Abstract
In contrast to microbially triggered inflammation, mechanisms promoting sterile inflammation remain poorly understood. Damage-associated molecular patterns (DAMPs) are considered key inducers of sterile inflammation following cell death, but the relative contribution of specific DAMPs, including high–mobility group box 1 (HMGB1), is ill defined. Due to the postnatal lethality of Hmgb1-knockout mice, the role of HMGB1 in sterile inflammation and disease processes in vivo remains controversial. Here, using conditional ablation strategies, we have demonstrated that epithelial, but not bone marrow–derived, HMGB1 is required for sterile inflammation following injury. Epithelial HMGB1, through its receptor RAGE, triggered recruitment of neutrophils, but not macrophages, toward necrosis. In clinically relevant models of necrosis, HMGB1/RAGE-induced neutrophil recruitment mediated subsequent amplification of injury, depending on the presence of neutrophil elastase. Notably, hepatocyte-specific HMGB1 ablation resulted in 100% survival following lethal acetaminophen intoxication. In contrast to necrosis, HMGB1 ablation did not alter inflammation or mortality in response to TNF- or FAS-mediated apoptosis. In LPS-induced shock, in which HMGB1 was considered a key mediator, HMGB1 ablation did not ameliorate inflammation or lethality, despite efficient reduction of HMGB1 serum levels. Our study establishes HMGB1 as a bona fide and targetable DAMP that selectively triggers a neutrophil-mediated injury amplification loop in the setting of necrosis.
Introduction
Inflammation is a key contributor to many disease processes. Whereas ligand-receptor systems regulating microbial-induced inflammation have been characterized in detail in the past 2 decades, the molecular regulation of sterile inflammation remains poorly understood. Sterile inflammation typically occurs in response to cell death (1) and often leads to detrimental exacerbation of the initial insult. It has been proposed that endogenous molecules released from dying cells, termed damage-associated molecular patterns (DAMPs), exert an essential role in this danger response (1–5). However, the role of specific DAMPs in disease processes in vivo and the mechanisms through which they regulate sterile inflammation remain ill-defined. Defining DAMPs that trigger sterile inflammation in response to cell death is of high relevance, as excessive inflammation often results in collateral damage and contributes significantly to morbidity and mortality (6). Accordingly, pharmacologic targeting of DAMPs may be of substantial therapeutic benefit — across organs — in settings that involve significant tissue injury. A large number of candidates including heat-shock proteins (HSPs), mitochondrial DNA, hyaluronan, formyl peptides, ATP, uric acid, S100B, and high–mobility group box 1 (HMGB1) have been suggested to act as DAMPs (5, 7), but only a few of these molecules have a confirmed DAMP function in vivo. As such, HSPs were once considered key proinflammatory DAMPs, but have recently been suggested to dampen immune responses (8). Likewise, HMGB1 has been suggested to amplify inflammation rather than directly triggering it (9–11). Much of the uncertainty about the role of DAMPs in sterile inflammation arises from a lack of clean genetic approaches that allow assessment of the contribution of single DAMPs to relevant disease settings in vivo. As such, there are virtually no mouse models in which candidate DAMPs have been knocked out. At the same time, most DAMP receptors (for which knockouts exist) are promiscuous and recognize multiple endogenous and bacterial ligands, making it difficult to sort out the contribution of specific DAMPs to relevant disease processes.
HMGB1 is a key DAMP candidate with proposed functions in the regulation of inflammation following tissue injury, LPS-induced shock, and UV irradiation (3, 4, 12, 13). In the healthy organism, HMGB1 functions as a DNA-binding protein that, without apparent sequence specificity, induces bends in the DNA helix, allowing interactions between DNA and proteins such as p53, NF-κB, homeobox-containing proteins, recombinases, and steroid hormone receptors (14). Although mice with global Hmgb1 deletion die shortly after birth (15), several recent studies have shown that intracellular HMGB1 is not required for cell homeostasis and organ function in the healthy adult organism (16–18). During tissue injury or sepsis, HMGB1 is passively released from necrotic cells and actively secreted by inflammatory cells, with signature posttranslational modifications that are characteristic of the respective release mechanism (19). In addition, the oxidation status of critical cysteine residues, which is largely determined by the physicochemical properties of the wound microenvironment, modulates the biological effects of extracellular HMGB1 (20). Thus, the function of the molecule is believed to be highly context dependent. It has been suggested that extracellular HMGB1 is a key mediator of sterile inflammation after necrosis and of LPS-induced lethality (13). Similar to many other DAMPs, the role of HMGB1 as a relevant promoter of inflammation and disease processes remains controversial: (a) the early postnatal lethality of Hmgb1-knockout mice (15) has precluded studying HMGB1 functions and its contributions to disease processes in vivo by genetic approaches; (b) HMGB1 has been suggested to act as an amplifier of TLR-dependent inflammation rather than a primary trigger of inflammation. In particular, contaminating DAMPs have been suggested to be responsible for proinflammatory effects of recombinant HMGB1, thus challenging its function as a true DAMP (10, 11). To date, virtually all functions of HMGB1 have been assessed using pharmacologic HMGB1 inhibition, recombinant HMGB1, or cell lines. Of note, previous septic shock studies, in which HMGB1 was shown to mediate lethality (13), have demonstrated major differences between experimental approaches using TNF- and IL-1–neutralizing antibodies, which prevented LPS-induced lethality (21–24), and genetic approaches, in which TNF-, TNFR-, and IL-1R–deficient mice displayed normal sensitivity to LPS-induced septic shock (25–27). Subsequent clinical studies that spanned more than a decade did not find clear benefits of TNF neutralization (28), and this approach has subsequently been abandoned as a therapeutic strategy in septic shock. These findings emphasize the importance of genetic approaches in understanding the contribution of inflammatory mediators to disease processes such as septic shock and tissue injury.
Here, we studied the role of HMGB1 in sterile and LPS-induced inflammation using conditional HMGB1 ablation in a recently generated transgenic mouse (16). We demonstrate that HMGB1 exerts an essential role in recruiting neutrophils to sites of necrotic tissue injury, resulting in sterile inflammation, injury amplification, and decreased survival. Surprisingly, HMGB1 did not promote inflammation or death in LPS-induced shock. Together, these data implicate HMGB1 as a potential therapeutic target in the setting of tissue necrosis to interrupt the HMGB1-triggered amplification loop that follows organ injury.
Results
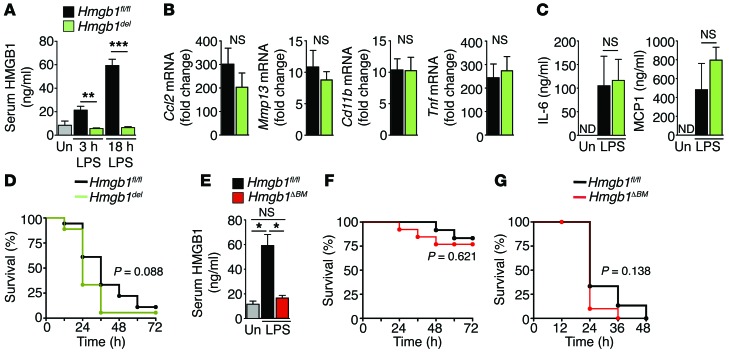
HMGB1 does not mediate LPS-induced lethal shock.
One of the most striking functions of extracellular HMGB1 in the regulation of inflammation has been demonstrated in LPS-mediated septic shock, an often lethal syndrome that affects 750,000 patients annually in the US alone and accounts for 10% of all ICU admissions (29). A previous study demonstrated that HMGB1 acts as a late mediator of LPS-mediated shock, with a neutralizing HMGB1 antibody increasing survival rates in LPS-treated mice from 0% to 70% (13). To study the contribution of HMGB1 to LPS-mediated shock, we used conditional ablation of HMGB1 using a recently generated transgenic mouse (16). This well-characterized mouse model does not display altered gene transcription, cellular homeostasis, or organ function after Hmgb1 deletion in adulthood (16), making it ideally suited to investigate HMGB1 contributions to inflammatory disease processes in vivo. Using Mx1-Cre as an inducible and highly efficient strategy to delete Hmgb1 from IFN-responsive cells (30), we achieved complete inhibition of the LPS-induced increase in serum HMGB1 at early and late time points (Figure 1A) as well as almost complete loss of HMGB1 expression in the liver, bone marrow (BM), and macrophages (Supplemental Figure 1, A–C; full uncut gels are shown in the Supplemental material). Despite this efficient reduction of LPS-induced serum HMGB1, we observed no influence on LPS-induced inflammatory gene expression or cytokine secretion and even observed a trend toward slightly increased mortality (Figure 1, B–D), in contrast therefore with results from previous antibody-based studies demonstrating that HMGB1 neutralization was protective against LPS-induced death (13, 31). These data were further confirmed by crossing Vav1-Cre–transgenic mice, which display Cre-mediated recombination in hematopoietic cells (32), with Hmgb1fl/fl (Hmgb1ΔBM) mice, resulting in almost complete loss of HMGB1 expression levels in BM, spleen, and F4/80-positive hepatic macrophages, but no significant reduction in overall HMGB1 levels in the liver (Supplemental Figure 1, D and E). Deletion in endothelial cells, as previously reported for a different Vav1-Cre–transgenic mouse (33), occurred only in a minority of endothelial cells in several tested organs including the liver (20.9% ± 0.9%), kidneys (21.1% ± 9.2%), and heart (23.0% ± 3.0%) (Supplemental Figure 1, E and F). As with Mx1-Cre–mediated deletion, Vav1-Cre–mediated Hmgb1 deletion resulted in significantly reduced levels of circulating HMGB1 (Figure 1E), but did not significantly affect LPS-induced lethality at 2 different LPS doses (Figure 1, F and G). Taken together, these data suggest that HMGB1 does not constitute a key mediator of LPS-induced inflammation or lethal shock. Based on these findings, we investigated the hypothesis that HMGB1 operates as a regulator of inflammation in settings other than LPS-induced shock.
