Sir,
Riveira-Muñoz et al.1 recently reported that HIV-1 elite suppressors (ES), who maintain low viral loads without pharmacological intervention,2 also maintain higher levels of restriction factor SAMHD1 transcript than either healthy donors (HD) or viraemic progressors (VP) in PBMCs. This compelling finding applied to individuals with and without protective HLA-B alleles. As the authors astutely stated, however, increased SAMHD1 ‘may not be a cause but the consequence’ of suppression.1 Beyond this caveat, it is also important to determine the extent to which SAMHD1 up-regulation is found in suppressing individuals in disparate cohorts. Indeed, a previous quantification of multiple known or suspected restriction factors did not find different SAMHD1 levels in PBMCs of ES, VP and HD.3
We had conducted a similar study with a group of PBMC samples from HD (n = 8), a cohort of ES (n = 7) and VP (n = 7) and would like to add these data to the ongoing discussion. Our samples were described in an earlier publication in the context of microRNA expression.4 We used essentially the same methods as previously described.1,5 Total RNA was extracted from PBMCs with the Ambion mirVana microRNA kit and quantitative PCR results for SAMHD1 were normalized to the average of two housekeeping genes: glyceraldehyde 3-phosphate dehydrogenase (GAPDH), as used by Riveira-Muñoz et al.,1 as well as β-actin. Because SAMHD1 responds to type II and, in some cell types, type I IFNs,6,7 we also measured MX1, an indicator of type I IFN antiviral response.
All samples used in this study were de-identified prior to transfer, such that this research is not considered human subject research. There are no privacy concerns, as the authors have access to no identifying information. Samples had previously been obtained with appropriate consent under the approval of the Johns Hopkins Institutional Review Board.
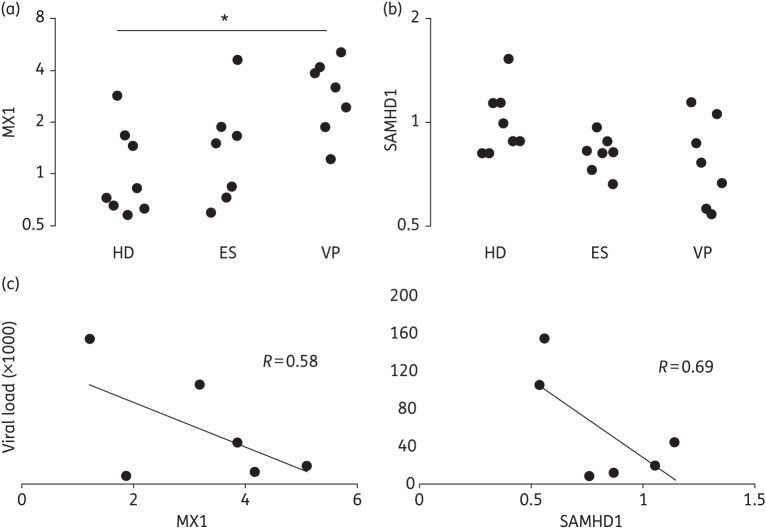
Most HD had low baseline levels of MX1, but several exhibited some degree of activation (Figure 1a). Activation was also seen in several ES. However, VP had the highest levels of MX1, on average >3-fold the HD average. Employing a Kruskal–Wallis test on the three groups, the group means were significantly different (P < 0.01) and post-tests with Bonferroni correction identified the VP–HD difference, but not the ES–HD difference, as significant (P < 0.017; Figure 1a).
Figure 1.
MX1 and SAMHD1 expression and correlation with viral load. HIV-1-positive VP, but not ES, displayed significantly higher levels of the type I IFN-responsive antiviral MX1 according to a Kruskal–Wallis test followed by post-tests (a); *P < 0.05. No differences were detected for retroviral restriction factor SAMHD1 (b). Within the VP group, there was an apparent trend towards negative correlation of both MX1 and SAMHD1 with viral load (c).
As for SAMHD1 expression, we did not observe the increase in ES PBMCs found in the cohort of Riveira-Muñoz et al.1 (Figure 1b). No ES had an SAMHD1 level higher than the average of the HD and there was little variability of SAMHD1 expression in the ES group. In fact, both ES and VP registered on average 20% less SAMHD1 transcript than HD. Despite the trend towards lower SAMHD1 expression in infected individuals, there was no significant difference between the groups (Figure 1b).
We found this lack of increased expression somewhat curious given the significantly higher IFN signalling in VP, as evidenced by increased MX1 levels, and our finding that SAMHD1 tracks with the type I IFN response in the brains of acutely retrovirus-infected animals (E. L. Buchanan, M. A. McAlexander and K. W. Witwer, unpublished results). Accordingly, we examined the relationship of MX1 and SAMHD1 in the three groups. Indeed, in HD and in VP, higher MX1 tended to coincide with higher SAMHD1 despite overall lower levels in VP, with R = 0.73 (VP) and R = 0.55 (HD). In contrast, there was no apparent correlation of MX1 and SAMHD1 in PBMCs from ES (R = 0.00).
