Abstract
Objectives
To identify β-lactamase genes in gut commensal Bacteroides species and to assess the impact of these enzymes, when carried by outer membrane vesicles (OMVs), in protecting enteric pathogens and commensals.
Methods
A deletion mutant of the putative class A β-lactamase gene (locus tag BT_4507) found in the genome of the human commensal Bacteroides thetaiotaomicron was constructed and a phenotypic analysis performed. A phylogenetic tree was built from an alignment of nine Bacteroides cephalosporinase protein sequences, using the maximum likelihood method. The rate of cefotaxime degradation after incubation with OMVs produced by different Bacteroides species was quantified using a disc susceptibility test. The resistance of Salmonella Typhimurium and Bifidobacterium breve to cefotaxime in liquid culture in the presence of B. thetaiotaomicron OMVs was evaluated by measuring bacterial growth.
Results
The B. thetaiotaomicron BT_4507 gene encodes a β-lactamase related to the CepA cephalosporinase of Bacteroides fragilis. OMVs produced by B. thetaiotaomicron and several other Bacteroides species, except Bacteroides ovatus, carried surface-associated β-lactamases that could degrade cefotaxime. β-Lactamase-harbouring OMVs from B. thetaiotaomicron protected Salmonella Typhimurium and B. breve from an otherwise lethal dose of cefotaxime.
Conclusions
The production of membrane vesicles carrying surface-associated β-lactamases by Bacteroides species, which constitute a major part of the human colonic microbiota, may protect commensal bacteria and enteric pathogens, such as Salmonella Typhimurium, against β-lactam antibiotics.
Keywords: β-lactamases, protective effect, gut microbiota, Salmonella
Introduction
The adult human gastrointestinal (GI) tract accommodates a bacterial community (the microbiota) comprising trillions of cells that carry out vital functions for human health. This association involves co-evolved beneficial human–microbiota interactions that are altered as a result of many environmental factors. In particular, the presence of antibiotics can disturb colonic metabolism and absorption of vitamins, and can alter susceptibility to infection.1,2 The human GI tract microbiota is dominated by two bacterial phyla, Firmicutes and Bacteroidetes.3 Species of the genus Bacteroides, which constitute ∼30% of all bacteria in the human GI tract, are among the most resistant anaerobes to antibiotics,4 including penicillins and broad-spectrum cephalosporins.5
During the 1980s, β-lactamase-producing strains of Bacteroides species were reported to protect penicillin-susceptible β-haemolytic streptococci from penicillin G in vivo, as shown by monitoring the formation of subcutaneous abscesses in mice co-infected by both organisms.6 Likewise, β-lactamases produced by Escherichia coli can protect Salmonella Typhimurium when both organisms are co-cultured in liquid broth in the presence of ampicillin.7 It was suggested in this study that the β-lactamase protected the Salmonella against ampicillin once it was exported in outer membrane vesicles (OMVs) produced and released into the medium by the E. coli strain. Schaar et al.8,9 later confirmed this idea by demonstrating that β-lactamases associated with OMVs produced by Moraxella catarrhalis protect Streptococcus pneumoniae and Haemophilus influenzae against amoxicillin, and that OMVs produced by H. influenzae could protect group A streptococci against amoxicillin. The possibility that OMVs produced by commensal Bacteroides species possess similar capabilities has not been considered.
In this study, we describe a novel cephalosporinase gene in the genome of a prominent member of the human GI tract microbiota, Bacteroides thetaiotaomicron. Homologues of this gene exist in most, if not all Bacteroides species. We also describe the protective effect of β-lactamase-carrying OMVs produced by B. thetaiotaomicron on the susceptibility of Salmonella Typhimurium and Bifidobacterium breve to β-lactam antibiotics.
Materials and methods
Bacterial strains and growth conditions
Bacteroides species and strains (Table 1) were grown under anaerobic conditions at 37°C in brain heart infusion (BHI) medium (Oxoid/Thermo Fisher, Basingstoke, UK) supplemented with 0.5 mg/L haemin (Sigma-Aldrich, St Louis, MO, USA) (BHI–haemin). Antibiotic resistance markers in B. thetaiotaomicron were selected using 1 mg/L tetracycline, 5 mg/L erythromycin or 200 mg/L gentamicin. E. coli strains GC10 (Sigma-Aldrich) transformed with pGH014-based plasmids (see below) and HB101(pRK2013) (DMSZ Collection, Braunschweig, Germany) were grown under agitation at 37°C in LB medium10 supplemented with 100 mg/L spectinomycin and 50 mg/L kanamycin, respectively. Electrocompetent E. coli cells were prepared and transformed by the method of Sambrook and Russell (2001).10 Salmonella enterica serovar Typhimurium ATCC 14028 was grown at 37°C in LB medium under agitation.
Table 1.
Bacteroides species and strains
| Strain | Origin | Locus taga | Combined disc method (mm)c | ESBL Etestd (cefotaxime/cefotaxime plus clavulanic acid in mg/L) |
|---|---|---|---|---|
| B. thetaiotaomicron VPI-5482 (NCTC 10582, Werner, 1970) | DSMZ | BT_4507 | 14 ± 0.8 | >16/1 |
| B. thetaiotaomicron GH221 | this work | ΔBT_4507 | no difference | 0.75/0.75 |
| B. dorei DSM 17855 | DSMZ | BACDOR_02757 | 9 ± 3.5 | 16/1 |
| B. fragilis NCTC 9343 (Garrod, 1955) | DSMZ | BF_1199 | 12.6 ± 1.7 | 16/0.75 |
| B. ovatus V975 | Whitehead and Hespell44 | BACOV975_02528b | 18 ± 0.8 | 16/0.19 |
| B. stercoris DMS 19555 | DSMZ | BACSTE_01456 | 7 ± 0.8 | >16/1 |
| B. xylanisolvens XB1A ATCC 43183 | DSMZ | GIB_3495 | 21.5 ± 0.5 | >16/0.125 |
aThe protein sequences were obtained from the National Center for Biotechnology Information (NCBI) protein databases.
bU. Wegmann, Institute of Food Research, Norwich, UK, personal communication.
cDifference between inhibition zones with cefpodoxime (10 μg) in the presence/absence of clavulanic acid (1 μg). The results shown are from three experiments performed independently.
dEtests were performed using strips containing cefotaxime and cefotaxime plus clavulanic acid.