Figure 1. HMGB1 does not mediate LPS-induced inflammation and lethality.
Hmgb1fl/flMx1-Creneg and Hmgb1fl/fl Mx1-Crepos mice (Hmgb1del) (A–D) as well as Hmgb1fl/fl Vav1-Creneg and Hmgb1fl/fl Vav1-Crepos mice (Hmgb1ΔBM) (E and F) were treated with a lethal dose of LPS (80 mg/kg i.v.). (A–C) HMGB1 serum levels (A) (n = 4 per group), hepatic inflammatory gene expression (B), and serum chemokine levels (C) were determined in Hmgb1fl/fl and Hmgb1del (n = 7 per group) mice by ELISA and qPCR, respectively. (D) Survival was determined in Hmgb1fl/fl and Hmgb1del (n = 18 per group) mice. (E and F) Hmgb1fl/fl and Hmgb1ΔBM mice were treated with LPS (30 mg/kg). HMGB1 serum levels were determined 18 hours after LPS challenge (E) (n = 4 per group). Survival was determined in Hmgb1fl/fl and Hmgb1ΔBM mice (n = 12 per group). (G) Hmgb1fl/fl and Hmgb1ΔBM mice were treated with LPS (80 mg/kg). Survival was determined in Hmgb1fl/fl (n = 15) and Hmgb1ΔBM mice (n = 10). *P < 0.05, **P < 0.01, and ***P < 0.001 by 1-way ANOVA followed by Tukey’s multiple comparisons test (A, C, and E), unpaired 2-tailed t test (B), and Mantel-Cox log-rank test (D, F, and G), respectively. Un, untreated; ND, nondetectable; NS, nonsignificant.
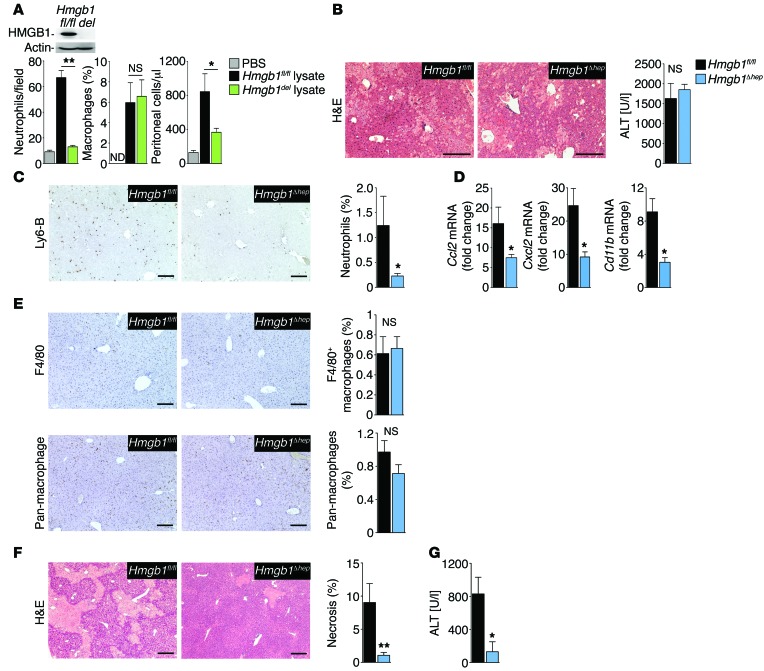
HMGB1 promotes neutrophil but not macrophage migration toward necrotic tissue.
In addition to active secretion from inflammatory cells, e.g., after stimulation with LPS, HMGB1 is also passively released from necrotic cells, consistent with the concept of a proinflammatory DAMP. We therefore tested the role of HMGB1 in host responses to injury using necrotic tissue from Hmgb1-deleted or Hmgb1fl/fl control mice. Neutrophils, the first responders to tissue injury, displayed a greater than 7-fold increase in migration toward Hmgb1fl/fl tissue lysates. Of note, Hmgb1 deletion almost completely mitigated the ability of tissue lysates to induce neutrophil migration (Figure 2A). In contrast to neutrophils, macrophage migration was not affected by Hmgb1 deletion of tissue lysates (Figure 2A). We made similar observations in vivo, where inflammatory cell infiltration was reduced after i.p. injection of Hmgb1-deleted liver lysates compared with injection of Hmgb1fl/fl lysates (Figure 2A). In summary, these data indicate that HMGB1 may be a key DAMP that triggers the migration of neutrophils but not macrophages toward necrotic tissue.
Figure 2. HMGB1 promotes neutrophil recruitment in vitro and in vivo.
(A) Neutrophil and macrophage migration toward Hmgb1-floxed and Hmgb1-deleted (Hmgb1del) liver extracts (induced by Mx1-Cre), determined in Boyden chambers (left 2 panels, n = 3 per group, with representative results from 3 separate isolations). Insert shows immunoblot confirming Hmgb1 deletion. Peritoneal inflammatory cell accumulation after i.p. injection of lysates from Hmgb1fl/fl (n = 7) and Hmgb1del (n = 8) livers. (B–E) Hmgb1fl/fl (n = 8) and Hmgb1Δhep (n = 9) mice were subjected to warm hepatic I/R and sacrificed 6 hours later. H&E staining (B, left panel) and serum ALT (B, right panel) demonstrate similar initial injury, whereas hepatic neutrophil infiltration (C) and hepatic expression of inflammatory genes (D) differed. Numbers of hepatic macrophages determined by F4/80 staining and staining with pan-macrophage antibody in Hmgb1fl/fl (n = 8) and Hmgb1Δhep (n = 9) mice (E). (F and G) Hepatic injury 24 hours after I/R injury in Hmgb1fl/fl (n = 11) and Hmgb1Δhep (n = 10) mice, determined by H&E staining (F) and serum ALT (G). *P < 0.05 and **P < 0.01 by 1-way ANOVA followed by Tukey’s multiple comparisons test (A) and unpaired 2-tailed t test (B–G). Scale bars: 200 μm.
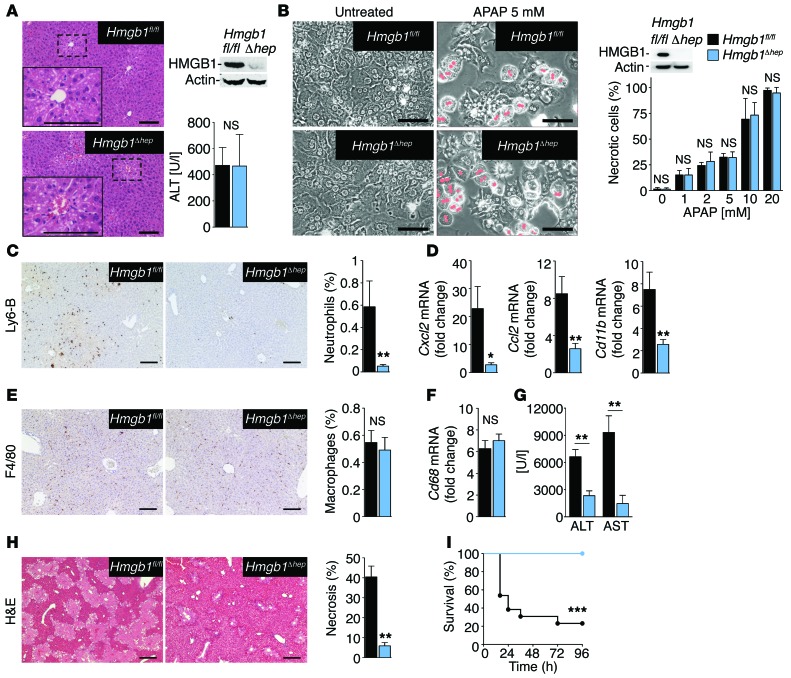
HMGB1 promotes neutrophil migration and injury amplification after necrosis in vivo.