Finally, we compared the gene expression data with the CD4+ count and viral load for each infected individual. For ES and VP, SAMHD1 was not correlated with the CD4+ T cell count (R = 0.01 and R = 0.06, respectively; data not shown). However, in VP, viral load appeared to be negatively correlated with both MX1 and SAMHD1 (R = 0.58 and R = 0.69, respectively; Figure 1c). This result suggests that, regardless of the nature of the link between type I IFN signalling and SAMHD1, increases in both tend to associate with viral control.
We acknowledge that the small number of samples and the relatively minor variation in the levels of SAMHD1 preclude firm statements about correlation. Nevertheless, we submit that three conclusions are warranted. First, reminiscent of Abdel-Mohsen et al.,3 the data are inconsistent with increased SAMHD1 mRNA levels in this cohort of ES. Second, it is possible that the lack of apparent correlation of SAMHD1 and IFN response in ES suggests a dampened response of SAMHD1 to IFN in cells from these individuals. Those in other cohorts might have a different response, however. Third, regardless of whether SAMHD1 plays a role in elite suppression, SAMHD1 levels in VP, while often lower than those of HD, appear to retain some sensitivity to IFN signalling and may be associated with lower viral load in VP.
In future research, it will be instructive to examine SAMHD1 and other restriction factors in specific cell types of ES and to include protein and post-translational modification-level tests. To exert an antiviral effect, the SAMHD1 transcript must be present first of all, so quantifying mRNA is certainly important. Beyond transcript and protein abundance, however, multimerization8 and phosphorylation9,10 determine SAMHD1 protein activity and the latter is clearly tied to IFN signalling.10
Funding
Funding was provided by the National Institute of Allergy and Infectious Diseases of the National Institutes of Health of the United States (R21AI102659 to K. W. W.).
Transparency declarations
We have no conflicts of interest to declare. The funding organization played no role in the design, execution, analysis or reporting of this research.
Author contributions
Study conception and design: K. W. W. Data acquisition: E. L. B. and M. A. M. Analysis and interpretation: E. L. B., M. A. M. and K. W. W. Drafting and/or revision of manuscript: K. W. W., E. L. B. and M. A. M.
Acknowledgements
We gratefully acknowledge Joel N. Blankson for providing de-identified PBMC samples.
References
- 1.Riveira-Muñoz E, Ruiz A, Pauls E, et al. Increased expression of SAMHD1 in a subset of HIV-1 elite controllers. J Antimicrob Chemother. 2014;69:3057–60. doi: 10.1093/jac/dku276. [DOI] [PubMed] [Google Scholar]
- 2.Buckheit RW, III, Salgado M, Martins KO, et al. The implications of viral reservoirs on the elite control of HIV-1 infection. Cell Mol Life Sci. 2013;70:1009–19. doi: 10.1007/s00018-012-1101-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Abdel-Mohsen M, Raposo RA, Deng X, et al. Expression profile of host restriction factors in HIV-1 elite controllers. Retrovirology. 2013;10:106. doi: 10.1186/1742-4690-10-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Witwer KW, Watson AK, Blankson JN, et al. Relationships of PBMC microRNA expression, plasma viral load, and CD4+ T cell count in HIV-1-infected elite suppressors and viremic patients. Retrovirology. 2012;9:5. doi: 10.1186/1742-4690-9-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sisk JM, Clements JE, Witwer KW. miRNA profiles of monocyte-lineage cells are consistent with complicated roles in HIV-1 restriction. Viruses. 2012;4:1844–64. doi: 10.3390/v4101844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lafuse WP, Brown D, Castle L, et al. Cloning and characterization of a novel cDNA that is IFN-γ-induced in mouse peritoneal macrophages and encodes a putative GTP-binding protein. J Leukoc Biol. 1995;57:477–83. doi: 10.1002/jlb.57.3.477. [DOI] [PubMed] [Google Scholar]
- 7.St Gelais C, de Silva S, Amie SM, et al. SAMHD1 restricts HIV-1 infection in dendritic cells (DCs) by dNTP depletion, but its expression in DCs and primary CD4+ T-lymphocytes cannot be upregulated by interferons. Retrovirology. 2012;9:105. doi: 10.1186/1742-4690-9-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hansen EC, Seamon KJ, Cravens SL, et al. GTP activator and dNTP substrates of HIV-1 restriction factor SAMHD1 generate a long-lived activated state. Proc Natl Acad Sci USA. 2014;111:E1843–51. doi: 10.1073/pnas.1401706111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.White TE, Brandariz-Nunez A, Valle-Casuso JC, et al. The retroviral restriction ability of SAMHD1, but not its deoxynucleotide triphosphohydrolase activity, is regulated by phosphorylation. Cell Host Microbe. 2013;13:441–51. doi: 10.1016/j.chom.2013.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cribier A, Descours B, Valadao AL, et al. Phosphorylation of SAMHD1 by cyclin A2/CDK1 regulates its restriction activity toward HIV-1. Cell Rep. 2013;3:1036–43. doi: 10.1016/j.celrep.2013.03.017. [DOI] [PubMed] [Google Scholar]