Electron microscopy
Samples were fixed for 1 h in 2.5% glutaraldehyde in 0.1 M piperazinediethanesulfonic acid (PIPES) buffer (pH 7.2). After washing with 0.1 M PIPES buffer, each sample was pipetted onto the centre of a small square of filter paper, which was folded and inserted into a critical-point drying capsule and dehydrated in a series of ethanol solutions (10, 20, 30, 40, 50, 60, 70, 80, 90 and 3 × 100%). Samples were critical-point dried in a Polaron E3000 drier (Quorum Technologies, Newhaven, UK) using liquid carbon dioxide as the transition fluid. The parcels were then carefully unfolded and the dry cells attached to sticky tabs mounted on scanning electron microscopy stubs by flicking the back of the filter paper in the direction of the stub. The samples were coated with gold in a high-resolution sputter-coater apparatus (Agar Scientific, Stansted, UK). Scanning electron microscopy was carried out using a Zeiss Supra 55 VP FEG SEM operating at 3 kV.
For negative staining, a drop of vesicle suspension was applied to a carbon-coated Formvar copper grid and left for 1 min before washing with five or six drops of 2% uranyl acetate solution in water. The excess stain was wicked off and the grids were left to dry thoroughly before viewing in the transmission electron microscope. The grids were examined and imaged in an Tecnai G2 20 Twin transmission electron microscope (FEI, Hillsboro, OR, USA) at 200 kV.
Oligonucleotide primers
The primers used are detailed in Table S1 (available as Supplementary data at JAC Online).
Construction of a BT_4507 deletion mutant
A 798 bp chromosomal DNA fragment upstream from BT_4507 and including the first 18 nucleotides of its 5′-end region was amplified by PCR using the primer pair BT4507_1 and BT4507_2. This product was then cloned into the SacI/BamHI site of the E. coli–Bacteroides suicide shuttle vector pGH014, consisting of plasmid pFD51611 with the tetracycline resistance gene tetQ from the Bacteroides plasmid pBT-212 inserted into the BamHI and SalI restriction sites. A 829 bp chromosomal DNA fragment downstream from BT_4507, including the last 46 nucleotides of the 3′-end region, was amplified by PCR using the primer pair BT4507_3 and BT4507_4 and was cloned into the SalI/PstI site of the pGH014-based plasmid. The resulting plasmid, containing the BT_4507::tetQ construct, was mobilized from E. coli GC10 into B. thetaiotaomicron by triparental filter mating,13 using E. coli HB101(pRK2013) as the helper strain. Transconjugants were selected on BHI–haemin agar containing 200 mg/L gentamicin and 1 mg/L tetracycline. Determination of susceptibility to either tetracycline or erythromycin was carried out to identify recombinants that were tetracycline resistant and erythromycin susceptible, after re-streaking transconjugant bacteria on LB agar containing tetracycline or both antibiotics. PCR analysis and sequencing were used to confirm the allelic exchange. A transconjugant, GH221, containing the BT_4507::tetQ construct inserted into the B. thetaiotaomicron chromosome was selected for further studies.
Overexpression of BT_4507 in the deletion mutant
To complement the B. thetaiotaomicron BT_4507 deletion mutant GH221, a derivative of the Bacteroides vector pGH043,14 was engineered for high-level expression of BT_4507. First, the primer pair Lactamase_F and Lactamase_EcoRI_R was used to amplify an 889 bp region encoding BT_4507 from B. thetaiotaomicron VPI-5482 genomic DNA. This BT_4507 fragment was digested with EcoRI before cloning into the NcoI (blunted)/EcoRI site of the Bacteroides expression vector pGH043, creating pGH092. To visualize the BT_4507 protein by fluorescence imaging, we made an in-frame fusion of the flavin mononucleotide-based fluorescent protein Pp1 with the C-terminus of BT_4507, using a 30-amino-acid linker. A PCR fragment of Pp1 was then obtained, using pGLOW-Pp1-stop (Evocatal GmbH, Dusseldorf, Germany) plasmid DNA as template. This fragment, attached to the linker by recombinant PCR, was cloned into the BsaAI/EcoRI site of pGH092, creating pGH095. To achieve overexpression, the ribosome-binding site region present in pGH095 was exchanged with the ribosome-binding site region of vector pGH090,14 by way of splice extension PCR. Initially, the primer pairs Reverse and 20-Lact_R, and 20-Lact_F and Lactamase_EcoRI_R were used, respectively, on templates pGH090 and pGH092. The resulting products were then used as templates for the splice PCR involving the primer pair Reverse and Lactamase_EcoRI_R. The 683 bp SphI, PshAI-digested PCR fragment was used to replace the corresponding 761 bp digest fragment of pGH095, creating pGH098. Transformation of the BT_4507 deletion mutant GH221 with pGH098 resulted in the creation of GH274.
Phylogenetic analysis
The evolutionary relationship of β-lactamases was inferred using the maximum likelihood method in the MEGA5 software tool.15 Amino acid sequences were aligned with PRANK,16 and highly variable regions removed from the dataset. The Whelan and Goldman model of evolution17 was used and initial tree(s) for the heuristic search were obtained automatically as follows. When the number of common sites was <100, or otherwise less than one-fourth of the total number of sites, the maximum parsimony method was used; otherwise the BIONJ method,18 with maximum composite likelihood (MCL) distance matrix, was used.19 To provide statistical support for each node on the tree, a consensus tree was generated from 1000 bootstrap datasets.
OMV isolation
Bacterial cultures (20 mL) were centrifuged at 5000 g for 15 min at 4°C and the supernatants were filtered through 0.22 μm pore-size polyethersulfone (PES) membranes (Sartorius, Goettingen, Germany) to remove debris and cells. Supernatants were concentrated by ultrafiltration (100 kDa molecular weight cut-off, Vivaspin 20, Sartorius) to 200 μL. The retentate was rinsed twice with 20 mL of PBS (pH 7.4) and concentrated to 200 μL (2 mg dry weight vesicles). OMV sterility was examined by checking for growth of any contaminating bacterial cells on BHI–haemin agar. OMV protein content was determined using the Total Protein Micro protein assay reagent kit (Sigma-Aldrich) after disruption by sonication. Alternatively, the dry weight of OMVs was determined after incubating OMV suspensions in a drying oven for 48 h. OMVs were also collected from the lower compartment of a Steritop filtration unit (Millipore, Billerica, USA) containing sterile BHI–haemin medium on a magnetic stir plate, after their diffusion through a 0.22 μm pore membrane from the upper compartment containing a growing B. thetaiotaomicron culture in BHI–haemin. The sterility of the BHI–haemin containing the vesicles was confirmed by plating OMV suspensions onto BHI–haemin agar immediately before collection.