To test the DAMP function of epithelial HMGB1 in vivo and its role in linking necrosis to neutrophil-mediated inflammatory responses in clinically relevant disease models, we subjected mice with an albumin-Cre–driven hepatocyte-specific Hmgb1 knockout (Hmgb1Δhep) to ischemia and reperfusion (I/R) injury. Of note, this hepatic epithelial–knockout mouse shows a virtually complete absence of HMGB1 in HNF4α-positive hepatocytes and cytokeratin-positive biliary epithelial cells, but preserves HMGB1 expression in other hepatic cell populations including F4/80-positive macrophages, CD31-positive endothelial cells, and desmin-positive hepatic stellate cells, as well as HMGB1 expression in other organs (Supplemental Figure 2, A–F). I/R injury is not only involved in the pathophysiology of a wide range of diseases and in complications of organ transplantation (34), but also represents a prototypical setting in which DAMPs elicit detrimental effects (6). Hmgb1Δhep mice displayed a profound reduction of infiltrating neutrophils and inflammatory gene expression following hepatic I/R injury, whereas the initial injury, as demonstrated by similar serum ALT levels and necrosis area, was similar to that seen in Hmgb1fl/fl mice (Figure 2, B–D). Similar to our in vitro data, Hmgb1 ablation in hepatocytes selectively affected neutrophils without impacting the number of F4/80-positive macrophages in the liver (Figure 2E). At later time points, the reduced inflammation translated into amelioration of liver injury (Figure 2, F and G), consistent with the notion that recruited neutrophils promote collateral damage and that HMGB1 is part of this neutrophil-mediated injury amplification loop. To confirm the relevance of HMGB1 as a disease-promoting DAMP in a second clinically relevant disease setting, we subjected Hmgb1Δhep and Hmgb1fl/fl mice to acetaminophen-induced liver injury. Acetaminophen intoxication induces HMGB1 release in mice (Supplemental Figure 3A) and humans (35) and represents the most common cause of acute liver failure in patients (36), with 70,000 annual hospital visits in the US alone (37). Similar to our findings in hepatic I/R injury, the initial acetaminophen-induced liver injury was similar between Hmgb1Δhep and Hmgb1fl/fl mice (Figure 3A). Despite this similar initial injury, we observed a strong reduction of early and late hepatic neutrophil infiltration and of proinflammatory gene expression in Hmgb1Δhep mice (Figure 3, C and D, and Supplemental Figure 3B). Consistent with our in vitro findings and observation in the I/R model, Hmgb1 deletion selectively affected neutrophil infiltration without altering the number of hepatic macrophages (Figure 3, E and F). Alterations in neutrophil recruitment and inflammation could not be attributed to differences in the metabolic activation of acetaminophen, a prerequisite for its toxicity, or to hepatocyte sensitivity to acetaminophen, since acetaminophen adducts, early hepatic GSH depletion, and Cyp2e1 expression (Supplemental Figure 3, C–F), as well as acetaminophen-induced death of primary hepatocytes (Figure 3B) were similar in Hmgb1fl/fl and Hmgb1Δhep mice. Since neutrophil infiltration preceded differences in chemokine expression between Hmgb1fl/fl and Hmgb1Δhep mice (Supplemental Figure 4A), the effects of HMGB1 on neutrophils appeared not to be directly caused by HMGB1-mediated modulation of chemokine expression. Accordingly, we found that neutrophil recruitment toward necrotic liver tissue was not altered by pharmacologic inhibition of CXCL2 or its receptor CXCR4 by neutralizing antibodies or AMD3100, respectively (Supplemental Figure 4B). Hence, the differences in chemokine levels between Hmgb1fl/fl and Hmgb1Δhep mice we observed at later time points were likely a consequence of different levels of neutrophil-mediated inflammation and liver injury. It is conceivable that these differences in chemokine expression additionally contribute to differences in neutrophil accumulation between Hmgb1fl/fl and Hmgb1Δhep mice at later stages. At later time points, we found that the decreased neutrophil infiltration in Hmgb1Δhep mice translated into attenuated liver injury, with striking reductions in serum ALT and AST (Figure 3G) and liver necrosis (Figure 3H). Of note, 100% of Hmgb1Δhep mice survived a lethal dose of acetaminophen versus only 22% of Hmgb1fl/fl mice (Figure 3I). Despite slight differences in kinetics, these data strikingly resemble our findings in the I/R model, confirming HMGB1 as a bona fide DAMP and a key trigger of an inflammation-mediated injury amplification loop.
Figure 3. HMGB1 mediates neutrophil recruitment, injury amplification, and lethality following acetaminophen-induced liver necrosis.
(A) Serum ALT and liver histology in Hmgb1fl/fl (n = 8) and Hmgb1Δhep (n = 7) mice 3 hours after injection of acetaminophen (300 mg/kg). (B) Acetaminophen-induced death in Hmgb1fl/fl and Hmgb1Δhep hepatocytes (n = 3 separate isolations per group). (C–H) Hepatic neutrophil recruitment (C), inflammatory gene expression (D), macrophage numbers (E and F), serum ALT and AST (G), and liver necrosis (H) 24 hours after acetaminophen (300 mg/kg) injection in Hmgb1fl/fl (n = 8) and Hmgb1Δhep (n = 8) mice. (I) Survival after a lethal dose of acetaminophen (500 mg/kg) in Hmgb1fl/fl and Hmgb1Δhep (n = 13 per group) mice. *P < 0.05, **P < 0.01, and ***P < 0.001 by 2-tailed unpaired t test (A–H) and Mantel-Cox log-rank test (I). Scale bars: 200 μm (A, C, E, and H) and 100 μm (B).
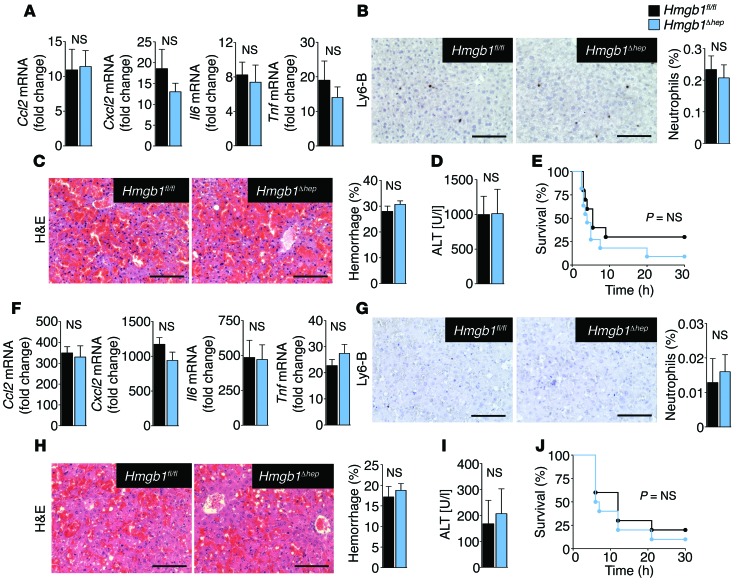
HMGB1 does not promote sterile inflammation or injury amplification after apoptosis.
To determine whether HMGB1 mediates inflammatory responses to all forms of cell death, we next tested its role in TNF- and FAS-mediated apoptosis. In striking contrast to our findings in necrotic injury, hepatic HMGB1 deficiency did not alter inflammation, injury, or survival in FAS- or TNF-mediated apoptotic liver injury (Figure 4, A–J), consistent with the notion that apoptosis is considered a low-inflammatory form of cell death (3). The unaltered hepatocyte injury in apoptotic injury models further corroborates our finding that HMGB1 status does not affect the intrinsic sensitivity of hepatocytes to cell death, thus emphasizing the cell-extrinsic nature of HMGB1-mediated inflammation in necrotic injury models.
Figure 4. HMGB1 does not modulate inflammation, injury, or survival in response to FAS- or TNF-induced apoptosis.
(A and B) Hepatic inflammatory gene expression (A) and neutrophil infiltration (B) 10 hours after injection of a sublethal dose of the FAS-agonistic antibody Jo2 (0.15 μg/g) in Hmgb1fl/fl (n = 6) and Hmgb1Δhep (n = 7) mice. (C) Hepatic H&E staining and hemorrhage 5 hours after a lethal dose of Jo2 (0.5 μg/g) in Hmgb1fl/fl and Hmgb1Δhep (n = 3 per group) mice. (D and E) Serum ALT after 5 hours (D) and survival (E) in Hmgb1fl/fl (n = 9) and Hmgb1Δhep (n = 10) mice after Jo2 injection (0.5 μg/g). (F–J) Mice were injected with 700 μg/kg D-Gal and 100 μg/kg LPS. Hepatic inflammatory gene expression (F), neutrophil infiltration (G), hemorrhage (H), and serum ALT (I) 6 hours later in Hmgb1fl/fl and Hmgb1Δhep (n = 5 per group) mice. (J) Survival of Hmgb1fl/fl and Hmgb1Δhep (n = 10 per group) mice. Statistical significance assessed by 2-tailed unpaired t test (A–D and F–I) and Mantel-Cox log-rank test (E and J). Scale bars: 200 μm (B, C, G, and H).
HMGB1 from different cellular compartments differentially affects the inflammatory response after necrosis and exhibits distinct posttranslational modifications.
