Abstract
A mutant of Escherichia coli is described that displays kirromycin resistance in a cell-free system by virtue of an altered elongation factor Tu (EF-Tu). In poly(U)-directed poly(Phe) synthesis the kirromycin resistance of the crystallized enzyme ranged between a factor of 80 and 700, depending on temperature. Similarly, kirromycin-induced EF-Tu GTPase activity uncoupled from ribosomes and aminoacyl-tRNA required correspondingly higher concentrations of the antibiotic. Resistance of EF-Tu to kirromycin is a consequence of a modified enzyme structure as indicated by its altered fingerprint pattern.
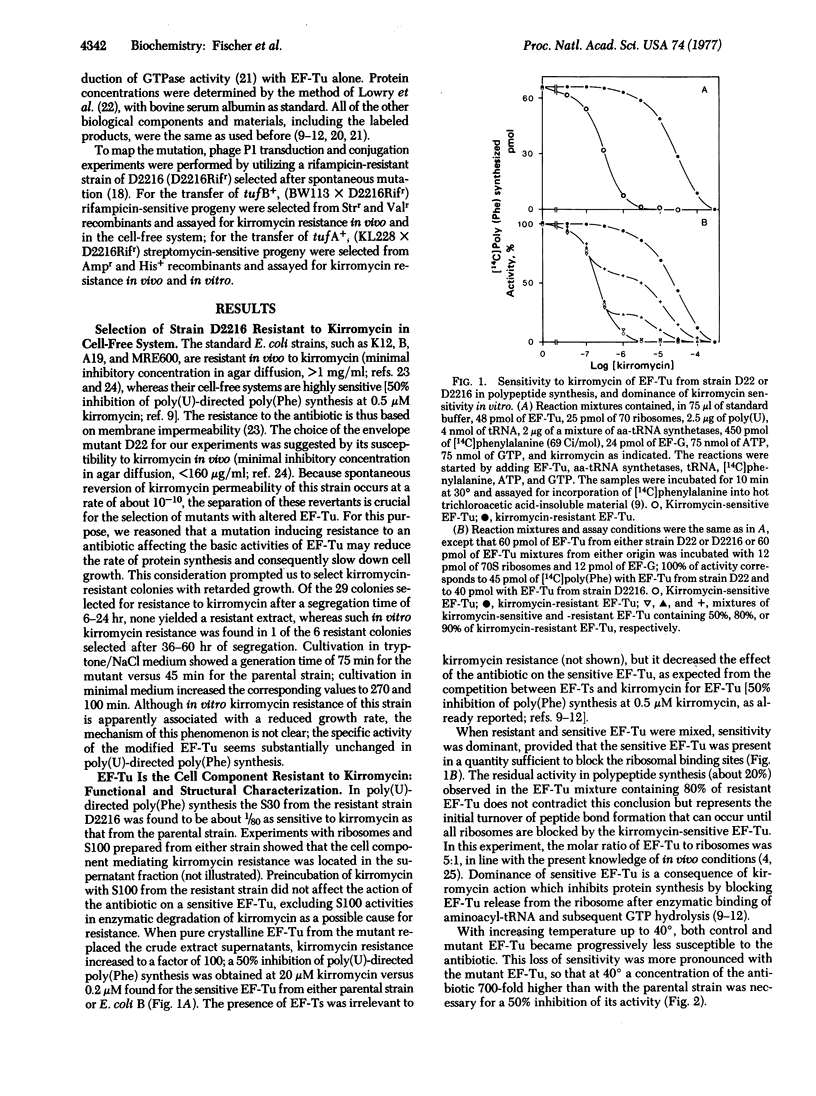
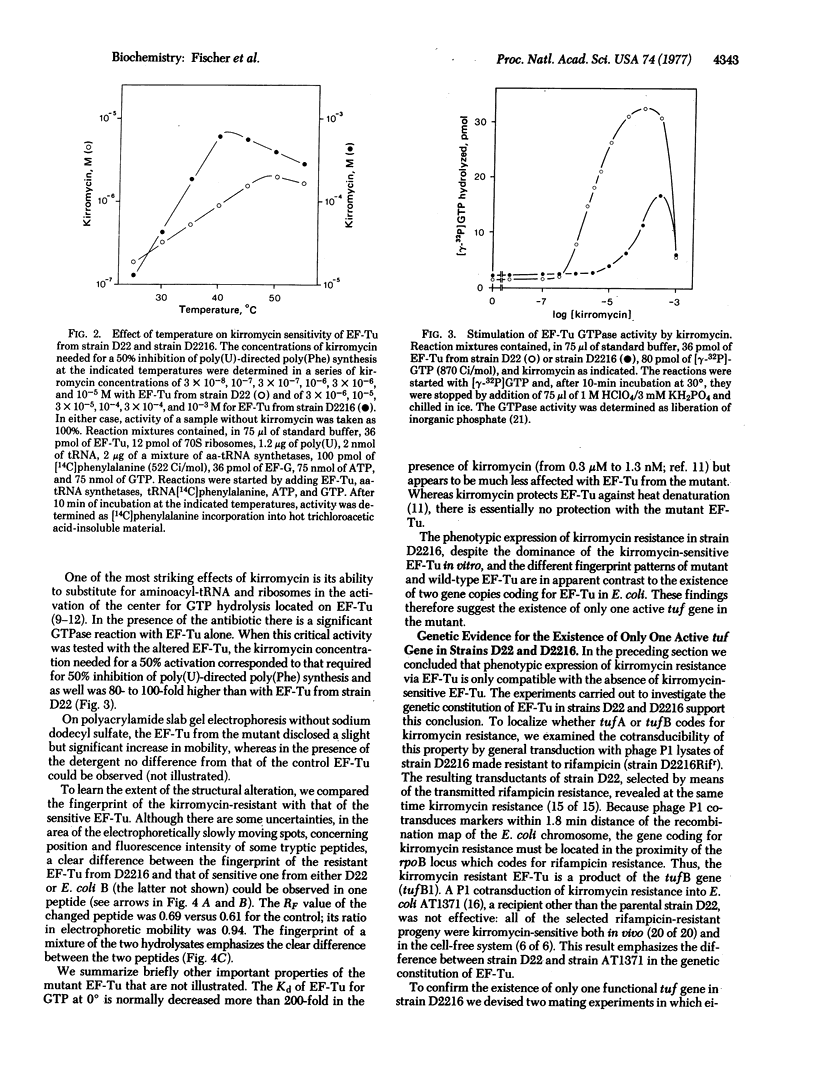
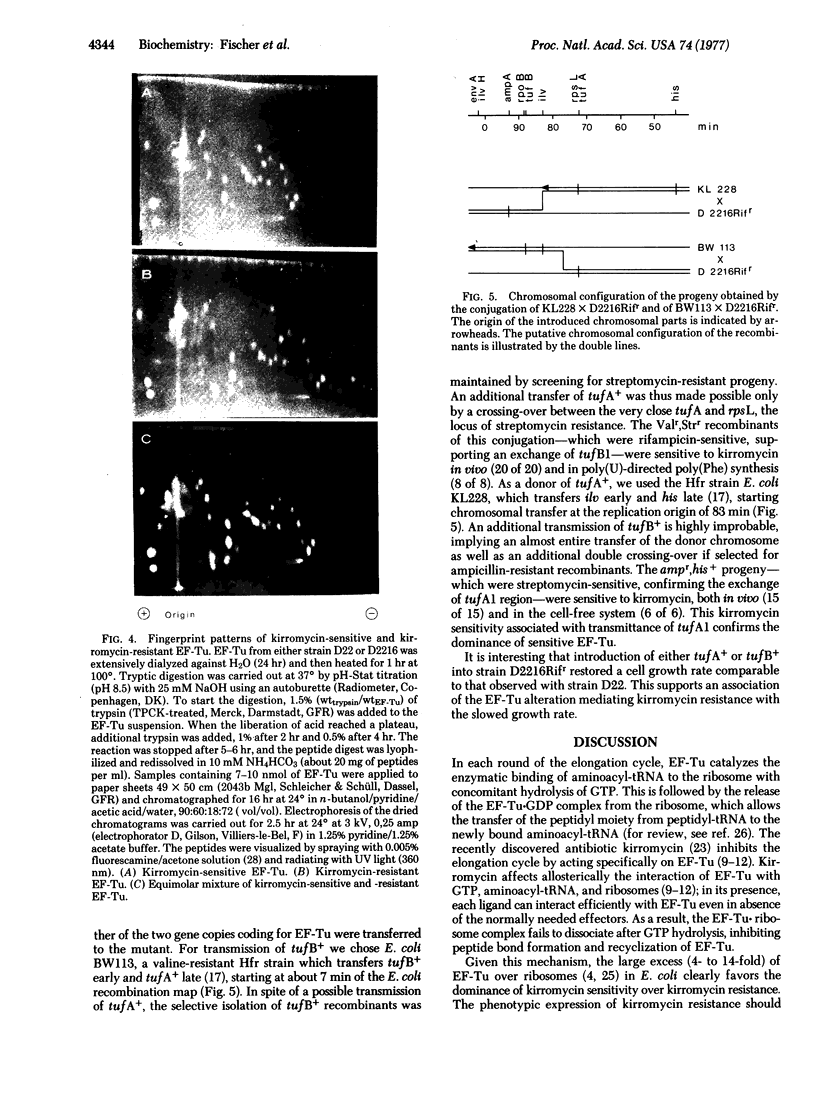
P1 transduction experiments showed that the kirromycin-resistant EF-Tu is coded by an altered tufB gene (tufB1). The known existence of two genes coding for EF-Tu would interfere with the recognition of a mutant altered in only one of those genes, if the mutation were recessive. Because kirromycin blocks EF-Tu release from the ribosome, kirromycin sensitivity is dominant, as shown by the failure of a mixed EF-Tu population to express resistance in vitro. Therefore, phenotypic expression of kirromycin resistance in vivo appears to be only possible if the EF-Tu mutant lacks an active tufA gene, a property likely to be inherited from the parental D22 strain. The observations that introduction of a tufA+ region makes the resistant strain sensitive to the antibiotic and that transduction of tufB1 into a recipient other than E. coli D22 yields kirromycin-sensitive progeny support these conclusions.
Keywords: protein biosynthesis, EF-Tu GTPase regulation, EF-Tu peptide pattern, tufB gene product, dominance of kirromycin sensitivity