Measurement of β-lactamase activity
Bacteroides species were grown in 20 mL of BHI–haemin for 16 h, centrifuged at 3500 g for 10 min and the periplasmic fraction was prepared according to the method described by Osborn et al.20 Briefly, the cell pellet was resuspended in 4 mL of fractionation buffer (30 mM Tris, 20% sucrose, 1 mM EDTA, pH 8) and incubated for 10 min at 20°C, then centrifuged for 10 min at 3000 g. This pellet was resuspended in 0.8 mL of ice-cold 5 mM MgSO4 and left on ice for 10 min, releasing an osmotic shock fluid, which was separated by centrifugation for 10 min at 3000 g, 4°C. OMV concentrations were adjusted according to the protein concentrations obtained after sonication.
To assess the BtCepA β-lactamase activity, 5 μL of protein extract (corresponding to 5 μg for the vesicles) was added to 95 μL of phosphate buffer (0.1 M phosphate/1 mM EDTA, pH 7.0) and assayed spectrophotometrically at 486 nm by hydrolysis of 50 mg/L nitrocefin according to the manufacturer's instructions (Calbiochem).
Enzyme activity at the surface of OMVs
A suspension of 250 μg of vesicles in 0.1 M phosphate/1 mM EDTA buffer (pH 7.0) was incubated for 5 min (for vesicles from B. thetaiotaomicron) and 1 h (for all other Bacteroides species) at 37°C in the presence of 100 mg/L proteinase K (Sigma-Aldrich). Proteinase K activity was stopped by addition of 1 mM phenylmethanesulfonyl fluoride (PMSF). To measure β-lactamase activity, 50 mg/L nitrocefin was added and changes in absorbance at 486 nm were measured. For BtMinpp activity, 1 mM inositol hexakisphosphate (InsP6) (Merck, Readington, NJ, USA) was added to the OMVs and the mix was incubated for 1 h at 37°C. Inositol phosphates were resolved by anion exchange chromatography as previously described.21 As a control, 25 mg/L of purified His-tagged BtMinpp was incubated21 with or without 100 mg/L proteinase K for 1 h at 37°C, with 1 mM PMSF added to stop the reaction. Degradation of 1 mM InsP6 was measured by HPLC (method described in Stentz et al.21) after incubation for 1 h at 37°C. BtMinpp activity was quantified as the ratio of the sum of the integrated peak areas of InsP5 products to the sum of the integrated InsP5 and InsP6 peaks.
β-Lactamase activity in OMVs from multiple Bacteroides species
For OMV production, the different Bacteroides species were grown in the presence of 10 mg/L cefotaxime to ensure optimized cephalosporinase expression. Vesicles corresponding to 10 μg of total protein obtained by sonication were added to 100 μL of a 10 mg/L cefotaxime solution. The antibiotic solution containing OMVs was incubated for 1 h at 37°C, with OMVs subsequently removed by filtration (Amicon Ultra-0.5 centrifugal filter device, 100 kDa, Millipore, Billerica, MA, USA), and 10 μL of filtrate (corresponding to 0.1 μg of cefotaxime) was applied to a disc previously placed on a Salmonella-inoculated soft agar overlay plate. The inhibition zones were measured after 16 h of incubation at 37°C.
Susceptibility tests
The susceptibility of Bacteroides to cefpodoxime and cefpodoxime plus clavulanic acid was tested using a cefpodoxime combination disc kit (Oxoid/Thermo Fisher, Basingstoke, UK) with the discs placed on BHI agar plates topped with a soft agar layer (0.75% agar) seeded with Bacteroides species. Double-ended Etest® strips (AB bioMérieux, Marcy-l'Étoile, France) containing gradients of cefotaxime (16–0.25 mg/L) at one end and cefotaxime (1–0.016 mg/L) plus clavulanic acid at the other end were used to measure susceptibility, on BHI agar. The different Bacteroides species used were pre-grown in the presence of 5 mg/L cefotaxime and the cells were rinsed with fresh BHI broth.
OMV–Salmonella and OMV–Bifidobacterium co-incubations
Cultures of S. enterica serovar Typhimurium strain ATCC 14028 and B. breve UCC2003,22 were grown for 16 h at 37°C under agitation in LB broth and anaerobically in BHI broth supplemented with 0.5% yeast extract (BHIY), respectively. Aliquots of 10 μL of 100-fold dilutions of these cultures were added to 10 mL of pre-warmed LB or BHIY, respectively. OMVs from a 20 mL culture of B. thetaiotaomicron (corresponding to 20 μg of total protein) were concentrated, rinsed and re-suspended in PBS. OMV concentrations were adjusted according to the protein concentrations measured after sonication. After addition of different concentrations of cefotaxime the Salmonella culture was incubated at 37°C with agitation (250 rpm) and the Bifidobacterium culture was incubated anaerobically while being stirred at 37°C. At varying times thereafter cells were plated for viable counts.
Results
BT_4507 encodes a β-lactamase that confers resistance to high levels of ampicillin
To determine whether OMVs produced by B. thetaiotaomicron harbour β-lactamase activity, we first identified the gene(s) responsible for the resistance of the bacterium to β-lactam antibiotics.
We began by measuring the resistance of B. thetaiotaomicron VPI-5482 to penicillins, observing high-level resistance to the penicillin derivative ampicillin (Table 2). We also noted that B. thetaiotaomicron grown in the presence of sublethal concentrations of ampicillin (10–25 mg/L) gave rise to large numbers of tangled filaments >150 μm long (Figure 1b), comprising non-septate cells 12.5 μm in length. In considering the distance between two nodes (Figure 1c), representing the site of cell septation in normal conditions, this dimension is >10-fold greater than that of non-treated cells (Figure 1a). Examples of filament formation in Bacteroides species grown with subinhibitory concentrations of β-lactam antibiotics have previously been reported.23 For E. coli and other Gram-negative aerobes,24,25 the binding of β-lactam antibiotics to PBP3, a key element of the cell-septation machinery, prevents cell division and leads to the formation of filamentous cells.
Table 2.
Resistance to ampicillin and related β-lactamase activity in B. thetaiotaomicron
| Strain | Genotypea | MICb | β-Lactamase activityc in the periplasm | β-Lactamase activityc associated with OMVsd |
||
|---|---|---|---|---|---|---|
| total activity | vesicle fraction | buffer | ||||
| GH196 | WT (pGH043) | 32 | 269 ± 8 | 33 ± 6 | 29 ± 5 | ND |
| GH266 | ΔBT_4507 (pGH043) | 1 | 0.1 ± 0.3 | ND | ND | ND |
| GH274 | ΔBT_4507 (pGH098) | 1024 | 12 415 ± 551 | 284 ± 14 | 271 ± 11 | 2.3 ± 0.5 |
ND, not detected.
apGH043, empty vector; pGH098, plasmid overexpressing BT_4507.
bConcentration of ampicillin in mg/L. The results are from two experiments performed independently.
cβ-Lactamase activity expressed in nmol of hydrolysed nitrocefin/mg of protein/min. The activity was assessed spectrophotometrically by hydrolysis of nitrocefin. The results shown are from three experiments performed independently.
dThe vesicles were incubated in phosphate buffer (0.1 M phosphate/1 mM EDTA, pH 7.0) for 1 h at 37°C and the total activity in the suspension was measured after sonication. Alternatively, the vesicles were removed by filtration and the activity was measured in the vesicle fraction and in the filtered buffer.
