Abstract
Triggering receptor expressed on myeloid cells (TREM)-1 is an orphan receptor implicated in innate immune activation. Inhibition of TREM-1 reduces sepsis in mouse models, suggesting a role for it in immune responses triggered by bacteria. However, the absence of an identified ligand has hampered a full understanding of TREM-1 function. We identified complexes between peptidoglycan recognition protein 1 (PGLYRP1) and bacterially derived peptidoglycan that constitute a potent ligand capable of binding TREM-1 and inducing known TREM-1 functions. Interestingly, multimerization of PGLYRP1 bypassed the need for peptidoglycan in TREM-1 activation, demonstrating that the PGLYRP1/TREM-1 axis can be activated in the absence of bacterial products. The role for PGLYRP1 as a TREM-1 activator provides a new mechanism by which bacteria can trigger myeloid cells, linking two known, but previously unrelated, pathways in innate immunity.
Introduction
Triggering receptor expressed on myeloid cells (TREM)-1 is a known proinflammatory receptor expressed on monocytes/macrophages and neutrophils (1, 2), although its exact mechanism of activation is unknown. TREM-1 expression is upregulated during bacterial infection, and antagonizing TREM-1 with an inhibitory peptide prevents mortality in mouse models of sepsis (3). In contrast with other TREM family members, which bind directly to bacterially derived components (4), no such link has been established for TREM-1. In the absence of a known ligand, TREM-1–induced responses have been studied using agonistic anti–TREM-1 Abs, demonstrating that TREM-1 cross-linking induces cellular activation, cytokine production, and amplification of inflammatory responses (1, 2). A more complete understanding of TREM-1 function, as well as its relevance to disease, requires the identification of the physiological ligand for TREM-1.
In this article, we describe the identification of peptidoglycan (PGN) recognition protein 1 (PGLYRP1) as a ligand for TREM-1. When multimerized or complexed with PGN, PGLYRP1 is able to activate TREM-1 and enhance cytokine production in human neutrophils and macrophages.
Materials and Methods
Abs and other reagents
Anti–TREM-1 (MAB1278), anti-PGLYRP1 polyclonal Ab, and isotype controls were from R&D Systems. Anti–TREM-1 (clone 9E2) (1) was purified from hybridoma supernatant. Anti-PGLYRP1 mAb was generated by immunizing mice with full-length human PGLYRP1. Anti-PGLYRP1 and anti-TNP (as isotype-matched control) were recombinantly expressed as human (h)IgG4 chimeric mAbs.
TREM-1–Fc, TREM-1 tetramer, and CD83 tetramer constructs, consisting of a CD33 leader peptide, followed by the extracellular domain of the corresponding protein (tandem domains for tetramers) fused to a mutated hIgG1 Fc with reduced Fc effector function (E216G, C220S, L234A, L235E, G237A, A330S, P331S), were expressed and purified from HEK293-6E cells.
Recombinant hPGLYRP1 was purchased from R&D Systems or made at Novo Nordisk. No major differences in potency were observed when comparing different sources of PGLYRP1. The following TLR ligands (TLRLs) were used: LPS, polyinosinic-polycytidylic acid (both from Sigma-Aldrich), PGN-SA, PGN-BS, PGN-ECndi, PGN-ECndss, FSL-1, Tri-DAP, M-TriDAP, and muramyl dipeptide (MDP) (all from InvivoGen). Lysozyme and mutanolysin were from Sigma-Aldrich. hM-CSF and hIL-4 were expressed and purified from Escherichia coli. All proteins were tested for endotoxins using the Limulus amebocyte lysate assay and confirmed to contain <1 EU/mg endotoxin.
Cells and cell lines
The BWZ.36/ TREM-1 reporter cell line was derived from BWZ.36 cells, which express an NFAT-driven LacZ reporter (5), by transfection with hTREM-1 and hDAP12. LacZ activation was measured using the Beta-Glo kit (Promega). Stimulation with 10 ng/ml PMA + 1 μM thapsigargin (both from Sigma-Aldrich) confirmed a functional NFAT reporter. For coculture stimulations, freshly purified human neutrophils were incubated overnight with the indicated stimulants before adding the TREM-1 reporter cells.
