Abstract
The gene encoding F-box protein FBXW7 is frequently mutated in many human cancers. Although most previous studies have focused on the tumor-suppressive capacity of FBXW7 in tumor cells themselves, we determined that FBXW7 in the host microenvironment also suppresses cancer metastasis. Deletion of Fbxw7 in murine BM-derived stromal cells induced accumulation of NOTCH and consequent transcriptional activation of Ccl2. FBXW7-deficient mice exhibited increased serum levels of the chemokine CCL2, which resulted in the recruitment of both monocytic myeloid-derived suppressor cells and macrophages, thereby promoting metastatic tumor growth. Administration of a CCL2 receptor antagonist blocked the enhancement of metastasis in FBXW7-deficient mice. Furthermore, in human breast cancer patients, FBXW7 expression in peripheral blood was associated with serum CCL2 concentration and disease prognosis. Together, these results suggest that FBXW7 antagonizes cancer development in not only a cell-autonomous manner, but also a non-cell-autonomous manner, and that modulation of the FBXW7/NOTCH/CCL2 axis may provide a potential approach to suppression of cancer metastasis.
Introduction
Metastasis is a major cause of death in cancer patients, and elucidation of the genes and mechanisms that underlie this process is expected to provide a basis for the development of new cancer treatments. Such mechanisms have remained poorly understood because of the complexity of metastasis, which includes detachment of cancer cells from a primary tumor followed by their invasion into surrounding tissue, entry into the circulatory system, and invasion and proliferation in distant organs. In addition to the genomic variation among malignant tumor cells, recent research has focused on the relationship between cancer and the host environment. BM-derived cells (BMDCs) — including T cells (1), B cells (2), granulocytic and monocytic myeloid-derived suppressor cells (G-MDSCs and Mo-MDSCs, respectively) (3–6), macrophages (7–10), BM-derived stromal cells (BMSCs) (11, 12), hematopoietic progenitor cells (HPCs) (13), and endothelial progenitor cells (EPCs) (14) — play pivotal roles in promoting metastasis, including facilitation of tumor cell growth and invasion as well as of angiogenesis (15).
Tumor cells and surrounding stromal cells secrete various growth factors, cytokines, and chemokines that promote cancer development (16, 17). Chemokines promote tumor development and progression in addition to recruiting immune cells to tumor sites. The chemokine CCL2 (also known as monocyte chemoattractant protein–1 [MCP-1]) regulates the recruitment of monocytes, macrophages, and other inflammatory cells to sites of inflammation through interaction with its receptor, CCR2 (18). CCL2 also contributes to the recruitment of monocytes/macrophages to sites of pulmonary metastasis in mice with breast cancer and then promotes tumor outgrowth (19). Systemic administration of neutralizing antibodies against CCL2 in mouse cancer models has resulted in marked attenuation of tumor growth, reduction in tumor blood vessel density, and inhibition of metastasis (19–23).
FBXW7 (also known as Fbw7, Sel-10, hCdc4, or hAgo) is the F-box protein component of an Skp1–Cul1–F-box protein–type (SCF-type) ubiquitin ligase, in which it functions as a receptor responsible for substrate recognition. Most of the substrates of FBXW7 are growth promoters, including c-MYC (24, 25), NOTCH (26–28), cyclin E (29–31), c-JUN (32, 33), KLF5 (34, 35), and mTOR (36), and FBXW7 is therefore thought to serve as a tumor suppressor. Analysis of FBXW7 in many primary human tumors revealed that approximately 6% of the tumors harbored mutations in this gene (37). Mutations were detected most frequently in cholangiocarcinoma (35%) and T cell acute lymphocytic leukemia (T-ALL; 31%). Notably, 43% of the identified mutations were found to be missense mutations that resulted in amino acid substitutions at key arginine residues (Arg465 and Arg479) within the WD40 domain that are responsible for substrate recognition, which suggests that defective degradation of FBXW7 substrates leads to tumorigenesis.
Prior findings in genetic analyses of mice in which Fbxw7 is conditionally deleted in various tissues collectively support a pivotal role for FBXW7 in suppression of tumorigenesis in vivo. Conditional inactivation of Fbxw7 in the T cell lineage of mice induced the development of thymic lymphoma as a result of excessive c-MYC accumulation (38). More than half of BM-specific FBXW7-deficient mice developed T-ALL within 16 weeks, manifesting marked accumulation of NOTCH1 and c-MYC proteins (39, 40). FBXW7-null mice harboring a mutation in the adenomatous polyposis coli (Apc) gene (Fbxw7Δ/Δ Apcmin/+ mice) showed an increase in both number and size of intestinal tumors, and a consequently reduced survival rate, compared with Apcmin/+ mice (41). These various observations thus suggest that FBXW7 is a potent tumor suppressor in mice as well as in humans.
In the present study, we show that FBXW7 expression in the host environment is a key determinant of cancer metastasis. Metastasis was found to be enhanced in mice lacking FBXW7 in BM compared with control mice. We characterized the mechanism underlying this enhancement of metastasis: deletion of Fbxw7 resulted in NOTCH accumulation and consequent activation of Ccl2 gene transcription in BMSCs. The increased production of CCL2 by these cells likely promoted the formation of metastatic niches through recruitment of Mo-MDSC and macrophages. Inhibition of CCL2/CCR2 signaling reduced the frequency of metastasis in the FBXW7-deficient mice. Our results thus suggest that the FBXW7/NOTCH/CCL2 pathway plays a central role in the regulation of cancer metastasis.
Results
Deletion of Fbxw7 in BM promotes cancer metastasis in mice.
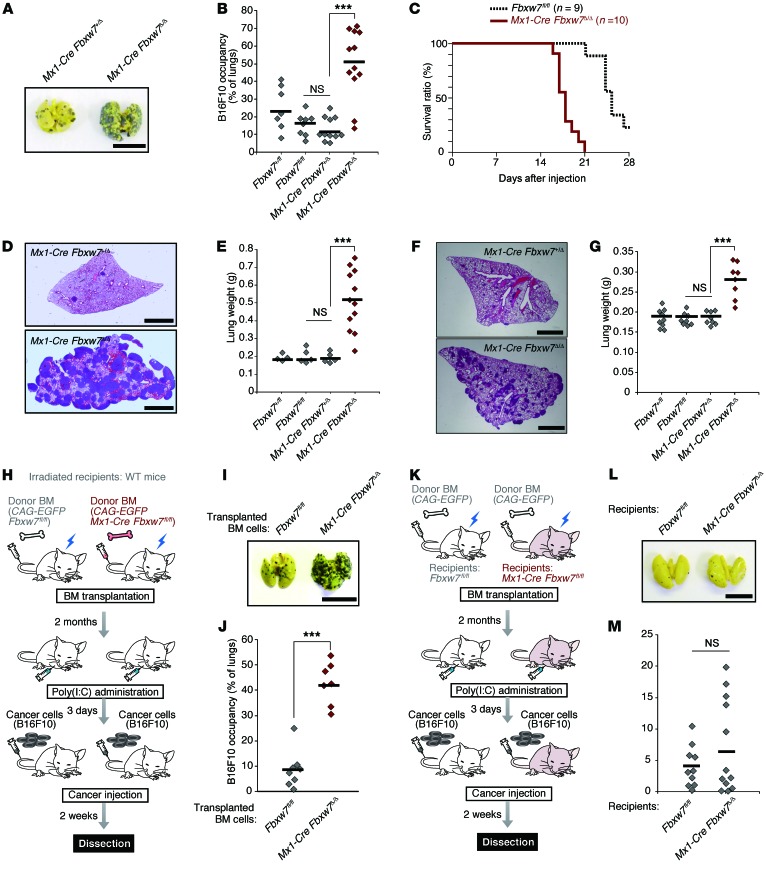
Most studies of FBXW7 have focused on its functions in tumor cells (42–44); little is known regarding the role of this protein in the host microenvironment with respect to tumor development. To investigate the role of FBXW7 in the host microenvironment, we transferred B16F10 melanoma cells into the tail vein of Mx1-Cre Fbxw7fl/fl mice that had been injected with polyinosinic:polycytidylic acid [poly(I:C)] to delete floxed Fbxw7 alleles selectively in BM (referred to hereafter as Mx1-Cre Fbxw7Δ/Δ mice). The frequency of metastasis of the melanoma cells to the lungs was markedly increased in Mx1-Cre Fbxw7Δ/Δ versus control mice (Figure 1, A and B), and this increased metastasis was accompanied by earlier death of the Mx1-Cre Fbxw7Δ/Δ mice (Figure 1C). Similar results were obtained when Lewis lung carcinoma (LLC) cells (Figure 1, D and E, and Supplemental Figure 1, A–C; supplemental material available online with this article; doi:10.1172/JCI78782DS1) or low–metastatic potential B16F1 melanoma cells (Figure 1, F and G, and Supplemental Figure 1, D–F) were injected into the tail vein of these mice. Thus, the level of FBXW7 in BM represents a key determinant of cancer metastasis in mice.
Figure 1. Fbxw7 deletion in BM promotes cancer metastasis in an intravenous tumor cell transplantation model.
(A and B) B16F10 cells were transplanted into Fbxw7+/fl (n = 7), Fbxw7fl/fl (n = 8), Mx1-Cre Fbxw7+/Δ (n = 11), and Mx1-Cre Fbxw7Δ/Δ (n = 12) mice. The gross appearance of the lungs (A) and their occupancy by tumor colonies (B) were examined. Horizontal bars in B indicate mean values. (C) Kaplan-Meier survival curves for Fbxw7fl/fl (n = 9) and Mx1-Cre Fbxw7Δ/Δ (n = 10) mice after injection of B16F10 cells. (D and E) LLC cells were transplanted into Fbxw7+/fl (n = 4), Fbxw7fl/fl (n = 5), Mx1-Cre Fbxw7+/Δ (n = 5), and Mx1-Cre Fbxw7Δ/Δ (n = 12) mice. (F and G) B16F1 cells were transplanted into Fbxw7+/fl (n = 9), Fbxw7fl/fl (n = 8), Mx1-Cre Fbxw7+/Δ (n = 8), and Mx1-Cre Fbxw7Δ/Δ (n = 8) mice. Lungs were subjected to H&E staining (D and F), and their gross weight was determined (E and G). (H–J) Metastasis assays performed in WT mice reconstituted with CAG-EGFP Fbxw7fl/fl (n = 8) or CAG-EGFP Mx1-Cre Fbxw7Δ/Δ (n = 7) donor BM. (K–M) Metastasis assays performed in Fbxw7fl/fl (n = 10) or Mx1-Cre Fbxw7Δ/Δ (n = 12) mice reconstituted with WT donor BM. Schematic representation (H and K), gross appearance of the lungs (I and L), and their occupancy by tumor colonies (J and M) are shown. Scale bars: 10 mm (A, I, and L); 2 mm (D and F). Horizontal bars in B, E, G, J, and M indicate means. ***P < 0.001, 1-way ANOVA and Bonferroni test (B, E, and G) or 2-tailed Student’s t test (J).
