Abstract
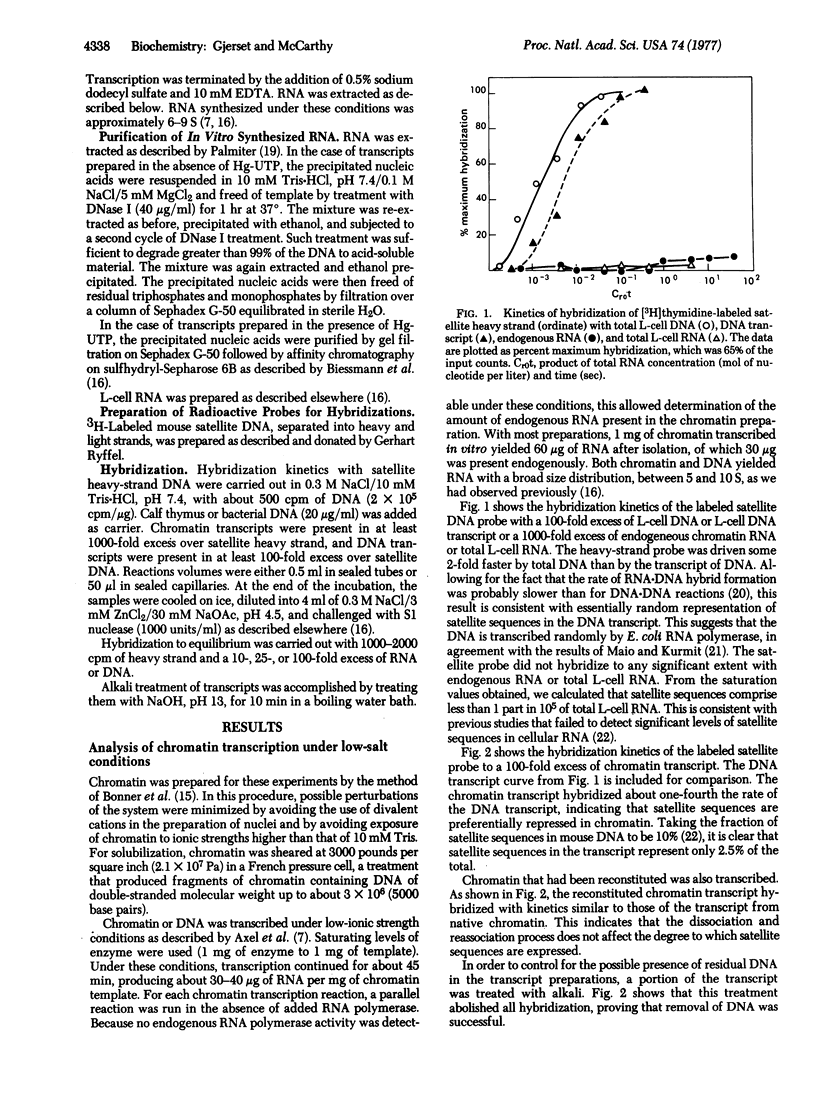
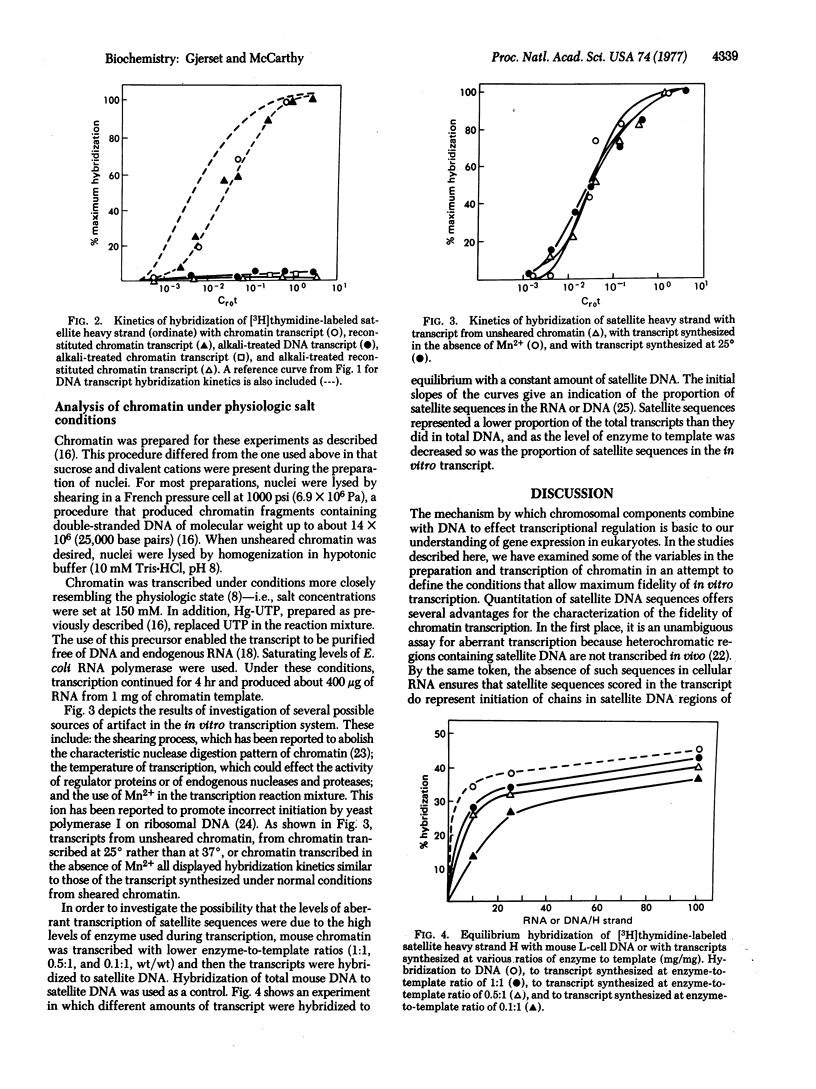
An attempt was made to elucidate some of the factors influencing the fidelity with which isolated chromatin from mouse L-cells is transcribed by RNA polymerase from Escherichia coli by analyzing the in vitro transcript for the presence of satellite sequences. These sequences are absent from cellular RNA and therefore reflect aberrant transcription. The results indicate that satellite sequences are underrepresented in chromatin transcripts relative to those of DNA. This selectivity is insensitive to many variables in procedures for the isolation and transcription of chromatin. However, lowering the ratio of enzyme to template further reduced the proportion of satellite sequences in the transcript. We conclude that a primary factor influencing the extent of aberrant transcription is the level of enzyme used. Under limiting enzyme conditions, an efficient selection against satellite sequences is observed. However, under conditions of enzyme excess, the enzyme initiates chains at weaker secondary promoters localized in regions of the chromatin containing satellite DNA.
Full text
PDF

Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Astrin S. M. Mapping of the SV40 specific sequences transcribed in vitro from chromatin of SV40 transformed cells. Biochemistry. 1975 Jun 17;14(12):2700–2704. doi: 10.1021/bi00683a022. [DOI] [PubMed] [Google Scholar]
- Axel R., Cedar H., Felsenfeld G. Synthesis of globin ribonucleic acid from duck-reticulocyte chromatin in vitro. Proc Natl Acad Sci U S A. 1973 Jul;70(7):2029–2032. doi: 10.1073/pnas.70.7.2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bekhor I., Kung G. M., Bonner J. Sequence-specific interaction of DNA and chromosomal protein. J Mol Biol. 1969 Jan;39(2):351–364. doi: 10.1016/0022-2836(69)90322-2. [DOI] [PubMed] [Google Scholar]
- Biessmann H., Gjerset R. A., Levy B., McCarthy B. J. Fidelity of chromatin transcription in vitro. Biochemistry. 1976 Oct 5;15(20):4356–4363. doi: 10.1021/bi00665a002. [DOI] [PubMed] [Google Scholar]
- Burgess R. R. A new method for the large scale purification of Escherichia coli deoxyribonucleic acid-dependent ribonucleic acid polymerase. J Biol Chem. 1969 Nov 25;244(22):6160–6167. [PubMed] [Google Scholar]
