Abstract
The use of genome-wide proteomic and RNA interference approaches has moved our understanding of signal transduction from linear pathways to highly integrated networks centered on core nodes. However, probing the dynamics of flow of information through such networks remains technically challenging. In particular, how the temporal dynamics of an individual pathway can elicit distinct outcomes in a single cell type and how multiple pathways may interact sequentially or synchronously to influence cell fate remain open questions in many contexts. The development of fluorescence-based reporters and optogenetic regulators of pathway activity enables the analysis of signaling in living cells and organisms with unprecedented spatiotemporal resolution and holds the promise of addressing these key questions. We present a brief overview of the evidence for the importance of temporal dynamics in cellular regulation, introduce these fluorescence-based tools, and highlight specific studies that leveraged these tools to probe the dynamics of information flow through signaling networks. In particular, we highlight two studies in Caenorhabditis elegans sensory neurons and cultured mammalian cells that demonstrate the importance of signal dynamics in determining cellular responses.
How Temporal Dynamics and Signal Crosstalk Influence Cell Behavior
Communication between cells is critical to both the development and homeostasis of multicellular organisms. Cells must respond appropriately to their surroundings by adopting a particular fate or adapting their behavior in response to challenges. How intercellular signaling defines cell fate or behavioral outcomes is therefore a critical question in cell and developmental biology. A simple explanation would be that a specific pathway is responsible for each specific fate or behavior. However, many years of studies have only identified a small number of core signaling pathways, for example, the epidermal growth factor (EGF) pathway, the fibroblast growth factor (FGF) pathway, the Hedgehog pathway, the cytokine Janus kinase/signal transducer and activator of transcription (JAK/STAT) pathway, the Notch pathway, the retinoic acid pathway, the transforming growth factor–β (TGF-β) pathway, the nuclear factor κB (NF-κB) pathway, the Hippo pathway, and the Wnt pathway [reviewed in (1)]. Thus, there appear to be insufficient individual signaling pathways to mediate the complexity in cellular outcomes and responses observed, suggesting that individual pathways can produce distinct responses or that signaling crosstalk is key to generating diverse outcomes or, as increasingly becoming apparent, a combination of the two.
Genomics approaches integrating protein-protein interaction data with large-scale RNA interference (RNAi) studies [for examples, (2–5)] have characterized complex signaling networks centered around core nodes, which have multiple upstream regulator and downstream effector connections. These networks suggest many possible points of crosstalk between pathways and ways in which a single pathway could produce different outcomes in different contexts. Differences in the expression of genes encoding network components may alter the flow of information in different cell types (6), and differences in the specific cell’s chromatin state may affect the accessibility of downstream transcriptional targets (7).
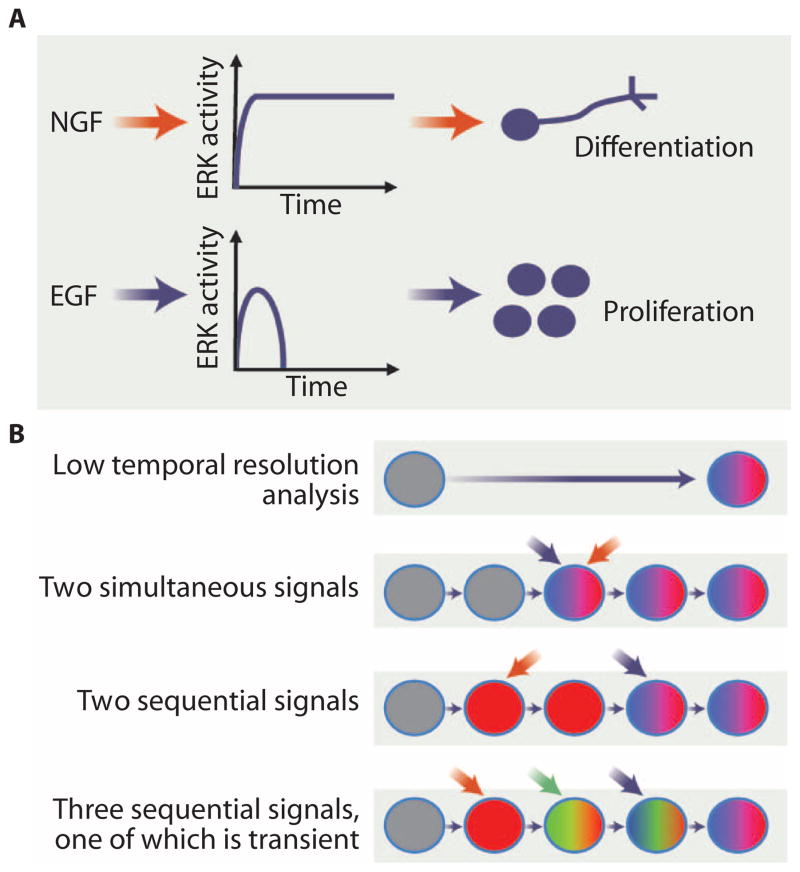
However, even in the same cell type, stimuli that activate the same pathways or hubs can, in some cases, produce different outcomes. In one classic example, nerve growth factor (NGF) and EGF both signal through the mitogen-activated protein kinase (MAPK) extracellular signal–regulated kinase (ERK) module but result in the differentiation and division of pheo-chromocytoma 12 (PC12) cells, respectively (8). A single signaling hub can therefore generate different outcomes in the same cell type depending on the input. Two potential explanations are that different upstream inputs may regulate different pathways in parallel, which both lead to ERK activation, or that the qualitative or quantitative nature of the input signal modulates the outcome of signaling through ERK. The duration of ERK activity differs depending on the stimulus and results in different outcomes. NGF stimulation results in sustained ERK activation and hence differentiation, whereas transient ERK activation in response to EGF results in proliferation (Fig. 1A) (9). Thus, the temporal dynamics of individual components in signaling networks can specify distinct cellular responses. Computational approaches can provide information and help formulate testable hypotheses regarding network wiring and dynamics. One such study used modular response analysis (10) to further study the PC12 response to EGF and NGF (11). This approach enabled the authors to identify different feedback responses downstream of ERK signaling, with NGF generating a positive feedback response and EGF negative feedback. Reversal of these feedback responses, such that NGF resulted in a negative feedback response and EGF positive feedback, was sufficient to reverse the responses to EGFand NGF. Analysis of this kind requires quantitative data and, although snapshots can be very informative, time series experiments or ideally tracking signaling in real time has the potential to offer greater insight (12).