To examine whether ablation of Fbxw7 specifically in BM was indeed responsible for the enhanced metastasis in Mx1-Cre Fbxw7Δ/Δ mice, we transplanted BM cells from Mx1-Cre Fbxw7fl/fl or control Fbxw7fl/fl mice that also harbor a transgene for enhanced green fluorescent protein (EGFP) under the control of the CAG promoter into irradiated C57BL/6 mice (Figure 1H). The recipient mice were subsequently injected with poly(I:C) to delete floxed alleles of Fbxw7; 3 days after injection, B16F10 or LLC cancer cells were transferred to these mice. Metastasis to the lungs was more pronounced in mice receiving CAG-EGFP Mx1-Cre Fbxw7fl/fl BM cells than in those receiving the control cells (Figure 1, I and J, and Supplemental Figure 1, G and H). In contrast, a reciprocal experiment revealed no such enhancement of metastasis in Mx1-Cre Fbxw7fl/fl mice subjected to transplantation with BM from CAG-EGFP mice and injected with poly(I:C) (Figure 1, K–M). These results confirmed that the loss of FBXW7 in BM is indeed responsible for the increased frequency of metastasis observed in Mx1-Cre Fbxw7Δ/Δ mice.
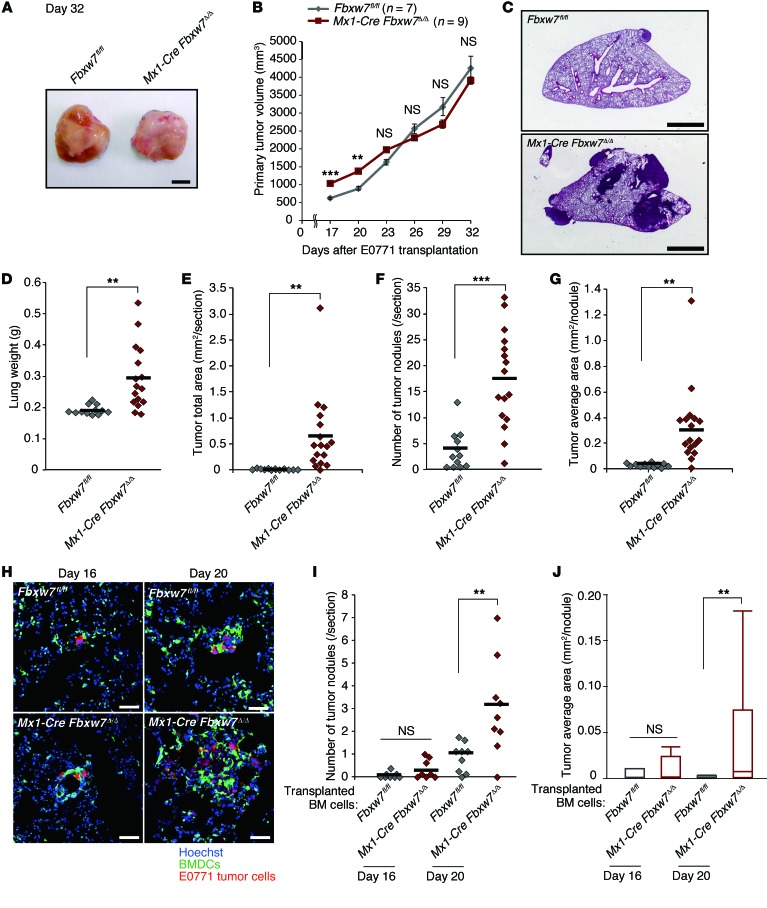
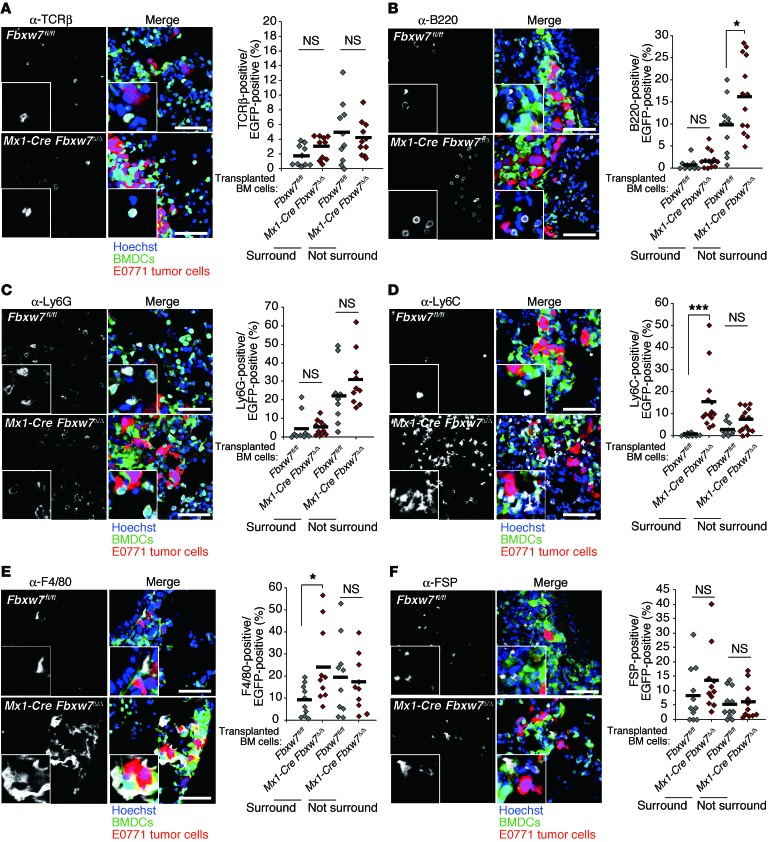
We also examined metastatic tumor growth in control and Mx1-Cre Fbxw7Δ/Δ mice after orthotopic transplantation of E0771 mouse breast cancer cells. Primary tumor growth was promoted in FBXW7-deficient mice on days 17 and 20, albeit not at later time points (Figure 2, A and B). Metastasis to the lungs was markedly enhanced in the mutant mice (Figure 2, C and D). In addition to lung weight, the total tumor area, number of tumor nodules, and average area per nodule in the lungs were greater for Mx1-Cre Fbxw7Δ/Δ mice than controls (Figure 2, D–G). We next monitored the progression of metastatic tumors in this model. Whereas we did not detect any tumor cells in the lungs at 12 days after cell transplantation, metastasis of E0771 cells was apparent in both control and Mx1-Cre Fbxw7Δ/Δ mice at 16 days (Figure 2, H–J). The number of tumor nodules and the average area per nodule did not differ between genotypes at 16 days after transplantation, but were significantly greater in Mx1-Cre Fbxw7Δ/Δ mice than in controls at 20 days. Although a premetastatic niche was previously shown to be formed by clusters of BMDCs (13), we found that such EGFP+ clusters were already present at day 0 (before E0771 cell transplantation) in the lungs of WT mice reconstituted with EGFP-labeled Mx1-Cre Fbxw7Δ/Δ or control BM cells (Supplemental Figure 2A). The number of these clusters did not change substantially with time after E0771 cell transplantation and did not differ between the genotypes (Supplemental Figure 2B). In contrast, the number of diffusely infiltrated BMDCs in the lungs was increased after tumor cell transplantation specifically in mice reconstituted with Mx1-Cre Fbxw7Δ/Δ BM cells (Supplemental Figure 2, A and C). Immunofluorescence analysis with antibodies against TCRβ (for T cells), B220 (for B cells), Ly6G (for G-MDSCs), Ly6C (for Mo-MDSCs), F4/80 (for monocytes/macrophages), fibroblast-specific protein (FSP; for stromal cells), MAC1 (for myeloid cells), c-KIT (for HPCs), and VE-cadherin (for EPCs) revealed that the number of Ly6C+, F4/80+, and MAC1+ cells increased among tumor-surrounding BMDCs, whereas only B220+ cells moderately increased in number among the nonsurrounding BMDCs (Figure 3, A–F, and Supplemental Figure 3, A–C). These results suggested that accumulation of Mo-MDSCs or of more differentiated macrophages might be responsible for the promotion of metastasis in Mx1-Cre Fbxw7Δ/Δ mice.
Figure 2. Fbxw7 deletion in BM promotes cancer metastasis in an orthotopic breast cancer transplantation model.
(A and B) E0771 cells were transplanted into the mammary fat pad of Fbxw7fl/fl (n = 11) and Mx1-Cre Fbxw7Δ/Δ (n = 13) mice. Primary tumor gross appearance after 32 days (A) and volume at the indicated times (B) are shown. (C–G) E0771 cells were transplanted into the mammary fat pad of Fbxw7fl/fl (n = 12) and Mx1-Cre Fbxw7Δ/Δ (n = 16) mice. After 32 days, lungs were subjected to H&E staining (C), and their gross weight was determined (D). Tumor total area (E), number of tumor nodules (F), and average tumor area (G) were calculated from the stained lung sections. (H–J) WT mice were reconstituted with BM cells of the indicated mice and subjected to orthotopic transplantation with tdTomato-labeled E0771 cells. Lungs were subjected to fluorescence microscopy for detection of BMDCs (green), tumor cells (red), and cell nuclei (Hoechst 33238) (H), and the number of tumor nodules (I) and average tumor area (J) were determined 16 (n = 8 per group) or 20 (n = 9 per group) days after tumor cell transplantation. Scale bars: 2 mm (C); 100 μm (H). Data in B are mean ± SEM; horizontal bars in D–G and I indicate means; box and whisker plots in J depict the smallest value, lower quartile, median, upper quartile, and largest value. **P < 0.01, ***P < 0.001, 2-tailed Student’s t test (B, D–G, I, and J).
Figure 3. Ly6C+ Mo-MDSCs and F4/80+ monocytes/macrophages accumulate in the microenvironment of metastatic tumors in the lungs of mice reconstituted with FBXW7-deficient BM cells.
Representative immunohistofluorescence staining (white) for TCRβ (A), B220 (B), Ly6G (C), Ly6C (D), F4/80 (E), and FSP (F) for lung sections from WT mice reconstituted with CAG-EGFP Fbxw7fl/fl (n = 10 [A, B, D, and E]; 9 [C]; 11 [F]) or CAG-EGFP Mx1-Cre Fbxw7Δ/Δ (n = 10 [A, C, and E]; 12 [B]; 14 [D]; 11 [F]) BM cells and subjected to orthotopic transplantation with tdTomato-labeled E0771 cells (20 days before analysis) as in Figure 2H. Intrinsic fluorescence of EGFP (green), tdTomato (red), and Hoechst 33258 (blue) was also imaged. Higher-magnification images (×2) are shown in the insets. Scale bars: 100 μm. The percentage of EGFP+ BMDCs positive for each marker in tumor-surrounding and nonsurrounding regions was quantified; horizontal bars indicate means. *P < 0.05, ***P < 0.001, 1-way ANOVA and Bonferroni test.