- Dale R. M., Martin E., Livingston D. C., Ward D. C. Direct covalent mercuration of nucleotides and polynucleotides. Biochemistry. 1975 Jun 3;14(11):2447–2457. doi: 10.1021/bi00682a027. [DOI] [PubMed] [Google Scholar]
- Flamm W. G., Walker P. M., McCallum M. Some properties of the single strands isolated from the DNA of the nuclear satellite of the mouse (Mus musculus). J Mol Biol. 1969 Mar 28;40(3):423–443. doi: 10.1016/0022-2836(69)90163-6. [DOI] [PubMed] [Google Scholar]
- Gottesfeld J. M., Garrard W. T., Bagi G., Wilson R. F., Bonner J. Partial purification of the template-active fraction of chromatin: a preliminary report. Proc Natl Acad Sci U S A. 1974 Jun;71(6):2193–2197. doi: 10.1073/pnas.71.6.2193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honjo T., Reeder R. H. Transcription of Xenopus chromatin by homologous ribonucleic acid polymerase: aberrant synthesis of ribosomal and 5S ribonucleic acid. Biochemistry. 1974 Apr 23;13(9):1896–1899. doi: 10.1021/bi00706a018. [DOI] [PubMed] [Google Scholar]
- Huang R. C., Huang P. C. Effect of protein-bound RNA associated with chick embryo chromatin on template specificity of the chromatin. J Mol Biol. 1969 Jan;39(2):365–378. doi: 10.1016/0022-2836(69)90323-4. [DOI] [PubMed] [Google Scholar]
- Hutton J. R., Wetmur J. G. Renaturation of bacteriophage phiX174 DNA-RNA hybrid: RNA length effect and nucleation rate constant. J Mol Biol. 1973 Jul 15;77(4):495–500. doi: 10.1016/0022-2836(73)90218-0. [DOI] [PubMed] [Google Scholar]
- Kleiman L., Huang R. C. Reconstitution of chromatin. The sequential binding of histones to DNA in the presence of salt and urea. J Mol Biol. 1972 Feb 28;64(1):1–8. doi: 10.1016/0022-2836(72)90317-8. [DOI] [PubMed] [Google Scholar]
- Maio J. J., Kurnit D. M. Transcription of mammalian satellite DNAs by homologous DNA-dependent RNA polymerases. Biochim Biophys Acta. 1974 May 31;349(3):305–319. doi: 10.1016/0005-2787(74)90118-x. [DOI] [PubMed] [Google Scholar]
- Marsh J. L., McCarthy B. J. Analysis of DNA-RNA hybridization data using the Scatchard plot. Biochem Biophys Res Commun. 1973 Dec 10;55(3):805–811. doi: 10.1016/0006-291x(73)91215-1. [DOI] [PubMed] [Google Scholar]
- Noll M., Thomas J. O., Kornberg R. D. Preparation of native chromatin and damage caused by shearing. Science. 1975 Mar 28;187(4182):1203–1206. doi: 10.1126/science.187.4182.1203. [DOI] [PubMed] [Google Scholar]