Fig. 1. The effects of pathway and input dynamics on cellular responses.
(A) Different inputs may generate different activation dynamics of a signal hub to produce different outcomes. For example, NGF and EGF signaling lead to sustained and transient ERK activity to result in differentiation or proliferation, respectively. (B) Different sequences of input stimuli can produce different outcomes. Analysis with low temporal resolution may detect the activation of two pathways (red and blue) in a process but not the temporal relationship between them. Transient activation of pathway (green) may be completely undetected. Credit: V. ALTOUNIAN/SCIENCE SIGNALING
In addition to temporally dynamic regulation of common pathway components, temporally regulated activity of multiple different pathways can also serve as a mechanism to control cell fate and behavior (Fig. 1B). Indeed, although a great deal is known about which pathways are involved in which processes, it is often unclear how they interact. Two classic examples of fate determination in development, the patterning of the Caenorhabditis elegans vulva and the Drosophila melanogaster photoreceptors, serve as paradigms of signal crosstalk. The three cell fates that form the nematode vulva differentiate from a group of initially equivalent cells. Elegant genetic and developmental biology studies revealed that patterning is achieved through the action of three pathways (13): an EGF morphogen signal (14), a lateral inhibition signal mediated by Notch signaling (15), and a Wnt signal that maintains the competence of the cells to respond to EGF (16). Although there is a qualitative understanding of the source of the stimuli and the sequence of the activity of the pathways (an inductive EGF signal followed by Notch-mediated lateral inhibition), a quantitative understanding of the relative importance of these signals for the robustness of vulval development has been the subject of computational modeling approaches [reviewed in (17)].
In the compound eye of Drosophila, each of the ommatidia contains eight different types of photoreceptor cell, along with cone cells, pigment cells, and cells that form bristles. Fate determination in the ommatidium depends on the spatiotemporal integration of Notch and EGF signaling to generate multiple distinct fates (18). Cone cell determination involves synchronous Notch and EGF signaling (19), whereas EGF induction of photoreceptor cells induces sequential expression of Delta that signals to adjacent cells through Notch to adopt the cone cell fate (20). These two examples show the complexity of response that can arise from the integration of just a few pathways. Studies of stem cell niches, such as the mammalian hair follicle and intestinal crypt and the Drosophila midgut and germ line, have identified complex regulation by multiple pathways (21–24). To dissect the relationships between pathways in these contexts, it will be important to obtain spatiotemporal information about pathway activity at sufficient resolution to determine, for example, which pathways are functioning sequentially or in parallel.
Time Scale of Signaling Dynamics
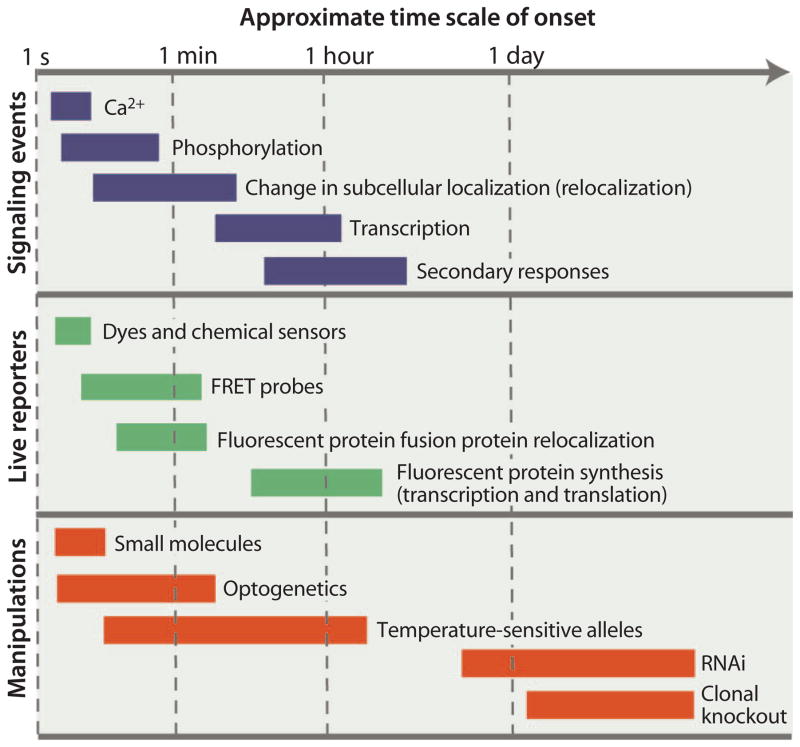
One of the challenges to studying signaling dynamics is the broad time scale (from seconds to days) over which the dynamic events can occur (Fig. 2). For example, calcium signals can occur within seconds, whereas transcriptional responses may take hours. Manipulation of individual pathways over a time scale of hours or days, as can be achieved with RNAi or induction of mutant clones, provides qualitative information about how the pathway affects cell fate and epistasis analysis can identify some relationships between pathways. However, primary signaling responses at the transcriptional level can occur on a time scale of minutes, so secondary responses and sequential pathway crosstalk may complicate interpretation of data from single, late time points as exemplified by the studies of Avraham et al. (25) and Housden et al. (26), each of which found complex secondary regulatory responses that influenced transcriptional responses.
Fig. 2. Qualitative signaling time scales.
The range of time scales over which signaling events occur and their reporters’ function. The response time within each class of reporter may vary; for example, a transcriptional reporter expressing superfolder GFP (sfGFP) will be detectable before one expressing GFP. Knockdown experiments or genetic knockouts may take many hours or days to take effect during which time secondary effects will occur. Credit: V. ALTOUNIAN/SCIENCE SIGNALING
Dissecting the relationships between pathways and the flow of information that regulates cell fate requires a level of temporal and spatial resolution that can best be achieved by either time-course experiments or ideally live imaging, but this requires appropriate tools to both track and regulate pathway activity at the appropriate time scale (Fig. 2). Here, we discuss developments in the tools available to observe and manipulate signaling with high spatiotemporal resolution, giving examples of the insights that these approaches can provide. We also highlight the potential application and extension of these approaches to address fundamental questions in signaling and regulation of cell fate.