The contribution of HMGB1 from different cellular sources, such as dying epithelial cells and HMGB1-secreting inflammatory cells, remains only poorly understood (4, 38). To investigate whether HMGB1 from BM-derived cells affects injury responses in addition to the observed role of epithelial HMGB1, we used the above-described Vav1-Cre–deleted Hmgb1ΔBM mice. In contrast to Hmgb1Δhep mice, Hmgb1ΔBM mice did not display altered hepatic neutrophil infiltration, inflammatory gene expression, or liver injury following necrosis (Supplemental Figure 5, A–D), thus emphasizing the key role of epithelial HMGB1 in triggering injury responses following necrotic cell death. In light of recent reports indicating that posttranslational modifications of HMGB1 modulate its bioactivity, we used proteomic mass spectrometry (MS) and tandem mass spectrometry (MS/MS) (39) to identify the presence of HMGB1 posttranslational modifications in the serum of acetaminophen-treated mice and to assign them to specific cellular sources. We observed the presence of 4 distinct isoforms of HMGB1 in Hmgb1fl/fl mice, all in high abundance, with molecular weights of 25,469 Da and 24,587 Da (fully reduced HMGB1) and 25,467 Da and 24,585 Da (disulfide HMGB1) — all of these isoforms being either nonacetylated or acetylated at the 2 NLS sites (ref. 40 and Supplemental Figure 6). One important difference between the Hmgb1Δhep and Hmgb1ΔBM mice was the complete absence of disulfide HMGB1 in the Hmgb1Δhep mice (Supplemental Figure 6). Since both Hmgb1Δhep and Hmgb1ΔBM mice lacked detectable circulating acetylated HMGB1 (Supplemental Figure 6), it can be concluded that acetylation alone is not sufficient to trigger neutrophil infiltration and that formation of disulphide HMGB1 in epithelial cells is likely crucial for this process.
HMGB1 promotes necrosis-induced sterile inflammation through RAGE.
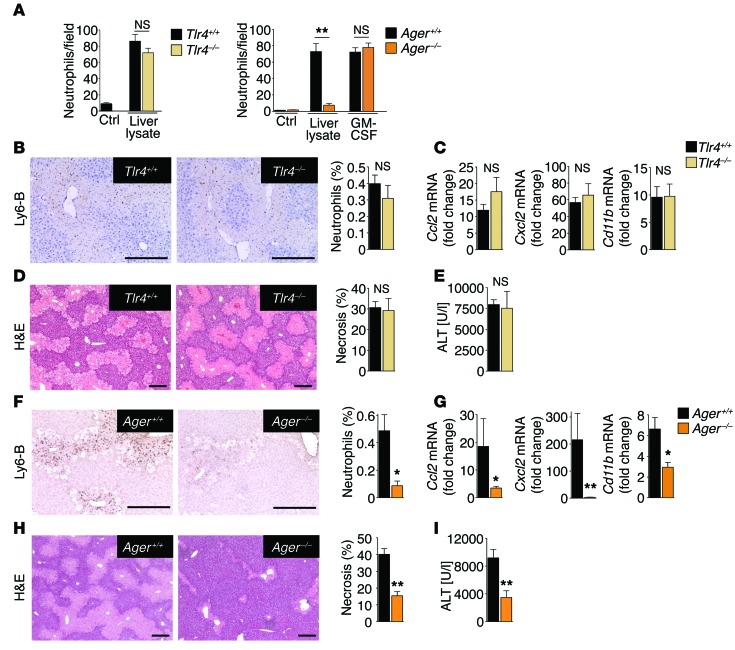
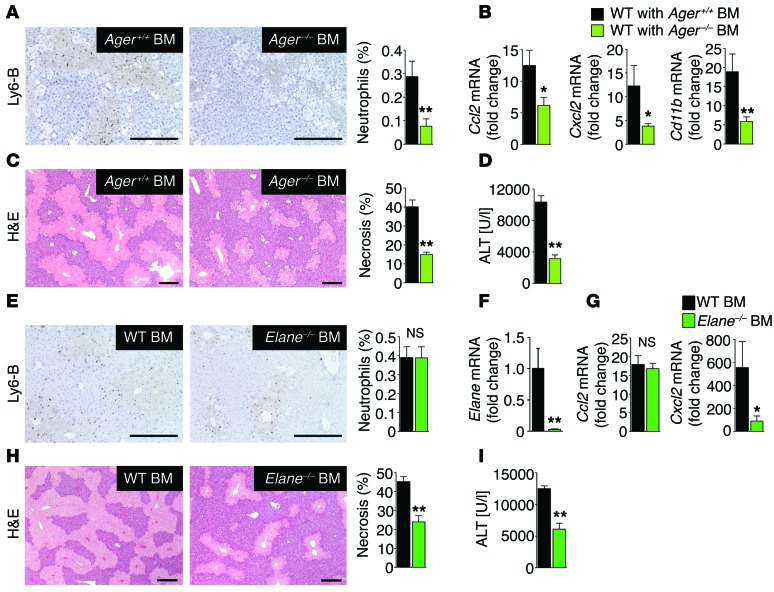
Having established epithelial HMGB1 as a disease-promoting DAMP in clinically relevant settings, we next aimed to identify the receptor through which HMGB1 recruits neutrophils and amplifies injury in vivo, focusing on Toll-like receptor 4 (TLR4) and the receptor for advanced glycation end-products (RAGE) as the best-established HMGB1 receptors (41). Whereas TLR4-deficient neutrophils displayed normal migration toward liver lysates, migration of RAGE-deficient neutrophils toward liver lysates, but not toward our positive control GM-CSF, was reduced by 80% (Figure 5A). To confirm these results in vivo and to test whether RAGE deficiency affects injury responses in a manner similar to that of HMGB1 ablation, we subjected WT, TLR4-deficient, and RAGE-deficient mice to a sublethal dose of acetaminophen. In line with the above in vitro findings, we did not detect differences in neutrophil infiltration, inflammation, or liver injury between WT and TLR4-deficient mice (Figure 5, B–E), but noted a profound reduction of these parameters in RAGE-deficient mice, similar to that observed in Hmgb1-deleted mice (Figure 5, F–I). Like Hmgb1Δhep hepatocytes, RAGE-deficient hepatocytes displayed normal acetaminophen metabolization and preserved sensitivity to acetaminophen-induced cell death (Supplemental Figure 7, A–C). To substantiate the hypothesis that RAGE expressed on BM-derived inflammatory cells and not other RAGE-expressing cell types is responsible for neutrophil infiltration, inflammation, and injury in response to necrosis, we generated RAGE BM–chimeric mice (Supplemental Figure 8, A and B). Following a sublethal dose of acetaminophen, chimeric mice with RAGE-deficient BM displayed strongly reduced neutrophil infiltration, inflammation, and late liver injury (Figure 6, A–D), similar to that seen in mice with global RAGE deficiency. Of note, macrophage depletion did not affect hepatic neutrophil recruitment after acetaminophen treatment (Supplemental Figure 9), confirming that the HMGB1-triggered neutrophil infiltration into the liver was not an indirect effect through RAGE-expressing macrophages. These findings are consistent with our observation that necrotic tissue could directly stimulate the migration of RAGE-expressing, but not RAGE-deficient, neutrophils in vitro (Figure 5A).
Figure 5. RAGE, but not TLR4, mediates neutrophil recruitment following necrosis.
(A) Migration of WT, Tlr4–/–, and RAGE-deficient (RAGE encoded by Ager) neutrophils (Ager–/– neutrophils, n ≥2 separate isolations per experiment) toward WT liver lysates. GM-CSF served as a positive control. (B–E) Analysis of hepatic neutrophil infiltration (B), inflammatory gene expression (C), H&E staining and necrosis quantification (D), and serum ALT concentrations (E) in WT (n = 9) and Tlr4–/– (n = 7) mice treated with acetaminophen (300 mg/kg). (F–I) Analysis of hepatic neutrophil infiltration (F), inflammatory gene expression (G), H&E staining and necrosis quantification (H), and serum ALT concentrations (I) in WT (n = 10) and Ager–/– (n = 9) mice treated with acetaminophen (300 mg/kg). *P < 0.05 and **P < 0.01 by unpaired 2-tailed t test. Scale bars: 200 μm.
Figure 6. RAGE-expressing neutrophils promote injury amplification after necrotic injury.
(A–D) Neutrophil infiltration (A), inflammatory gene expression (B), H&E staining and necrosis (C), and serum ALT concentrations (D) in chimeric mice with WT BM (n = 8) and Ager–/– BM (n = 10) treated with acetaminophen (300 mg/kg). (E–I) Chimeric mice with WT BM and Elane–/– BM (n = 12 per group) were treated with acetaminophen (300 mg/kg). Hepatic neutrophil infiltration was determined by staining for Ly6-B (E). BM reconstitution was confirmed by qPCR for Elane in spleens of BM-chimeric mice (F). Hepatic inflammatory gene expression was determined by qPCR (G). Liver injury was assessed by hepatic H&E staining and necrosis quantification (H), and serum ALT concentrations (I). *P < 0.05 and **P < 0.01 by unpaired 2-tailed t test. Scale bars: 200 μm.
HMGB1-recruited neutrophils amplify injury following necrosis.