Figure 1.
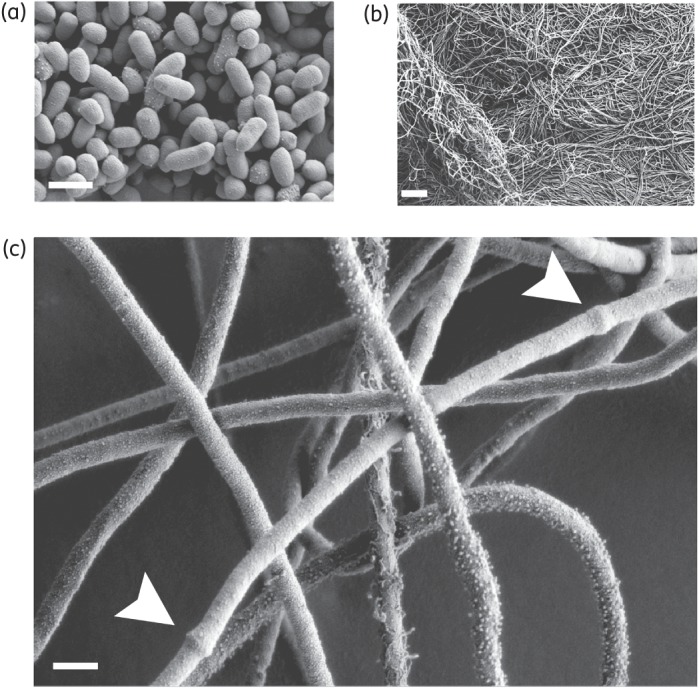
Scanning electron microscope image of B. thetaiotaomicron cells. (a) Non-treated cells. (b and c) Cells grown in the presence of 10 mg/L ampicillin. Scale bar, ∼10 μm (b) and 1 μm (a and c). The nodes, likely to represent the site of cell septation, are indicated with white arrows in (c).
The genome of B. thetaiotaomicron VPI-5482 (NCTC 10582)26 contains four putative β-lactamase genes: three putative class B3 metallo-β-lactamase (MBL) genes (locus tags BT_0822, BT_1146 and BT_1410) and a class A serine β-lactamase gene (BT_4507). There is, however, a significant risk in naming these gene products as class B3 MBLs because of their sequence and structural homology to e.g. metallo-glyoxylases II (based on pairwise sequence alignments using BLASTP,27 and protein structure alignments performed with 3D-JIGSAW28).29 In addition, because of the susceptibility of the strain and the genus Bacteroides as a whole to carbapenems, we concluded that MBLs were not a significant factor in resistance and focused our attention on the serine β-lactamase. The BT_4507-encoded β-lactamase protein contains a predicted secretion signal (http://www.cbs.dtu.dk/services/SignalP/), implying that this enzyme is potentially secreted into the periplasm and could subsequently be packaged into OMVs, as shown for other bacterial species.30 We therefore chose to study BT_4507 and constructed a deletion mutant, which lost resistance to ampicillin, further supporting the absence of other significant β-lactamase activities (Table 2). For ease of detection of β-lactamase activity, a B. thetaiotaomicron strain was engineered to overexpress BT_4507. Overexpression of BT_4507 in trans in the deleted mutant raised the MIC of ampicillin to levels 32-fold higher than for the WT strain (Table 2). Moreover, the β-lactamase activities measured in periplasmic protein extracts of the different variants correlated with the level of resistance to ampicillin (Table 2).
Identification of novel cepA-like chromosomal genes encoding cephalosporinases in Bacteroides species
A BLASTP search of the BT_4507-encoded protein sequence against the non-redundant protein database31 identified 273 sequences with significant alignments (E value ≤1 e−40 and a minimal number of 60 identical amino acids over the entire sequence length). These protein sequences derived mainly from bacteria of the Bacteroidetes phylum. Among them, CepA and CfxA are two class-A cephalosporinase variants from B. fragilis32,33 and CblA is a class A cephalosporinase from Bacteroides uniformis.34 A phylogenetic tree was constructed by alignment of nine Bacteroides representatives (Figure 2), with two GES-type β-lactamases from Enterobacteriaceae as an outgroup.35 BT_4507 belonged to the CepA clade, which includes CepA from B. fragilis. We shall therefore refer to the BT_4507-encoded enzyme as BtCepA.
Figure 2.
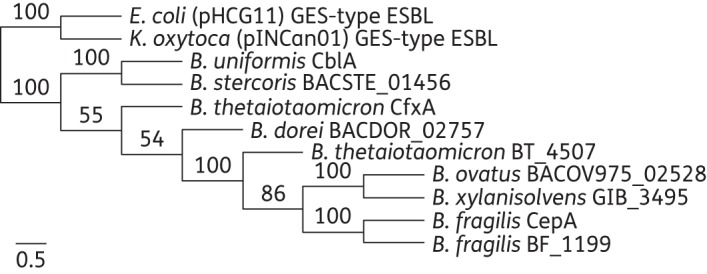
Phylogenetic tree derived from the alignment of 11 β-lactamase proteins from different bacterial species constructed using the maximum likelihood method. To provide statistical support for each node on the tree, a consensus tree was generated from 1000 bootstrap datasets. The tree is drawn to scale, with branch lengths measured as the number of substitutions per site. E., Escherichia; K., Klebsiella; B., Bacteroides; GES enzymes are plasmid-mediated types that are disseminated among Enterobacteriaceae.
To test whether the cephalosporinase homologues from reference Bacteroides listed in the phylogenetic tree were β-lactamases, we adapted two phenotypic confirmatory tests developed for Enterobacteriaceae ESBLs to Bacteroides.36 Resistance to third-generation cephalosporin antibiotics was measured and compared in the presence and absence of clavulanic acid, which inhibits class A β-lactamases (Table 1). Except for the B. thetaiotaomicron cepA deletion mutant, the zone size measured in the combined disc method using cefpodoxime (10 μg) increased by >5 mm when clavulanic acid (1 μg) was added. This result was confirmed using a cefotaxime-based Etest method (Table 1). Therefore, all Bacteroides tested contained at least one class A cephalosporinase gene.