Tools for Tracking Signaling in Real Time
A range of fluorescent probes and reporters have been developed that enable tracking of pathway activity in living cells and organisms. These molecular tools function at different levels in the pathways, from upstream ligands to transduction machinery and downstream transcriptional outputs, and on different time scales (Fig. 2). Some of these fluorescent tools are small molecules that can be introduced into cells to monitor the changes in concentration of various signaling components or changes in metabolic state or redox potential. Others are based on green fluorescent protein (GFP) and its variants, such as yellow fluorescent protein (YFP), cyan fluorescent protein (CFP), and enhanced GFP (eGFP). This second group requires fusion of the fluorescent protein to a sensor of the signal and then introduction of the fluorescent protein–tagged fusion protein into the cell or animal for imaging. These genetically encoded fluorescent reporters can be either fusions with a full-length signaling protein or fusion with a domain that is responsive to a signal [reviewed in (27)].
In various systems, researchers have exploited the mechanisms of signal transduction to provide live readouts of signaling. In some pathways, specific components show clear subcellular redistribution in response to pathway activation, and this change in subcellular localization can be visualized as a readout of pathway activity either qualitatively or quantitatively. For example, STAT translocates to the nucleus when activated by JAK, and tagging of different STAT isoforms with GFP combined with photobleaching has characterized distinct mechanisms of regulating nuclear import (28, 29). In another example, fusion of YFP to the yeast stress-responsive transcription factor Msn2 showed that the dynamics of its nuclear transport differ in response to different stimuli and thus explain the different transcriptional responses that occur (30). This system was the subject of another study that combined computational modeling with proteomics and imaging of GFP-tagged Msn2 to show that nuclear phosphorylation and export were key to its localization (31).
In other instances, dynamic protein-protein interactions or conformational changes of single proteins during pathway activity have been exploited for the development of genetically encoded fluorescence resonance energy transfer (FRET) probes. FRET involves the transfer of energy from an excited donor fluorophore to an acceptor fluorophore over a range of just 1 to 10 nm, allowing the quantification of the proximity of the fluorophores [reviewed in (32)]. Donor and acceptor fluorophores may be fused to different proteins to measure their interactions (intermolecular FRET) or to different domains of the same protein to measure conformational changes (intramolecular FRET). Intramolecular sensors for each of the guanosine triphosphatases (GTPases) Rac, RhoA, and Cdc42, for example, have been used to elucidate their roles in mechanotransduction in response to cyclic stretch (33, 34). Intermolecular FRET between YFP-ERK and CFP-MEK (MAPK kinase) demonstrated their direct interaction in the cytoplasm, and the loss of the FRET signal was used to quantify ERK translocation to the nucleus (35).
Tracking Ligands, Signal Transducers, and Transcriptional Responses
A comprehensive understanding of signaling dynamics requires tracking of signaling at all levels from the upstream ligands to the downstream transcriptional responses. At the ligand level, fluorescent tools have enabled the visualization of gradients that are key to cell fate determination. Many protein ligands can be directly visualized by fusion to fluorescent reporters, enabling quantification of diffusion dynamics. For example, fusion of GFP to the TGF-β ligands Nodal and Lefty enabled the measurement of their distributions and diffusivity during zebrafish development, providing evidence for reaction-diffusion models of patterning (36). Non-protein ligands, such as the hormone retinoic acid that plays essential roles in vertebrate development, cannot be fused to fluorescent proteins and require different approaches. Fusion of the retinoic acid–binding domains from the retinoic acid receptor (RAR) to FRET donor and acceptor pairs allowed imaging of the local concentration of retinoic acid by measuring shifts in FRETefficiency caused by conformational changes in response to retinoic acid binding (37). This allowed visualization of the gradient responsible for the anterior-posterior patterning of the vertebrate hindbrain.
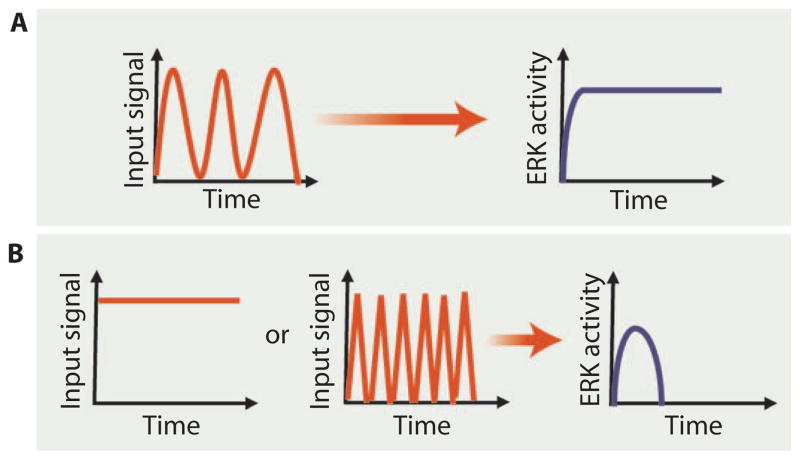
The conserved Ras–MAPK kinase kinase (MAPKKK)–MAPK–ERK module lies downstream of many extracellular signals, and its activation can result in a range of cellular responses depending on the specific signal. Thus, quantitatively tracking ERK activity in living cells has the potential to reveal how dynamics contribute to these divergent responses. Staining for ERK localization or phosphorylation in fixed samples gives a readout of the activity of the module at single time points (38), but fluorescence-based approaches enable tracking of activity in living cells. A range of FRET systems have been developed to measure ERK activity in cultured cells (39, 40) and, by optimizing the brightness of the fluorophores and FRET efficiency, even in vivo, where signal detection can be difficult (41). Tomida and colleagues used a single-molecule Y-Pet enhanced CFP–based intramolecular FRET sensor to assess ERK activation dynamics in a single C. elegans NaCl-responsive sensory neuron invivo (Fig. 3) (41). In addition to tracking ERK dynamics, the authors used a microfluidic chamber to precisely temporally control NaCl concentrations so that the effects of different input dynamics could be assessed. They found that the frequency of stimulation was critical to the extent of ERK activation: Maximal sustained activation was achieved only by cyclic stimulation and rest with a periodicity of around 40 s (Fig. 3A). Either sustained stimulation or more rapid cycles of stimulation and rest failed to generate a sustained response (Fig. 3B) (41). This study highlights the importance of input dynamics as opposed to simply the nature or concentration of the stimulus in controlling the activity of the pathway. The authors proposed that the nature of the response may allow the sensory neuron to filter out both continuous signals and transient noise and thus to respond to relevant changes in the environment, providing a direct link between signaling dynamics and function.