To directly link neutrophil infiltration to amplification of liver injury, we investigated whether inhibition of neutrophil function ameliorates acetaminophen-induced injury. We tested the role of neutrophils in injury amplification using chimeric mice with BM-specific deficiency of neutrophil elastase (Elane), a protease that is essential for neutrophil effector functions but not for neutrophil migration (42). Mice with Elane–/– BM displayed profoundly reduced necrosis and ALT levels after acetaminophen challenge despite intact hepatic neutrophil recruitment (Figure 6, E–I), thus confirming that neutrophils in injured tissue amplify already existing injury. To further determine whether this HMGB1-dependent neutrophil-mediated injury amplification pathway could be pharmacologically exploited, we tested the effects of the HMGB1 inhibitor glycyrrhizin (12, 43). This inhibitor is a natural component of liquorice and has been widely used for the treatment of liver disease in Asia and Europe (44), but its mechanisms of action are not understood. Glycyrrhizin markedly attenuated acetaminophen-induced hepatic inflammation and injury (Supplemental Figure 10, A–C) to almost the same degree as that of genetic HMGB1 ablation. Moreover, treatment with glycyrrhizin 2 hours after acetaminophen intoxication also reduced liver injury, albeit to a lesser extent than did pretreatment (Supplemental Figure 10, D and E), suggesting the possibility of pharmacologic prevention of HMGB1-mediated injury amplification.
Discussion
Using conditional HMGB1 ablation, our study could for the first time to our knowledge reliably characterize the contributory role of HMGB1 to inflammatory processes driven by LPS and necrosis, the 2 proposed key functions of HMGB1. Surprisingly, our results revealed no role of HMGB1 in the promotion of LPS-induced lethal shock. Although we cannot fully explain differences in a previous study by Tracey et al., which demonstrated a 70% reduction of lethality by HMGB1-neutralizing antibodies (13), our approach virtually eliminated LPS-induced serum HMGB1 by 2 genetic approaches and therefore effectively rules out a key role of HMGB1 in promoting LPS-induced lethality. Similar differences between pharmacologic and genetic approaches have been observed in studies of TNF in septic shock in mice (21–27), and subsequent studies in humans have failed to reproduce therapeutic effects of TNF neutralization seen in mice (28). A recent study by Yanai et al., using conditional ablation of HMGB1 exons 2–4 in myeloid cells also reported no protection from LPS-induced lethality by Hmgb1 deletion (18). However, in contrast to our study, Yanai et al. reported only marginally lowered levels of serum HMGB1 levels, thus precluding conclusions about the functional contribution of extracellular HMGB1 to LPS-induced shock (18). In our study, there was even a trend toward slightly increased LPS-induced lethality in mice with Mx1-Cre–mediated Hmgb1 deletion, but not in mice with Vav1-Cre–mediated deletion (at 2 different doses), indicating a possible but only minor role for HMGB1 in protection from LPS-mediated septic shock, as suggested by Yanai et al. (18). Rather than being a key mediator of LPS-mediated lethality, our study instead suggests that HMGB1 functions as a neutrophil-recruiting DAMP following necrosis. Its key role in controlling neutrophil infiltration and inflammation following necrosis was observed in 2 clinically relevant necrosis models and was further highlighted by the complete prevention of acetaminophen-induced lethality in Hmgb1Δhep mice. In conjunction with the strong reduction of inflammation and injury by the HMGB1 inhibitor glycyrrhizin and a key role of HMGB1 in promoting the migration of human neutrophils toward necrotic tissue (Supplemental Figure 11), our results suggest that HMGB1 might serve as a therapeutic target for diseases in which cell death is a key disease driver. Our post-treatment data for glycyrrhizin also suggest that targeting HMGB1 may have to be a truly preventive approach, i.e., before cell death occurs (e.g., as treatment in chronic diseases) or given at very early time points before HMGB1-induced neutrophil recruitment has started. In contrast to the striking effects of epithelial HMGB1 ablation, we observed no contribution of HMGB1 in BM-derived cells following necrosis, suggesting that HMGB1 fulfills other functions in this compartment. Our results also underpin the notion that DAMPs such as HMGB1 are critical in response to necrosis but not apoptosis — as HMGB1 ablation did not alter TNF- or FAS-induced injury responses or lethality.
Our study further highlights the role of RAGE as the receptor target for HMGB1, promoting neutrophil migration to necrotic tissue. We observed the requirement of RAGE not only in neutrophil migration toward necrotic tissue in vitro, but also in the acetaminophen model in vivo. The contribution of RAGE on non–BM-derived cell populations to acetaminophen-induced neutrophil migration was excluded using BM-chimeric mice. Likewise, we did not observe a role for TLR4, another receptor target of HMGB1, in inflammatory responses to necrotic injury, nor in the migration of neutrophils toward necrotic tissue in vitro. Although liver-resident macrophages typically do not increase following liver injury, BM-derived macrophages often migrate to sites of injury (45). However, we did not observe effects of epithelial Hmgb1 deletion on macrophage migration in vitro, nor on the number of hepatic macrophages in vivo, suggesting that the HMGB1/RAGE axis selectively modulates neutrophil migration toward injured tissue. Moreover, our in vitro migration assays and in vivo data, including experiments in which mice were depleted of macrophages, demonstrate that the HMGB1/RAGE axis provides a direct link between necrotic cells and neutrophils without requirement for macrophage-mediated signals. Using Elane–/– BM–chimeric mice, our study firmly established injury amplification by HMGB1-recruited neutrophils. Our study focused primarily on pathways through which necrotic tissue triggers neutrophil recruitment and sterile inflammation and did not determine in detail effector pathways through which neutrophils amplify tissue injury. The incomplete inhibition of liver injury in mice lacking Elane is consistent with previous studies that demonstrate the essential role of additional pathways such as proteinase 3 in neutrophil activation (46). Moreover, recent studies have also highlighted the role of additional DAMP receptors such as CLEC12A in the negative regulation of inflammation in response to cell death responses (47), suggesting that these may counteract the proinflammatory effects of HMGB1. Future studies need to further investigate the functional interactions between pro- and antiinflammatory DAMP receptors on neutrophils following necrosis and determine the effectors through which HMGB1-recruited neutrophils exacerbate tissue injury. These are likely to not only include neutrophil proteases and ROS, but possibly also neutrophil extracellular traps, a key effector of neutrophils in infection (48).
Before being known as a DAMP, HMGB1 was described as a nuclear protein with intracellular functions, such as bending of DNA, to facilitate DNA-protein interactions (14). We previously did not find evidence for a requirement of intracellular HMGB1 in the regulation of gene expression, organ function, and viability after tissue-specific ablation of exons 2–4 of Hmgb1 (16), consistent with the lack of a spontaneous phenotype in mice with conditional HMGB1 ablation from other groups (17, 18). Our results indicate that intracellular functions of HMGB1 are likely restricted to development and/or specific cell types and that the main function of HMGB1 in epithelial cells lies in its role as a danger signal in the extracellular space in the context of tissue injury. Consistent with these findings, we also do not observe a role of intracellular HMGB1 in epithelial injury responses: (a) in our 2 necrosis models, initial injury of epithelial cells is similar and only diverges at later time points after differences in inflammation and neutrophil infiltration have occurred; (b) in the acetaminophen models, we found APAP metabolization and similar APAP-induced death of primary hepatocytes in floxed and Hmgb1-deleted mice; (c) the sensitivity to FAS- and TNF-induced injury was not altered by HMGB1 status. Consistent with these findings, Yanai et al. found no effect of Hmgb1 deletion on early liver injury after hepatic I/R (18). However, their study did not further investigate DAMP functions of HMGB1, such as neutrophil infiltration, inflammation, and late liver injury (18). In contrast to our study and that of Yanai et al., conditional ablation of HMGB1 exons 2–3 resulted in worsened hepatic injury, even at early time points after I/R, as well as in worsened pancreatitis (17, 49). Of note, the targeting strategy in these studies appears to affect mitochondrial quality and function, which are likely to affect the injury response (17, 49), whereas our approach avoids this potential confounder (16). Our data indicate that HMGB1 isoforms differ between epithelial cells and BM-derived cells and suggest disulfide HMGB1 as a key mediator whose absence in epithelial cells is correlated with reduced neutrophil recruitment and injury. While our data demonstrate the hepatocyte-specific origin of disulfide HMGB1 following injury, it is also conceivable that HMGB1 oxidation to its disulfide isoform within hepatocytes requires hepatic recruitment of neutrophils, a key source of ROS. Importantly, disulfide HMGB1 has recently been shown to exert cytokine-like properties, whereas all-thiol HMGB1 exerts chemotactic activity via CXCL12 heterodimerization and CXCR4 activation (20). Since the chemotactic activity of HMGB1-containing liver extracts toward neutrophils was not mediated via CXCR4, it needs to be investigated whether fully reduced HMGB1 (which was present in Hmgb1Δhep mice, albeit at reduced levels) or disulfide HMGB1 (which was absent in Hmgb1Δhep mice) mediates this chemotactic activity in neutrophils.