- Palmiter R. D. Magnesium precipitation of ribonucleoprotein complexes. Expedient techniques for the isolation of undergraded polysomes and messenger ribonucleic acid. Biochemistry. 1974 Aug 13;13(17):3606–3615. doi: 10.1021/bi00714a032. [DOI] [PubMed] [Google Scholar]
- Paul J., Gilmour R. S., Affara N., Birnie G., Harrison P., Hell A., Humphries S., Windass J., Young B. The globin gene: structure and expression. Cold Spring Harb Symp Quant Biol. 1974;38:885–890. doi: 10.1101/sqb.1974.038.01.090. [DOI] [PubMed] [Google Scholar]
- Paul J., Gilmour R. S. Organ-specific restriction of transcription in mammalian chromatin. J Mol Biol. 1968 Jul 14;34(2):305–316. doi: 10.1016/0022-2836(68)90255-6. [DOI] [PubMed] [Google Scholar]
- Reeder R. H. Transcription of chromatin by bacterial RNA polymerase. J Mol Biol. 1973 Oct 25;80(2):229–241. doi: 10.1016/0022-2836(73)90169-1. [DOI] [PubMed] [Google Scholar]
- Smith K. D., Church R. B., McCarthy B. J. Template specificity of isolated chromatin. Biochemistry. 1969 Nov;8(11):4271–4277. doi: 10.1021/bi00839a007. [DOI] [PubMed] [Google Scholar]
- Smith M. M., Huang R. C. Transcription in vitro of immunoglobulin kappa light chain genes in isolated mouse myeloma nuclei and chromatin. Proc Natl Acad Sci U S A. 1976 Mar;73(3):775–779. doi: 10.1073/pnas.73.3.775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein G., Park W., Thrall C., Mans R., Stein J. Regulation of cell cycle stage-specific transcription of histone genes from chromatin by non-histone chromosomal proteins. Nature. 1975 Oct 30;257(5529):764–767. doi: 10.1038/257764a0. [DOI] [PubMed] [Google Scholar]
- Tan C. H., Miyagi M. Specificity of transcription of chromatin in vitro. J Mol Biol. 1970 Jun 28;50(3):641–653. doi: 10.1016/0022-2836(70)90090-2. [DOI] [PubMed] [Google Scholar]
- Van Keulen H., Planta R. J., Retèl J. Structure and transcription specificity of yeast RNA polymerase A. Biochim Biophys Acta. 1975 Jun 16;395(2):179–190. doi: 10.1016/0005-2787(75)90157-4. [DOI] [PubMed] [Google Scholar]
- Weintraub H., Groudine M. Chromosomal subunits in active genes have an altered conformation. Science. 1976 Sep 3;193(4256):848–856. doi: 10.1126/science.948749. [DOI] [PubMed] [Google Scholar]
- Wilson G. N., Steggles A. W., Kantor J. A., Nienhuis A. W., Anderson W. F. Cell-free transcription of mammalian chromatin. Quantitative measurement of newly synthesized globin messenger RNA sequences. J Biol Chem. 1975 Nov 25;250(22):8604–8613. [PubMed] [Google Scholar]