HEK293-6E cells expressing a membrane-bound PGLYRP1 were generated by transient transfection with a construct encoding PGLYRP1 fused to the intracellular and transmembrane domain of the type II receptor MDL-1 and with DAP12 (Fig. 3B). In Fig. 2B, the charged lysine residue in the transmembrane domain was changed to leucine to eliminate the need for DAP12 cotransfection. Binding of TREM-1–Fc to PGLYRP1 cells was analyzed by flow cytometry, using anti-human IgG-PE as secondary Ab (Jackson ImmunoResearch) and gating on live cells.
FIGURE 3.
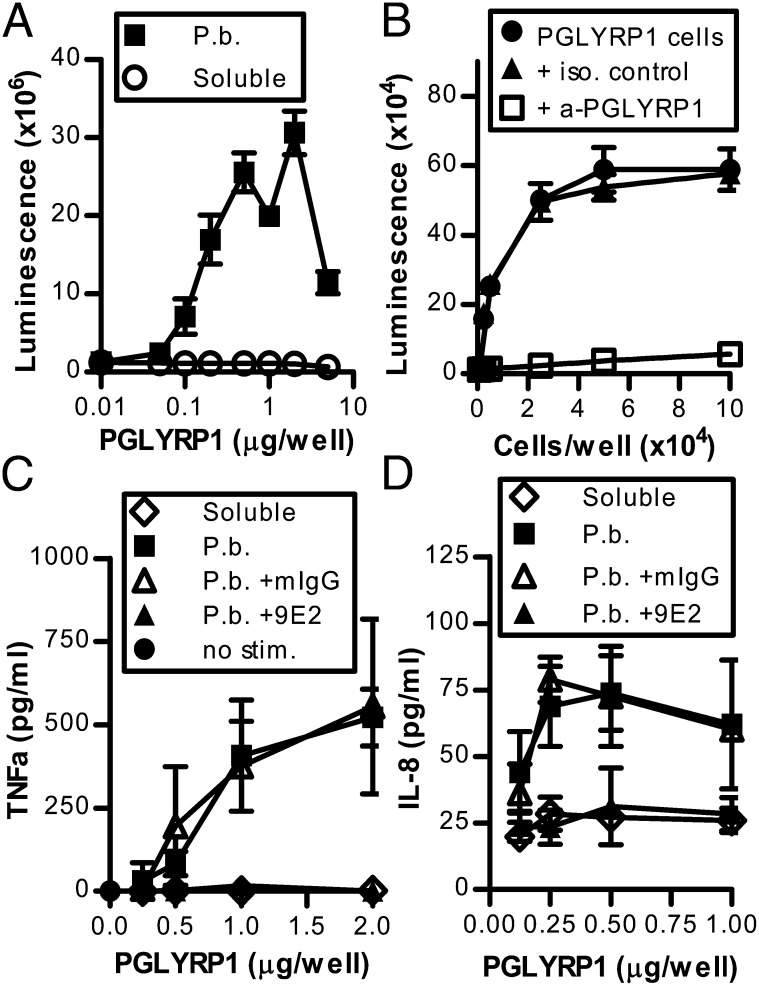
PGLYRP1 induces TREM-1 activation independently of PGN. (A) Stimulation of the TREM-1 reporter cell line with soluble or pb PGLYRP1 in the absence of PGN. Values are mean ± SD of duplicate samples. Data are representative of three independent experiments. (B) Stimulation of the TREM-1 reporter cell line with HEK293 cells expressing membrane-bound PGLYRP1. Values are mean ± SD of duplicate samples. Data are representative of three independent experiments. (C) Human macrophages stimulated with soluble or pb PGLYRP1 and inhibited with anti–TREM-1 9E2 or isotype control. Data show measurements of TNF-α in culture supernatants (mean ± SD of triplicate samples) and are representative of three independent experiments. (D) Purified neutrophils stimulated with soluble or pb PGLYRP1 and inhibited with anti–TREM-1 9E2 or isotype control. Data show measurements of IL-8 in culture supernatants (mean ± SD of triplicate samples) and are representative of two independent experiments.