Fig. 3. Responding to input signal frequency.
(A) Only a specific frequency of oscillating input produced a sustained ERK response in a specific sensory neuron in C. elegans. (B) Sustained input signals or signals that oscillated too frequently produced a transient response. Credit: V. ALTOUNIAN/SCIENCE SIGNALING
Similar to ligands, not all signaling molecules are proteins and some of the most rapidly responding pathways involve ions, such as calcium, or ligand-activated transcription factors, such as the RAR or nuclear steroid hormone receptors. Small-molecule sensors of calcium concentrations, such as fura-2, were among the first developed (42) and, along with the more recently developed genetically encoded reporters (43), have enabled extensive investigation into the dynamics of calcium signaling in cells and in vivo (44).
Frequently, effects on cell fate and initiation of secondary responses are mediated by changes in gene expression, and the effects on cell fate can require variable time scales. Tracking transcriptional responses with high temporal resolution is therefore key to dissecting signal crosstalk and linking signaling to cell fate because primary transcriptional responses may influence secondary responses. In cell culture, real-time reverse transcription polymerase chain reaction, a method to quantitatively detect mRNA abundance, and RNAseq (a method for transcriptome analysis) time-course experiments have been used successfully to separate primary and secondary signaling responses on a scale of tens of minutes, but are not easily applied in vivo in which multiple cell types receive distinct signals and respond differently (25, 26). Reporter constructs driving the expression of fluorescent proteins, such as TOPFlash for Wnt signaling (45), allow live imaging of expression in vivo with excellent spatial resolution but, depending on the folding speed and stability of the reporter protein, may create a delay in detecting the response or in detecting the termination of the response. Studies on somite formation in vertebrates, which depends on a Notch-based oscillating clock, have highlighted the possibilities of destabilized fluorescent reporters, in which the fluorescent protein is fused to a tag that promotes degradation of the fusion protein and hence reduces its half-life, for real-time imaging (46). Oscillations in Notch signaling occur with ~1 hour between peak and trough; thus, conventional GFP-based reporters persist longer than the duration of a single cycle (47). Oscillations in Notch signaling in zebrafish were visualized in real time with destabilized luciferase and YFP reporters for Notch signaling components Hes1 and Lunatic fringe, respectively (47, 48). Similar short-period oscillations in Notch signaling also regulate fate in neural stem cells (49) and embryonic stem cells (50), suggesting that this type of temporal signaling could be of broad importance (51). An alternative approach to fluorescent protein–based reporters is the fluorescence-based detection of mRNA, which can be achieved by incorporating MS2 RNA stem-loops, which can be bound by MS2 protein, into a transcriptional reporter construct. Constitutive expression of nuclear-localized MS2-YFP reveals activity of the promoter as spots of intense fluorescence against a background of MS2-YFP that is not bound to regions of active transcription (52).
Manipulating Input Dynamics
Although a range of tools has already been developed to provide live readouts of pathway activity, the tools necessary for high-resolution temporal manipulation of pathway components have been comparatively lacking. In vivo studies generally rely on single perturbations, such as overexpression, systemic or local administration of agonists, or knockdown or knockout of pathway components. All of these approaches require a time scale of hours to days (Fig. 2) (53), during which time secondary responses involving feedback, autocrine signaling, or paracrine signaling can replace, mask, modify, or compensate for primary responses. Temperature-sensitive forms of proteins may offer quicker manipulation but are not available for all pathways and have additional caveats related to the effects of heat shock on the system (53). The long duration involved also prevents fine-tuning of pathway stimulation to probe the role of dynamic inputs.
In some cases, the nature of the stimulus readily allows manipulation. For example, the work on ERK activation by Tomida and colleagues relied on environmental NaCl and used a microfluidic device to provide precise control (41). Dynamic studies of mechanotransduction have exploited equipment, such as optical tweezers, for in vitro cell studies (34). For organisms, such as yeast, or cells that can be grown in culture, agonists or antagonists of a pathway that can be rapidly washed on and off enable reversible, dynamic manipulation of pathway activity. Direct regulation of the nuclear transport dynamics of the yeast transcription factor Msn2 was achieved in this way with a small-molecule inhibitor of its regulator protein kinase A, demonstrating that differences in nuclear transport dynamics modulate transcriptional output (30). Similarly, the importance of p53 dynamics in the outcome of DNA damage responses in a breast cancer cell line was assessed using a small-molecule inhibitor of a p53 regulator (54).
Most of the techniques used in the previous examples are not readily applicable to studies in intact tissues or readily extended to a broad range of pathways. The development of optogenetics, the use of genetically encoded light-responsive proteins to enable light to regulate cellular functions, has opened the possibility of high-resolution pathway manipulation (55). A recent study combined optogenetic manipulation with fluorescence-based reporters to interrogate how dynamic inputs produce distinct signaling dynamics through the Ras-MAPKKK-MAPKK-ERK pathway (referred to as Ras-ERK module) (56). Toettcheret al. used a reversible optogenetic switch to activate and deactivate the Ras-ERK module with a temporal resolution of just a few minutes (56). This switch is based on the Phy-PIF system: The Phy protein can be photoconverted between two conformational states, only one of which interacts with PIF (57). PIF was fused to the catalytic domain of a RasGEF, the activity of which depends on membrane localization, and co-expressed with a membrane-localized form of Phy. Stimulation with red light triggers the interaction of PIFand Phy, resulting in membrane localization of the RasGEF and hence activation of Ras. Exposure to far red light blocks the interaction and deactivated RasGEF. By coupling this reversible optogenetic system with a blue fluorescent protein (BFP)–tagged ERK, the input stimulus of Ras activity was experimentally controlled and the output response of ERK activity was tracked with high spatiotemporal resolution. The dynamic signaling data were then combined with proteomics to assess the downstream response to different patterns of temporal activation, which revealed two distinct responses that depended on the signal duration. Differential dynamic activation of a common module (Ras-MAPKKK-MAPK-ERK) is therefore sufficient to generate discrete responses.