We also characterized cells in the peripheral blood of mice at various times from 2 days before to 32 days after tumor cell transplantation. The frequency of MAC1+Ly6G–Ly6C+ Mo-MDSCs and MAC1+F4/80+CD115+ monocytes/macrophages in peripheral blood increased in Mx1-Cre Fbxw7Δ/Δ versus control mice before tumor cell transplantation, whereas the frequency of MAC1+Ly6G+Ly6C+ immature MDSCs did not differ between the genotypes at this time (Supplemental Figure 2, D–F). However, the frequency of these latter cells in peripheral blood increased transiently — to a greater extent in Mx1-Cre Fbxw7Δ/Δ mice than in control mice — between days 16 and 24. In contrast, the frequency of MAC1+Ly6G–Ly6C+ Mo-MDSCs in BM did not differ between genotypes at day 0 or day 20 (Supplemental Figure 2, G and H). Collectively, these results suggested that the increased infiltration of BMDCs during the early phase of metastasis to the lungs in Mx1-Cre Fbxw7Δ/Δ mice might represent increased migration or recruitment of Mo-MDSCs and macrophages, rather than differentiation of these cells in BM.
Serum chemokine levels are increased in mice lacking FBXW7 in BM.
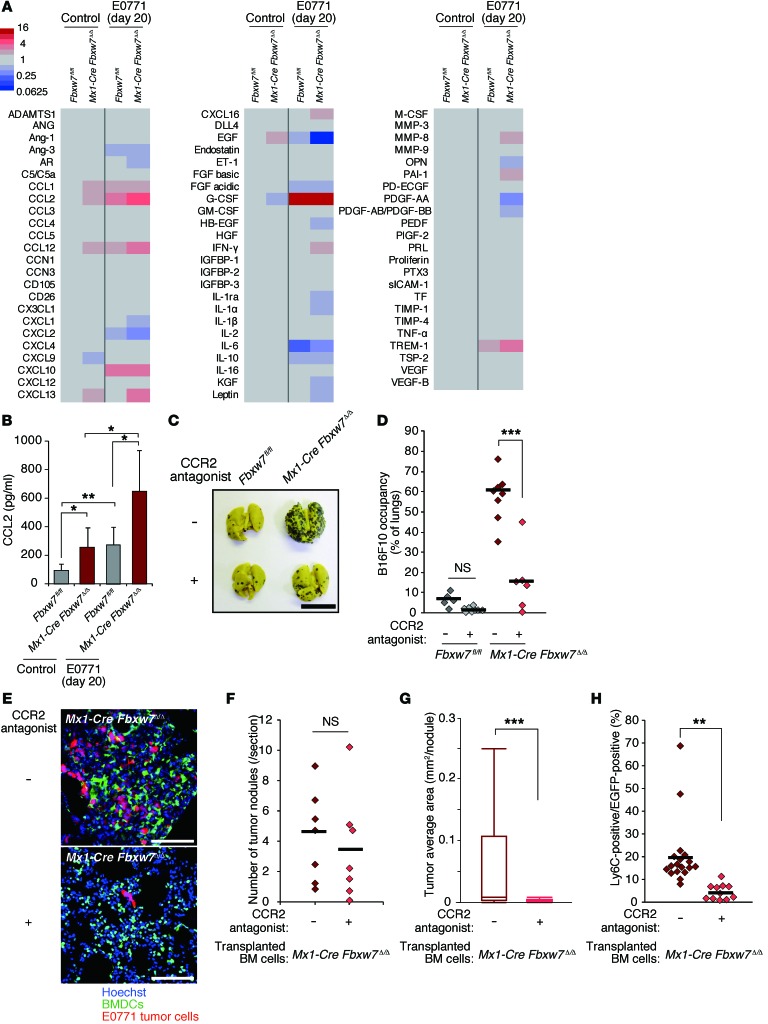
To explore the mechanism responsible for this increased mobilization of Mo-MDSCs and macrophages induced by FBXW7 deficiency, we examined the serum concentrations of various cytokines before and after E0771 cell transplantation. Cytokine array analysis revealed that the levels of CCL2, CCL12 (also known as MCP-5), and the chemokine CXCL13 (also known as B lymphocyte chemoattractant [BLC]) were increased more than 2-fold in Mx1-Cre Fbxw7Δ/Δ versus control mice both before and after E0771 cell transplantation (Figure 4A and Supplemental Figure 4). Both CCL2 and CCL12 induce the migration of monocytes/macrophages by binding to their common receptor, CCR2. CCL12 is mainly secreted from macrophages, but Fbxw7 ablation in the macrophages of LysM-Cre Fbxw7fl/fl mice did not affect metastasis frequency (Supplemental Figure 5, A and B), which suggests that CCL12 is not largely responsible for the promotion of metastasis. We thus focused on CCL2 and found greater serum CCL2 concentrations in Mx1-Cre Fbxw7Δ/Δ mice compared with control mice both before and after E0771 cell transplantation (Figure 4B).
Figure 4. Increased production of CCL2 promotes metastasis in FBXW7-deficient mice.
(A) Serum cytokine levels in Fbxw7fl/fl and Mx1-Cre Fbxw7Δ/Δ mice after orthotopic transplantation of tdTomato-labeled E0771 cells or in nontransplant controls. Optical densities for cytokines are indicated according to the color scale. See also Supplemental Figure 5. (B) Serum concentration of CCL2, determined by ELISA, in the indicated mice transplanted with tdTomato-labeled E0771 cells. Data are mean ± SD (n = 4 per group). (C and D) Gross appearance of the lungs (C) and their occupancy by visible B16F10 tumor colonies (D) for Fbxw7fl/fl (n = 5 [not treated]; 8 [treated]) and Mx1-Cre Fbxw7Δ/Δ (n = 8 [not treated]; 6 [treated]) mice injected with B16F10 cells and treated or not with the CCR2 antagonist propagermanium. (E–H) WT mice were reconstituted with CAG-EGFP Mx1-Cre Fbxw7Δ/Δ BM cells, subjected to orthotopic transplantation with tdTomato-labeled E0771 cells, and treated or not with propagermanium (n = 7). At 20 days after tumor cell transplantation, lungs were subjected to fluorescence microscopy (E), and number of tumor nodules (F) and average tumor area (G) were determined. (H) The percentage of EGFP+ BMDCs positive for Ly6C in the tumor-bearing lungs of mice as in E–G was determined by immunohistofluorescence analysis. Scale bars: 10 mm (C); 100 μm (E). Horizontal bars in D, F, and H indicate means; box and whisker plots in G depict the smallest value, lower quartile, median, upper quartile, and largest value. *P < 0.05, **P < 0.01, ***P < 0.001, 1-way ANOVA and Bonferroni test (B and D) or 2-tailed Student’s t test (F–H).
To examine whether the enhanced metastasis apparent in FBXW7-deficient mice is dependent on the CCL2/CCR2 pathway, we treated Mx1-Cre Fbxw7Δ/Δ mice with propagermanium, a CCR2 antagonist. The extent of B16F10 or E0771 cell metastasis in Mx1-Cre Fbxw7Δ/Δ mice was significantly attenuated by propagermanium administration (Figure 4, C–H). The frequency of EGFP+Ly6C+ cells in the lungs of WT mice reconstituted with EGFP-labeled Mx1-Cre Fbxw7Δ/Δ BM cells and injected with E0771 cells was also significantly reduced by propagermanium treatment (Figure 4H). Unexpectedly, the number of tumor nodules in the lungs was not affected by propagermanium treatment, whereas the size of each nodule was markedly reduced in the treated mice (Figure 4, F and G). Collectively, these results suggested that the increased production of CCL2 by FBXW7-deficient BMDCs promotes the growth of tumors that have already metastasized to the lungs.
The Ccl2 gene is activated in FBXW7-deficient BMSCs.
We next investigated which cells of the BM are responsible for the metastasis promotion and increased serum CCL2 concentration observed in FBXW7-deficient mice. For these experiments, we used mice deficient in FBXW7 in different lineages. LysM-Cre Fbxw7–/Δ mice, in which the Cre gene is activated to delete floxed Fbxw7 alleles only in the granulocyte-macrophage lineage, did not show an increase in metastasis frequency compared with control mice (Supplemental Figure 5, A and B). Furthermore, neither Lck-Cre Fbxw7Δ/Δ nor Cd19-Cre Fbxw7Δ/Δ mice (Fbxw7 deletion specific to T and B cells, respectively) showed enhancement of metastasis like that in Mx1-Cre Fbxw7Δ/Δ mice (Supplemental Figure 5, C–F). These results suggested that neither myeloid (granulocyte and macrophage) nor lymphoid (T and B cell) lineages are responsible for the promotion of metastasis induced by BM Fbxw7 ablation.
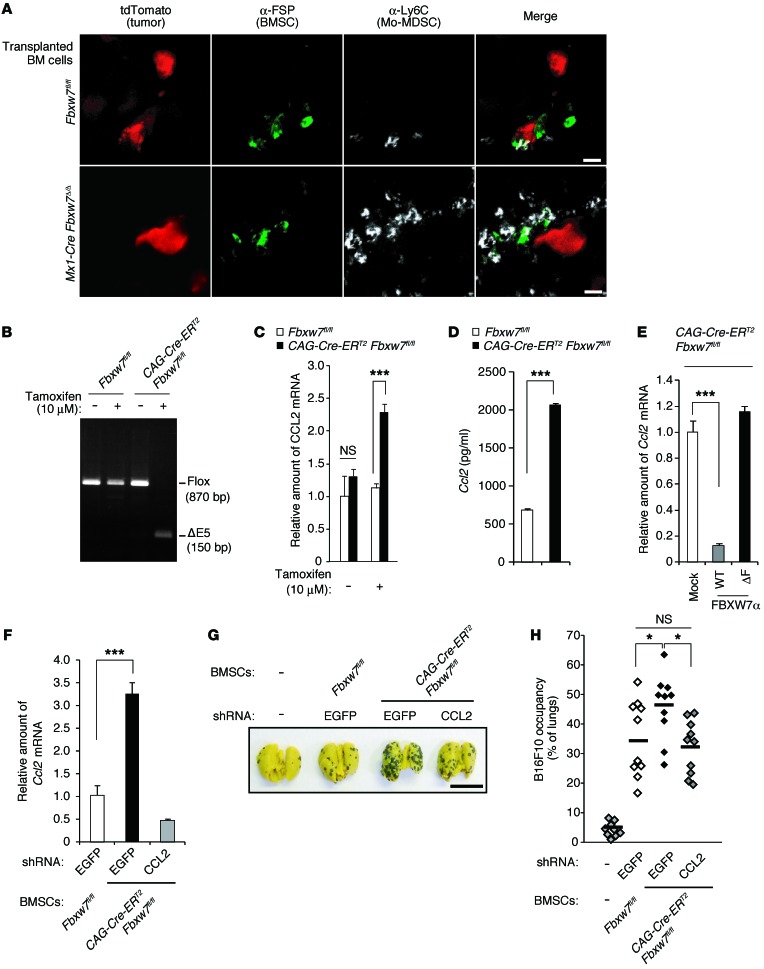
Neither the serum level of CCL2 nor the frequency of MAC1+F4/80+ monocytes/macrophages in peripheral blood differed between LysM-Cre Fbxw7–/Δ and control mice (Supplemental Figure 6, A and B), which suggests that monocytes/macrophages are not the major source of CCL2 produced in response to Fbxw7 loss. We found that FSP+ BMSCs colocalized with metastatic tumor cells and Mo-MDSCs in the lungs of mice after orthotopic transplantation of E0771 cells (Figure 5A). Given that BMSCs were previously shown to secrete CCL2 and to contribute to the emigration of monocytes from BM (45, 46), we hypothesized that BMSCs might be a major source of CCL2 production in our models. To test this hypothesis, we isolated BMSCs from BM of CAG-Cre-ERT2 Fbxw7fl/fl (or control Fbxw7fl/fl) mice and treated them with 10 μM tamoxifen to induce deletion of floxed Fbxw7 alleles (Figure 5B). The abundance of Ccl2 mRNA was increased in CAG-Cre-ERT2 Fbxw7Δ/Δ BMSCs compared with control cells (Figure 5C). The amount of CCL2 released from CAG-Cre-ERT2 Fbxw7Δ/Δ BMSCs into the culture medium was also substantially greater than that released from control cells (Figure 5D). Furthermore, the introduction of WT Fbxw7a cDNA into CAG-Cre-ERT2 Fbxw7Δ/Δ BMSCs resulted in a marked decrease in the abundance of Ccl2 mRNA, whereas introduction of cDNA for a mutant form of Fbxw7a that lacks the F-box domain (ΔF) had no such effect (Figure 5E), which suggests that FBXW7 negatively regulates CCL2 production in BMSCs.
