJMoa1112304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gaidzik VI, Paschka P, Späth D, et al. TET2 mutations in acute myeloid leukemia (AML): results from a comprehensive genetic and clinical analysis of the AML study group. J Clin Oncol. 2012;30(12):1350–1357. doi: 10.1200/JCO.2011.39.2886. [DOI] [PubMed] [Google Scholar]
- 40.Weissmann S, Alpermann T, Grossmann V, et al. Landscape of TET2 mutations in acute myeloid leukemia. Leukemia. 2012;26(5):934–942. doi: 10.1038/leu.2011.326. [DOI] [PubMed] [Google Scholar]
- 41.Hart DO, Raha T, Lawson ND, Green MR. Initiation of zebrafish haematopoiesis by the TATA-box-binding protein-related factor Trf3. Nature. 2007;450(7172):1082–1085. doi: 10.1038/nature06349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lin H, Grosschedl R. Failure of B-cell differentiation in mice lacking the transcription factor EBF. Nature. 1995;376(6537):263–267. doi: 10.1038/376263a0. [DOI] [PubMed] [Google Scholar]
- 43.Pongubala JM, Northrup DL, Lancki DW, et al. Transcription factor EBF restricts alternative lineage options and promotes B cell fate commitment independently of Pax5. Nat Immunol. 2008;9(2):203–215. doi: 10.1038/ni1555. [DOI] [PubMed] [Google Scholar]
- 44.Yamasaki N, Miyazaki K, Nagamachi A, et al. Identification of Zfp521/ZNF521 as a cooperative gene for E2A-HLF to develop acute B-lineage leukemia. Oncogene. 2010;29(13):1963–1975. doi: 10.1038/onc.2009.475. [DOI] [PubMed] [Google Scholar]
- 45.Adams S, van der Laan LJ, Vernon-Wilson E, et al. Signal-regulatory protein is selectively expressed by myeloid and neuronal cells. J Immunol. 1998;161(4):1853–1859. [PubMed] [Google Scholar]
- 46.Fujioka Y, Matozaki T, Noguchi T, et al. A novel membrane glycoprotein, SHPS-1, that binds the SH2-domain-containing protein tyrosine phosphatase SHP-2 in response to mitogens and cell adhesion. Mol Cell Biol. 1996;16(12):6887–6899. doi: 10.1128/mcb.16.12.6887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kharitonenkov A, Chen Z, Sures I, Wang H, Schilling J, Ullrich A. A family of proteins that inhibit signalling through tyrosine kinase receptors. Nature. 1997;386(6621):181–186. doi: 10.1038/386181a0. [DOI] [PubMed] [Google Scholar]
- 48.Seiffert M, Cant C, Chen Z, et al. Human signal-regulatory protein is expressed on normal, but not on subsets of leukemic myeloid cells and mediates cellular adhesion involving its counterreceptor CD47. Blood. 1999;94(11):3633–3643. [PubMed] [Google Scholar]
- 49.Barclay AN, Brown MH. The SIRP family of receptors and immune regulation. Nat Rev Immunol. 2006;6(6):457–464. doi: 10.1038/nri1859. [DOI] [PubMed] [Google Scholar]
- 50.Chan KS, Espinosa I, Chao M, et al. Identification, molecular characterization, clinical prognosis, and therapeutic targeting of human bladder tumor-initiating cells. Proc Natl Acad Sci USA. 2009;106(33):14016–14021. doi: 10.1073/pnas.0906549106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chao MP, Alizadeh AA, Tang C, et al. Therapeutic antibody targeting of CD47 eliminates human acute lymphoblastic leukemia. Cancer Res. 2011;71(4):1374–1384. doi: 10.1158/0008-5472.CAN-10-2238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Edris B, Weiskopf K, Volkmer AK, et al. Antibody therapy targeting the CD47 protein is effective in a model of aggressive metastatic leiomyosarcoma. Proc Natl Acad Sci USA. 2012;109(17):6656–6661. doi: 10.1073/pnas.1121629109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Theocharides AP, Jin L, Cheng PY, et al. Disruption of SIRPα signaling in macrophages eliminates human acute myeloid leukemia stem cells in xenografts. J Exp Med. 2012;209(10):1883–1899. doi: 10.1084/jem.20120502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Willingham SB, Volkmer JP, Gentles AJ, et al. The CD47-signal regulatory protein alpha (SIRPa) interaction is a therapeutic target for human solid tumors. Proc Natl Acad Sci USA. 2012;109(17):6662–6667. doi: 10.1073/pnas.1121623109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Beyer M, Thabet Y, Müller RU, et al. Repression of the genome organizer SATB1 in regulatory T cells is required for suppressive function and inhibition of effector differentiation. Nat Immunol. 2011;12(9):898–907. doi: 10.1038/ni.2084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cai S, Lee CC, Kohwi-Shigematsu T. SATB1 packages densely looped, transcriptionally active chromatin for coordinated expression of cytokine genes. Nat Genet. 2006;38(11):1278–1288. doi: 10.1038/ng1913. [DOI] [PubMed] [Google Scholar]