In addition to demonstrating the importance of temporal dynamics in signaling, Toettcher and colleagues (56) also highlighted the importance of high temporal resolution for dissecting signaling crosstalk. Because the method enabled input manipulation and output monitoring within 10-min intervals, a secondary response was revealed that other manipulation approaches, such as RNAi, would have merged with primary response. Activation of the Ras-ERK module for >1 hour triggers activation of STAT3, and analysis of cocultures of differently labeled cells showed that this was a paracrine effect in which sustained ERK signaling in one cell population induces cytokines that trigger JAK-STAT signaling in the other cells. The ERK-signaling cells themselves were refractory to the secondary cytokine signal, and STATactivation was probably only detected in the initial analysis of a single population of cells because of the variable amounts of the optogenetic components in each cell (that is, the population was not homogeneous). Nevertheless, this study shows not only that dynamically manipulating and tracking pathway activity can reveal the importance of signaling dynamics in controlling cellular behavior, but studies with high temporal resolution can also resolve the temporal relationships between pathways, thereby revealing crosstalk among pathways.
Future Directions in Understanding Information Flow and Signal Crosstalk
Clearly, signaling dynamics themselves are critical regulators of cell responses and may be useful as pharmacological targets (58, 59). The examples outlined here emphasize that the ability to manipulate and monitor signaling dynamics at high temporal resolution is key to revealing both the nature of dynamic signaling and its importance for information transfer and cell responses. The approach of Toettcher and colleagues could potentially be applied to look at multiple tiers of different pathways and quantify the flow of information to identify key points of regulation because there are many points of interaction and molecular relocalization events that could form the basis for optogenetic tools and fluorescence-based reporters (56). Development of tools to stimulate single or multiple pathways at different levels with precise temporal control coupled with dynamic reporters of pathway output at downstream levels, including transcriptional targets, will enable systematic dissection of information flow through specific pathways and offer new insights into how information is transmitted.
Although some studies look at intact organisms or tissues, such as the work of Tomida and colleagues (41) and intravital imaging studies of fluorescently labeled cells (60), an ongoing challenge is extending these approaches to the complex signaling that occurs in intact tissues from developing or adult organisms or in living multicellular organisms to couple the dynamics to regulation of cell fate, organismal behavior, and physiology in vivo. In vivo systems are further complicated by secondary signaling events that occur between different cell types, but the ability to apply these tools in vivo should reveal these downstream cell-cell signaling events and how they influence development or cellular behavior. Cells may be exposed to many signals simultaneously or may experience a precise temporal pattern of stimuli, and multiple pathways may be active with signaling occurring between multiple cell types. To what extent these pathways function synchronously or sequentially is not easily resolved by approaches based on the study of fixed samples using reporters with low temporal resolution. The ability to monitor signaling events in space and time in live tissues can show both individual pathway dynamics and the temporal relationships between pathways that may represent signal crosstalk. Many of the fluorescent reporters also provide spatial information, and when combined with high temporal resolution, these tools can be used to study interactions between different cell types in tissues. Even in an apparently homogeneous population of a single cell type, analysis at single-cell resolution is important, because averaging across cell populations may mask heterogeneity in responses and responsiveness (61). Again, extension of the published approaches to study different pathway components at similar resolution will be critical.
The ability to monitor signaling in vivo is limited by the brightness of the currently available fluorescent proteins, but brighter fluorescent protein variants continue to be developed. Improved FRET sensors are also being developed that are brighter and have improved dynamic range. One study, for example, generated libraries of RhoA and ERK biosensors (62) and showed that the improved ERK biosensor revealed ERK activity at single-cell resolution in zebrafish embryos in vivo. Improved FRET pairs with increased brightness and sensitivity to subtle changes will also facilitate the detection of signaling events with high temporal and spatial resolution (63).
Tools to monitor transcriptional responses with high temporal resolution are further limited by the speed with which the fluorescent proteins fold and are degraded. The development of brighter and faster-folding fluorescent proteins, such as sfGFP, across the color spectrum should lead to the development of more sensitive transcriptional reporters for simultaneously tracking the activity of multiple pathways (64, 65). Although destabilized reporters, such as those for the Notch pathway (46) and the JAK-STAT pathway (66), have improved the temporal resolution of detecting some transcriptional regulatory events, these generally rely on synthetic promoters containing multiple binding sites for the pathway-regulated transcription factors, which is not reflective of the native genetic regulatory mechanisms. Genome engineering with transcription activator–like effector nucleases (TALENs), zinc finger nucleases (ZFNs), and clustered regularly interspaced short palindromic repeats (CRISPR)–Cas9 (67–69) will make the targeting of reporters to endogenous loci to reflect the endogenous transcriptional regulation tractable. However, when placed in a native chromosomal context, these reporters may only be transcribed at low levels, which would present technical challenges in their detection.
The rapidly expanding field of optogenetics is likely to continue to offer new tools for pathway manipulation. Using light to trigger the association of a TALE DNA binding domain with a range of transcriptional effectors or chromatin modifiers, for example, has provided optogenetic control of transcription and chromatin states, respectively (70).
The continued application and development of fluorescence-based tools holds the promise of both visualizing and manipulating pathway activity with high spatiotemporal resolution, offering new insights into how pathways signal individually and in combination to regulate cell fate and behavior in culture, ex vivo, and in vivo.
Acknowledgments
We thank B. Housden for helpful comments.
Funding: D.P.D. is supported by the Human Frontier Science Program. Work in the Perrimon laboratory is supported by the Howard Hughes Medical Institute and the NIH.