FIGURE 2.
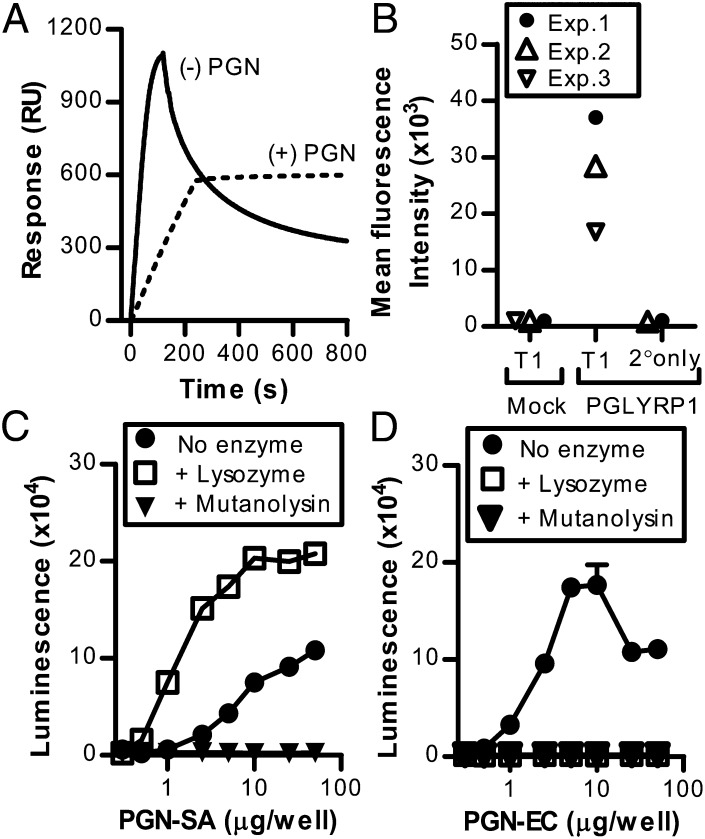
Role of PGN in the TREM-1–PGLYRP1 interaction. (A) Surface plasmon resonance analysis of PGLYRP1 binding to immobilized TREM-1 in the presence or absence of PGN-EC. Values are response units (RU). Shown is one experiment. (B) Flow cytometry analysis of TREM-1–Fc or secondary Ab–alone binding to either mock- or membrane-bound PGLYRP1-transfected cells. Values are single samples from three independent experiments. Stimulation of the TREM-1 reporter cell line with soluble PGLYRP1 in the presence of PGN-SA (C) or PGN-EC (D) pretreated with the digestive enzymes mutanolysin or lysozyme. Values are mean ± SD of triplicate samples. Data are representative of two independent experiments.
Primary human cells were purified from whole blood (Danish Blood Donor Core [Ethical approval no. H-D-2007-0055] or Astarte Biologics), from buffy coats (Danish Blood Donor Core [Ethical approval no. H-D-2008-113]), or from leukopheresis (Research Blood Components).
Cellular assays
Primary neutrophils were purified by centrifugation on Ficoll-Paque (Pharmacia), followed by sedimentation in 4% dextran (Sigma-Aldrich). Remaining erythrocytes were lysed using 0.2% NaCl. TLRLs were used at 10 μg/ml (PGN-SA, FSL-1, TriDAP, MDP, M-TriDAP), 1 μg/ml (polyinosinic-polycytidylic acid), or 100 ng/ml (LPS). After overnight culture, supernatants were analyzed for IL-8 using BioPlex (Bio-Rad).