While our study characterized the role of the HMGB1/RAGE axis using the liver as a model system, in which necrotic injury is accompanied by massive neutrophil infiltration and often results in life-threatening acute liver failure, we did not study its contribution to disease processes in other organs or in other contexts. Our data showing reduced neutrophil migration toward HMGB1-deleted cardiac tissue lysates (Supplemental Figure 12) suggest, in principle, that the neutrophil-recruiting HMGB1/RAGE axis may be operational in other tissues. Since other DAMPs such as formyl-peptides and SAP130 and their corresponding receptors (FPR-1 and Mincle, respectively) may mediate neutrophil recruitment in settings such as heat-induced liver injury (50) or irradiation-induced thymocyte death (51), the relative contribution of specific DAMPs needs to be further studied and compared between different injury types and organs. Moreover, the role of HMGB1 released from immune cells, e.g., undergoing pyroptosis or NETosis during infection, is likely to differ substantially from HMGB1 released during necrosis (52). In this regard, the benefits of HMGB1-mediated neutrophil recruitment in sterile inflammation and LPS-induced shock need to be better understood. It could be speculated that HMGB1 recruits neutrophils not only to promote phagocytosis of cellular debris, but also to constitute a preemptive strike against secondary infection of necrotic tissue — at the price of increased tissue injury. Likewise, the increase in HMGB1 secretion after LPS — rather than mediating lethal complications of LPS — may be involved in recruiting or activating effector cells in order to fight bacterial infections, as was recently suggested by Yanai et al. (18). Finally, cell death is a key driver of many disease processes including cancer, suggesting that DAMPs such as HMGB1 provide a link between epithelial injury and cancer in the setting of chronic injury.
Methods
Animals.
Mice were maintained on a 12-hour dark/12-hour light cycle with free access to food and water unless otherwise indicated. Six- to 8-week-old mice were used for all experiments. Hmgb1fl/fl mice and MHC-Cre mice have been previously described (16). C57BL6 mice, albumin-Cre mice, Mx1-Cre mice, Vav1-Cre mice, and TLR4-knockout mice were obtained from The Jackson Laboratory. RAGE-knockout mice were a gift of Ann-Marie Schmidt (New York University, New York, New York, USA). Mx1-Cre activation for deletion of Hmgb1 was induced by 3 i.p. injections of poly(I:C) (10 mg/kg; GE Healthcare), given every other day (30).
Models of LPS-induced inflammation and mortality.
Four weeks after administration of poly-(I:C), Hmgb1fl/fl and Hmgb1del mice were injected i.v. with LPS (80 mg/kg) dissolved in saline to induce systemic inflammation and lethal shock. Serum was collected after 3 and 18 hours to determine HMGB1 serum levels by ELISA (IBL International). For some experiments, mice were sacrificed after 18 hours to collect tissue. For survival analysis, animals were injected with 80 mg/kg LPS and monitored without further manipulation. To determine the contribution of HMGB1 from BM-derived cells, Vav1-Cre–negative Hmgb1fl/fl and Vav1-Cre–positive Hmgb1fl/fl mice (Hmgb1ΔBM) were treated accordingly, without prior exposure to poly-(I:C).
Liver injury models.
Mice were injected i.v. with hamster anti–mouse anti–CD95 antibody (10 μg/mouse, clone Jo2, catalog 554254; BD Biosciences) or with 700 μg/kg D-galactosamine i.p. followed by 100 μg/kg LPS i.v. (both from Sigma-Aldrich) 2 hours later. Mice were sacrificed 4 hours after the LPS injection or monitored for survival. Acetaminophen intoxication was induced in overnight-starved male mice by i.p. injection of 300 mg/kg (sublethal dose) or 500 mg/kg (lethal dose) acetaminophen (Sigma-Aldrich), dissolved in sterile warm saline. For some experiments, mice were pretreated with either clondroate liposomes or PBS liposomes (200 μl/mouse, obtained from Nico van Rooijen, Vrije Universiteit, Amsterdam, The Netherlands), followed by treatment with acetaminophen 96 hours later. For some experiments, mice were pretreated with 50 mg/kg glycyrrhizic acid dissolved in saline (catalog 50531; Sigma-Aldrich). For ischemia/reperfusion experiments, male mice were starved overnight and subjected to warm partial hepatic ischemia as previously described (53). Briefly, after midline abdominal incision, the liver hilum was exposed, and the hepatic artery, portal vein, and bile duct were occluded using a microvascular clamp (Fine Science Tools) proximally to caudate hepatic branches to avoid portal venous congestion. Rapid blanching of the respective liver lobes indicated ischemia. Body temperature was monitored with a UV thermometer (Fisher Scientific) and maintained at 37°C ± 1°C with heat pads and warm lamps. After 60 minutes of ischemia, the clamp was removed, and reperfusion of all segments was visually confirmed. In the event of incomplete reperfusion, the animal was excluded from further analysis.
BM transplantation.
BM transplantation (BMT) experiments were performed as previously described (54). Briefly, 5 × 106 BM cells from donor animals were injected i.v. into lethally irradiated (2 × 6 Gy) recipients. Successful BMT in RAGE chimeric mice was confirmed via GFP fluorescence as described (55), taking advantage of GFP expression in Ager–/– mice. Successful BMT in Elane chimeric mice was confirmed by quantitative PCR (qPCR) in spleens. APAP treatment was performed 8 weeks after BMT.
Serum transaminase, chemokine, and HMGB1 measurements.
Serum ALT and AST measurements were performed using ALT/GPT and AST/GOT kits (both from Thermo Scientific) according to the manufacturer’s instructions and calculated using a standard curve. IL-6 and MCP-1 ELISA (both from R&D Systems) and HMGB1 ELISA (IBL International) were performed according to the manufacturer’s instructions.
Chemotaxis assays.
To assess the effects of HMGB1 on neutrophil migration toward necrotic tissue, HMGB1-containing and HMGB1-deficient liver extracts were prepared according the following procedure: Hmgb1fl/fl Mx1-Cre–positive and Mx1-Cre–negative mice were injected 3 times with poly-(I:C), and livers were harvested 10 days later. Livers were lysed in sterile PMN buffer (PBS containing glucose and BSA) and subjected to 5 freeze-thaw cycles, followed by centrifugation. For in vitro chemotaxis assays, neutrophils were isolated from the peripheral blood of healthy donors using a histopaque gradient as previously described (56). For additional experiments involving WT and Ager–/– neutrophils, neutrophils were isolated from the BM of WT or Ager–/– mice as previously described (57). For some experiments, neutrophils and liver extracts were pretreated with either the CXCR4 inhibitor AMD3100 (Sigma-Aldrich) or anti–mouse CXCL2 antibody (AF-452-NA; R&D Systems). Neutrophil chemotaxis through 3-μm inserts (BD) coated with laminin, fibronectin, or fibrinogen was determined in Boyden chambers, with 1 × 106 neutrophils placed in the upper chamber and 30 mg liver extract in the lower chamber, both in sterile PMN buffer. Migration was quantified after 3 hours. Chemotaxis of macrophages was performed in an analogous manner using BM-derived macrophages, isolated as previously described (58), using 8-μm-pore-size filters. For in vivo chemotaxis, 30 mg lysed liver tissue was injected i.p. into recipient mice. After 20 hours, the peritoneum of the recipient mice was lavaged with 4 ml sterile PBS, and cells were counted using a hemocytometer.
RNA extraction, qPCR, Western blotting, and immunohistochemistry.
RNA from snap-frozen tissues was column purified (Roche Diagnostics). Following reverse transcription, qPCR was performed as previously described using primer-probe pairs (Applied Biosystems), with normalization to 18S and relative quantification by the standard curve method (59). Electrophoresis of protein extracts and subsequent blotting were performed as previously described (54). Blots were incubated with rabbit antibody against HMGB1 at a dilution of 1:1,000 (ab18256; Abcam) and rabbit anti–acetaminophen protein adducts (a gift of L. Pohl, NIH, Bethesda, Maryland, USA) at a dilution of 1:1,000 to 1:5,000 and visualized by chemiluminescence (Thermo Scientific). Blots were reprobed with mouse antibodies against β-actin (A5441; Sigma-Aldrich) or GADPH (G9225; Sigma-Aldrich). Immunohistochemical staining was performed on paraffin-embedded liver sections after 10% formalin fixation using a primary antibody against Ly-6B.2 (clone MCA771G; AbD Serotec) and anti-F4/80 (clone CI:A3-1, catalog MCA497R; AbD Serotec), followed by biotinylated anti–rabbit or anti–rat IgG, respectively, and developed with DAB peroxidase substrate (Vector Laboratories). HMGB1 immunohistochemistry was performed using rabbit anti–HMGB1 (ab18256; Abcam). To identify HMGB1 in specific cell populations, hepatocytes were identified by HNF4α staining (using anti-HNF4α antibody SC-6556; Santa Cruz Biotechnology Inc.), hepatic stellate cells by desmin staining (using Thermo Scientific Lab Vision anti–desmin RB-9014-P antibody), macrophages (using clone CI:A3-1 from AbD Serotec for F4/80 or pan-macrophage antibody ab56297 from Abcam), endothelial cells by endomucin staining (using anti-endomucin antibody sc-65495; Santa Cruz Biotechnology Inc.), and by fluorescent secondary antibodies as previously described (60), followed by confocal microscopy on a Nikon A1R MP confocal microscope (Nikon Instruments) with a ×40 oil immersion lens.
Hepatocyte isolation.
Mouse hepatocytes were isolated as previously described via 2-step collagenase perfusion (59). Hepatocytes were allowed to attach for 90 minutes on collagen-coated plates in RPMI containing 10% FBS (Gibco), followed by overnight starvation in serum-free medium before experiments. Cells were harvested for protein analysis 3 hours after APAP exposure, and cell death was quantified by assessing the percentage of PI-positive nuclei 24 hours after APAP exposure.