Figure 5. Increased Ccl2 gene expression in FBXW7-deficient BMSCs promotes cancer metastasis.
(A) Representative immunohistofluorescence staining of Ly6C and FSP in lung sections from WT mice reconstituted with indicated BM cells and subjected to orthotopic transplantation with tdTomato-labeled E0771 cells (20 days before analysis). (B) Genomic PCR analysis of BMSCs from the indicated mice incubated in the absence or presence of 10 μM tamoxifen. The positions of amplified fragments corresponding to floxed and exon 5–deleted (ΔE5) Fbxw7 alleles are indicated. (C) Relative abundance of Ccl2 mRNA in BMSCs from the indicated mice. (D) Concentration of CCL2 released into culture supernatants from the indicated BMSCs. (E) Relative abundance of Ccl2 mRNA in CAG-Cre-ERT2 Fbxw7Δ/Δ BMSCs infected with retroviruses encoding WT or ΔF mutant forms of FBXW7α or with the empty virus (Mock). (F) Relative abundance of Ccl2 mRNA in BMSCs from Fbxw7fl/fl or CAG-Cre-ERT2 Fbxw7fl/fl mice treated with tamoxifen and subjected to RNAi with shRNA vectors targeting EGFP (control) or CCL2. (G and H) Gross appearance of the lungs (G) and their occupancy by visible B16F10 colonies (H) for WT mice 2 weeks after injection both with B16F10 cells and with BMSCs isolated from the indicated mice and treated as in F (n = 10 per group). Scale bars: 10 μm (A), 10 mm (G). Data are mean ± SD (C–F); horizontal bars in H indicate means. *P < 0.05, ***P < 0.001, 1-way ANOVA and Bonferroni test (C, E, F, and H) or 2-tailed Student’s t test (D).
To examine whether the increased CCL2 production by BMSCs is responsible for the promotion of cancer metastasis in FBXW7-deficient mice, we depleted CAG-Cre-ERT2 Fbxw7Δ/Δ BMSCs of CCL2 by shRNA-mediated RNAi (Figure 5F) and then transferred these cells, together with B16F10 melanoma cells, into recipient mice via the tail vein. BMSCs were detected in many tissues, such as BM and lungs, even 4 months after transplantation (47). The extent of lung metastasis in recipient mice was increased by Fbxw7Δ/Δ versus control BMSCs when coinjected with melanoma cells (Figure 5, G and H). However, this effect of Fbxw7 deletion was abolished by depletion of CCL2 in BMSCs, which suggests that the increased production of CCL2 by FBXW7-deficient BMSCs contributes to the promotion of metastasis.
NOTCH accumulation in FBXW7-deficient BMSCs promotes metastasis by increasing CCL2 production.
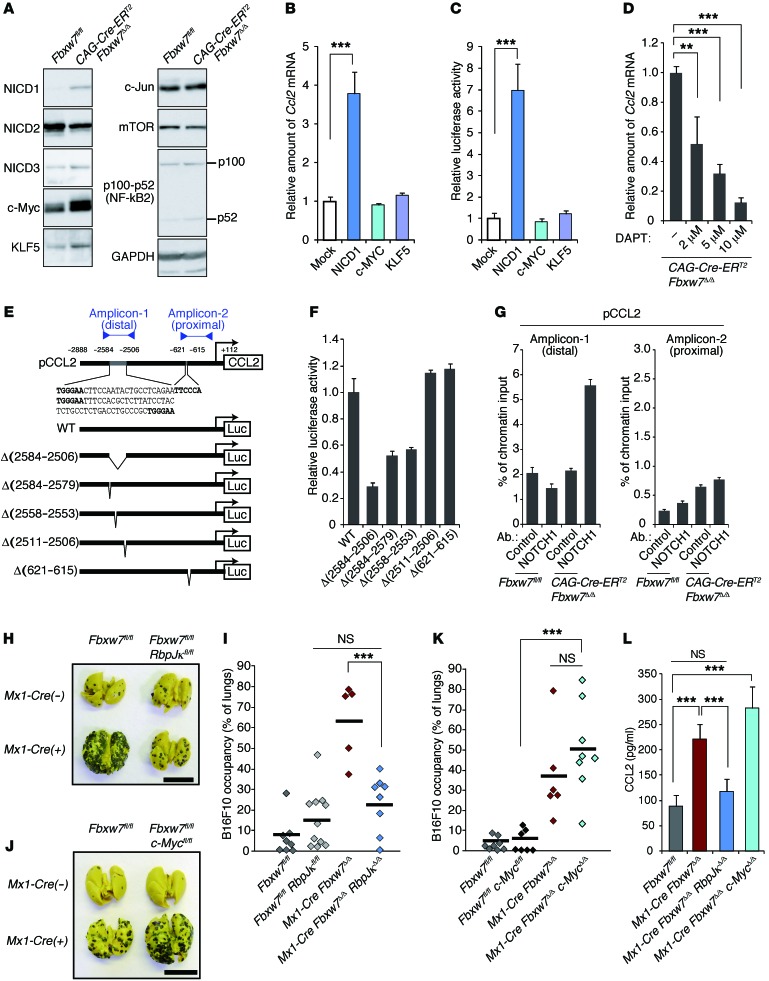
Immunoblot analysis revealed that, among the FBXW7 substrates examined, NOTCH1 intracellular domain (NICD1), c-MYC, and KLF5 accumulated at high levels in FBXW7-deficient BMSCs (Figure 6A). Forced expression of NICD1 in WT BMSCs resulted in a marked increase both in the abundance of Ccl2 mRNA and in the activity of the Ccl2 gene promoter, whereas that of c-MYC or KLF5 had no such effects (Figure 6, B and C). Inhibition of NOTCH signaling in FBXW7-deficient BMSCs by exposure to the γ-secretase inhibitor N-[N-(3,5-difluorophenacetyl)-L-alanyl]-S-phenylglycine t-butyl ester (DAPT) resulted in a concentration-dependent reduction in the amount of Ccl2 mRNA (Figure 6D). The promoter of the mouse Ccl2 gene contains 4 consensus sequences for NOTCH binding (Figure 6E and ref. 48), and a luciferase reporter assay with WT and mutant forms of this promoter indicated that the first 2 upstream elements are required for full promoter activity (Figure 6F). This finding was consistent with the results of ChIP analysis showing that NICD1 was associated with the distal region of the Ccl2 gene promoter in Fbxw7Δ/Δ BMSCs (Figure 6G). Together, these observations suggested that the NOTCH/CCL2 pathway in BMSCs contributes to the promotion of metastasis in FBXW7-deficient mice.
Figure 6. NOTCH accumulation in FBXW7-deficient BMSCs promotes CCL2 expression and cancer metastasis.
(A) Immunoblot analysis of FBXW7 substrates in the indicated BMSCs. (B) Relative abundance of Ccl2 mRNA in WT BMSCs infected with retroviruses encoding NICD1, c-MYC, or KLF5. (C) Luciferase assay for the Ccl2 gene in BMSCs infected with retroviruses for NICD1, c-MYC, or KLF5. (D) Relative abundance of Ccl2 mRNA in CAG-Cre-ERT2 Fbxw7Δ/Δ BMSCs incubated with DAPT. (E) WT and mutant forms of the mouse Ccl2 gene promoter fused to the firefly luciferase gene. Consensus binding sequences for NOTCH–RBP-Jκ are shown in bold. Proximal and distal amplicons in G are indicated. (F) Luciferase assay for the Ccl2 gene in CAG-Cre-ERT2 Fbxw7Δ/Δ BMSCs. (G) ChIP analysis of the Ccl2 gene promoter. Immunoprecipitation was performed with antibodies against NOTCH1 or with control IgG. (H and I) Intravenous transplantation with B16F10 cells for Fbxw7fl/fl (n = 8), Fbxw7fl/fl RbpJκfl/fl (n = 11), Mx1-Cre Fbxw7Δ/Δ (n = 5), and Mx1-Cre Fbxw7Δ/Δ RbpJκΔ/Δ (n = 8) mice. (J and K) Intravenous transplantation with B16F10 cells for Fbxw7fl/fl (n = 8), Fbxw7fl/fl c-Mycfl/fl (n = 7), Mx1-Cre Fbxw7Δ/Δ (n = 6), and Mx1-Cre Fbxw7Δ/Δ c-MycΔ/Δ (n = 8) mice. Gross appearance of the lungs (H and J) and their occupancy by B16F10 colonies (I and K) are shown. (L) Serum concentration of CCL2, determined by ELISA. Scale bars: 10 mm (H and J). Data are mean ± SD (n = 3) (B–D, F, G, and L); horizontal bars in I and K indicate means. **P < 0.01, ***P < 0.001, 1-way ANOVA and Bonferroni test (B–D, I, K, and L).
We further evaluated this notion by genetic analyses. Additional ablation of RBP-Jκ, an essential cofactor for NOTCH-dependent transactivation, markedly attenuated the enhanced metastasis observed in Mx1-Cre Fbxw7Δ/Δ mice (Figure 6, H and I). In contrast, inactivation of c-MYC in Mx1-Cre Fbxw7Δ/Δ mice had no such effect (Figure 6, J and K). Additional ablation of RBP-Jκ, but not that of c-MYC, also reduced the serum concentration of CCL2 to the control level in Mx1-Cre Fbxw7Δ/Δ mice (Figure 6L).
Low FBXW7 expression in the host microenvironment is associated with poor prognosis in breast cancer patients.