The cephalosporinase enzymes inferred to be the products of these genes were isolated, purified and extensively characterized from crude extracts of clinical isolates of B. thetaiotaomicron, B. fragilis and Bacteroides vulgatus by Sato et al.37 three decades ago. The susceptibility profile of B. thetaiotaomicron VPI-5482 is unexceptional for a Bacteroides species, with susceptibility to carbapenems, co-amoxiclav and cefoxitin (data not shown), but resistance to ampicillin, amoxicillin, cefotaxime and cefpodoxime.
B. thetaiotaomicron OMVs are naturally produced and carry BtCepA on their surface
We recently established that the phosphatase BtMinpp is exported and active in OMVs produced by B. thetaiotaomicron.21 However, one of the first steps of the standard protocol used for OMV isolation involves centrifugation of a bacterial culture in order to separate bacterial cells from the soluble fraction containing the vesicles. This centrifugation can cause cell surface damage, giving a potential for artefacts.38 Therefore, to ensure that OMVs isolated after cell centrifugation are produced during bacterial cell growth and are not derived from ruptured and damaged cells, we developed an alternative procedure in which OMVs were collected from a growing bacterial culture after their diffusion through a membrane filter into a compartment containing sterile medium (see the Materials and methods section). The OMVs obtained were visualized by electron microscopy (Figure 3a) and were indistinguishable from those generated by the standard centrifugation protocol, confirming that they are produced by living cells and not by, e.g. centrifugal shear forces.
Figure 3.
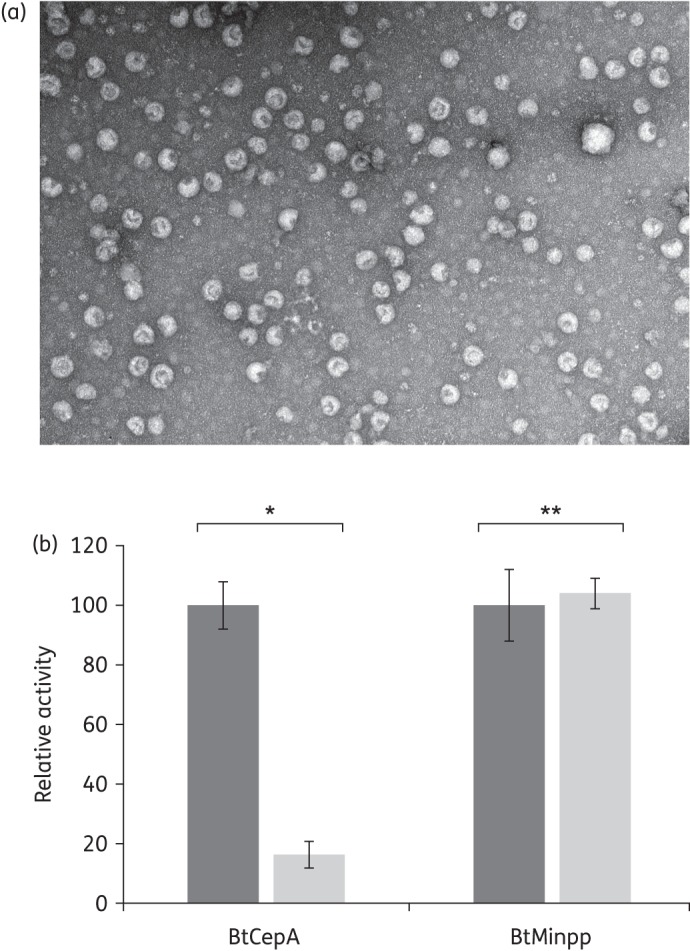
OMVs are produced in vivo by B. thetaiotaomicron and display BtCepA on their surface. (a) Electron microscopic photograph of OMVs collected from a sterile compartment after their diffusion through a 0.22 μm membrane from a compartment containing a B. thetaiotaomicron culture (see the Materials and methods section). Scale bar, ∼100 nm. (b) BtCepA and BtMinpp activities measured after treatment of OMVs with proteinase K. The relative activity is the ratio of the activity measured after proteinase K treatment compared with the activity measured without treatment. Dark grey bars, no proteinase K pre-treatment; light grey bars, proteinase K pre-treatment. *P < 0.0001; **P = 0.69.
We next addressed whether BtCepA is exported to OMVs. Assessment of the β-lactamase activity in B. thetaiotaomicron OMV protein extracts confirmed that BtCepA is associated with OMVs (Table 2). To localize BtCepA, OMV preparations were treated with proteinase K to digest any BtCepA associated with the outer surface of the vesicles. Only 5 min of proteinase K treatment was required for OMVs to lose 84% of their enzymatic activity when compared with non-treated OMVs (Figure 3b). Control experiments showed that activity of the BtMinpp phosphatase was not affected by proteinase K, confirming its location inside the vesicles. This result strongly suggests that a large fraction of BtCepA is exposed on the surface of the vesicles.
To ensure that the β-lactamase activity measured in solutions containing OMVs was not the result of enzyme release from OMVs into the milieu, a control experiment was run using vesicles isolated from B. thetaiotaomicron over-expressing BtCepA. No activity was detected in the milieu after 1 h of incubation (Table 2), consistent with the degradation of exogenous β-lactam antibiotics by OMVs occurring primarily at their surface and to a lesser extent within the vesicles.
Cephalosporinases are exported on the surface of OMVs released by other Bacteroides species
Based on the results obtained with BtCepA, we anticipated that cephalosporinases from other Bacteroides species would also be exported to OMVs. Accordingly, we investigated the capacity for isolated vesicles from Bacteroides dorei, B. fragilis, Bacteroides ovatus, Bacteroides stercoris and Bacteroides xylanisolvens to degrade β-lactam antibiotics present in the external milieu. OMVs were incubated in the presence of the third-generation cephalosporin cefotaxime and the degradation of the antibiotic was measured by applying the antibiotic solution, once OMVs had been removed, to a blank filter paper disc on a plate seeded with Salmonella as the indicator organism (Figure 4). S. enterica serovar Typhimurium has previously been used as an indicator strain in studies showing a protective effect by ampicillin-resistant E. coli.7
Figure 4.