REFERENCES AND NOTES
- 1.Perrimon N, Pitsouli C, Shilo BZ. Signaling mechanisms controlling cell fate and embryonic patterning. Cold Spring Harb Perspect Biol. 2012;4:a005975. doi: 10.1101/cshperspect.a005975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Friedman AA, Tucker G, Singh R, Yan D, Vinayagam A, Hu Y, Binari R, Hong P, Sun X, Porto M, Pacifico S, Murali T, Finley RL, Asara JM, Berger B, Perrimon N. Proteomic and functional genomic landscape of receptor tyrosine kinase and Ras to extracellular signal–regulated kinase signaling. Sci Signal. 2011;4:rs10. doi: 10.1126/scisignal.2002029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Couzens AL, Knight JDR, Kean MJ, Teo G, Weiss A, Dunham WH, Lin ZY, Bagshaw RD, Sicheri F, Pawson T, Wrana JL, Choi H, Gingras AC. Protein interaction network of the mammalian Hippo pathway reveals mechanisms of kinase-phosphatase interactions. Sci Signal. 2013;6:rs15. doi: 10.1126/scisignal.2004712. [DOI] [PubMed] [Google Scholar]
- 4.Kwon Y, Vinayagam A, Sun X, Dephoure N, Gygi SP, Hong P, Perrimon N. The Hippo signaling pathway interactome. Science. 2013;342:737–740. doi: 10.1126/science.1243971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang W, Li X, Huang J, Feng L, Dolinta KG, Chen J. Defining the protein–protein interaction network of the human Hippo pathway. Mol Cell Proteomics. 2014;13:119–131. doi: 10.1074/mcp.M113.030049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kiel C, Verschueren E, Yang JS, Serrano L. Integration of protein abundance and structure data reveals competition in the ErbB signaling network. Sci Signal. 2013;6:ra109. doi: 10.1126/scisignal.2004560. [DOI] [PubMed] [Google Scholar]
- 7.Wong LY, Hatfield JK, Brown MA. Ikaros sets the potential for Th17 lineage gene expression through effects on chromatin state in early T cell development. J Biol Chem. 2013;288:35170–35179. doi: 10.1074/jbc.M113.481440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chao MV. Growth factor signaling: Where is the specificity? Cell. 1992;68:995–997. doi: 10.1016/0092-8674(92)90068-n. [DOI] [PubMed] [Google Scholar]
- 9.Marshall CJ. Specificity of receptor tyrosine kinase signaling: Transient versus sustained extracellular signal-regulated kinase activation. Cell. 1995;80:179–185. doi: 10.1016/0092-8674(95)90401-8. [DOI] [PubMed] [Google Scholar]
- 10.Bruggeman FJ, Westerhoff HV, Hoek JB. Modular response analysis of cellular regulatory networks. J Theor Biol. 2002;218:507–520. [PubMed] [Google Scholar]
- 11.Santos SDM, Verveer PJ, Bastiaens PIH. Growth factor-induced MAPK network topology shapes Erk response determining PC-12 cell fate. Nat Cell Biol. 2007;9:324–330. doi: 10.1038/ncb1543. [DOI] [PubMed] [Google Scholar]
- 12.Sontag E, Kiyatkin A, Kholodenko BN. Inferring dynamic architecture of cellular networks using time series of gene expression, protein and metabolite data. Bioinformatics. 2004;20:1877–1886. doi: 10.1093/bioinformatics/bth173. [DOI] [PubMed] [Google Scholar]
- 13.Kenyon C. A perfect vulva every time: Gradients and signaling cascades in C. elegans. Cell. 1995;82:171–174. doi: 10.1016/0092-8674(95)90302-x. [DOI] [PubMed] [Google Scholar]
- 14.Hill RJ, Sternberg PW. The gene lin-3 encodes an inductive signal for vulval development in C. elegans. Nature. 1992;358:470–476. doi: 10.1038/358470a0. [DOI] [PubMed] [Google Scholar]
- 15.Sternberg PW. Lateral inhibition during vulval induction in Caenorhabditis elegans. Nature. 1988;335:551–554. doi: 10.1038/335551a0. [DOI] [PubMed] [Google Scholar]
- 16.Myers TR, Greenwald I. Wnt signal from multiple tissues and lin-3/EGF signal from the gonad maintain vulval precursor cell competence in Caenorhabditis elegans. Proc Natl Acad Sci USA. 2007;104:20368–20373. doi: 10.1073/pnas.0709989104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Félix MA, Barkoulas M. Robustness and flexibility in nematode vulva development. Trends Genet. 2012;28:185–195. doi: 10.1016/j.tig.2012.01.002. [DOI] [PubMed] [Google Scholar]
- 18.Nagaraj R, Banerjee U. The little R cell that could. Int J Dev Biol. 2004;48:755–760. doi: 10.1387/ijdb.041881rn. [DOI] [PubMed] [Google Scholar]
- 19.Flores GV, Duan H, Yan H, Nagaraj R, Fu W, Zou Y, Noll M, Banerjee U. Combinatorial signaling in the specification of unique cell fates. Cell. 2000;103:75–85. doi: 10.1016/s0092-8674(00)00106-9. [DOI] [PubMed] [Google Scholar]
- 20.Tsuda L, Nagaraj R, Zipursky SL, Banerjee U. An EGFR/Ebi/Sno pathway promotes Delta expression by inactivating Su(H)/SMRTER repression during inductive notch signaling. Cell. 2002;110:625–637. doi: 10.1016/s0092-8674(02)00875-9. [DOI] [PubMed] [Google Scholar]
- 21.Blanpain C, Fuchs E. Epidermal stem cells of the skin. Annu Rev Cell Dev Biol. 2006;22:339–373. doi: 10.1146/annurev.cellbio.22.010305.104357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Clevers H. The intestinal crypt, a prototype stem cell compartment. Cell. 2013;154:274–284. doi: 10.1016/j.cell.2013.07.004. [DOI] [PubMed] [Google Scholar]
- 23.de Cuevas M, Matunis EL. The stem cell niche: Lessons from the Drosophila testis. Development. 2011;138:2861–2869. doi: 10.1242/dev.056242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jiang H, Edgar BA. Intestinal stem cells in the adult Drosophila midgut. Exp Cell Res. 2011;317:2780–2788. doi: 10.1016/j.yexcr.2011.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Avraham R, Sas-Chen A, Manor O, Steinfeld I, Shalgi R, Tarcic G, Bossel N, Zeisel A, Amit I, Zwang Y, Enerly E, Russnes HG, Biagioni F, Mottolese M, Strano S, Blandino G, Børresen-Dale AL, Pilpel Y, Yakhini Z, Segal E, Yarden Y. EGF decreases the abundance of microRNAs that restrain oncogenic transcription factors. Sci Signal. 2010;3:ra43. doi: 10.1126/scisignal.2000876. [DOI] [PubMed] [Google Scholar]