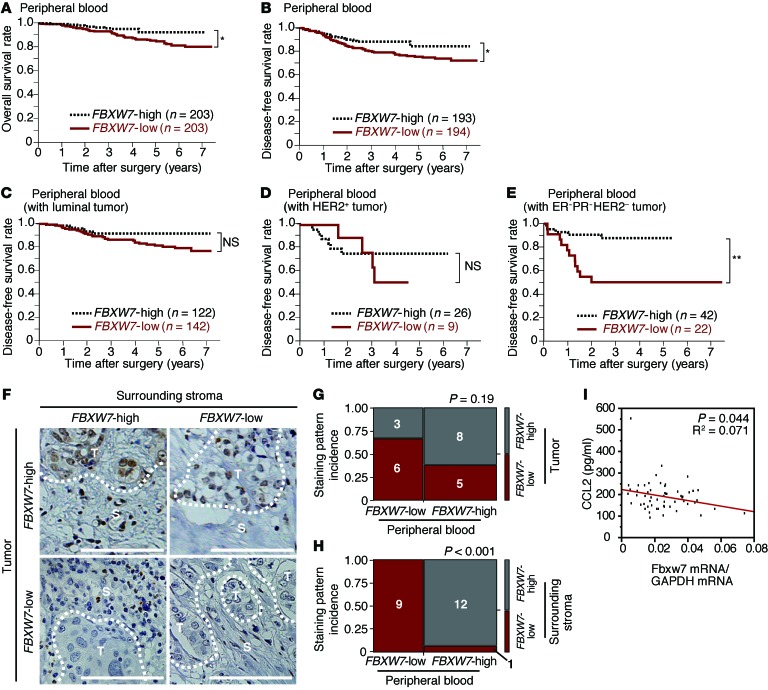
To extend our observations in mice to humans, we measured the abundance of FBXW7 mRNA in peripheral blood of breast cancer specimens and examined the relationship between FBXW7 mRNA abundance and prognosis. The prognosis of individuals with low FBXW7 expression in peripheral blood was significantly poorer than that of those in the corresponding high-FBXW7 group (Figure 7, A and B), consistent with our model. The difference in prognosis between the low- and high-expression groups was even more pronounced when the analysis was restricted to patients with tumors triple-negative for estrogen receptor (ER), progesterone receptor (PR), and HER2 (Figure 7, C–E) or to those with tumors of high histological grade or low stage (Supplemental Figure 7, A–G). The frequency of CD45– circulating tumor cells that expressed CD326 (also known as EpCAM) was at most 0.003% of total blood mononuclear cells (Supplemental Figure 7H), which suggests that the contribution of such cells to the total abundance of FBXW7 mRNA in peripheral blood is negligible.
Figure 7. Clinical relevance of FBXW7 expression in breast cancer patients.
(A and B) Kaplan-Meier curves for overall (A) and disease-free (B) survival of breast cancer patients (n = 406) classified according to the abundance of FBXW7 mRNA in peripheral blood. (C–E) Kaplan-Meier curves for disease-free survival of breast cancer patients with luminal (C), HER2+ (D), or triple-negative ER–PR–HER2– (E) tumors classified according to the abundance of FBXW7 mRNA in peripheral blood. *P < 0.05, **P < 0.01, log-rank test. (F) Representative immunohistochemical staining patterns for FBXW7 in breast cancer patients: positive in both primary tumor cells and stroma, positive in tumor cells only, positive in stroma only, or negative in both tumor cells and stroma. T, tumor cells; S, surrounding stromal cells. Scale bars: 100 μm. (G and H) Mosaic plots summarizing the abundance of FBXW7 mRNA in peripheral blood and FBXW7 expression in tumor cells (G) and surrounding stroma (H) for the indicated numbers of breast cancer patients (n = 22). P values for the association between these parameters were calculated by χ2 test. (I) Correlation between the abundance of FBXW7 mRNA in peripheral blood and the serum CCL2 concentration in breast cancer patients (n = 57).
Immunohistochemical analysis of FBXW7 in primary tumor lesions revealed that the abundance of FBXW7 mRNA in peripheral blood was not significantly correlated with FBXW7 expression in tumor cells, but was highly correlated with that in surrounding stromal cells (Figure 7, F–H). These results suggested that the abundance of FBXW7 mRNA in peripheral blood is a marker for FBXW7 expression in stromal cells that surround tumor cells. We also observed a negative correlation between FBXW7 mRNA abundance in peripheral blood and serum CCL2 concentration (Figure 7I). High serum levels of CCL2 were associated with poor prognosis in breast cancer patients (S. Akiyoshi and K. Mimori, unpublished observation). These data support our concept that FBXW7 ablation gives rise to increased CCL2 production, facilitating metastatic tumor growth (Figure 8).
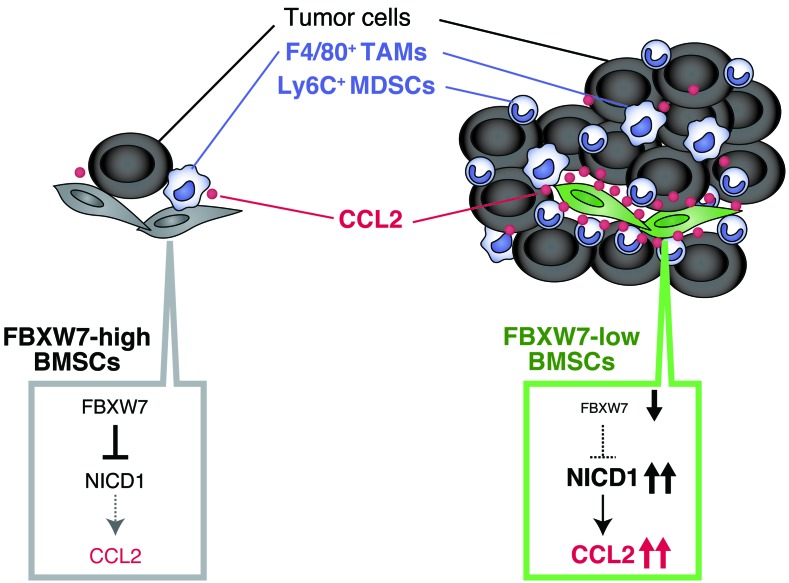
Figure 8. Promotion of cancer metastasis by loss of FBXW7 in the host environment.
Loss of FBXW7 in BMSCs results in accumulation of NICD1 and increased secretion of CCL2, which in turn promotes recruitment of Mo-MDSCs and macrophages. These cells then promote the growth of tumors that have already colonized the lungs. TAM, tumor-associated macrophage.
Discussion
The FBXW7 gene is a potent tumor suppressor, as evidenced by the many corresponding mutations associated with human cancer (37) as well as by the tumor formation observed in conditional knockout mice (38, 39, 41). The anticancer function of FBXW7 is thought to be mediated by specific ubiquitylation both of growth-promoting oncoproteins — such as c-MYC (24, 25), NOTCH (26, 27, 49), cyclin E (29–31), c-JUN (32, 33), KLF5 (34, 35), and mTOR (36) — and of the antiapoptotic molecule MCL-1 (50, 51). Most previous studies have focused on the tumor-suppressive role of FBXW7 in tumor cells themselves. The putative tumor suppressor C/EBPδ was shown to inhibit FBXW7 expression and to promote mammary tumor metastasis through attenuation of FBXW7-dependent degradation of mTOR in tumor cells (52). We have now discovered what we believe to be a new aspect of FBXW7 function in tumor suppression — namely, its role in the host environment to suppress cancer metastasis. Our data also provide mechanistic insight into the suppression of cancer metastasis by FBXW7. We found that NOTCH, one of the major substrates of FBXW7, activated transcription of the gene encoding CCL2, one of the most well-characterized chemokines with respect to cancer development. Our human clinical data, showing that reduced FBXW7 expression in peripheral blood was associated with a poor prognosis in breast cancer patients, appeared to be consistent with our experimental results in mice. We therefore propose that the FBXW7/NOTCH/CCL2 axis in the host environment limits cancer metastasis.
CCL2 is thought to be secreted from both cancer cells and noncancer cells in the tumor environment. In a xenograft model in which human cancer cells were transplanted into mice, administration of antibodies specific for human or mouse CCL2 inhibited tumor growth and metastasis, which supports the notion that CCL2 secreted from both the tumor and the host environment plays a key role in tumor development (19, 21). It is likely that the FBXW7/NOTCH1/CCL2 axis in cancer cells also plays a key role in metastasis. We showed here that the level of FBXW7 expression in peripheral blood was related to prognosis in breast cancer patients. These results thus suggest that constitutional variability in FBXW7 expression might be an important determinant of prognosis. We propose that the level of FBXW7 expression is a potentially powerful prognostic marker for cancer patients in general, and that targeting the CCL2/CCR2 system might prove a rational approach for preventing cancer metastasis. Indeed, we found that the CCR2 antagonist propagermanium had a marked inhibitory effect on cancer metastasis in mice. Propagermanium is currently administered clinically for the treatment of individuals with hepatitis B virus infection, and its long-term safety has been well demonstrated. Our results suggest that evaluation of this drug for its ability to inhibit cancer metastasis in humans is warranted.
We found that the number of tumor nodules in the lungs and the average area per nodule were greater in FBXW7-deficient mice than in controls subjected to orthotopic transplantation of breast cancer cells. Unexpectedly, however, treatment with propagermanium did not affect the number of tumor nodules, although the size of each nodule and the frequency of associated Mo-MDSCs were markedly reduced. Collectively, these results suggest that CCL2-dependent infiltration of Mo-MDSCs in the lungs influences the growth of established metastatic tumors rather than the recruitment of tumor cells to the lungs, which might be regulated by other factors, such as CXCL13, triggering receptor expressed on myeloid cells 1 (TREM1); epidermal growth factor, CXCL2 (also known as macrophage inflammatory protein 2α [MIP-2α]); and PDGF-AA. The serum levels of these proteins were increased or decreased more than 2-fold in Mx1-Cre Fbxw7Δ/Δ mice compared with controls after E0771 cell transplantation. Further studies are needed to elucidate the molecular mechanisms responsible for the increase in tumor cell engraftment in FBXW7-deficient mice.
In the present study, infiltration of Mo-MDSCs and macrophages into metastatic tumor lesions was promoted by the increased production of chemokines such as CCL2 in FBXW7-deficient mice. We found that BMSCs likely represent a major source of CCL2 production, although a possible contribution of other BMDCs cannot be excluded. In humans, CCL2 produced from other cell types — such as fibroblasts, endothelial cells, and smooth muscle cells — is also thought to promote cancer metastasis (53–56). Our identification of FBXW7 and NOTCH as upstream regulators of CCL2 expression provides both important insight into the mechanism by which production of this chemokine is regulated and a basis for the development of new strategies for cancer treatment.
Methods
Analysis of human clinical specimens.