Primary monocytes were purified using RosetteSep (STEMCELL Technologies) and differentiated into macrophages by culturing for 6 d with 50 ng/ml hM-CSF, followed by 1 d of culture with 40 ng/ml hIL-4. Macrophages were recovered using 5 mM EDTA (Life Technologies), reseeded, and cultured overnight with either plate-bound (pb) PGLYRP1 or 3 μg/ml PGN-BS plus 1 μg/ml PGLYRP1, and the supernatants were analyzed for TNF-α using BioPlex (Bio-Rad) or ELISA (R&D Systems). All monocyte/macrophage incubations were done under hypoxic conditions (2% O2) to enhance TREM-1 expression, as previously described for monocyte differentiation to dendritic cells (6). TREM-1 upregulation was confirmed by flow cytometric analysis.
XL immunoprecipitation mass spectrometry
XL immunoprecipitation mass spectrometry (IPMS) was done, essentially as described previously (7), using TREM-1 tetramer as bait and CD83 tetramer as control protein to immunoprecipitate proteins from neutrophil lysates. Precipitated proteins were trypsin digested and analyzed by liquid chromatography tandem mass spectrometry using an LTQ Orbitrap XL mass spectrometer (Thermo Scientific). The data were searched using SEQUEST-Sorcerer engine, and protein IDs were filtered with a false discovery rate of 1%.
Surface plasmon resonance analysis
Assays were run in 1× HBS-P, 25°C, at 20–30 μl/min on a Biacore T200 (GE Healthcare). TREM-1 tetramer (4641.1 RU) was coupled to a CM5 chip, and 150 nM PGLYRP1 was injected over the surface in the presence or absence of 10 μg/ml PGN-ECndss. As a negative control for nonspecific binding, a neutralized CM5 surface was run in parallel with the injected reagents and was subtracted to obtain the depicted binding data.
Results and Discussion
PGN-stimulated neutrophils activate TREM-1 via PGLYRP1
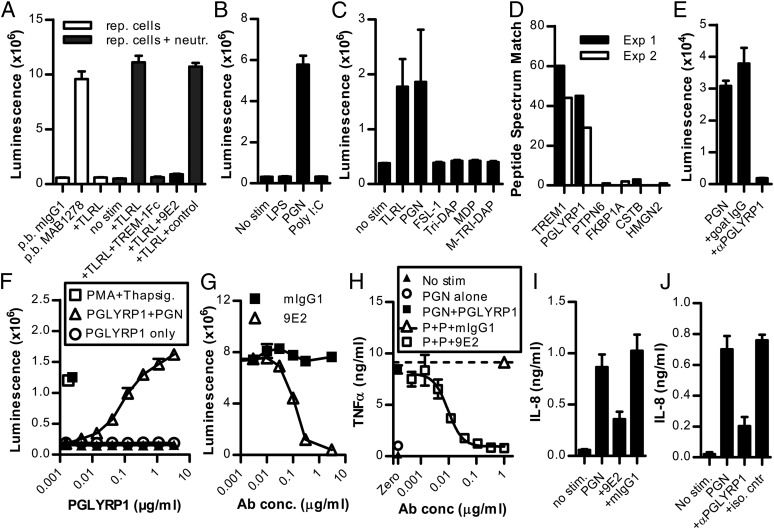
An endogenous ligand for TREM-1 was suggested to exist on neutrophils activated by bacteria or TLRLs (8). Extending this work, we tested TLRL-stimulated human neutrophils for their ability to activate a TREM-1–expressing reporter cell line. As shown in Fig. 1A, neutrophils stimulated with a TLRL mixture triggered TREM-1 to a similar extent as did an agonistic anti–TREM-1 mAb, whereas stimulation with only TLRL did not induce TREM-1 activation. The neutrophil-induced activity was specific to TREM-1 because it could be blocked by both TREM-1–Fc fusion protein and the anti–TREM-1 mAb 9E2. Although this anti–TREM-1 mAb was used previously by other investigators as a pb Ab to stimulate TREM-1 (1), we consistently find that it behaves as an antagonist when soluble. Among the components in the TLRL mixture, only PGN induced the neutrophil-mediated stimulation of TREM-1 (Fig. 1B), whereas other TLR or NOD1/2 ligands were ineffective (Fig. 1C). This suggested that the role of PGN in neutrophil-mediated TREM-1 activation was not simply a requirement for concurrent TLR2/NOD1/2 stimulation but specifically supported ligand induction or function.