- 26.Housden BE, Fu AQ, Krejci A, Bernard F, Fischer B, Tavaré S, Russell S, Bray SJ. Transcriptional dynamics elicited by a short pulse of notch activation involves feed-forward regulation by E(spl)/Hes genes. PLOS Genet. 2013;9:e1003162. doi: 10.1371/journal.pgen.1003162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Giepmans BNG, Adams SR, Ellisman MH, Tsien RY. The fluorescent toolbox for assessing protein location and function. Science. 2006;312:217–224. doi: 10.1126/science.1124618. [DOI] [PubMed] [Google Scholar]
- 28.McBride KM, Banninger G, McDonald C, Reich NC. Regulated nuclear import of the STAT1 transcription factor by direct binding of importin-α. EMBO J. 2002;21:1754–1763. doi: 10.1093/emboj/21.7.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen HC, Reich NC. Live cell imaging reveals continuous STAT6 nuclear trafficking. J Immunol. 2010;185:64–70. doi: 10.4049/jimmunol.0903323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hao N, O’Shea EK. Signal-dependent dynamics of transcription factor translocation controls gene expression. Nat Struct Mol Biol. 2012;19:31–39. doi: 10.1038/nsmb.2192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sunnåker M, Zamora-Sillero E, Dechant R, Ludwig C, Busetto AG, Wagner A, Stelling J. Automatic generation of predictive dynamic models reveals nuclear phosphorylation as the key Msn2 control mechanism. Sci Signal. 2013;6:ra41. doi: 10.1126/scisignal.2003621. [DOI] [PubMed] [Google Scholar]
- 32.Jares-Erijman EA, Jovin TM. FRET imaging. Nat Biotechnol. 2003;21:1387–1395. doi: 10.1038/nbt896. [DOI] [PubMed] [Google Scholar]
- 33.Aoki K, Matsuda M. Visualization of small GTPase activity with fluorescence resonance energy transfer-based biosensors. Nat Protoc. 2009;4:1623–1631. doi: 10.1038/nprot.2009.175. [DOI] [PubMed] [Google Scholar]
- 34.Liu B, Kim TJ, Wang Y. Live cell imaging of mechanotransduction. J R Soc Interface. 2010;7(Suppl 3):S365–S375. doi: 10.1098/rsif.2010.0042.focus. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Burack WR, Shaw AS. Live cell imaging of ERK and MEK: Simple binding equilibrium explains the regulated nucleocytoplasmic distribution of ERK. J Biol Chem. 2005;280:3832–3837. doi: 10.1074/jbc.M410031200. [DOI] [PubMed] [Google Scholar]
- 36.Müller P, Rogers KW, Jordan BM, Lee JS, Robson D, Ramanathan S, Schier AF. Differential diffusivity of Nodal and Lefty underlies a reaction-diffusion patterning system. Science. 2012;336:721–724. doi: 10.1126/science.1221920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shimozono S, Iimura T, Kitaguchi T, Higashijima SI, Miyawaki A. Visualization of an endogenous retinoic acid gradient across embryonic development. Nature. 2013;496:363–366. doi: 10.1038/nature12037. [DOI] [PubMed] [Google Scholar]
- 38.Friedman A, Perrimon N. A functional RNAi screen for regulators of receptor tyrosine kinase and ERK signalling. Nature. 2006;444:230–234. doi: 10.1038/nature05280. [DOI] [PubMed] [Google Scholar]
- 39.Sato M, Kawai Y, Umezawa Y. Genetically encoded fluorescent indicators to visualize protein phosphorylation by extracellular signal-regulated kinase in single living cells. Anal Chem. 2007;79:2570–2575. doi: 10.1021/ac062171d. [DOI] [PubMed] [Google Scholar]
- 40.Harvey CD, Ehrhardt AG, Cellurale C, Zhong H, Yasuda R, Davis RJ, Svoboda K. A genetically encoded fluorescent sensor of ERK activity. Proc Natl Acad Sci USA. 2008;105:19264–19269. doi: 10.1073/pnas.0804598105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tomida T, Oda S, Takekawa M, Iino Y, Saito H. The temporal pattern of stimulation determines the extent and duration of MAPK activation in a Caenorhabditis elegans sensory neuron. Sci Signal. 2012;5:ra76. doi: 10.1126/scisignal.2002983. [DOI] [PubMed] [Google Scholar]
- 42.Grynkiewicz G, Poenie M, Tsien RY. A new generation of Ca2+ indicators with greatly improved fluorescence properties. J Biol Chem. 1985;260:3440–3450. [PubMed] [Google Scholar]
- 43.Palmer AE, Tsien RY. Measuring calcium signaling using genetically targetable fluorescent indicators. Nat Protoc. 2006;1:1057–1065. doi: 10.1038/nprot.2006.172. [DOI] [PubMed] [Google Scholar]
- 44.Reiff DF, Ihring A, Guerrero G, Isacoff EY, Nakai J, Borst A. In vivo performance of genetically encoded indicators of neural activity in flies. J Neurosci. 2006;25:4766–4778. doi: 10.1523/JNEUROSCI.4900-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Veeman MT, Slusarski DC, Kaykas A, Louie SH, Moon RT. Zebrafish prickle, a modulator of noncanonical Wnt/Fz signaling, regulates gastrulation movements. Curr Biol. 2003;13:680–685. doi: 10.1016/s0960-9822(03)00240-9. [DOI] [PubMed] [Google Scholar]
- 46.Soroldoni D, Oates AC. Live transgenic reporters of the vertebrate embryo’s Segmentation Clock. Curr Opin Genet Dev. 2011;21:600–605. doi: 10.1016/j.gde.2011.09.006. [DOI] [PubMed] [Google Scholar]