All clinical results in this study are from retrospective studies. Peripheral blood specimens were obtained from 406 Japanese women with breast cancer who underwent surgery between 2000 and 2005 at the National Kyushu Cancer Center. All patients were clearly identified as having breast cancer based on clinicopathologic findings, and none underwent chemotherapy or radiotherapy before surgery. Collection of peripheral blood through a venous catheter for the measurement of FBXW7 mRNA was performed immediately before surgery with the patients under general anesthesia. The initial 1.0 ml of peripheral blood was discarded to avoid contamination by skin cells; the second 1.0 ml was mixed with 4.0 ml Isogen-LS (Nippon Gene) for extraction of total RNA.
Mice.
Fbxw7fl/fl mice, homozygous for the floxed Fbxw7 allele (38), were crossed with Mx1-Cre transgenic mice (provided by K. Rajewsky, University of Cologne, Cologne, Germany; ref. 57), and deletion of the floxed allele in the resulting offspring was induced by intraperitoneal injection (6 total injections on alternate days) with 20 μg poly(I:C) (Calbiochem) per gram of body weight. Deletion of exon 5 of the floxed Fbxw7 allele was confirmed by PCR analysis of genomic DNA as previously described (38). Fbxw7fl/fl mice were also crossed with RbpJκfl/fl mice (provided by T. Honjo, Kyoto University, Kyoto, Japan; ref. 58) or c-Mycfl/fl mice (provided by I.M. de Alborán, Centro Nacional de Biotecnologia/Consejo Superior de Investigaciones Científicas [CNB/CSIC], Madrid, Spain; ref. 59) or with Lck-Cre (60), LysM-Cre (61), Cd19-Cre (62), CAG-Cre-ERT2 (63), or CAG-EGFP (provided by M. Okabe, Osaka University, Osaka, Japan; ref. 64) transgenic mice.
BM transplantation.
C57BL/6 or other recipient mice (8 weeks of age) were irradiated with a lethal dose (11 Gy) of γ rays and injected via the tail vein with BM cells (2.0 × 106 in 100 μl PBS) isolated from 8-week-old CAG-EGFP, CAG-EGFP Fbxw7fl/fl, or CAG-EGFP Mx1-Cre Fbxw7fl/fl mice. At 2 months after transplantation, recipients were injected with poly(I:C) as described above to delete floxed Fbxw7 alleles; at 3 days after the final poly(I:C) injection, animals were injected with B16F10, LLC, or E0771 cells as described below. Recipient peripheral blood cells were examined for chimerism by flow cytometry each month after BM transplantation as well as at the time of lung dissection.
Assay of tumor metastasis.
Suspensions of B16F10 (2.0 × 105), B16F1 (2.0 × 105), or LLC (5.0 × 105) cells in PBS were injected into the tail vein of 8- to 11-week-old host mice. After 2 weeks, the animals were killed, and the lungs were removed and fixed in Bouin’s solution or 4% paraformaldehyde. Lung occupancy by visible B16F10 tumor colonies was analyzed using NIH ImageJ. E0771 cells (5.0 × 105) were injected subcutaneously into the mammary fat pad. Tumor volume (in mm3) was measured with calipers and calculated as (w2 × l)/2. For stable expression of tdTomato, E0771 cells were infected with a lentivirus encoding MYC epitope–tagged tdTomato for 2 days. Mice were fed normal chow without or with supplementation with 0.005% propagermanium (3-oxygemylpropinic acid polymer; provided by Sanwa Kagaku Kenkyusho Co.) beginning 1 day before cancer cell injection. Experiments were randomized, and investigators were blinded during experiments in the animal studies.
Cell culture.
B16F10 (provided by Cell Resource Center of Tohoku University), LLC (provided by Cell Resource Center of Tohoku University), B16F1 (provided by S. Okano, Kyushu University, Fukuoka, Japan), and E0771 (CH3 BioSystems) cells were maintained in RPMI 1640 medium (Sigma-Aldrich) supplemented with 10% fetal bovine serum (Invitrogen), 1 mM sodium pyruvate, 100 U/ml penicillin, 100 μg/ml streptomycin, 2 mM l-glutamine, and 10 ml/l nonessential amino acids (Gibco). BMSCs were isolated from BM collected from the tibia and femur of 8- to 10-week-old mice, and they were cultured in RPMI 1640 medium supplemented with 10% fetal bovine serum, 10% horse serum, 2 mM l-glutamine, 100 U/ml penicillin, and 100 μg/ml streptomycin. Nonadherent cells were removed after 24 hours, and adherent cells were maintained with replenishment of the medium every 3 days. BMSCs were treated with 10 μM tamoxifen (Sigma-Aldrich) for 2 days in order to delete floxed Fbxw7 alleles. They were also treated with DAPT (Calbiochem) for 2 days to inhibit NOTCH signaling. MEFs were prepared from embryos at embryonic day 13.5 and maintained as previously described (65), and they were treated with 2 μM tamoxifen (Sigma-Aldrich) for 2 days in order to delete floxed Fbxw7 alleles.
Flow cytometry.
For sorting of peripheral blood cells of breast cancer patients, we obtained 6 ml heparinized peripheral blood from 4 patients with recurrent breast cancer and metastasis. Mononuclear cells were isolated from the blood by Ficoll (GE Healthcare) density centrifugation at 500 g for 25 minutes at 4°C. Erythrocytes were lysed with 1× BD Pharm Lyse buffer (BD Biosciences). The isolated cells were then stained with antibodies against CD45 (clone HI100, Sony Biotechnology) and CD326 (clone 9C4, Sony Biotechnology) for analysis using Cell Sorter SH800 (Sony Biotechnology). For analysis of mouse BM and peripheral blood, erythrocytes were lysed with hemolysis buffer (0.14 M NH4Cl and 0.01 M Tris-HCl at pH 7.5), and the remaining cells were stained with antibodies against F4/80 (clone BM8, eBioscience), CD115 (clone AFS98, eBioscience), MAC1 (clone M1/70, eBioscience), Ly6G (clone 1A8, BD Biosciences), and Ly6C (clone AL-21, BD Biosciences). The stained cells were analyzed with a FACSCalibur flow cytometer (BD).
Histological, immunohistochemical, and immunohistofluorescence analyses.
For H&E staining, tissue was fixed in Bouin’s solution, embedded in paraffin, cut into serial sections (4 μm thickness), and stained as described previously (66). For immunohistochemical analysis, breast tissue microarray slides obtained from Kyushu University Beppu Hospital were stained with antibodies against FBXW7 (clone 3D1, Abnova) using an Envision immunostaining system (DAKO). The sections were counterstained with hematoxylin. Immunohistochemical staining intensity of breast cancer regions and surrounding stroma was scored as negative (low) or positive (high). For immunohistofluorescence analysis, tissue was fixed with 4% paraformaldehyde in 0.1 M phosphate buffer, embedded in 30% sucrose overnight, sectioned (15 μm thickness) with a cryostat, and stained as described previously (66). Antibodies against TCRβ (clone H57-597), B220 (clone RA3-6B2), MAC1 (clone M1/70), and c-Kit (clone 2B8) were from eBioscience; antibodies against Ly6C (clone AL-21), Ly6G (clone 1A8), and VE-cadherin (clone 11D4.1) were from BD Biosciences; antibodies against F4/80 (clone A3-1) were from Serotec; and antibodies against FSP (clone D9F9D) were from Cell Signaling Technology. Immune complexes were detected with secondary antibodies labeled with Alexa Fluor 633 or Alexa Fluor 405 (Molecular Probes), each at a dilution of 1:2,000. The sections were mounted in Fluoromount (Diagnostic BioSystems) and examined with a laser-scanning confocal microscope (LSM700, Carl Zeiss).
Immunoblot analysis.
Total protein extracts were prepared from BMSCs with lysis buffer (50 mM Tris-HCl, pH 7.5; 150 mM NaCl; 0.5% Triton X-100; 10 mM NaF; 10 mM Na4P2O7; 0.4 mM Na3VO4; 0.4 mM EDTA; 20 μg/ml leupeptin; 10 μg/ml aprotinin; 1 mM phenylmethylsulfonyl fluoride). The extracts (20 μg protein) were subjected to immunoblot analysis as previously described (67). Antibodies against NOTCH3 (clone M-20) and KLF5 (clone H-300) were obtained from Santa Cruz Biotechnology; antibodies against cleaved NOTCH1 (clone D1E11), NOTCH2 (clone D76A6), c-JUN (clone 60A8), p100-p52 (catalog no. 4882), and mTOR (clone 7C10) were from Cell Signaling Technology; antibodies against c-MYC were from Abcam; and antibodies against GAPDH (loading control) were from BD Biosciences.
Retroviral expression system.
Complementary DNA encoding hemagglutinin epitope–tagged mouse NICD1, mouse c-MYC, mouse KLF5, or FLAG epitope–tagged mouse FBXW7α (or its ΔF mutant) were subcloned into pMX-puro (provided by T. Kitamura, University of Tokyo, Tokyo, Japan), and the resulting vectors were used to transfect Plat E cells (68) and thereby generate recombinant retroviruses. BMSCs were infected with recombinant retroviruses and subjected to selection in medium containing puromycin (10 μg/ml). Cells stably expressing each recombinant protein were pooled for experiments.
RNAi.
Construction of shRNA vectors and RNAi were performed as described previously (69). The sequence targeted for mouse Ccl2 was 5′-GGTATTCCCTTTCATGAATAC-3′. An RNAi vector for EGFP was used as a control.
RT and real-time PCR analysis.
Total RNA (1 μg), isolated from mouse cells using Isogen (Nippon Gene), was subjected to RT with a QuantiTect Reverse Transcription Kit (Qiagen), and the resulting cDNA was subjected to real-time PCR analysis with SYBR Green PCR Master Mix and specific primers in a StepOnePlus Real-Time PCR System (Applied Biosystems). PCR primer sequences were as follows: Ccl2 sense, 5′-CAGCAGCAGGTGTCCCAAAG-3′; Ccl2 antisense, 5′-TGTCTGGACCCATTCCTTCTTG-3′; Rps18 sense, 5′-GAGGACCTGGAGAGGCTGAAG-3′; Rps18 antisense, 5′-CTGCGGCCAGTGGTCTTG-3′. The amount of Ccl2 mRNA was normalized to that of Rps18 mRNA. For human clinical specimens, total RNA (2.7 μg) isolated from cells using Isogen-LS (Nippon Gene) was subjected to RT with Moloney leukemia virus reverse transcriptase (BRL), and the resulting cDNA was subjected to real-time PCR analysis with SYBR-Green I dye and specific primers in a LightCycler system (Roche Applied Science). Amplification was monitored as described previously (70). PCR primer sequences were as follows: Fbxw7 sense, 5′-CCTCCAGGAATGGCTAAAAA-3′; Fbxw7 antisense, 5′-AAGAGTTCATCTAAAGCAAGCAA-3′; Gapdh sense, 5′-AGCCACATCGCTCAGACAC-3′; Gapdh antisense, 5′-GCCCAATACGACCAAATCC-3′. The amount of Fbxw7 mRNA was normalized to that of Gapdh mRNA.
Antibody array and ELISA.