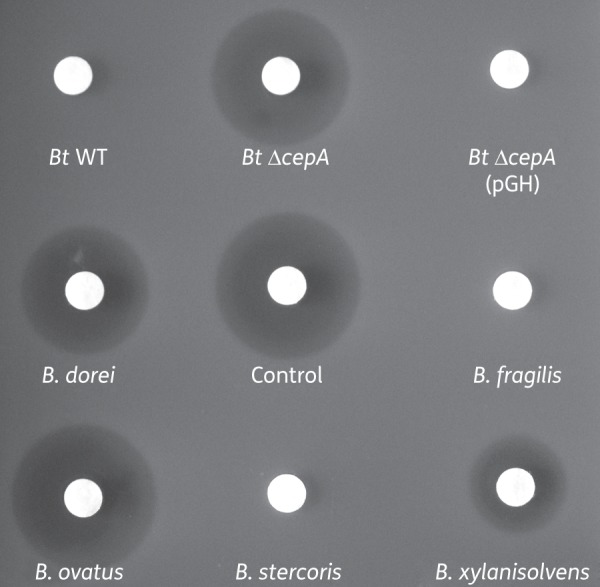
OMVs produced by Bacteroides spp. degrade cefotaxime. Antibiotic disc susceptibility test using Salmonella Typhimurium. The discs were loaded with 10 μL of a 10 mg/L cefotaxime solution that had been incubated for 1 h at 37°C with OMVs from different Bacteroides species. The discs were then placed onto Salmonella-inoculated agar plates. The inhibition zones were read after 16 h of incubation at 37°C. Control, 0.1 μg cefotaxime.
OMVs from B. fragilis, B. stercoris and B. thetaiotaomicron showed the highest capacity to degrade cefotaxime, with no visible inhibition of Salmonella growth remaining (Figure 4). In contrast, a zone of inhibition similar to that with untreated antibiotic was observed with B. ovatus OMVs, indicating that no β-lactamase activity was present in/on these OMVs despite expression of an active cephalosporinase (Table 1). These data suggest that, under laboratory conditions, the cephalosporinase produced by B. ovatus may be excluded from OMVs, as previously shown for various outer membrane proteins in E. coli,30 or, alternatively, does not bind to the surface of OMVs.
Using the proteinase K procedure applied to B. thetaiotaomicron OMVs (see above), 80%, 86% and 93% of the β-lactamase activity is exposed on the surface of B. fragilis, B. stercoris and B. xylanisolvens OMVs, respectively. The activity from B. dorei OMVs was very weak, making evaluation of the surface-exposed fraction difficult.
The clear reduction in the cefotaxime inhibition zone following exposure to OMVs from most Bacteroides species strongly suggests that these OMVs have the capability to serve as antibiotic degradation machineries in the GI tract of animals, acting remotely and independently of their parental bacterial cells.
B. thetaiotaomicron OMVs protect Salmonella Typhimurium and B. breve against β-lactam antibiotics
To confirm that OMVs carrying cephalosporinases can protect bacteria from β-lactam activity, B. thetaiotaomicron OMVs were added to growing cultures of Salmonella Typhimurium (Figure 5a) or to those of the human commensal intestinal bacterium B. breve (Figure 5b) in the presence or absence of cefotaxime. Vesicles produced by the ΔBtcepA deletion mutant were used as a negative control. As expected, cefotaxime had a dramatic effect on the viability of Salmonella Typhimurium cells when grown in the presence of vesicles lacking BtcepA (Figure 5a). By contrast, addition of BtCepA-loaded vesicles allowed the salmonellae to grow normally in the presence of the lowest cefotaxime concentration (1 mg/L) and at a slower rate for the highest concentration (10 mg/L). The same phenomenon was observed when ampicillin was used (data not shown). Because of its slower growth rate, B. breve appeared more resistant to cefotaxime than Salmonella Typhimurium over time (Figure 5b). However, the presence of B. thetaiotaomicron BtCepA-loaded vesicles in the medium allowed the cells to grow until saturation of the culture at 24 h, whereas most of the cells were killed after 24 h when the culture was supplemented with ΔBtcepA vesicles. Thus, β-lactamase-loaded OMVs from Bacteroides protected Salmonella Typhimurium and B. breve against β-lactam antibiotics.
Figure 5.
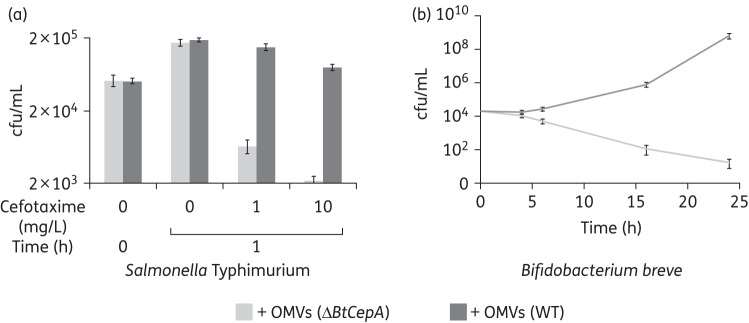
Killing curves of Salmonella Typhimurium (a) and B. breve (b) in liquid broth in the presence of B. thetaiotaomicron OMVs produced either by the ΔBtcepA mutant (light grey) or the WT strain (dark grey). The final concentration of cefotaxime added to the culture at time zero is indicated.
Discussion
Our study focuses on the identification of cephalosporinase genes in human gut Bacteroides species and on the distribution of their product. We find that, in addition to being secreted to the periplasm of bacterial cells, these cephalosporinases also associate with OMVs produced by these bacteria; the results are consistent with a role for these OMVs in degrading β-lactam antibiotics remotely from their parental cells.
Whereas Gram-positive β-lactamase-producing organisms largely liberate their enzymes into the surrounding milieu, the β-lactamases of Gram-negative bacteria are usually considered to be confined to the periplasmic space of the cells, acting in concert with the diffusion barrier of the outer membrane.39 The outer membrane of E. coli constitutes a diffusion barrier for both penicillins and cephalosporins;40 it thereby slows the influx of antibiotic molecules and is one of the major factors that determine the degree of antibiotic resistance of the organism.41 In this report, we show that part of cephalosporinase BtCepA becomes associated with membrane vesicles and this BtCepA–OMV association efficiently degrades substrate β-lactam antibiotics present in the surrounding medium. The extracellular degradation of β-lactam antibiotics is attributable to the OMVs only, since no leakage of the enzyme into the medium could be detected (Table 2). Degradation experiments with proteinase K showed that BtCepA was exposed on the surface of the vesicles rather than being sequestered within their lumen (Figure 3b). This is of interest since surface-exposed enzymes, exposed to the full antibiotic concentration, can achieve swifter substrate turnover than periplasmic enzymes in hydrolysing β-lactams in the surrounding medium40 and, by extension, would be more efficient than enzymes located inside OMVs.