Glutathione measurements.
Glutathione measurements were performed using a glutathione assay kit according to the manufacturer’s instructions (Cayman Chemicals).
Analysis of HMGB1 by electrospray ionization liquid chromatography MS/MS.
All chemicals and solvents used were of the highest available grade (Sigma-Aldrich). Samples were precleared with 50 μl protein G sepharose beads for 1 hour at 4°C. HMGB1 was immunoprecipitated with 5 μg rabbit anti–HMGB1 (ab18256; Abcam) for 16 hours at 4°C as previously described (61). For the analysis of HMGB1 posttranslational modifications, free thiol groups within HMGB1 were alkylated for 90 minutes with 10 mM iodoacetamide at 4°C. Cysteine residues in disulfide bonds were then reduced with 30 mM dithiothreitol at 4°C for 1 hour followed by alkylation of newly exposed thiol groups with 90 mM NEM at 4°C for 10 minutes as previously described (62). Samples were subjected to trypsin (Promega) or GluC (New England BioLabs Inc.) digestion according to the manufacturer’s instructions and desalted using C18 Ziptips (EMD Millipore). Characterization of acetylated or phosphorylated lysine residues and redox modifications on cysteine residues within HMGB1 were determined using an AB SCIEX QTRAP 5500 (AB SCIEX) equipped with a NanoSpray II source by in-line liquid chromatography using a U3000 HPLC System connected to a 180 μm × 20 mm nanoACQUITY UPLC C18 trap column and a 75 μm × 15 cm nanoACQUITY UPLC BEH130 C18 column (Waters) via reducing unions. A gradient from 0.05% TFA (v/v) to 50% ACN/0.08% TFA (v/v) over a 40-minute period was applied at a flow rate of 200 nl/minute (63). The ion spray potential was set at 2,200 to 3,500 V, the nebulizer gas at 19, and the interface heater at 150°C.
Statistics.
All data are expressed as the mean ± SEM. For comparison of 2 groups, an unpaired 2-tailed t or Mann-Whitney U test was used. Differences in survival were compared using a log-rank (Mantel-Cox) test. A P value of less than 0.05 was considered statistically significant.
Study approval.
All animal procedures were performed with Columbia University IACUC approval and were in accordance with the Guide for the Care and Use of Laboratory Animals (National Academies Press. 8th edition. Revised 2011). Isolatio of neutrophils from the blood of volunteers was performed with Columbia University IRB approval.
Supplementary Material
Acknowledgments
This study was supported by NIH grants 5R01DK76920 and 1U01AA021912 (to R.F. Schwabe). P. Huebener was supported by a fellowship from the German Research Foundation (Hu 1953/1-1). J.M. Caviglia was supported by NIH grant 1K22CA178098.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information:J Clin Invest. 2015;125(2):539–550. doi:10.1172/JCI76887.
References
- 1.Kono H, Rock KL. How dying cells alert the immune system to danger. Nat Rev Immunol. 2008;8(4):279–289. doi: 10.1038/nri2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Matzinger P. The danger model: a renewed sense of self. Science. 2002;296(5566):301–305. doi: 10.1126/science.1071059. [DOI] [PubMed] [Google Scholar]
- 3.Scaffidi P, Misteli T, Bianchi ME. Release of chromatin protein HMGB1 by necrotic cells triggers inflammation. Nature. 2002;418(6894):191–195. doi: 10.1038/nature00858. [DOI] [PubMed] [Google Scholar]
- 4.Lotze MT, Tracey KJ. High-mobility group box 1 protein (HMGB1): nuclear weapon in the immune arsenal. Nat Rev Immunol. 2005;5(4):331–342. doi: 10.1038/nri1594. [DOI] [PubMed] [Google Scholar]
- 5.Zitvogel L, Kepp O, Kroemer G. Decoding cell death signals in inflammation and immunity. Cell. 2010;140(6):798–804. doi: 10.1016/j.cell.2010.02.015. [DOI] [PubMed] [Google Scholar]
- 6.Chen GY, Nunez G. Sterile inflammation: sensing and reacting to damage. Nat Rev Immunol. 2010;10(12):826–837. doi: 10.1038/nri2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kaczmarek A, Vandenabeele P, Krysko DV. Necroptosis: the release of damage-associated molecular patterns and its physiological relevance. Immunity. 2013;38(2):209–223. doi: 10.1016/j.immuni.2013.02.003. [DOI] [PubMed] [Google Scholar]
- 8.Broere F, van der Zee R, van Eden W. Heat shock proteins are no DAMPs, rather ‘DAMPERs’. Nat Rev Immunol. 2011;11(8):565. doi: 10.1038/nri2873-c1. [DOI] [PubMed] [Google Scholar]
- 9.Bianchi ME. HMGB1 loves company. J Leukoc Biol. 2009;86(3):573–576. doi: 10.1189/jlb.1008585. [DOI] [PubMed] [Google Scholar]
- 10.Hreggvidsdottir HS, et al. The alarmin HMGB1 acts in synergy with endogenous and exogenous danger signals to promote inflammation. J Leukoc Biol. 2009;86(3):655–662. doi: 10.1189/jlb.0908548. [DOI] [PubMed] [Google Scholar]
- 11.Tsan MF. Heat shock proteins and high mobility group box 1 protein lack cytokine function. J Leukoc Biol. 2011;89(6):847–853. doi: 10.1189/jlb.0810471. [DOI] [PubMed] [Google Scholar]
- 12.Bald T, et al. Ultraviolet-radiation-induced inflammation promotes angiotropism and metastasis in melanoma. Nature. 2014;507(7490):109–113. doi: 10.1038/nature13111. [DOI] [PubMed] [Google Scholar]
- 13.Wang H, et al. HMG-1 as a late mediator of endotoxin lethality in mice. Science. 1999;285(5425):248–251. doi: 10.1126/science.285.5425.248. [DOI] [PubMed] [Google Scholar]
- 14.Bianchi ME, Agresti A. HMG proteins: dynamic players in gene regulation and differentiation. Curr Opin Genet Dev. 2005;15(5):496–506. doi: 10.1016/j.gde.2005.08.007. [DOI] [PubMed] [Google Scholar]
- 15.Calogero S, et al. The lack of chromosomal protein Hmg1 does not disrupt cell growth but causes lethal hypoglycaemia in newborn mice. Nat Genet. 1999;22(3):276–280. doi: 10.1038/10338. [DOI] [PubMed] [Google Scholar]
- 16.Huebener P, et al. High-mobility group box is dispensable for autophagy, mitochondrial quality control and organ function in vivo. Cell Metab. 2014;19(1):1–9. doi: 10.1016/j.cmet.2013.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang H, et al. Hepatocyte specific HMGB1 deletion worsens the injury in liver ischemia/reperfusion: A role for intracellular HMGB1 in cellular protection. Hepatology. 2014;59(5):1984–1997. doi: 10.1002/hep.26976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yanai H, et al. Conditional ablation of HMGB1 in mice reveals its protective function against endotoxemia and bacterial infection. Proc Natl Acad Sci U S A. 2013;110(51):20699–20704. doi: 10.1073/pnas.1320808110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang H, Antoine DJ, Andersson U, Tracey KJ. The many faces of HMGB1: molecular structure-functional activity in inflammation, apoptosis, and chemotaxis. J Leukoc Biol. 2013;93(6):865–873. doi: 10.1189/jlb.1212662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Venereau E, et al. Mutually exclusive redox forms of HMGB1 promote cell recruitment or proinflammatory cytokine release. J Exp Med. 2012;209(9):1519–1528. doi: 10.1084/jem.20120189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Beutler B, Milsark IW, Cerami AC. Passive immunization against cachectin/tumor necrosis factor protects mice from lethal effect of endotoxin. Science. 1985;229(4716):869–871. doi: 10.1126/science.3895437. [DOI] [PubMed] [Google Scholar]
- 22.Tracey KJ, et al. Anti-cachectin/TNF monoclonal antibodies prevent septic shock during lethal bacteraemia. Nature. 1987;330(6149):662–664. doi: 10.1038/330662a0. [DOI] [PubMed] [Google Scholar]
- 23.Ohlsson K, Bjork P, Bergenfeldt M, Hageman R, Thompson RC. Interleukin-1 receptor antagonist reduces mortality from endotoxin shock. Nature. 1990;348(6301):550–552. doi: 10.1038/348550a0. [DOI] [PubMed] [Google Scholar]
- 24.Silva AT, Bayston KF, Cohen J. Prophylactic and therapeutic effects of a monoclonal antibody to tumor necrosis factor-alpha in experimental gram-negative shock. J Infect Dis. 1990;162(2):421–427. doi: 10.1093/infdis/162.2.421. [DOI] [PubMed] [Google Scholar]
- 25.Pasparakis M, Alexopoulou L, Episkopou V, Kollias G. Immune and inflammatory responses in TNFα-deficient mice: a critical requirement for TNFα in the formation of primary B cell follicles, follicular dendritic cell networks and germinal centers, and in the maturation of the humoral immune response. J Exp Med. 1996;184(4):1397–1411. doi: 10.1084/jem.184.4.1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rothe J, Mackay F, Bluethmann H, Zinkernagel R, Lesslauer W. Phenotypic analysis of TNFR1-deficient mice and characterization of TNFR1-deficient fibroblasts in vitro. Circ Shock. 1994;44(2):51–56. [PubMed] [Google Scholar]