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bachmann B. J., Low K. B., Taylor A. L. Recalibrated linkage map of Escherichia coli K-12. Bacteriol Rev. 1976 Mar;40(1):116–167. doi: 10.1128/br.40.1.116-167.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blumenthal T., Landers T. A., Weber K. Bacteriophage Q replicase contains the protein biosynthesis elongation factors EF Tu and EF Ts. Proc Natl Acad Sci U S A. 1972 May;69(5):1313–1317. doi: 10.1073/pnas.69.5.1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown S., Blumenthal T. Function and structure in ribonucleic acid phage Qbeta ribonucleic acid replicase. Effect of inhibitors of EF-Tu on ribonucleic acid synthesis and renaturation of active enzyme. J Biol Chem. 1976 May 10;251(9):2749–2753. [PubMed] [Google Scholar]
- Chinali G., Wolf H., Parmeggiani A. Effect of kirromycin on elongation factor Tu. Location of the catalytic center for ribosome-elongation-factor-Tu GTPase activity on the elongation factor. Eur J Biochem. 1977 May 2;75(1):55–65. doi: 10.1111/j.1432-1033.1977.tb11503.x. [DOI] [PubMed] [Google Scholar]
- DAVIS B. D., MINGIOLI E. S. Mutants of Escherichia coli requiring methionine or vitamin B12. J Bacteriol. 1950 Jul;60(1):17–28. doi: 10.1128/jb.60.1.17-28.1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furano A. V. Content of elongation factor Tu in Escherichia coli. Proc Natl Acad Sci U S A. 1975 Dec;72(12):4780–4784. doi: 10.1073/pnas.72.12.4780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobson G. R., Rosenbusch J. P. Abundance and membrane association of elongation factor Tu in E. coli. Nature. 1976 May 6;261(5555):23–26. doi: 10.1038/261023a0. [DOI] [PubMed] [Google Scholar]
- Jaskunas S. R., Lindahl L., Nomura M. Identification of two copies of the gene for the elongation factor EF-Tu in E. coli. Nature. 1975 Oct 9;257(5526):458–462. doi: 10.1038/257458a0. [DOI] [PubMed] [Google Scholar]