- 57.Stam RW, Schneider P, Hagelstein JA, et al. Gene expression profiling-based dissection of MLL translocated and MLL germline acute lymphoblastic leukemia in infants. Blood. 2010;115(14):2835–2844. doi: 10.1182/blood-2009-07-233049. [DOI] [PubMed] [Google Scholar]
- 58.Franzoni A, Passon N, Fabbro D, Tiribelli M, Damiani D, Damante G. Histone post-translational modifications associated to BAALC expression in leukemic cells. Biochem Biophys Res Commun. 2012;417(2):721–725. doi: 10.1016/j.bbrc.2011.12.013. [DOI] [PubMed] [Google Scholar]
- 59.Zhang H, Zhou Y, Rui Y, et al. Association between the polymorphism rs3217927 of CCND2 and the risk of childhood acute lymphoblastic leukemia in a Chinese population. PLoS ONE. 2014;9(4):e95059. doi: 10.1371/journal.pone.0095059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ishikawa F, Yasukawa M, Lyons B, et al. Development of functional human blood and immune systems in NOD/SCID/IL2 receptor gamma chain(null) mice. Blood. 2005;106(5):1565–1573. doi: 10.1182/blood-2005-02-0516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shultz LD, Lyons BL, Burzenski LM, et al. Human lymphoid and myeloid cell development in NOD/LtSz-scid IL2R gamma null mice engrafted with mobilized human hemopoietic stem cells. J Immunol. 2005;174(10):6477–6489. doi: 10.4049/jimmunol.174.10.6477. [DOI] [PubMed] [Google Scholar]
- 62.Ishikawa F, Yoshida S, Saito Y, et al. Chemotherapy-resistant human AML stem cells home to and engraft within the bone-marrow endosteal region. Nat Biotechnol. 2007;25(11):1315–1321. doi: 10.1038/nbt1350. [DOI] [PubMed] [Google Scholar]
- 63.Castor A, Nilsson L, Astrand-Grundström I, et al. Distinct patterns of hematopoietic stem cell involvement in acute lymphoblastic leukemia. Nat Med. 2005;11(6):630–637. doi: 10.1038/nm1253. [DOI] [PubMed] [Google Scholar]
- 64.Cox CV, Evely RS, Oakhill A, Pamphilon DH, Goulden NJ, Blair A. Characterization of acute lymphoblastic leukemia progenitor cells. Blood. 2004;104(9):2919–2925. doi: 10.1182/blood-2004-03-0901. [DOI] [PubMed] [Google Scholar]
- 65.Saito Y, Kitamura H, Hijikata A, et al. Identification of therapeutic targets for quiescent, chemotherapy-resistant human leukemia stem cells. Sci Transl Med. 2010;2(17):ra9. doi: 10.1126/scitranslmed.3000349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Iwasaki T, Takeda Y, Maruyama K, et al. Deletion of tetraspanin CD9 diminishes lymphangiogenesis in vivo and in vitro. J Biol Chem. 2013;288(4):2118–2131. doi: 10.1074/jbc.M112.424291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Nielsen PJ, Lorenz B, Müller AM, et al. Altered erythrocytes and a leaky block in B-cell development in CD24/HSA-deficient mice. Blood. 1997;89(3):1058–1067. [PubMed] [Google Scholar]
- 68.Takai T, Ono M, Hikida M, Ohmori H, Ravetch JV. Augmented humoral and anaphylactic responses in Fc gamma RII-deficient mice. Nature. 1996;379(6563):346–349. doi: 10.1038/379346a0. [DOI] [PubMed] [Google Scholar]
- 69.Kristiansen G, Denkert C, Schlüns K, Dahl E, Pilarsky C, Hauptmann S. CD24 is expressed in ovarian cancer and is a new independent prognostic marker of patient survival. Am J Pathol. 2002;161(4):1215–1221. doi: 10.1016/S0002-9440(10)64398-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kristiansen G, Winzer KJ, Mayordomo E, et al. CD24 expression is a new prognostic marker in breast cancer. Clin Cancer Res. 2003;9(13):4906–4913. [PubMed] [Google Scholar]
- 71.Nishida H, Yamazaki H, Yamada T, et al. CD9 correlates with cancer stem cell potentials in human B-acute lymphoblastic leukemia cells. Biochem Biophys Res Commun. 2009;382(1):57–62. doi: 10.1016/j.bbrc.2009.02.123. [DOI] [PubMed] [Google Scholar]
- 72.Bretz NP, Salnikov AV, Perne C, et al. CD24 controls Src/STAT3 activity in human tumors. Cell Mol Life Sci. 2012;69(22):3863–3879. doi: 10.1007/s00018-012-1055-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Maloney DG. Anti-CD20 antibody therapy for B-cell lymphomas. N Engl J Med. 2012;366(21):2008–2016. doi: 10.1056/NEJMct1114348. [DOI] [PubMed] [Google Scholar]
- 74.Grupp SA, Kalos M, Barrett D, et al. Chimeric antigen receptor-modified T cells for acute lymphoid leukemia. N Engl J Med. 2013;368(16):1509–1518. doi: 10.1056/NEJMoa1215134. [DOI] [PMC free article] [PubMed] [Google Scholar]