- 47.Masamizu Y, Ohtsuka T, Takashima Y, Nagahara H, Takenaka Y, Yoshikawa K, Okamura H, Kageyama R. Real-time imaging of the somite segmentation clock: Revelation of unstable oscillators in the individual presomitic mesoderm cells. Proc Natl Acad Sci USA. 2006;103:1313–1318. doi: 10.1073/pnas.0508658103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Aulehla A, Wiegraebe W, Baubet V, Wahl MB, Deng C, Taketo M, Lewandoski M, Pourquié O. A β-catenin gradient links the clock and wavefront systems in mouse embryo segmentation. Nat Cell Biol. 2008;10:186–193. doi: 10.1038/ncb1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shimojo H, Ohtsuka T, Kageyama R. Oscillations in notch signaling regulate maintenance of neural progenitors. Neuron. 2008;58:52–64. doi: 10.1016/j.neuron.2008.02.014. [DOI] [PubMed] [Google Scholar]
- 50.Kobayashi T, Mizuno H, Imayoshi I, Furusawa C, Shirahige K, Kageyama R. The cyclic gene Hes1 contributes to diverse differentiation responses of embryonic stem cells. Genes Dev. 2009;23:1870–1875. doi: 10.1101/gad.1823109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kageyama R, Niwa Y, Shimojo H, Kobayashi T, Ohtsuka T. Ultradian oscillations in Notch signaling regulate dynamic biological events. Curr Top Dev Biol. 2010;92:311–331. doi: 10.1016/S0070-2153(10)92010-3. [DOI] [PubMed] [Google Scholar]
- 52.Larson DR, Fritzsch C, Sun L, Meng X, Lawrence DS, Singer RH. Direct observation of frequency modulated transcription in single cells using light activation. Elife. 2013;2:e00750. doi: 10.7554/eLife.00750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shogren-Knaak MA, Alaimo PJ, Shokat KM. Recent advances in chemical approaches to the study of biological systems. Annu Rev Cell Dev Biol. 2001;17:405–433. doi: 10.1146/annurev.cellbio.17.1.405. [DOI] [PubMed] [Google Scholar]
- 54.Purvis JE, Karhohs KW, Mock C, Batchelor E, Loewer A, Lahav G. p53 dynamics control cell fate. Science. 2012;336:1440–1444. doi: 10.1126/science.1218351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Toettcher JE, Voigt CA, Weiner OD, Lim WA. The promise of optogenetics in cell biology: Interrogating molecular circuits in space and time. Nat Methods. 2011;8:35–38. doi: 10.1038/nmeth.f.326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Toettcher JE, Weiner OD, Lim WA. Using optogenetics to interrogate the dynamic control of signal transmission by the Ras/Erk module. Cell. 2013;155:1422–1434. doi: 10.1016/j.cell.2013.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Levskaya A, Weiner OD, Lim WA, Voigt CA. Spatiotemporal control of cell signalling using a light-switchable protein interaction. Nature. 2009;461:997–1001. doi: 10.1038/nature08446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Purvis JE, Lahav G. Encoding and decoding cellular information through signaling dynamics. Cell. 2013;152:945–956. doi: 10.1016/j.cell.2013.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Behar M, Barken D, Werner SL, Hoffmann A. The dynamics of signaling as a pharmacological target. Cell. 2013;155:448–461. doi: 10.1016/j.cell.2013.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mempel TR, Scimone ML, Mora JR, von Andrian UH. In vivo imaging of leukocyte trafficking in blood vessels and tissues. Curr Opin Immunol. 2004;16:406–417. doi: 10.1016/j.coi.2004.05.018. [DOI] [PubMed] [Google Scholar]
- 61.Canham MA, Sharov AA, Ko MSH, Brickman JM. Functional heterogeneity of embryonic stem cells revealed through translational amplification of an early endodermal transcript. PLOS Biol. 2010;8:e1000379. doi: 10.1371/journal.pbio.1000379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fritz RD, Letzelter M, Reimann A, Martin K, Fusco L, Ritsma L, Ponsioen B, Fluri E, Schulte-Merker S, van Rheenen J, Pertz O. A versatile toolkit to produce sensitive FRET biosensors to visualize signaling in time and space. Sci Signal. 2013;6:rs12. doi: 10.1126/scisignal.2004135. [DOI] [PubMed] [Google Scholar]
- 63.Day RN, Davidson MW. Fluorescent proteins for FRET microscopy: Monitoring protein interactions in living cells. Bioessays. 2012;34:341–350. doi: 10.1002/bies.201100098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pédelacq JD, Cabantous S, Tran T, Terwilliger TC, Waldo GS. Engineering and characterization of a superfolder green fluorescent protein. Nat Biotechnol. 2006;24:79–88. doi: 10.1038/nbt1172. [DOI] [PubMed] [Google Scholar]
- 65.Khmelinskii A, Keller PJ, Bartosik A, Meurer M, Barry JD, Mardin BR, Kaufmann A, Trautmann S, Wachsmuth M, Pereira G, Huber W, Schiebel E, Knop M. Tandem fluorescent protein timers for in vivo analysis of protein dynamics. Nat Biotechnol. 2012;30:708–714. doi: 10.1038/nbt.2281. [DOI] [PubMed] [Google Scholar]
- 66.Bach EA, Ekas LA, Ayala-Camargo A, Flaherty MS, Lee H, Perrimon N, Baeg GH. GFP reporters detect the activation of the Drosophila JAK/STAT pathway in vivo. Gene Expr Patterns. 2007;7:323–331. doi: 10.1016/j.modgep.2006.08.003. [DOI] [PubMed] [Google Scholar]
- 67.Joung JK, Sander JD. TALENs: A widely applicable technology for targeted genome editing. Nat Rev Mol Cell Biol. 2013;14:49–55. doi: 10.1038/nrm3486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Carroll D. Genome engineering with zinc-finger nucleases. Genetics. 2011;188:773–782. doi: 10.1534/genetics.111.131433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pennisi E. The CRISPR craze. Science. 2013;341:833–836. doi: 10.1126/science.341.6148.833. [DOI] [PubMed] [Google Scholar]
- 70.Konermann S, Brigham MD, Trevino AE, Hsu PD, Heidenreich M, Cong L, Platt RJ, Scott DA, Church GM, Zhang F. Optical control of mammalian endogenous transcription and epigenetic states. Nature. 2013;500:472–476. doi: 10.1038/nature12466. [DOI] [PMC free article] [PubMed] [Google Scholar]