Although our findings are consistent with a large fraction of BtCepA being associated with the membrane of OMVs and being exposed on the surface of the vesicles, no transmembrane region was predicted in BtCepA using the TMHMM server (http://www.cbs.dtu.dk/services/TMHMM-2.0/), suggesting that BtCepA is unlikely to be embedded in the outer membrane. It is also unlikely that BtCepA is a lipoprotein, since Bacteroides β-lactamases give discrete and sharp bands by isoelectric focusing analysis,42 instead of smearing, as expected for lipoproteins. BtCepA and other Bacteroides cephalosporinases perhaps may electrostatically associate with surface phosphates of vesicle phospholipids, but further analysis of their exact localization and attachment is needed, in both bacterial cells and OMVs, to confirm or refute this hypothesis and to determine exactly how these proteins anchor to the surface of OMVs.
It is well established that antibiotic usage strongly affects the intestinal microbiota.2,43 For example, Pérez-Cobas et al.2 recently reported that combined intravenous therapy with ampicillin/sulbactam and cefazolin caused significant microbiota disturbance in the human colon. Hence, we speculate that Bacteroides species in the human GI tract microbiota have the potential to protect Salmonella and possibly other pathogens and commensal microorganisms, such as the probiotic B. breve, by yielding large numbers of cephalosporinase-coated OMVs. More generally, the presence of cephalosporinase-coated OMVs in the colon could contribute to the maintenance of a balanced microbiota protecting against the adverse effects of antibiotic treatments. It remains unclear whether members of the genus have specifically evolved to do this during the antibiotic era or whether, as seems more likely, the association of these cephalosporins, which are inherent to most members of the genus, and the OMVs is essentially fortuitous. Notably, the B. fragilis NCTC 9343 strain, which produces β-lactamase-coated vesicles, was deposited with a culture collection as early as 1955, suggesting that the behaviour is not a very recent development.
Funding
This work was supported by institutional grants from the Biotechnology and Biological Sciences Research Council (BBSRC) (BB/J004529/1, S. R. C.).
Transparency declarations
None to declare.
Supplementary data
Table S1 is available as Supplementary data at JAC Online (http://jac.oxfordjournals.org).
Acknowledgements
We would like to thank Marine Rouyer for her technical assistance, Dr Isabelle Hautefort for helping with the manuscript and Dr Udo Wegmann for scientific discussions. We are also grateful to Dr Lindsay Hall and Cristina Alcon for providing B. breve UCC2003.
References
- 1.Jernberg C, Löfmark S, Edlund C, et al. Long-term impacts of antibiotic exposure on the human intestinal microbiota. Microbiology. 2010;156:3216–23. doi: 10.1099/mic.0.040618-0. [DOI] [PubMed] [Google Scholar]
- 2.Pérez-Cobas AE, Gosalbes MJ, Friedrichs A, et al. Gut microbiota disturbance during antibiotic therapy: a multi-omic approach. Gut. 2013;62:1591–601. doi: 10.1136/gutjnl-2012-303184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Turnbaugh PJ, Hamady M, Yatsunenko T, et al. A core gut microbiome in obese and lean twins. Nature. 2009;457:480–4. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Krieg NR, Ludwig W, Euzéby J, et al. Phylum XIV. Bacteroidetes phyl. nov. In: Krieg NR, Staley JT, Brown DR, editors. Bergey's Manual of Systematic Bacteriology 2nd Ed. Vol. 4. New York: Springer Science & Business Media: 2010. pp. 25–470. [Google Scholar]
- 5.Wexler HM, Finegold SM. Current susceptibility patterns of anaerobic bacteria. Yonsei Med J. 1998;39:495–501. doi: 10.3349/ymj.1998.39.6.495. [DOI] [PubMed] [Google Scholar]
- 6.Brook I, Pazzaglia G, Coolbaugh JC, et al. In-vivo protection of group A β-haemolytic streptococci from penicillin by β-lactamase-producing Bacteroides species. J Antimicrob Chemother. 1983;12:599–606. doi: 10.1093/jac/12.6.599. [DOI] [PubMed] [Google Scholar]
- 7.Perlin MH, Clark DR, McKenzie C, et al. Protection of Salmonella by ampicillin-resistant Escherichia coli in the presence of otherwise lethal drug concentrations. Proc Biol Sci. 2009;276:3759–68. doi: 10.1098/rspb.2009.0997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schaar V, Nordström T, Mörgelin M, et al. Moraxella catarrhalis outer membrane vesicles carry β-lactamase and promote survival of Streptococcus pneumoniae and Haemophilus influenzae by inactivating amoxicillin. Antimicrob Agents Chemother. 2011;55:3845–53. doi: 10.1128/AAC.01772-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schaar V, Uddbäck I, Nordström T, et al. Group A streptococci are protected from amoxicillin-mediated killing by vesicles containing β-lactamase derived from Haemophilus influenzae. J Antimicrob Chemother. 2014;69:117–20. doi: 10.1093/jac/dkt307. [DOI] [PubMed] [Google Scholar]
- 10.Sambrook J, Russell DW. Molecular Cloning: A Laboratory Manual. NY, USA: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- 11.Smith CJ, Rollins LA, Parker AC. Nucleotide sequence determination and genetic analysis of the Bacteroides plasmid, pBI143. Plasmid. 1995;34:211–22. doi: 10.1006/plas.1995.0007. [DOI] [PubMed] [Google Scholar]
- 12.Tancula E, Feldhaus MJ, Bedzyk LA, et al. Location and characterization of genes involved in binding of starch to the surface of Bacteroides thetaiotaomicron. J Bacteriol. 1992;174:5609–16. doi: 10.1128/jb.174.17.5609-5616.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shoemaker NB, Getty C, Gardner JF, et al. Tn4351 transposes in Bacteroides spp. and mediates the integration of plasmid R751 into the Bacteroides chromosome. J Bacteriol. 1986;165:929–36. doi: 10.1128/jb.165.3.929-936.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wegmann U, Horn N, Carding SR. Defining the Bacteroides ribosomal binding site. Appl Environ Microbiol. 2013;79:1980–9. doi: 10.1128/AEM.03086-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tamura K, Peterson D, Peterson N, et al. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–9. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Löytynoja A, Goldman N. webPRANK: a phylogeny-aware multiple sequence aligner with interactive alignment browser. BMC Bioinformatics. 2010;11:579. doi: 10.1186/1471-2105-11-579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Whelan S, Goldman N. A general empirical model of protein evolution derived from multiple protein families using a maximum-likelihood approach. Mol Biol Evol. 2001;18:691–9. doi: 10.1093/oxfordjournals.molbev.a003851. [DOI] [PubMed] [Google Scholar]