The serum concentrations of chemokines and cytokines were analyzed using a Proteome Profiler kit (catalog nos. ARY006 and ARY015, R&D Systems). CCL2 levels in mouse or human serum and in mouse BMSC or MEF culture supernatants were also measured by ELISA (Ready-SET-Go kit; eBioscience). For measurement of CCL2 release by BMSCs or MEFs, cells (1 × 104 per well in 24-well plates) were cultured for 48 hours.
Luciferase reporter assay.
The promoter region of mouse Ccl2 and its deletion mutants were subcloned into pGL2-Basic (Promega), which encodes firefly luciferase. BMSCs were seeded (2 × 104 per well in 24-well plates) 24 hours before transfection with promoter constructs (0.25 μg) and the internal control vector pRL-TK (0.25 μg; Promega) for Renilla luciferase using the FuGENE HD reagent (Promega). Luciferase activities were measured using a Dual-Luciferase Reporter Assay System (Promega) and a Lumat LB9507 luminometer (EG&G Berthold) at 48 hours after transfection. Firefly luciferase activity was normalized to that of Renilla luciferase.
ChIP.
BMSCs (2 × 107) were fixed for 5 minutes with 0.5% formaldehyde in RPMI1640 medium. Fixation was terminated by addition of glycine to a final concentration of 0.125 M, and cells were then washed with ice-cold PBS, lysed with 2 ml lysis buffer (5 mM Hepes-NaOH, pH 8.0; 200 mM KCl; 1 mM CaCl2; 1.5 mM MgCl2; 5% sucrose; and 0.5% Nonidet P-40) and exposed to micrococcal nuclease (New England Biolabs) to yield chromatin fragments consisting of 1–5 nucleosomes. The nucleosomes were incubated for 6 hours at 4°C with 2 μg of antibodies against NOTCH1 (clone C-20, Santa Cruz Biotechnology) conjugated to 20 μl Dynabeads–Protein G. The immunoprecipitated material was washed, chromatin was eluted, and the crosslinks were reversed. The DNA fragments were purified by phenol-chloroform extraction followed by precipitation with isopropanol, then used as a template for real-time PCR analysis with the SYBR Select PCR system (Applied Biosystems). PCR primer sequences were as follows: Ccl2 (distal) sense, 5′-GCTCACATTCCAGCTAAATATCTCT-3′; Ccl2 (distal) antisense, 5′-GAGTTATTGTCTGTTTCCCTCTCA-3′; Ccl2 (proximal) sense, 5′-TTACTGGGGGTCCTTTCCCA-3′; Ccl2 (proximal) antisense, 5′-GGAGTGGCTCTGCTTTCACT-3′. The extent of chromatin enrichment was normalized to input.
Statistics.
Quantitative data were subjected to statistical analysis as indicated. A P value less than 0.05 was considered statistically significant.
Study approval.
All animal experiments were approved by the IACUC of Kyushu University, and animal care was in accordance with institutional guidelines. All clinical samples were approved for analysis by the ethical committees of the National Hospital Organization Kyushu Cancer Center (no. 2001-15; October 16, 2001) and Kyushu University (no. 302; February 9, 2006). Written informed consent was obtained from all patients with cancers analyzed in this study.
Supplementary Material
Acknowledgments
We thank T. Honjo for RbpJκfl/fl mice; I.M. de Alborán for c-Mycfl/fl mice; K. Rajewsky for Mx1-Cre transgenic mice; M. Okabe for CAG-EGFP transgenic mice; S. Okano for B16F1 melanoma cells; T. Kitamura for pMX-puro; Sanwa Kagaku Kenkyusho Co. for propagermanium; A. Kataoka for collection of BM and peripheral blood from breast cancer patients; S. Tokunaga for comments on statistical analysis; Y. Tateishi, S. Yamamura, K. Watanabe, N. Nishimura, K. Tsunematsu, A. Niihara, M. Tanaka, K. Motomura, and N. Kinoshita for technical assistance; members of our laboratories for comments on the manuscript; and A. Ohta for help with manuscript preparation. This work was supported in part by a grant from the Project for Development of Innovative Research on Cancer Therapeutics (P-DIRECT).
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information:J Clin Invest. 2015;125(2):621–635. doi:10.1172/JCI78782.
References
- 1.Fridman WH, Pages F, Sautes-Fridman C, Galon J. The immune contexture in human tumours: impact on clinical outcome. Nat Rev Cancer. 2012;12(4):298–306. doi: 10.1038/nrc3245. [DOI] [PubMed] [Google Scholar]
- 2.de Visser KE, Korets LV, Coussens LM. De novo carcinogenesis promoted by chronic inflammation is B lymphocyte dependent. Cancer Cell. 2005;7(5):411–423. doi: 10.1016/j.ccr.2005.04.014. [DOI] [PubMed] [Google Scholar]
- 3.Murdoch C, Muthana M, Coffelt SB, Lewis CE. The role of myeloid cells in the promotion of tumour angiogenesis. Nat Rev Cancer. 2008;8(8):618–631. doi: 10.1038/nrc2444. [DOI] [PubMed] [Google Scholar]
- 4.Youn JI, Nagaraj S, Collazo M, Gabrilovich DI. Subsets of myeloid-derived suppressor cells in tumor-bearing mice. J Immunol. 2008;181(8):5791–5802. doi: 10.4049/jimmunol.181.8.5791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Khaled YS, Ammori BJ, Elkord E. Myeloid-derived suppressor cells in cancer: recent progress and prospects. Immunol Cell Biol. 2013;91(8):493–502. doi: 10.1038/icb.2013.29. [DOI] [PubMed] [Google Scholar]
- 6.Talmadge JE, Gabrilovich DI. History of myeloid-derived suppressor cells. Nat Rev Cancer. 2013;13(10):739–752. doi: 10.1038/nrc3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bingle L, Brown NJ, Lewis CE. The role of tumour-associated macrophages in tumour progression: implications for new anticancer therapies. J Pathol. 2002;196(3):254–265. doi: 10.1002/path.1027. [DOI] [PubMed] [Google Scholar]
- 8.Mosser DM, Edwards JP. Exploring the full spectrum of macrophage activation. Nat Rev Immunol. 2008;8(12):958–969. doi: 10.1038/nri2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Qian BZ, Pollard JW. Macrophage diversity enhances tumor progression and metastasis. Cell. 2010;141(1):39–51. doi: 10.1016/j.cell.2010.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Siveen KS, Kuttan G. Role of macrophages in tumour progression. Immunol Lett. 2009;123(2):97–102. doi: 10.1016/j.imlet.2009.02.011. [DOI] [PubMed] [Google Scholar]
- 11.Karnoub AE, et al. Mesenchymal stem cells within tumour stroma promote breast cancer metastasis. Nature. 2007;449(7162):557–563. doi: 10.1038/nature06188. [DOI] [PubMed] [Google Scholar]
- 12.Hanahan D, Coussens LM. Accessories to the crime: functions of cells recruited to the tumor microenvironment. Cancer Cell. 2012;21(3):309–322. doi: 10.1016/j.ccr.2012.02.022. [DOI] [PubMed] [Google Scholar]
- 13.Kaplan RN, et al. VEGFR1-positive haematopoietic bone marrow progenitors initiate the pre-metastatic niche. Nature. 2005;438(7069):820–827. doi: 10.1038/nature04186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gao D, et al. Endothelial progenitor cells control the angiogenic switch in mouse lung metastasis. Science. 2008;319(5860):195–198. doi: 10.1126/science.1150224. [DOI] [PubMed] [Google Scholar]
- 15.Joyce JA, Pollard JW. Microenvironmental regulation of metastasis. Nat Rev Cancer. 2009;9(4):239–252. doi: 10.1038/nrc2618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Balkwill F. Cancer and the chemokine network. Nat Rev Cancer. 2004;4(7):540–550. doi: 10.1038/nrc1388. [DOI] [PubMed] [Google Scholar]
- 17.Mukaida N, Baba T. Chemokines in tumor development and progression. Exp Cell Res. 2011;318(2):95–102. doi: 10.1016/j.yexcr.2011.10.012. [DOI] [PubMed] [Google Scholar]
- 18.Van Coillie E, Van Damme J, Opdenakker G. The MCP/eotaxin subfamily of CC chemokines. Cytokine Growth Factor Rev. 1999;10(1):61–86. doi: 10.1016/S1359-6101(99)00005-2. [DOI] [PubMed] [Google Scholar]
- 19.Qian BZ, et al. CCL2 recruits inflammatory monocytes to facilitate breast-tumour metastasis. Nature. 2011;475(7355):222–225. doi: 10.1038/nature10138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Salcedo R, et al. Human endothelial cells express CCR2 and respond to MCP-1: direct role of MCP-1 in angiogenesis and tumor progression. Blood. 2000;96(1):34–40. [PubMed] [Google Scholar]
- 21.Loberg RD, et al. Targeting CCL2 with systemic delivery of neutralizing antibodies induces prostate cancer tumor regression in vivo. Cancer Res. 2007;67(19):9417–9424. doi: 10.1158/0008-5472.CAN-07-1286. [DOI] [PubMed] [Google Scholar]
- 22.Zhu X, Fujita M, Snyder LA, Okada H. Systemic delivery of neutralizing antibody targeting CCL2 for glioma therapy. J Neurooncol. 2011;104(1):83–92. doi: 10.1007/s11060-010-0473-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lu X, Kang Y. Chemokine (C-C motif) ligand 2 engages CCR2+ stromal cells of monocytic origin to promote breast cancer metastasis to lung and bone. J Biol Chem. 2009;284(42):29087–29096. doi: 10.1074/jbc.M109.035899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Welcker M, et al. The Fbw7 tumor suppressor regulates glycogen synthase kinase 3 phosphorylation-dependent c-Myc protein degradation. Proc Natl Acad Sci U S A. 2004;101(24):9085–9090. doi: 10.1073/pnas.0402770101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yada M, et al. Phosphorylation-dependent degradation of c-Myc is mediated by the F-box protein Fbw7. EMBO J. 2004;23(10):2116–2125. doi: 10.1038/sj.emboj.7600217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hubbard EJ, Wu G, Kitajewski J, Greenwald I. sel-10, a negative regulator of lin-12 activity in Caenorhabditis elegans, encodes a member of the CDC4 family of proteins. Genes Dev. 1997;11(23):3182–3193. doi: 10.1101/gad.11.23.3182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gupta-Rossi N, et al. Functional interaction between SEL-10, an F-box protein, and the nuclear form of activated Notch1 receptor. J Biol Chem. 2001;276(37):34371–34378. doi: 10.1074/jbc.M101343200. [DOI] [PubMed] [Google Scholar]
- 28.Oberg C, et al. The Notch intracellular domain is ubiquitinated and negatively regulated by the mammalian Sel-10 homolog. J Biol Chem. 2001;276(38):35847–35853. doi: 10.1074/jbc.M103992200. [DOI] [PubMed] [Google Scholar]