FIGURE 1.
Neutrophils activate TREM-1 via PGLYRP1 and PGN. (A) Stimulation of TREM-1 reporter cells with pb anti–TREM-1 (MAB1278) or with neutrophils plus TLRL mix, in the presence or absence of TREM-1–Fc protein, anti–TREM-1 9E2, or matched IgG control. Values are mean ± SD of triplicate samples. Data are representative of >10 independent experiments. (B and C) TREM-1 reporter cells stimulated with neutrophils plus individual TLRL. Values are mean ± SD of triplicate samples. Shown is one experiment out of two (B) or three (C) independent experiments. (D) The top six peptides identified by XL IPMS with TREM-1 tetramer bound to PGN-treated neutrophils. Values are the spectral peptide counts from two independent experiments. (E) Stimulation of TREM-1 reporter cells with PGN-treated neutrophils and inhibition with anti-PGLYRP1 or control Ab. Values are mean ± SD of duplicate samples. Data are representative of six independent experiments. (F) Stimulation of BWZ.36/TREM-1 (open symbols) or parental BWZ.36 (filled symbols) reporter cells with soluble PGLYRP1 plus PGN-EC. Values are mean ± SD of quadruplicate samples. Data are representative of three independent experiments. (G) Blockade of PGLYRP1+PGN-induced TREM-1 reporter cell activation by anti–TREM-1 9E2. Values are mean ± SD of triplicate samples. Data are representative of five independent experiments. (H) Measurement of TNF-α in supernatants from macrophages stimulated with PGN-BS plus soluble PGLYRP1 and inhibited with anti-TREM-1 9E2 or isotype control. Values are mean ± SD of duplicate samples. Data are representative of >10 independent experiments. (I and J) Measurement of IL-8 in supernatants from purified neutrophils stimulated with PGN-SA. Inhibition with anti–TREM-1 9E2 (I) or anti-PGLYRP1 mAb (J). Values are mean ± SD of triplicate samples. Data are representative of four (I) or two (J) independent experiments.
To identify the putative neutrophil-expressed TREM-1 ligand, soluble TREM-1 or a control protein were cross-linked to neutrophils cultured with PGN. Resulting complexes were isolated by immunoprecipitation and identified by mass spectrometry in two independent experiments (XL IPMS) to contain PGLYRP1 peptides (Fig. 1D). In line with this, an anti-PGLYRP1 Ab could specifically block the neutrophil-mediated activation of the TREM-1 reporter cell line (Fig. 1E), supporting the conclusion that PGLYRP1 is the TREM-1–stimulating protein on neutrophils.
PGLYRP1 forms a TREM-1 ligand complex with PGN
PGLYRP1 is an antibacterial protein found in neutrophil tertiary granules, and it belongs to a family of PGN-binding proteins (PGRPs) that is highly conserved among insects and mammals (9). In Drosophila, PGRPs function as pattern-recognition proteins that bind bacterial PGN and activate Toll and Imd pathways to initiate immune responses (10). Mammalian PGRPs have primarily been described as effector molecules with direct bactericidal activity (9). Although PGLYRP1 was reported to be involved in neutrophil production of reactive oxygen species (11), as well as modulation of immune responses in vivo (12), the underlying mechanism has remained undefined. Using TREM-1 reporter cells we found that soluble PGLYRP1 could stimulate TREM-1, but only in the presence of PGN (Fig. 1F), and this could be blocked by anti–TREM-1 (Fig. 1G). This suggests that neutrophil-released PGLYRP1, when present with PGN, constitutes a functional ligand for TREM-1.