- 27.Peschon JJ, et al. TNF receptor-deficient mice reveal divergent roles for p55 and p75 in several models of inflammation. J Immunol. 1998;160(2):943–952. [PubMed] [Google Scholar]
- 28.Fisher CJ, et al. Treatment of septic shock with the tumor necrosis factor receptor:Fc fusion protein. The Soluble TNF Receptor Sepsis Study Group. N Engl J Med. 1996;334(26):1697–1702. doi: 10.1056/NEJM199606273342603. [DOI] [PubMed] [Google Scholar]
- 29.Angus DC, van der Poll T. Severe sepsis and septic shock. N Engl J Med. 2013;369(9):840–851. doi: 10.1056/NEJMra1208623. [DOI] [PubMed] [Google Scholar]
- 30.Kuhn R, Schwenk F, Aguet M, Rajewsky K. Inducible gene targeting in mice. Science. 1995;269(5229):1427–1429. doi: 10.1126/science.7660125. [DOI] [PubMed] [Google Scholar]
- 31.Yang H, et al. Reversing established sepsis with antagonists of endogenous high-mobility group box 1. Proc Natl Acad Sci U S A. 2004;101(1):296–301. doi: 10.1073/pnas.2434651100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.de Boer J, et al. Transgenic mice with hematopoietic and lymphoid specific expression of Cre. Eur J Immunol. 2003;33(2):314–325. doi: 10.1002/immu.200310005. [DOI] [PubMed] [Google Scholar]
- 33.Georgiades P, et al. VavCre transgenic mice: a tool for mutagenesis in hematopoietic and endothelial lineages. Genesis. 2002;34(4):251–256. doi: 10.1002/gene.10161. [DOI] [PubMed] [Google Scholar]
- 34.Eltzschig HK, Eckle T. Ischemia and reperfusion — from mechanism to translation. Nat Med. 2011;17(11):1391–1401. doi: 10.1038/nm.2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Antoine DJ, et al. Mechanistic biomarkers provide early and sensitive detection of acetaminophen-induced acute liver injury at first presentation to hospital. Hepatology. 2013;58(2):777–787. doi: 10.1002/hep.26294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bernal W, Wendon J. Acute liver failure. N Engl J Med. 2013;369(26):2525–2534. doi: 10.1056/NEJMra1208937. [DOI] [PubMed] [Google Scholar]
- 37.Heard KJ. Acetylcysteine for acetaminophen poisoning. N Engl J Med. 2008;359(3):285–292. doi: 10.1056/NEJMct0708278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bonaldi T, et al. Monocytic cells hyperacetylate chromatin protein HMGB1 to redirect it towards secretion. EMBO J. 2003;22(20):5551–5560. doi: 10.1093/emboj/cdg516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ge X, et al. High mobility group box-1 (HMGB1) participates in the pathogenesis of alcoholic liver disease (ALD). J Biol Chem. 2014;289(33):22672–22691. doi: 10.1074/jbc.M114.552141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Antoine DJ, Harris HE, Andersson U, Tracey KJ, Bianchi ME. A systematic nomenclature for the redox states of high mobility group box (HMGB) proteins. Mol Med. 2014;20:135–137. doi: 10.2119/molmed.2014.00022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Andersson U, Tracey KJ. HMGB1 is a therapeutic target for sterile inflammation and infection. Annu Rev Immunol. 2011;29:139–162. doi: 10.1146/annurev-immunol-030409-101323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Belaaouaj A, et al. Mice lacking neutrophil elastase reveal impaired host defense against gram negative bacterial sepsis. Nat Med. 1998;4(5):615–618. doi: 10.1038/nm0598-615. [DOI] [PubMed] [Google Scholar]
- 43.Mollica L, et al. Glycyrrhizin binds to high-mobility group box 1 protein and inhibits its cytokine activities. Chem Biol. 2007;14(4):431–441. doi: 10.1016/j.chembiol.2007.03.007. [DOI] [PubMed] [Google Scholar]
- 44.Manns MP, et al. Glycyrrhizin in patients who failed previous interferon alpha-based therapies: biochemical and histological effects after 52 weeks. J Viral Hepat. 2012;19(8):537–546. doi: 10.1111/j.1365-2893.2011.01579.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zigmond E, et al. Infiltrating monocyte-derived macrophages and resident kupffer cells display different ontogeny and functions in acute liver injury. J Immunol. 2014;193(1):344–353. doi: 10.4049/jimmunol.1400574. [DOI] [PubMed] [Google Scholar]
- 46.Kessenbrock K, et al. Proteinase 3 and neutrophil elastase enhance inflammation in mice by inactivating antiinflammatory progranulin. J Clin Invest. 2008;118(7):2438–2447. doi: 10.1172/JCI34694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Neumann K, et al. Clec12a is an inhibitory receptor for uric acid crystals that regulates inflammation in response to cell death. Immunity. 2014;40(3):389–399. doi: 10.1016/j.immuni.2013.12.015. [DOI] [PubMed] [Google Scholar]
- 48.Yipp BG, et al. Infection-induced NETosis is a dynamic process involving neutrophil multitasking in vivo. Nat Med. 2012;18(9):1386–1393. doi: 10.1038/nm.2847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kang R, et al. Intracellular Hmgb1 inhibits inflammatory nucleosome release and limits acute pancreatitis in mice. Gastroenterology. 2014;146(4):1097–1107. doi: 10.1053/j.gastro.2013.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.McDonald B, et al. Intravascular danger signals guide neutrophils to sites of sterile inflammation. Science. 2010;330(6002):362–366. doi: 10.1126/science.1195491. [DOI] [PubMed] [Google Scholar]
- 51.Yamasaki S, Ishikawa E, Sakuma M, Hara H, Ogata K, Saito T. Mincle is an ITAM-coupled activating receptor that senses damaged cells. Nat Immunol. 2008;9(10):1179–1188. doi: 10.1038/ni.1651. [DOI] [PubMed] [Google Scholar]
- 52.Nystrom S, et al. TLR activation regulates damage-associated molecular pattern isoforms released during pyroptosis. EMBO J. 2013;32(1):86–99. doi: 10.1038/emboj.2012.328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tsung A, et al. The nuclear factor HMGB1 mediates hepatic injury after murine liver ischemia-reperfusion. J Exp Med. 2005;201(7):1135–1143. doi: 10.1084/jem.20042614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Seki E, et al. TLR4 enhances TGF-β signaling and hepatic fibrosis. Nat Med. 2007;13(11):1324–1332. doi: 10.1038/nm1663. [DOI] [PubMed] [Google Scholar]
- 55.Pusterla T, et al. Receptor for advanced glycation endproducts (RAGE) is a key regulator of oval cell activation and inflammation-associated liver carcinogenesis in mice. Hepatology. 2013;58(1):363–373. doi: 10.1002/hep.26395. [DOI] [PubMed] [Google Scholar]
- 56.Li Y, Karlin A, Loike JD, Silverstein SC. A critical concentration of neutrophils is required for effective bacterial killing in suspension. Proc Natl Acad Sci U S A. 2002;99(12):8289–8294. doi: 10.1073/pnas.122244799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Swamydas M, Lionakis MS. Isolation, purification and labeling of mouse bone marrow neutrophils for functional studies and adoptive transfer experiments. J Vis. 2013;Exp(77):e50586. doi: 10.3791/50586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Davis BK. Isolation, culture, and functional evaluation of bone marrow-derived macrophages. Methods Mol Biol. 2013;1031:27–35. doi: 10.1007/978-1-62703-481-4_3. [DOI] [PubMed] [Google Scholar]
- 59.Dapito DH, et al. Promotion of hepatocellular carcinoma by the intestinal microbiota and TLR4. Cancer Cell. 2012;21(4):504–516. doi: 10.1016/j.ccr.2012.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mederacke I, et al. Fate tracing reveals hepatic stellate cells as dominant contributors to liver fibrosis independent of its aetiology. Nat Commun. 2013;4:2823. doi: 10.1038/ncomms3823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Antoine DJ, et al. High-mobility group box-1 protein and keratin-18, circulating serum proteins informative of acetaminophen-induced necrosis and apoptosis in vivo. Toxicol Sci. 2009;112(2):521–531. doi: 10.1093/toxsci/kfp235. [DOI] [PubMed] [Google Scholar]
- 62.Yang H, et al. Redox modification of cysteine residues regulates the cytokine activity of high mobility group box-1 (HMGB1). Mol Med. 2012;18:250–259. doi: 10.2119/molmed.2011.00389. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 63.Antoine DJ, et al. Molecular forms of HMGB1 and keratin-18 as mechanistic biomarkers for mode of cell death and prognosis during clinical acetaminophen hepatotoxicity. J Hepatol. 2012;56(5):1070–1079. doi: 10.1016/j.jhep.2011.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.