- 18.Gascuel O. BIONJ: an improved version of the NJ algorithm based on a simple model of sequence data. Mol Biol Evol. 1997;14:685–95. doi: 10.1093/oxfordjournals.molbev.a025808. [DOI] [PubMed] [Google Scholar]
- 19.Tamura K, Dudley J, Nei M, et al. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–9. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 20.Osborn MJ, Gander JE, Parisi E, et al. Mechanism of assembly of the outer membrane of Salmonella typhimurium: site of synthesis of lipopolysaccharide. J Biol Chem. 1972;247:3973–86. [PubMed] [Google Scholar]
- 21.Stentz R, Osborne S, Horn N, et al. A bacterial homolog of a eukaryotic inositol phosphate signaling enzyme mediates cross-kingdom dialog in the mammalian gut. Cell Rep. 2014;6:646–56. doi: 10.1016/j.celrep.2014.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fanning S, Hall LJ, van Sinderen D. Bifidobacterium breve UCC2003 surface exopolysaccharide production is a beneficial trait mediating commensal-host interaction through immune modulation and pathogen protection. Gut Microbes. 2012;3:420–5. doi: 10.4161/gmic.20630. [DOI] [PubMed] [Google Scholar]
- 23.Guss SP, Bawdon RE. Effects of sub-minimum inhibitory concentrations of cephalosporins on Bacteroides species. J Antimicrob Chemother. 1984;13:633–5. doi: 10.1093/jac/13.6.633. [DOI] [PubMed] [Google Scholar]
- 24.Spratt BG. Distinct penicillin binding proteins involved in the division, elongation, and shape of Escherichia coli K12. Proc Natl Acad Sci USA. 1975;72:2999–3003. doi: 10.1073/pnas.72.8.2999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Buijs J, Dofferhoff AS, Mouton JW, et al. Concentration-dependency of β-lactam-induced filament formation in Gram-negative bacteria. Clin Microbiol Infect. 2008;14:344–9. doi: 10.1111/j.1469-0691.2007.01940.x. [DOI] [PubMed] [Google Scholar]
- 26.Xu J, Bjursell MK, Himrod J, et al. A genomic view of the human-Bacteroides thetaiotaomicron symbiosis. Science. 2003;299:2074–6. doi: 10.1126/science.1080029. [DOI] [PubMed] [Google Scholar]
- 27.Altschul SF, Gish W, Miller W, et al. Basic local alignment search tool. J Mol Biol. 1990;215:403–10. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 28.Bates PA, Kelley LA, MacCallum RM, et al. Enhancement of protein modelling by human intervention in applying the automatic programs 3D-JIGSAW and 3D-PSSM. Proteins. 2001;45(Suppl 5):39–46. doi: 10.1002/prot.1168. [DOI] [PubMed] [Google Scholar]
- 29.Bebrone C. Metallo-β-lactamases (classification, activity, genetic organization, structure, zinc coordination) and their superfamily. Biochem Pharmacol. 2007;74:1686–701. doi: 10.1016/j.bcp.2007.05.021. [DOI] [PubMed] [Google Scholar]
- 30.Kulp A, Kuehn MJ. Biological functions and biogenesis of secreted bacterial outer membrane vesicles. Annu Rev Microbiol. 2010;64:163–84. doi: 10.1146/annurev.micro.091208.073413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Benson DA, Boguski MS, Lipman DJ, et al. GenBank. Nucleic Acids Res. 1997;25:1–6. doi: 10.1093/nar/25.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rogers MB, Parker AC, Smith CJ. Cloning and characterization of the endogenous cephalosporinase gene, cepA, from Bacteroides fragilis reveals a new subgroup of Ambler class A β-lactamases. Antimicrob Agents Chemother. 1993;37:2391–400. doi: 10.1128/aac.37.11.2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.García N, Gutiérrez G, Lorenzo M, et al. Genetic determinants for cfxA expression in Bacteroides strains isolated from human infections. J Antimicrob Chemother. 2008;62:942–7. doi: 10.1093/jac/dkn347. [DOI] [PubMed] [Google Scholar]
- 34.Smith CJ, Bennett TK, Parker AC. Molecular and genetic analysis of the Bacteroides uniformis cephalosporinase gene, cblA, encoding the species-specific 3-lactamase. Antimicrob Agents Chemother. 1994;38:1711–5. doi: 10.1128/aac.38.8.1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Barrios H, Garza-Ramos U, Ochoa-Sanchez LE, et al. A plasmid-encoded class 1 integron contains GES-type extended-spectrum β-lactamases in Enterobacteriaceae clinical isolates in Mexico. Antimicrob Agents Chemother. 2012;56:4032–4. doi: 10.1128/AAC.05980-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing: Twenty-second Informational Supplement M100-S22. Wayne, PA, USA: CLSI; 2012. [Google Scholar]
- 37.Sato K, Matsuura Y, Miyata K, et al. Characterization of cephalosporinases from Bacteroides fragilis, Bacteroides thetaiotaomicron and Bacteroides vulgatus. J Antibiot (Tokyo) 1983;36:76–85. doi: 10.7164/antibiotics.36.76. [DOI] [PubMed] [Google Scholar]
- 38.Peterson BW, Sharma PK, van der Mei HC, et al. Bacterial cell surface damage due to centrifugal compaction. Appl Environ Microbiol. 2012;78:120–5. doi: 10.1128/AEM.06780-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zimmermann W, Rosselet A. Function of the outer membrane of Escherichia coli as a permeability barrier to β-lactam antibiotics. Antimicrob Agents Chemother. 1977;12:368–72. doi: 10.1128/aac.12.3.368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu W, Nikaido H. Contribution of the cell-surface-associated enzyme in the Zimmermann-Rosselet assay of outer membrane permeability of β-lactam antibiotics. Antimicrob Agents Chemother. 1991;35:177–9. doi: 10.1128/aac.35.1.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Datta N, Kontomichalou P. Penicillinase synthesis controlled by infectious R factors in Enterobacteriaceae. Nature. 1965;208:239–41. doi: 10.1038/208239a0. [DOI] [PubMed] [Google Scholar]
- 42.Appelbaum PC, Philippon A, Jacobs MR, et al. Characterization of β-lactamases from non-Bacteroides fragilis group Bacteroides spp. belonging to seven species and their role in β-lactam resistance. Antimicrob Agents Chemother. 1990;34:2169–76. doi: 10.1128/aac.34.11.2169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Blaser M. Antibiotic overuse: stop the killing of beneficial bacteria. Nature. 2011;476:393–4. doi: 10.1038/476393a. [DOI] [PubMed] [Google Scholar]
- 44.Whitehead TR, Hespell RB. The genes for three xylan-degrading activities from Bacteroides ovatus are clustered in a 3.8-kilobase region. J Bacteriol. 1990;172:2408–12. doi: 10.1128/jb.172.5.2408-2412.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