PGN is a macromolecular component in the cell wall of all bacteria and is known to bind PGLYRP1 (13). Most myeloid cells express the PGN-sensing receptors TLR2 and NOD1/2. Although TREM-1 does not bind bacterial components directly (4), our findings suggest that TREM-1 is a bacteria-sensing receptor on myeloid cells, exploiting PGLYRP1 for specific recognition of PGN. Thus, the presence of PGLYRP1 can regulate whether PGN is able to induce TREM-1 activation. In support of this, we found that the addition of PGLYRP1 to PGN-stimulated primary human macrophages increased TNF-α secretion >7-fold; notably, this response was blocked by anti–TREM-1 (Fig. 1H).
Neutrophils, like macrophages, express both TLRs and TREM-1. When neutrophils degranulate upon stimulation, granule proteins, such as PGLYRP1, are released (11). We hypothesized that PGLYRP1 released by PGN-stimulated neutrophils binds PGN to form ligand complexes capable of activating adjacent TREM-1–expressing cells, thus enhancing the response. Indeed, when we examined IL-8 responses from PGN-stimulated neutrophils, blockade of either TREM-1 (Fig. 1I) or PGLYRP1 (Fig. 1J) partially inhibited IL-8 release, confirming a contribution of both PGLYRP1 and TREM-1 to PGN-induced responses in neutrophils. The finding that PGLYRP1 combined with PGN triggers TREM-1 provides a novel mechanism for how bacteria induce TREM-1 activation, and it may provide an explanation for why TREM-1 blockade reduces mortality in mouse models of sepsis (3).
Multimeric PGN is required for PGN-PGLYRP1–mediated TREM-1 activation
To further investigate the role of PGN in PGLYRP1-mediated TREM-1 activation, we examined the interaction of PGN, TREM-1, and PGLYRP1. Using surface plasmon resonance analysis, we found that PGLYRP1 bound transiently to immobilized TREM-1 (Fig. 2A). Interestingly, despite PGN being critical for soluble PGLYRP1 activating TREM-1 (Fig. 1F), PGN only had a stabilizing effect, and it was not essential for the interaction (Fig. 2A). This PGN independence was confirmed using flow cytometry, showing TREM-1–Fc binding to HEK293 cells expressing a membrane-bound PGLYRP1 but not to mock-transfected cells (Fig. 2B). Thus, PGLYRP1 and TREM-1 can bind each other, and although PGN can enhance the binding, it is not essential for the interaction. No direct binding of PGN to TREM-1 was observed (data not shown), arguing against PGN being a direct ligand for TREM-1.
Because PGN is a complex macromolecule, we hypothesized that PGLYRP1 might require multimerization to stimulate TREM-1 and that PGN functions as a scaffold enabling this. To address this, we tested enzymatically digested PGN together with PGLYRP1 in the reporter cell assay. In agreement with the hypothesis, the ability of PGN to support PGLYRP1-mediated TREM-1 activation was eliminated by mutanolysin treatment, which fully degrades both E. coli–derived PGN (PGN-ECndi) and Staphylococcus aureus–derived PGN (PGN-SA), whereas lysozyme treatment results in full degradation of PGN-ECndi but not PGN-SA (Fig. 2C, 2D) (14). Furthermore, the PGN subunit MDP could not substitute for PGN in PGLYRP1-mediated TREM-1 activation (data not shown). Lysozyme digestion consistently increased the PGN-SA–mediated PGLYRP1–TREM-1 signal (Fig. 2C). Although not fully understood, we observed that fractionation of PGN species by size impacts the ability of the PGN to support PGLYRP1–TREM-1 activation (data not shown). Lysozyme may result in a partial digestion of PGN-SA, thus affecting the signaling strength.
PGLYRP1 alone can stimulate TREM-1 on myeloid cells
To further address the role of PGN during PGLYRP1–TREM-1 signaling, we constructed different multimeric formats of PGLYRP1. We found that pb PGLYRP1 alone (Fig. 3A), as well as HEK293 cells expressing PGLYRP1 on the cell surface (Fig. 3B), was able to activate TREM-1. Thus, although stimulation with soluble PGLYRP1 alone did not induce a signal (Fig. 1F), PGLYRP1 presented in either a pb form or anchored to a cell surface was a potent stimulant of TREM-1.