- LOWRY O. H., ROSEBROUGH N. J., FARR A. L., RANDALL R. J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951 Nov;193(1):265–275. [PubMed] [Google Scholar]
- Low B. Rapid mapping of conditional and auxotrophic mutations in Escherichia coli K-12. J Bacteriol. 1973 Feb;113(2):798–812. doi: 10.1128/jb.113.2.798-812.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas-Lenard J. Protein biosynthesis. Annu Rev Biochem. 1971;40:409–448. doi: 10.1146/annurev.bi.40.070171.002205. [DOI] [PubMed] [Google Scholar]
- Lupker J. H., Verschoor G. J., de Rooij F. W., Rörsch A., Bosch L. An Escherichia coli mutant with an altered elongation factor Tu. Proc Natl Acad Sci U S A. 1974 Feb;71(2):460–463. doi: 10.1073/pnas.71.2.460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monner D. A., Jonsson S., Boman H. G. Ampicillin-resistant mutants of Escherichia coli K-12 with lipopolysaccharide alterations affecting mating ability and susceptibility to sex-specific bacteriophages. J Bacteriol. 1971 Aug;107(2):420–432. doi: 10.1128/jb.107.2.420-432.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Normark S., Boman H. G., Matsson E. Mutant of Escherichia coli with anomalous cell division and ability to decrease episomally and chromosomally mediated resistance to ampicillin and several other antibiotics. J Bacteriol. 1969 Mar;97(3):1334–1342. doi: 10.1128/jb.97.3.1334-1342.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen S., Blumenthal R. M., Reeh S., Russell L. B., Lemaux P., Laursen R. A., Nagarkatti S., Friesen J. D. A mutant of Escherichia coli with an altered elongation factor Tu. Proc Natl Acad Sci U S A. 1976 May;73(5):1698–1701. doi: 10.1073/pnas.73.5.1698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen S., Reeh S. V. Analysis of the proteins synthesized in ultraviolet light-irradiated Escherichia coli following infection with the bacteriophages lambdadrifd 18 and lambdadfus-3. Mol Gen Genet. 1976 Mar 30;144(3):339–343. doi: 10.1007/BF00341733. [DOI] [PubMed] [Google Scholar]