- 29.Koepp DM, et al. Phosphorylation-dependent ubiquitination of cyclin E by the SCFFbw7 ubiquitin ligase. Science. 2001;294(5540):173–177. doi: 10.1126/science.1065203. [DOI] [PubMed] [Google Scholar]
- 30.Moberg KH, Bell DW, Wahrer DC, Haber DA, Hariharan IK. Archipelago regulates Cyclin E levels in Drosophila and is mutated in human cancer cell lines. Nature. 2001;413(6853):311–316. doi: 10.1038/35095068. [DOI] [PubMed] [Google Scholar]
- 31.Strohmaier H, et al. Human F-box protein hCdc4 targets cyclin E for proteolysis and is mutated in a breast cancer cell line. Nature. 2001;413(6853):316–322. doi: 10.1038/35095076. [DOI] [PubMed] [Google Scholar]
- 32.Nateri AS, Riera-Sans L, Da Costa C, Behrens A. The ubiquitin ligase SCFFbw7 antagonizes apoptotic JNK signaling. Science. 2004;303(5662):1374–1378. doi: 10.1126/science.1092880. [DOI] [PubMed] [Google Scholar]
- 33.Wei W, Jin J, Schlisio S, Harper JW, Kaelin WG., Jr The v-Jun point mutation allows c-Jun to escape GSK3-dependent recognition and destruction by the Fbw7 ubiquitin ligase. Cancer Cell. 2005;8(1):25–33. doi: 10.1016/j.ccr.2005.06.005. [DOI] [PubMed] [Google Scholar]
- 34.Liu N, et al. The Fbw7/human CDC4 tumor suppressor targets proproliferative factor KLF5 for ubiquitination and degradation through multiple phosphodegron motifs. J Biol Chem. 2010;285(24):18858–18867. doi: 10.1074/jbc.M109.099440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhao D, Zheng HQ, Zhou Z, Chen C. The Fbw7 tumor suppressor targets KLF5 for ubiquitin-mediated degradation and suppresses breast cell proliferation. Cancer Res. 2010;70(11):4728–4738. doi: 10.1158/0008-5472.CAN-10-0040. [DOI] [PubMed] [Google Scholar]
- 36.Mao JH, et al. FBXW7 targets mTOR for degradation and cooperates with PTEN in tumor suppression. Science. 2008;321(5895):1499–1502. doi: 10.1126/science.1162981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Akhoondi S, et al. FBXW7/hCDC4 is a general tumor suppressor in human cancer. Cancer Res. 2007;67(19):9006–9012. doi: 10.1158/0008-5472.CAN-07-1320. [DOI] [PubMed] [Google Scholar]
- 38.Onoyama I, et al. Conditional inactivation of Fbxw7 impairs cell-cycle exit during T cell differentiation and results in lymphomatogenesis. J Exp Med. 2007;204(12):2875–2888. doi: 10.1084/jem.20062299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Matsuoka S, et al. Fbxw7 acts as a critical fail-safe against premature loss of hematopoietic stem cells and development of T-ALL. Genes Dev. 2008;22(8):986–991. doi: 10.1101/gad.1621808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Thompson BJ, et al. The SCFFBW7 ubiquitin ligase complex as a tumor suppressor in T cell leukemia. J Exp Med. 2007;204(8):1825–1835. doi: 10.1084/jem.20070872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Babaei-Jadidi R, et al. FBXW7 influences murine intestinal homeostasis and cancer, targeting Notch, Jun, and DEK for degradation. J Exp Med. 2011;208(2):295–312. doi: 10.1084/jem.20100830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nakayama KI, Nakayama K. Ubiquitin ligases: cell-cycle control and cancer. Nat Rev Cancer. 2006;6(5):369–381. doi: 10.1038/nrc1881. [DOI] [PubMed] [Google Scholar]
- 43.Welcker M, Clurman BE. FBW7 ubiquitin ligase: a tumour suppressor at the crossroads of cell division, growth and differentiation. Nat Rev Cancer. 2008;8(2):83–93. doi: 10.1038/nrc2290. [DOI] [PubMed] [Google Scholar]
- 44.Wang Z, et al. Tumor suppressor functions of FBW7 in cancer development and progression. FEBS Lett. 2012;586(10):1409–1418. doi: 10.1016/j.febslet.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Molloy AP, et al. Mesenchymal stem cell secretion of chemokines during differentiation into osteoblasts, and their potential role in mediating interactions with breast cancer cells. Int J Cancer. 2009;124(2):326–332. doi: 10.1002/ijc.23939. [DOI] [PubMed] [Google Scholar]
- 46.Shi C, et al. Bone marrow mesenchymal stem and progenitor cells induce monocyte emigration in response to circulating toll-like receptor ligands. Immunity. 2011;34(4):590–601. doi: 10.1016/j.immuni.2011.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Allers C, et al. Dynamic of distribution of human bone marrow-derived mesenchymal stem cells after transplantation into adult unconditioned mice. Transplantation. 2004;78(4):503–508. doi: 10.1097/01.TP.0000128334.93343.B3. [DOI] [PubMed] [Google Scholar]
- 48.Ling PD, Rawlins DR, Hayward SD. The Epstein-Barr virus immortalizing protein EBNA-2 is targeted to DNA by a cellular enhancer-binding protein. Proc Natl Acad Sci U S A. 1993;90(20):9237–9241. doi: 10.1073/pnas.90.20.9237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ross DA, Kadesch T. The notch intracellular domain can function as a coactivator for LEF-1. Mol Cell Biol. 2001;21(22):7537–7544. doi: 10.1128/MCB.21.22.7537-7544.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Inuzuka H, et al. SCFFBW7 regulates cellular apoptosis by targeting MCL1 for ubiquitylation and destruction. Nature. 2011;471(7336):104–109. doi: 10.1038/nature09732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wertz IE, et al. Sensitivity to antitubulin chemotherapeutics is regulated by MCL1 and FBW7. Nature. 2011;471(7336):110–114. doi: 10.1038/nature09779. [DOI] [PubMed] [Google Scholar]
- 52.Balamurugan K, et al. The tumour suppressor C/EBPδ inhibits FBXW7 expression and promotes mammary tumour metastasis. EMBO J. 2010;29(24):4106–4117. doi: 10.1038/emboj.2010.280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cushing SD, et al. Minimally modified low density lipoprotein induces monocyte chemotactic protein 1 in human endothelial cells and smooth muscle cells. Proc Natl Acad Sci U S A. 1990;87(13):5134–5138. doi: 10.1073/pnas.87.13.5134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Standiford TJ, Kunkel SL, Phan SH, Rollins BJ, Strieter RM. Alveolar macrophage-derived cytokines induce monocyte chemoattractant protein-1 expression from human pulmonary type II-like epithelial cells. J Biol Chem. 1991;266(15):9912–9918. [PubMed] [Google Scholar]
- 55.Lu Y, et al. PTHrP-induced MCP-1 production by human bone marrow endothelial cells and osteoblasts promotes osteoclast differentiation and prostate cancer cell proliferation and invasion in vitro. Int J Cancer. 2007;121(4):724–733. doi: 10.1002/ijc.22704. [DOI] [PubMed] [Google Scholar]
- 56.Silzle T, et al. Tumor-associated fibroblasts recruit blood monocytes into tumor tissue. Eur J Immunol. 2003;33(5):1311–1320. doi: 10.1002/eji.200323057. [DOI] [PubMed] [Google Scholar]
- 57.Kuhn R, Schwenk F, Aguet M, Rajewsky K. Inducible gene targeting in mice. Science. 1995;269(5229):1427–1429. doi: 10.1126/science.7660125. [DOI] [PubMed] [Google Scholar]
- 58.Tanigaki K, et al. Regulation of αβ/γδ T cell lineage commitment and peripheral T cell responses by Notch/RBP-J signaling. Immunity. 2004;20(5):611–622. doi: 10.1016/S1074-7613(04)00109-8. [DOI] [PubMed] [Google Scholar]
- 59.de Alboran IM, et al. Analysis of C-MYC function in normal cells via conditional gene-targeted mutation. Immunity. 2001;14(1):45–55. doi: 10.1016/S1074-7613(01)00088-7. [DOI] [PubMed] [Google Scholar]
- 60.Wolfer A, Wilson A, Nemir M, MacDonald HR, Radtke F. Inactivation of Notch1 impairs VDJβ rearrangement and allows pre-TCR-independent survival of early αβ lineage thymocytes. Immunity. 2002;16(6):869–879. doi: 10.1016/S1074-7613(02)00330-8. [DOI] [PubMed] [Google Scholar]
- 61.Clausen BE, Burkhardt C, Reith W, Renkawitz R, Forster I. Conditional gene targeting in macrophages and granulocytes using LysMcre mice. Transgenic Res. 1999;8(4):265–277. doi: 10.1023/A:1008942828960. [DOI] [PubMed] [Google Scholar]
- 62.Rickert RC, Roes J, Rajewsky K. B lymphocyte-specific, Cre-mediated mutagenesis in mice. Nucleic Acids Res. 1997;25(6):1317–1318. doi: 10.1093/nar/25.6.1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hayashi S, McMahon AP. Efficient recombination in diverse tissues by a tamoxifen-inducible form of Cre: a tool for temporally regulated gene activation/inactivation in the mouse. Dev Biol. 2002;244(2):305–318. doi: 10.1006/dbio.2002.0597. [DOI] [PubMed] [Google Scholar]
- 64.Okabe M, Ikawa M, Kominami K, Nakanishi T, Nishimune Y. ‘Green mice’ as a source of ubiquitous green cells. FEBS Lett. 1997;407(3):313–319. doi: 10.1016/S0014-5793(97)00313-X. [DOI] [PubMed] [Google Scholar]
- 65.Nakayama K, et al. Mice lacking p27Kip1 display increased body size, multiple organ hyperplasia, retinal dysplasia, and pituitary tumors. Cell. 1996;85(5):707–720. doi: 10.1016/S0092-8674(00)81237-4. [DOI] [PubMed] [Google Scholar]
- 66.Nakayama K, et al. Targeted disruption of Skp2 results in accumulation of cyclin E and p27Kip1, polyploidy and centrosome overduplication. EMBO J. 2000;19(9):2069–2081. doi: 10.1093/emboj/19.9.2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kamura T, et al. Degradation of p57Kip2 mediated by SCFSkp2-dependent ubiquitylation. Proc Natl Acad Sci U S A. 2003;100(18):10231–10236. doi: 10.1073/pnas.1831009100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Morita S, Kojima T, Kitamura T. Plat-E: an efficient and stable system for transient packaging of retroviruses. Gene Ther. 2000;7(12):1063–1066. doi: 10.1038/sj.gt.3301206. [DOI] [PubMed] [Google Scholar]
- 69.Kamura T, et al. Cytoplasmic ubiquitin ligase KPC regulates proteolysis of p27Kip1 at G1 phase. Nat Cell Biol. 2004;6(12):1229–1235. doi: 10.1038/ncb1194. [DOI] [PubMed] [Google Scholar]
- 70.Nagahara H, et al. Clinicopathologic and biological significance of kallikrein 6 overexpression in human gastric cancer. Clin Cancer Res. 2005;11(19 pt 1):6800–6806. doi: 10.1158/1078-0432.CCR-05-0943. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.