As seen with TREM-1 reporter cells, human macrophages responded to pb PGLYRP1 in a TREM-1–dependent manner (Fig. 3C), although the response was lower than the one to PGN plus PGLYRP1 (Fig. 1H). The lower response most likely reflects the synergy between TREM-1 and a concurrent TLR stimulation (1, 2), but it also may be caused by differences in the PGLYRP1–TREM-1 interaction when artificially immobilizing PGLYRP1 on a plastic surface. Human neutrophils stimulated with pb PGLYRP1 produced both IL-8 (Fig. 3D) and superoxide (data not shown), consistent with published data using pb agonistic anti–TREM-1 (2). PGLYRP1-induced responses in both macrophages and neutrophils could be inhibited with the anti–TREM-1 mAb, confirming the role of TREM-1 (Fig. 3C, 3D). Taken together, these data support the hypothesis that multimeric presentation of PGLYRP1 is sufficient for TREM-1 activation in myeloid cells and raise the possibility that TREM-1 activation also occurs in the absence of infections.
In conclusion, we present strong evidence that PGLYRP1 is a functional ligand for TREM-1. PGLYRP1 is a well-known neutrophil granule protein with antibacterial properties (9) that is present at readily detectable levels in plasma from healthy individuals (15) (data not shown). Our results show that PGLYRP1 must be multimerized to become a functional TREM-1 ligand. This can be achieved in the presence of PGN, thus providing a likely explanation for how TREM-1 activation is induced during bacterial infection. The need for cross-linking of PGLYRP1 also could be a safeguard ensuring that TREM-1 on circulating myeloid cells is not inappropriately stimulated. Interestingly, PGLYRP1 was shown to be present in neutrophil extracellular traps (16), suggesting another potential mechanism for PGLYRP1 multimerization and TREM-1 activation and opening new areas of research into the role of TREM-1 in disease.
Acknowledgments
We thank L. Madsen, B. Davidsen, B. Breuning, B. Stegmann, P. Birn, C. Wiberg, N. Rasmussen, T.-Y. Chao, Y. Luo, Z. Yang, T. Dong, Z. Guan, C. Hauskins, and A. Yabannavar for technical expertise and protein production. We also thank B. Ursø, F. Ramsdell, J. Peschon, P. Kong, H. Stennicke, and S. Read for discussions and scientific input during this work.
Footnotes
- h
- human
- IPMS
- immunoprecipitation mass spectrometry
- MDP
- muramyl dipeptide
- pb
- plate-bound
- PGLYRP1
- PGN recognition protein 1
- PGN
- peptidoglycan
- PGRP
- PGN-binding protein
- TLRL
- TLR ligand
- TREM
- triggering receptor expressed on myeloid cells.
Disclosures
The findings described in this article are part of a patent application submitted by Novo Nordisk A/S. All authors are or have been employed by Novo Nordisk A/S.