- Sander G., Marsh R. C., Voigt J., Parmeggiani A. A comparative study of the 50S ribosomal subunit and several 50S subparticles in EF-T-and EF-G-dependent activities. Biochemistry. 1975 May 6;14(9):1805–1814. doi: 10.1021/bi00680a001. [DOI] [PubMed] [Google Scholar]
- Travers A. A., Kamen R. I., Schleif R. F. Factor necessary for ribosomal RNA synthesis. Nature. 1970 Nov 21;228(5273):748–751. doi: 10.1038/228748a0. [DOI] [PubMed] [Google Scholar]
- Vandekerckhove J., Van Montagu M. Sequence analysis of fluorescamine-stained peptides and proteins purified on a nanomole scale. Application to proteins of bacteriophage MS2. Eur J Biochem. 1974 May 2;44(1):279–288. doi: 10.1111/j.1432-1033.1974.tb03483.x. [DOI] [PubMed] [Google Scholar]
- Weissbach H., Redfield B., Kaback H. R. Nucleotide binding by Escherichia coli membranes and solubilized membrane proteins. Arch Biochem Biophys. 1969 Dec;135(1):66–74. doi: 10.1016/0003-9861(69)90517-7. [DOI] [PubMed] [Google Scholar]
- Wolf H., Chinali G., Parmeggiani A. Kirromycin, an inhibitor of protein biosynthesis that acts on elongation factor Tu. Proc Natl Acad Sci U S A. 1974 Dec;71(12):4910–4914. doi: 10.1073/pnas.71.12.4910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf H., Chinali G., Parmeggiani A. Mechanism of the inhibition of protein synthesis by kirromycin. Role of elongation factor Tu and ribosomes. Eur J Biochem. 1977 May 2;75(1):67–75. doi: 10.1111/j.1432-1033.1977.tb11504.x. [DOI] [PubMed] [Google Scholar]
- Wolf H., Zähner H. Stoffwechselprodukte von Mikroorganismen. 99. Kirromycin. Arch Mikrobiol. 1972;83(2):147–154. [PubMed] [Google Scholar]