References
- 1.Bleharski J. R., Kiessler V., Buonsanti C., Sieling P. A., Stenger S., Colonna M., Modlin R. L. 2003. A role for triggering receptor expressed on myeloid cells-1 in host defense during the early-induced and adaptive phases of the immune response. J. Immunol. 170: 3812–3818. [DOI] [PubMed] [Google Scholar]
- 2.Radsak M. P., Salih H. R., Rammensee H. G., Schild H. 2004. Triggering receptor expressed on myeloid cells-1 in neutrophil inflammatory responses: differential regulation of activation and survival. J. Immunol. 172: 4956–4963. [DOI] [PubMed] [Google Scholar]
- 3.Gibot S., Kolopp-Sarda M. N., Béné M. C., Bollaert P. E., Lozniewski A., Mory F., Levy B., Faure G. C. 2004. A soluble form of the triggering receptor expressed on myeloid cells-1 modulates the inflammatory response in murine sepsis. J. Exp. Med. 200: 1419–1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cannon J. P., O’Driscoll M., Litman G. W. 2012. Specific lipid recognition is a general feature of CD300 and TREM molecules. Immunogenetics 64: 39–47. [DOI] [PubMed] [Google Scholar]
- 5.Sanderson S., Shastri N. 1994. LacZ inducible, antigen/MHC-specific T cell hybrids. Int. Immunol. 6: 369–376. [DOI] [PubMed] [Google Scholar]
- 6.Bosco M. C., Pierobon D., Blengio F., Raggi F., Vanni C., Gattorno M., Eva A., Novelli F., Cappello P., Giovarelli M., Varesio L. 2011. Hypoxia modulates the gene expression profile of immunoregulatory receptors in human mature dendritic cells: identification of TREM-1 as a novel hypoxic marker in vitro and in vivo. Blood 117: 2625–2639. [DOI] [PubMed] [Google Scholar]
- 7.Brandt C. S., Baratin M., Yi E. C., Kennedy J., Gao Z., Fox B., Haldeman B., Ostrander C. D., Kaifu T., Chabannon C., et al. 2009. The B7 family member B7-H6 is a tumor cell ligand for the activating natural killer cell receptor NKp30 in humans. J. Exp. Med. 206: 1495–1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gibot S., Buonsanti C., Massin F., Romano M., Kolopp-Sarda M. N., Benigni F., Faure G. C., Béné M. C., Panina-Bordignon P., Passini N., Lévy B. 2006. Modulation of the triggering receptor expressed on the myeloid cell type 1 pathway in murine septic shock. Infect. Immun. 74: 2823–2830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dziarski R., Gupta D. 2010. Review: Mammalian peptidoglycan recognition proteins (PGRPs) in innate immunity. Innate Immun 16: 168–174. [DOI] [PubMed] [Google Scholar]
- 10.Kurata S. 2014. Peptidoglycan recognition proteins in Drosophila immunity. Dev. Comp. Immunol. 42: 36–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dziarski R., Platt K. A., Gelius E., Steiner H., Gupta D. 2003. Defect in neutrophil killing and increased susceptibility to infection with nonpathogenic gram-positive bacteria in peptidoglycan recognition protein-S (PGRP-S)-deficient mice. Blood 102: 689–697. [DOI] [PubMed] [Google Scholar]
- 12.Park S. Y., Jing X., Gupta D., Dziarski R. 2013. Peptidoglycan recognition protein 1 enhances experimental asthma by promoting Th2 and Th17 and limiting regulatory T cell and plasmacytoid dendritic cell responses. J. Immunol. 190: 3480–3492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kumar S., Roychowdhury A., Ember B., Wang Q., Guan R., Mariuzza R. A., Boons G. J. 2005. Selective recognition of synthetic lysine and meso-diaminopimelic acid-type peptidoglycan fragments by human peptidoglycan recognition proteins Iα and S. J. Biol. Chem. 280: 37005–37012. [DOI] [PubMed] [Google Scholar]
- 14.Bera A., Herbert S., Jakob A., Vollmer W., Götz F. 2005. Why are pathogenic staphylococci so lysozyme resistant? The peptidoglycan O-acetyltransferase OatA is the major determinant for lysozyme resistance of Staphylococcus aureus. Mol. Microbiol. 55: 778–787. [DOI] [PubMed] [Google Scholar]
- 15.Liu T., Qian W.-J., Gritsenko M. A., Xiao W., Moldawer L. L., Kaushal A., Monroe M. E., Varnum S. M., Moore R. J., Purvine S. O., et al. Inflammation and the Host Response to Injury Large Scale Collaborative Research Programm 2006. High dynamic range characterization of the trauma patient plasma proteome. Mol. Cell. Proteomics 5: 1899–1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cho J. H., Fraser I. P., Fukase K., Kusumoto S., Fujimoto Y., Stahl G. L., Ezekowitz R. A. B. 2005. Human peptidoglycan recognition protein S is an effector of neutrophil-mediated innate immunity. Blood 106: 2551–2558. [DOI] [PMC free article] [PubMed] [Google Scholar]